Abstract
RNA splicing and epigenetic gene mutations are the most frequent genetic lesions found in patients with myelodysplastic neoplasm (MDS). About 25% of patients present concomitant mutations in such pathways, suggesting a cooperative role in MDS pathogenesis. Importantly, mutations in the splicing factor ZRSR2 frequently associate with alterations in the epigenetic regulator TET2. However, the impact of these concurrent mutations in hematopoiesis and MDS remains unclear. Using CRISPR/Cas9 genetically engineered mice, we demonstrate that Zrsr2m/mTet2−/− promote MDS with reduced penetrance. Animals presented peripheral blood cytopenia, splenomegaly, extramedullary hematopoiesis, and multi-lineage dysplasia, signs consistent with MDS. We identified a myelo-erythroid differentiation block accompanied by an expansion of LT-HSC and MPP2 progenitors. Transplanted animals presented a similar phenotype, thus indicating that alterations were cell-autonomous. Whole-transcriptome analysis in HSPC revealed key alterations in ribosome, inflammation, and migration/motility processes. Moreover, we found the MAPK pathway as the most affected target by mRNA aberrant splicing. Collectively, this study shows that concomitant Zrsr2 mutation and Tet2 loss are sufficient to initiate MDS in mice. Understanding this mechanistic interplay will be crucial for the identification of novel therapeutic targets in the spliceosome/epigenetic MDS subgroup.
Subject terms: Myelodysplastic syndrome, Haematopoietic stem cells, Cancer genetics
Introduction
Genetic mutations in splicing factors are observed in circa 50% of human myelodysplastic neoplasm (MDS) and about 45% of cases carry mutations in epigenetic regulators. Importantly, a subgroup of patients (25%) presents mutations in both splicing factors and epigenetic regulators, indicating possible cooperation among both functional categories in the pathomechanism of the disease.
The splicing factor ZRSR2 is mutated in about 5% of patients with MDS and appears associated with mutations in the epigenetic regulator TET2 [1]. ZRSR2 is implicated in the minor spliceosome and acts on U2-type introns (99.7% of introns in the human genome) as well as on U12-type introns (only present in approximately <0.3% of introns of all human genes) [2]. Mutations in ZRSR2 are very diverse and comprise nonsense, frameshift, missense, and splice-site mutations. While different hotspots are affected in other mutated splicing factors, mutations in ZRSR2 distribute across the entire coding region leading to protein loss-of-function [3]. ZRSR2 mutations affect predominantly males and cause abnormal splicing via intron retention of U12-dependent introns [4–7]. Apart from MDS, ZRSR2 mutations have also been reported in other hematological malignancies including chronic myelomonocytic leukemia (CMML) [3], myeloproliferative neoplasms (MPN) [3], and blastic plasmacytoid dendritic cell neoplasm (BPCDN) [8]. Functionally, shRNA-mediated ZRSR2 silencing in AML cell lines induced aberrant retention of U12-type introns, diminished cell growth, and altered myeloid differentiation [4]. Recently, two in vivo Zrsr2 knockout mouse models have been developed. Constitutive germline deletion of Zrsr2 in mice demonstrated that Zrsr2 is dispensable for hematopoietic development and that absence of a major phenotype is due to a Zrsr1 compensatory mechanism [6]. In contrast, Zrsr2 hematopoietic conditional knockout mice resulted in enhanced hematopoietic stem cell (HSC) self-renewal in vitro and in vivo [7].
On the other hand, TET2 promotes DNA demethylation by oxidizing the methyl group to 5-hydroxymethylcytosine. TET2 is the most frequently mutated gene in MDS (25–35%) [9], and biallelic TET2 gene inactivation is frequently observed in myeloid neoplasms [10]. In addition, TET2 mutations are highly prevalent in clonal hematopoiesis of indeterminate potential [11], suggesting that TET2 lesions are early driver events with the potential to predispose for further malignant transformation. TET2 is involved in the epigenetic control of gene regulation, HSC self-renewal, and myeloid lineage commitment [12]. Deletion of Tet2 in mice leads to sporadic development of MDS/CMML-like disease [13–16] and co-expression of mutated Tet2 with additional oncogenic mutations, including Asxl1, Ezh2, Jak2, c-Kit, Flt3, Bcor, Sf3b1, and AML1::ETO, promotes disease in mice [12, 17, 18]. A recent work explored the co-occurrence of Zrsr2 and Tet2 mutations in the context of BPDCN [8]. They reported that the combined inactivation of Zrsr2 and Tet2 expands common myeloid progenitors (CMP), granulocyte-monocyte progenitors (GMP), and megakaryocyte-erythroid progenitors (MEP) compartments in in vivo transplantation experiments. To the best of our knowledge, no studies have explored the concurrence of Zrsr2 and Tet2 in MDS.
To investigate the functional role of Zrsr2 and Tet2 mutations in the context of hematopoiesis and MDS, we generated a novel mutant Zrsr2 allele, either alone or in combination with Tet2 deficiency. Here, we show that concurrent mutations in Zrsr2 and Tet2 promote MDS via dysregulation of gene expression and alternative splicing. We detect a disruption in myelo-erythroid differentiation and an expansion of LT-HSC and MPP2 compartments. Notably, 20–25% of double-mutant mice developed a more severe phenotype characterized by cytopenias (thrombocytopenia with or without anemia), splenomegaly with extramedullary hematopoiesis, and multi-lineage dysplasia, signs reminiscent of MDS. Moreover, we identify global gene expression changes and aberrant mRNA splicing of key biological pathways. Taken together, we demonstrate that mutations in Zrsr2 and Tet2 promote MDS with reduced penetrance.
Materials and methods
Animals
Novel Zrsr2 mutant mice were created by CRISPR/Cas9 technology. In particular, exon 10 was targeted and a 17-nucleotide deletion was introduced, which generated a frameshift mutation [19]. Further details are provided in Supplementary information. Mice were then crossed to obtain homozygous mutants (Zrsr2m/m and Zrsr2m/y) and wild-type (Zrsr2+/+ and Zrsr2+/y) animals. Both female and male Zrsr2 mutant mice were used for experiments. To simplify nomenclature, we referred to all of them with the female genotypes. To generate animals carrying alterations in both Zrsr2 and Tet2, Zrsr2 mutant mice were crossed with a Tet2 KO line previously described [20]. Both female and male double mutant animals were used for experiments. Mice were genotyped by standard PCR using Zrsr2 and Tet2 specific primers (Supplementary Table 1). CD45.1 mice, used as recipients in transplantation experiments, were purchased from Charles Rivers (Wilmington, MA, USA). Pilot preliminary experiments were performed for sample size estimates. No animals were excluded from the analysis. Regarding randomization, in all cases where multiple experimental groups were analyzed, mice were not randomly distributed but rather allocated in such a way that each group was evenly matched with regard to age range and frequency of mice of each sex. Investigators were not blinded to the mouse genotypes. This study was approved by the Regional Valencian Ministry of Agriculture (permit 2017/VSC/PEA/00200) and the Ethics Committee at IIS La Fe (permit 2016/0756).
Flow cytometry analysis and cell sorting
Mice were sacrificed, autopsied, and bones (femurs, tibias, and cristae), spleen, and thymus were collected for further analysis. Red blood cells were lysed with Gey’s Solution (NH4Cl 8.3 g/L, NaHCO3 1.0 g/L, EDTA 37 mg/L) and RBC-depleted cells were stained using monoclonal antibodies for 15 min at 4 °C. PB samples were lysed with BD FACS™ Lysing Solution (BD, Franklin Lakes, NJ, USA) and stained with monoclonal antibodies for 15 min at 4 °C. Lin−Sca-1+c-Kit+ (LSK) cells were sorted from a pool of 3 mice for whole transcriptome analysis and transplantation assays. BM cells were lysed with Gey’s solution, stained with anti-mouse CD117 MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany), and enriched using LS columns (Miltenyi Biotec). c-Kit+ cells were subsequently stained with monoclonal antibodies for cell separation. Please refer to the Supplementary Information for additional details on antibodies and marker combinations.
In vivo transplantation assays
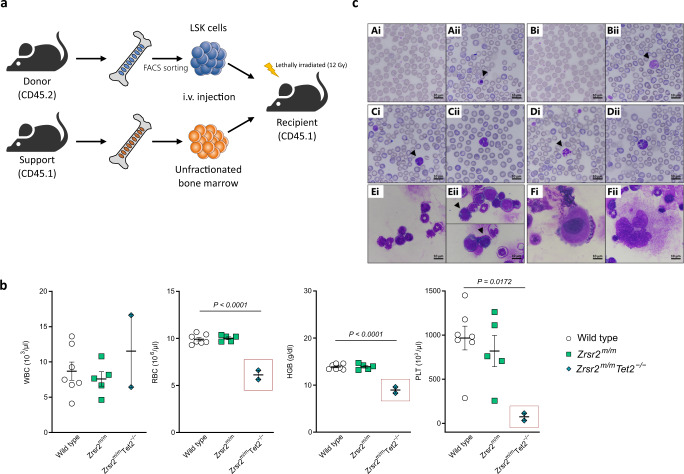
For HSPC transplantations, LSK cells were FACS-sorted from a pool of 3 donor mice, and 8000 LSK cells (CD45.2) were transplanted along with 2 × 105 support unfractionated bone marrow cells (CD45.1) into each lethally irradiated (12 Gray, split dose) mouse (CD45.1). Recipient mice were given enrofloxacin (Bayer, Leverkusen, Germany) in the drinking water for 4 weeks post-irradiation. PB engraftment was monitored monthly by flow cytometry.
Gene expression analysis by RT-qPCR
Total RNA was extracted by RNeasy Mini Kit (QIAGEN, Hilden, Germany) from 10 × 106 bone marrow or spleen cells previously lysed in RLT buffer (QIAGEN) supplemented with 1% β-mercaptoethanol (Sigma-Aldrich, St. Louis, MO, USA) and stored at −80 °C. RNA was treated with DNA-free DNase Treatment and Removal Reagents (Ambion, Austin, TX, USA). DNase-treated total RNA (1 µg) was reverse transcribed with oligo(dT) and TaqMan Reverse Transcription Reagents (Thermo Fisher Scientific, Waltham, MA, USA). The resulting cDNA was diluted tenfold before use. Real-time quantitative PCR was performed in technical triplicates using AceQ SYBR qPCR Master Mix (Vazyme Biotech Co., Nanjing, China) and the corresponding primers (Supplementary Table 1) using a ViiA 7 Real-Time PCR System (Thermo Fisher Scientific). All RT–qPCR analyses were performed on an Applied Biosystems QuantStudio Software V1.3 (Thermo Fisher Scientific). Primer set efficiency was firstly calculated in a standard curve experiment. Relative gene expression levels were calculated using the comparative CT method. Since target and housekeeping primer set efficiencies differed >10%, we applied the corrective formula described in [21] to calculate the relative expression ratio. Gene expression levels were then normalized to that of Hprt.
RNA sequencing
LSK cells (50,000 per sample) from pools of WT (n = 4), Zrsr2m/m (n = 5), Tet2−/− (n = 5), and Zrsr2m/mTet2−/− (n = 6) were sorted into RLT buffer supplemented with 1% β-mercaptoethanol. Total RNA extraction was performed using an RNEasy Micro kit (QIAGEN). DNA was then removed using on-column DNAse treatment. RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). Samples with an RNA Integrity Number value ≥7 were selected for further analysis. Low input directional RNA-seq libraries were prepared using NEBNext Ultra Directional RNA Library Prep Kit for Illumina (Illumina Inc., Cat N°. 7420, San Diego, CA, USA). Purified libraries were paired-end sequenced on the Illumina NovaSeq 6000 (Novogene, Beijing, China).
The quality of raw RNA-seq data was assessed in fastq data files using Fastp. Reads were mapped to the reference mouse genome GRCm38 (mm10), with genome annotations from GENCODE vM24, using the splicing aware mapping software STAR v2.5 [22]. Quantification of reads was calculated with HTSeq v0.6.1 [23]. Differential gene expression was determined using DESeq2 v2_1.6.3 [24] with adjusted P value ≤ 0.05, log2FoldChange ≥ 0. Differential alternative splicing was assessed using rMATS v3.2.1 [25]. This approach categorizes splicing events into five groups: retained intron (RI), skipped exon (SE), alternative 5′ splice site (A5SS), alternative 3′ splice site (A3SS), and mutually exclusive exons (MXE). Splicing events associated with each pair-wise comparison were identified based on false discovery rate (FDR) ≤ 0.05. Functional enrichment within lists of differentially spliced genes was identified with KEGG [26] and the enrichment cut-off p value was set as adjusted P value ≤ 0.05. RNA-seq data have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-10481.
The protein–protein interaction network between MAPK mis-spliced targets and related proteins was retrieved from STRING (http://string-db.org) [27]. The minimum required interaction score was set in high confidence (0.700). Network type was set in full STRING network (the edges indicate both functional and physical protein associations).
Results
Development of a germline Zrsr2 mutant mouse
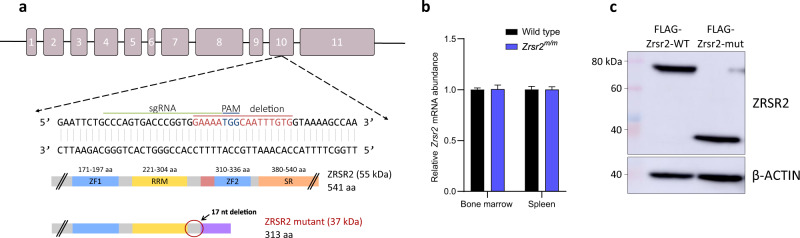
To gain insight into the role of Zrsr2 in normal and malignant hematopoiesis, we generated a mutant allele targeting Zrsr2 in vivo by CRISPR/Cas9 technology (Fig. 1a). This approach generated a 17-nucleotide deletion in exon 10 of Zrsr2 DNA sequence (Supplementary Fig. 1a), which resulted in a frameshift mutation. Consequently, the mutated Zrsr2 allele lacks the second zinc finger domain and the serine-rich domain (Fig. 1a). Seventeen nucleotide deletion was confirmed in different hematopoietic tissues (Supplementary Fig. 1b). Mutant bone marrow and spleen cells expressed levels of Zrsr2 mRNA equivalent to WT controls, suggesting that the mutated allele escapes the nonsense-mediated mRNA decay (Fig. 1b). In addition, 17-nucleotide deletion, in turn, caused skipping of exon 10, spliced event that was confirmed by RT-PCR in cDNA from LSK cells (Supplementary Fig. 1c). Due to difficulties to detect ZRSR2 protein in murine tissue, we tagged the WT and truncated mutant protein with FLAG and over-expressed it in HEK293T cells. Both WT and mutant ZRSR2 forms were detected, indicating that the truncated protein product is stable (Fig. 1c). Hemizygous Zrsr2m/y or homozygous Zrsr2m/m mice were mated with Tet2 knockout mice to produce Zrsr2m/mTet2−/− double mutant mice. Zrsr2 mutant mice had a normal lifespan (24 months) while double mutant mice died at earlier ages (average 14 months). Zrsr2m/m females were infertile.
Fig. 1. Generation of a germline Zrsr2 allele.
a Upper panel: Zrsr2 gene consists of eleven exons (boxes). Middle panel: CRISPR/Cas9 construction targeting Zrsr2 exon 10. Lines indicate gRNA sequence (green), PAM motif (blue), and the resulting 17-nucleotides deletion (red). Lower panel: Diagram of WT and truncated ZRSR2 protein with their functional domains: ZF (Zinc Finger), RRM (RNA recognition motif), SR (Serine/Arginine rich domain). Mutant Zrsr2m/m mice expressed a mutated short ZRSR2 protein containing a ZF domain and the RRM domain. The numbers indicate amino acids encoded by WT and mutant protein as predicted in silico. b Relative mRNA abundance of Zrsr2 in bone marrow and spleen determined by RT-qPCR. Data normalized to Hprt. BM: WT (n = 9), Zrsr2m/m (n = 10); SPL: WT (n = 6), Zrsr2m/m (n = 6). c Western blot of FLAG-tagged WT and truncated ZRSR2 protein overexpression in HEK293T cells. Proteins were detected using an anti-FLAG antibody in total cell lysates. Data from two independent experiments.
Zrsr2 mutation and Tet2 loss expand LT-HSC and MPP2 compartments and impair myelo-erythroid progenitor differentiation in steady-state
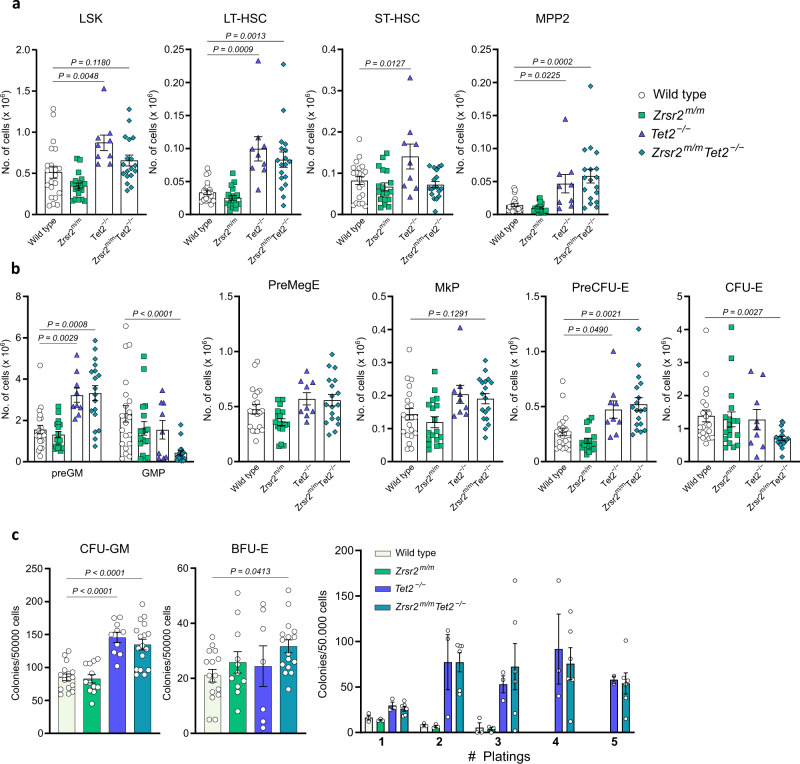
We analyzed the stem and myelo-erythroid progenitor compartments at 12-13 weeks. Fractionation of the HSC (LSK) compartment using CD150 and CD48 expression showed expansion of some phenotypic HSC subsets, such as the long-term HSC (LT-HSC, LSK CD150+CD48−) [28] and the multipotent MPP2 progenitors (LSK CD150+CD48+, erythroid, and megakaryocyte biased progenitors) [29, 30] (Fig. 2a, Supplementary Fig. 2a). At the progenitor level, we observed a significant increase in the early precursors of granulo-monocytic (PreGM) and erythroid (PreCFU-E) progenitors, an increase in megakaryocytic progenitors (MkP) and a decrease in the more committed granulo-monocytic (GMP), and erythroid (CFU-E) progenitors, suggestive of a differentiation block at the early stages of myelo-erythroid lineage commitment [31] (Fig. 2b, Supplementary Fig. 2b). These phenotypic alterations were accompanied by an increase in GM-CFU and BFU-E colony-forming units in Zrsr2m/mTet2−/− mice (Fig. 2c). Zrsr2m/mTet2−/− myeloid progenitors exhibited properties of self-renewal as evidenced by sustained CFU activity in serial replating assays (Fig. 2c). Overall, we observed a general skew towards myeloid lineage commitment at expense of the B-lymphoid lineage (Supplementary Fig. 2c). No effects on T lymphopoiesis were observed in single or double mutant mice (Supplementary Fig. 2d).
Fig. 2. Zrsr2 mutation and Tet2 loss expand LT-HSC and impair myelo-erythroid differentiation.
a Flow cytometry analysis of the LSK compartment from 3-months-old WT (n = 21), Zrsr2m/m (n = 18), Tet2−/− (n = 9), and Zrsr2m/mTet2−/− mice (n = 18). Representative dot plots are shown in Supplementary Fig. 2. b Flow cytometry analysis of myelo-erythroid progenitors from 3-months-old WT (n = 21), Zrsr2m/m (n = 18), Tet2−/− (n = 9), and Zrsr2m/mTet2−/− (n = 18) mice. Representative dot plots are shown in Supplementary Fig. 2. c Left and middle panel: GM-CFU and BFU-E forming capacity of unfractionated BM from 3-month-old WT (n = 18), Zrsr2m/m (n = 12), Tet2−/− (n = 10), and Zrsr2m/m Tet2−/− (n = 17) mice. Right panel: Serial replating capacity of unfractionated BM from 3-months-old WT (n = 3), Zrsr2m/m (n = 3), Tet2−/− (n = 3), and Zrsr2m/m Tet2−/− (n = 6) mice cultured under myeloid conditions. Data represent mean ± SEM.
Zrsr2 mutation and Tet2 loss result in MDS
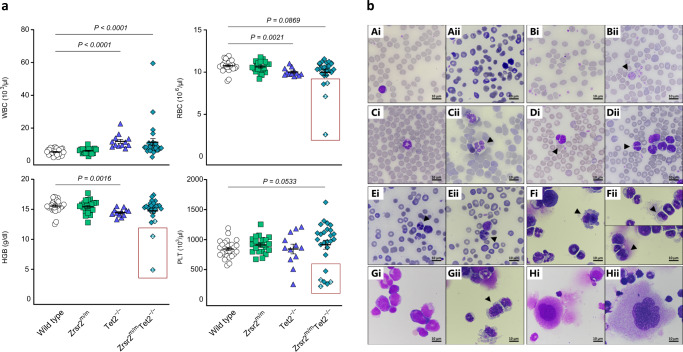
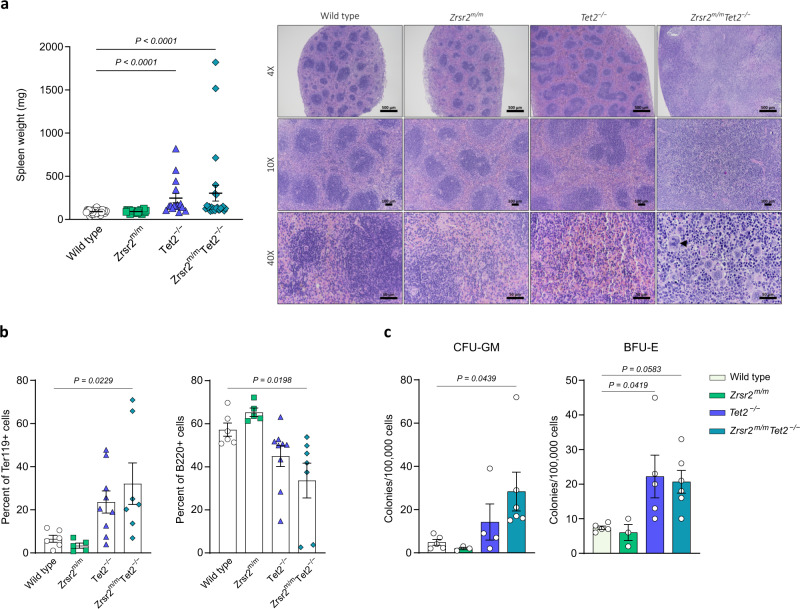
Notably, within the cohort of Zrsr2m/mTet2−/− mice analyzed, a subset of mice (25%, 6 out of 24) showed a more aggressive hematological phenotype. These mice presented cytopenias (thrombocytopenia with or without anemia) (Fig. 3a). Evaluation of peripheral blood smears and bone marrow cytospins indicated that Zrsr2m/mTet2−/− mice presented multi-lineage dysplastic features (Fig. 3b). We observed signs of dyserythropoiesis including RBC anisocytosis, polychromasia, and the presence of circulating nucleated red cells (Fig. 3b/Aii). In addition, we observed erythrophagocytosis and binucleated erythroid precursors in the bone marrow (Fig. 3b/Fi, Fii). Megakaryocytic dysplasia included platelet anisocytosis, the presence of abnormally large platelets in peripheral blood (Fig. 3B/Bii), and giant megakaryocytes in the bone marrow (Fig. 3b/Hii). Myeloid dysplasia was present in the form of neutrophil hypersegmentation (Fig. 3b/Cii), granulocyte hyposegmentation with pseudo Pelger-Huët anomalies (Fig. 3b/Di, Dii), nuclear fragmentation and karyorrhexis of neutrophils, and abnormal vacuolization in myeloid precursors (Fig. 3b/Gii). Macroscopic analysis of Zrsr2m/mTet2−/− spleens showed splenomegaly (Fig. 4a). Histopathological analysis of splenic sections demonstrated disruption of red and white pulp in Zrsr2m/mTet2−/− mice (Fig. 4a). A closer examination revealed infiltration by megakaryocyte and erythroid (Ter119+) cells and a decrease in B cells (B220+) as assessed by flow cytometry (Fig. 4b). Signs of extramedullary hematopoiesis were evidenced by the presence of GM-CFU and BFU-E activity in the spleen of Zrsr2m/mTet2−/− mice (Fig. 4c). Overall, a subset of Zrsr2m/mTet2−/− mice showed features consistent with myelodysplasia [32].
Fig. 3. Concomitant Zrsr2 mutation and Tet2 loss result in PB cytopenias and multilineage dysplasia.
a Hematological parameters from 3-month-old WT (n = 28), Zrsr2m/m (n = 21), Tet2−/− (n = 12), and Zrsr2m/mTet2−/− mice (n = 26). WBC: white blood cells, RBC: red blood cells, HGB: hemoglobin, PLT: platelets. Boxes show mice presenting cytopenias. b Peripheral blood smears and bone marrow cytospins from 3-months-old WT and Zrsr2m/mTet2−/− mice stained with May-Grünwald Giemsa. Normal RBC from WT (Ai) and dysplastic RBC from Zrsr2m/mTet2−/− mice (Aii) showing RBC anisocytosis, polychromasia, and presence of nucleated erythrocytes. Normal platelets from WT (Bi) and dysplastic platelets from Zrsr2m/mTet2−/− mice (Bii) showing anisocytosis and giant size. Normal neutrophil from WT (Ci), hypersegmented neutrophil (Cii) and hyposegmented neutrophils with pseudo Pelger-Huët anomalies (Di–Dii) from Zrsr2m/mTet2−/− mice. Nuclear fragmentation and karyorrhexis of granulocytes (Ei-Eii). Erythrophagocytosis (Fi) and binucleated erythroid precursors (Fii) in BM of Zrsr2m/mTet2−/− mice. Normal myeloid precursors from WT (Gi) and dysplastic myeloid precursors with cytoplasm vacuolization from Zrsr2m/mTet2−/− mice (Gii). Normal megakaryocyte in WT (Hi) and giant megakaryocyte in Zrsr2m/mTet2−/− mice (Hii). Dysplastic features are indicated with arrows.
Fig. 4. Concomitant Zrsr2 mutation and Tet2 loss result in splenomegaly with extramedullary hematopoiesis.
a Left panel: Spleen weight from 3-months-old WT (n = 25), Zrsr2m/m (n = 21), Tet2−/− (n = 9), and Zrsr2m/mTet2−/− (n = 24) mice. Right panel: Spleen sections from 3-months-old WT (n = 3), Zrsr2m/m (n = 2), Tet2−/− (n = 3), and Zrsr2m/mTet2−/− (n = 6) mice stained with H&E. The arrow indicates presence of megakaryocytes with dysplastic changes. b Percentage of erythroid (Ter119+) and B cells (B220+) in spleen of 3-months-old WT (n = 6), Zrsr2m/m (n = 5), Tet2−/− (n = 9), and Zrsr2m/mTet2−/− (n = 7) mice. c GM-CFU and BFU-E forming capacity of spleen cells from 3-month-old WT (n = 5), Zrsr2m/m (n = 3), Tet2−/− (n = 5), and Zrsr2m/mTet2−/− (n = 6) mice. Data represent mean ± SEM.
Since MDS is a disease of the elderly, we also analyzed the impact of mutations in aged control (24 months), Zrsr2m/m (24 months), and Zrsr2m/mTet2−/− mice (18 months). Old Zrsr2m/m mice presented anemia and one out three cases had leukocytosis, thrombocytopenia, and splenomegaly with disrupted spleen histology. Zrsr2m/mTet2−/− mice displayed anemia and thrombocytopenia (Supplementary Fig. 3a), and exhibited erythroid and myeloid dysplasia (Supplementary Fig. 3b). This was accompanied by splenomegaly with disrupted splenic architecture (Supplementary Fig. 3c, d). Notably, aged Zrsr2m/mTet2−/− mice were analyzed at earlier time-points (18 months) due to evident signs of illness (reduced mobility, hunched posture, ruffled fur, weight loss, and white foot paws). Interestingly, 1 out of 6 aged Zrsr2m/mTet2−/− mice showed signs suggestive of progression to secondary AML, such as an increase of blasts in PB smears and myeloperoxidase (MPO) staining in spleen and liver sections (Supplementary Fig. 3e–g).
Zrsr2 mutation and Tet2 loss in HSPC enhance BM repopulation capacity and cause cell-autonomous hematopoietic alterations
Since there is evidence of HSPC involvement in MDS, we evaluated hematopoietic stem cell function in Zrsr2 mutant and Zrsr2m/mTet2−/− mice. We performed transplantation assays using 8000 LSK cells (CD45.2) along with 2 × 105 support unfractionated bone marrow cells (CD45.1) (Fig. 5a). 8–12 weeks after transplantation, the reconstitution of the recipient (CD45.1) hematopoietic system by the donor cells was verified by flow cytometric analysis of the peripheral blood. We observed that Zrsr2m/mTet2−/− cells displayed an increased repopulation capacity and retained multi-lineage reconstitution ability (Supplementary Fig. 4a). In the donor-derived CD45.2+ fraction, we detected an increase in erythroid-megakaryocytic biased MPP2 progenitors and a decrease in PreMegE, suggestive of a differentiation block along the erythroid lineage at an earlier step (Supplementary Fig. 4b, c). As occurred in steady-state, we also observed an accumulation of PreGM progenitors and a decrease in GMP (Supplementary Fig. 4c). LT-HSC were reduced, suggesting a push of LT-HSC towards more committed HSPC in a transplantation context. Further, Zrsr2m/mTet2−/− mice presented cytopenias (anemia and thrombocytopenia) (Fig. 5b) and myeloid and erythroid dysplasia (Fig. 5c). Remarkably, transplanted Zrsr2m/mTet2−/− animals were moribund at the time of sacrifice (15 months post-transplantation).
Fig. 5. Combined Zrsr2-Tet2 mutations in HSPC cause cell-autonomous hematopoietic alterations.
a Experimental design for transplantation. Lethally irradiated CD45.1 recipient mice were co-transplanted with 8.000 LSK (CD45.2, isolated from each genotype) and 200.000 support unfractionated bone marrow (CD45.1) cells. Peripheral blood was analyzed monthly by flow cytometry after transplantation. b Hematological parameters in transplanted animals at 15 months post-transplantation. WT (n = 7), Zrsr2m/m (n = 5), and Zrsr2m/mTet2−/− (n = 2). Boxes show mice presenting cytopenias. c May-Grünwald Giemsa staining of peripheral blood smears and bone marrow cytospins in transplanted animals at 15 months post-transplantation. Normal RBC from WT (Ai) and dysplastic RBC from Zrsr2m/mTet2−/− (Aii), showing RBC anisocytosis, polychromasia, and a nucleated RBC. Normal platelets from WT (Bi) and giant platelet from Zrsr2m/mTet2−/− mice (Bii). Hyposegmented granulocytes (Ci-Cii) and granulocytes with nuclear fragmentation and karyorrhexis (Di–Dii) from Zrsr2m/mTet2−/− mice. Normal progenitors from WT (Ei) and binucleated erythroid precursors from Zrsr2m/mTet2−/− mice (Eii). Megakaryocyte from WT (Fi) and enlarged megakaryocyte from Zrsr2m/mTet2−/− mice (Fii). (A-F) Data represent mean ± SEM and come from three independent experiments.
Whole transcriptome sequencing in HSPC shows an alteration of lineage-specific hematopoietic genes and an enrichment of inflammatory pathways
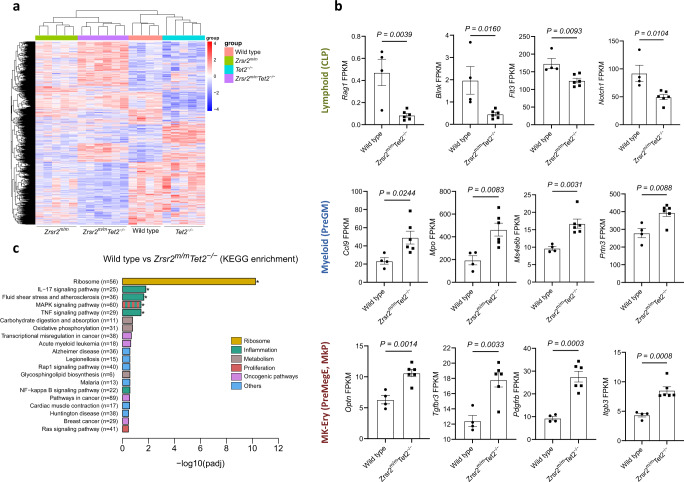
Next, we performed RNA sequencing experiments to get insight into the molecular changes of Zrsr2m/mTet2−/− HSPC. Transcriptome analysis resulted in distinct gene expression signatures (Fig. 6a). Differential gene expression analysis in pair-wise comparisons against WT control by DESEq2 [24] yielded 571 differentially expressed genes (DEG) in Zrsr2 mutant, 1203 DEG in Tet2−/− and 2952 DEG in Zrsr2m/mTet2−/− LSK cells (adjusted P-value ≤ 0.05). To evaluate the impact of the combined dysfunction of Zrsr2 and Tet2 on DNA methylation, we performed global epigenomic profiling of WT, Tet2−/−, and Zrsr2m/mTet2−/− mice using Infinium Mouse Methylation Bead Chip array (285K, Illumina). Principal component analyses (PCA) revealed that Zrsr2m/mTet2−/− and Tet2−/− hematopoietic cells had distinct epigenomic profiles from those of WT (Supplementary Fig. 5a). We observed that 8747 CpG, corresponding to 531 murine genes, were found differentially methylated in Zrsr2m/mTet2−/− cells in comparison to WT cells (Supplementary Table 2). Next, we performed a Venn diagram analysis to identify the overlapping genes in both transcriptomic and epigenomic analyses. We detected that circa 3% of DEG presented alterations in DNA methylation levels (Supplementary Fig. 5b, Supplementary Table 4a). Within Zrsr2m/mTet2−/− LSK, 1327 genes were up-regulated and 1625 genes were down-regulated. Molecularly, Zrsr2m/mTet2−/− LSK were primed towards a myeloid fate since transcripts related to common lymphoid progenitors (Rag1, Blnk, Flt3) and the Notch1 pathway were down-regulated (Fig. 6b) while transcripts associated to preGM (Ccl9, Mpo, Ms4a6b, Prtn3) (Fig. 6b), preMegE (Optn, Tgfbr3), and MkP (Pdgfrb, Itgb3) [33] were up-regulated (Fig. 6b). Interestingly, gene expression changes in Blnk, Mpo, Tgfbr3 are potentially deregulated by DNA methylation changes (Supplementary Table 4b). This suggests that myeloid and megakaryocyte-erythroid priming originates in the immature HSPC in concordance with the increased proportion of erythroid-megakaryocytic biased progenitors (MPP2) within the LSK compartment. Enrichment analysis of DEG using GO indicated that genes up-regulated in Zrsr2m/mTet2−/− LSK were associated to ribosome, inflammation, and cell migration/motility processes (Supplementary Fig. 5c). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis [26] in Zrsr2m/mTet2−/− LSK showed up enrichment in ribosome function and pro-inflammatory cytokine pathways, such as those mediated by IL-17, TNF-α, and mitogen-activated protein kinase (MAPK) signaling (Fig. 6c). MDS is frequently associated with deregulated inflammatory signaling profiles in BM [34]. On this basis, we conducted a cytokine multiplex assay and found that mice displaying a more aggressive MDS phenotype presented higher pro-inflammatory cytokines levels (IFN-γ, IL-6, IL-10, and TNF-α), suggesting a contributing role of inflammation in Zrsr2m/mTet2−/− MDS phenotype (Supplementary Fig. 6).
Fig. 6. Gene expression dysregulation in Zrsr2m/mTet2−/− LSK cells leads to transcriptional myeloid priming and enrichment of inflammatory pathways.
a Heatmap visualizing the differentially expressed genes (DEG) identified in WT, Zrsr2m/m, Tet2−/−, and Zrsr2m/mTet2−/− LSK cells from 3-months-old mice (red, up-regulated genes; blue, down-regulated genes). n = 4-6 replicates/genotype. b Gene expression levels of CLP, PreGM, and PreMegE and MkP associated genes. c KEGG enrichment analysis of DEG in LSK from WT vs Zrsr2m/mTet2−/− mice.
Aberrant mRNA splicing in Zrsr2m/mTet2−/− mice target the MAPK protein family
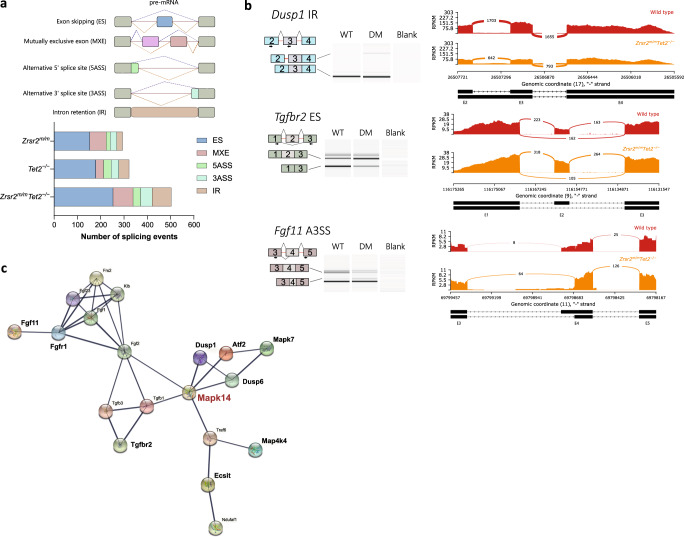
To determine alternative splicing patterns in WT, Zrsr2m/m, Tet2−/−, and Zrsr2m/mTet2−/− LSK cells we employed rMATS [25]. Alternative spliced events were categorized into five major types: skipped exon (SE), MXE, A5SS, A3SS, and RI. Using pair‐wise comparisons against WT control, we identified 294 significant differentially spliced events in Zrsr2m/m, 322 in Tet2−/−, and 503 in Zrsr2m/mTet2−/− (P value ≤ 0.01, FDR ≤ 0.05) (Fig. 7a, Supplementary Table 3). We found that 253 alternative spliced genes were uniquely present in Zrsr2m/mTet2−/− cells, implying that the combined alteration of Zrsr2 and Tet2 cooperatively promoted mis-splicing of these transcripts (Supplementary Fig. 7a). KEGG pathway enrichment analysis was performed on differentially spliced genes (Supplementary Fig. 7b). We found that the MAPK kinase family and the Fanconi anemia pathway were the most affected targets by mRNA aberrant splicing in Zrsr2m/mTet2−/−. Importantly, a total of nine genes related to the MAPK pathway were identified as mis-spliced, from which we validated 3, Dusp1, Tgfbr2, and Fgf11 (Fig. 7b). Strikingly, we observed that these targets converged into a particular kinase, the MAPK14 (Fig. 7c). We also identified and corroborated exon skipping events in Per1 and Ttbk2 (in Zrsr2m/mTet2−/−), and Frrs1 (in Zrsr2m/m) transcripts, involved in circadian regulation, cell migration/motility, and iron metabolism, respectively (Supplementary Fig. 7c). Remarkably, mis-splicing of these targets was also found in human ZRSR2 and ZRSR2-TET2 co-mutated samples (unpublished observations).
Fig. 7. Aberrant alternative splicing in Zrsr2m/mTet2−/− LSK cells targets the MAPK pathway.
a Number of significant aberrant splicing events in Zrsr2m/m, Tet2−/−, and Zrsr2m/m Tet2−/− LSK cells (P value ≤ 0.01, FDR ≤ 0.05). b Validation of relevant aberrant splicing events identified by RNA-seq using RT- PCR. Sashimi plots showing an IR event in Dusp1, an ES event in Tgfbr2, and an A3SS event in Fgf11. c Protein-protein interactions among MAPK-related targets were retrieved from STRING. Line thickness represents interaction confidence and edges indicate both functional and physical protein associations. Relevant mis-spliced targets identified in this study (in bold) converge in the MAPK14 (red).
Discussion
Here we show that mutations of Zrsr2 and Tet2, genes commonly co-mutated in human MDS, resulted in impairment of myelo-erythroid differentiation and the development of thrombocytopenia and anemia, multi-lineage dysplasia, and splenomegaly with extramedullary hematopoiesis. These features, manifested with incomplete penetrance, are compatible with MDS [32]. Further, Zrsr2 and Tet2 mutations were associated with an expansion of LT-HSC and MPP2 compartments and an increase of CFUs, with CFU-GM cells acquiring self-renewal abilities. Moreover, the phenotype observed in steady-state mice was reproduced in transplanted mice, thus indicating that these alterations were cell-autonomous. Altogether, Zrsr2 and Tet2 mutations cause a phenotype that recapitulates critical features of myelodysplasia.
Prior studies exploring in vivo Zrsr2 function in hematopoiesis provided somewhat contrasting results. Zrsr2 constitutive knockout mice exhibited normal myeloid development and regular repopulating ability [6]. Data from mice with hematopoietic-specific conditional deletion of Zrsr2 showed that Zrsr2 loss promotes HSC self-renewal [7]. In our study, young adult mice expressing truncated ZRSR2 (Zrsr2m/m) exhibited normal myeloid development and regular repopulating ability, similar to the phenotype described in Zrsr2 constitutive knockout [6]. However, old Zrsr2m/m mice did display several hematological alterations, such as reduced RBC and hemoglobin levels akin to patients with isolated ZRSR2 mutations showing refractory macrocytic anemias [35]. One aspect for which our model differs from previous studies is that we did not generate a knockout but a stable truncated protein. Many Zrsr2 mutations observed in the clinics are likely analogous to the Zrsr2 mutant allele that we describe here in that they generate a loss of the C-terminal region [3]. Other models of MDS based on mutations in recurrently mutated genes have been described. Sf3b1+/K700ETet2−/− mice [18] showed similar characteristics to the ones we report here, such as anemia, expansion of the LT-HSC compartment, and erythroid and megakaryocyte dysplasia. Other studies recapturing MDS/MPN-like disease also showed splenomegaly with extramedullary hematopoiesis [36]. Loss of Tet2 itself promotes myeloid-biased hematopoiesis and HSC self-renewal [15]. Correspondingly, we show that concurrent deletion of Tet2 enhanced myeloid-biased differentiation, as reflected by increased MPP2 progenitors and increased myelo-erythroid lineage priming in LSK HSPC. Further, Tet2 loss confers growth advantage and self-renewal capacity to Zrsr2 mutant progenitors, as evidenced by clonogenic and serial replating assays. This increased output of colonies has also been shown in an Asxl1−/−Tet2−/− model [37]. In the transplantation setting, Zrsr2m/mTet2−/− cells showed higher repopulation capacity than Zrsr2 mutant and WT cells, in accordance with the clonal advantage typical of MDS HSC. In addition, we observed an increased reconstitution to megakaryocytic-erythroid biased MPP2 progenitors, LSK, and PreGM in mice transplanted with Zrsr2m/mTet2−/− cells. Similarly, previous work in bone marrow chimeras transplanted with Zrsr2 and Tet2 double knockout cells reported an increase in LSK and myeloid progenitors (CMP, GMP, and MEP) [8].
Deregulated DNA methylation is one of the hallmarks of MDS [38], although in our study only circa 3% of genes that were differentially expressed in Zrsr2m/mTet2−/− LSK may potentially be regulated by an epigenetic mechanism such as DNA methylation. Importantly, transcriptional profiling followed by GO enrichment analysis indicated that genes upregulated in Zrsr2m/mTet2−/− LSK were associated with ribosome, inflammation, and cell migration/motility. Interestingly, alterations in ribosome biogenesis are associated with MDS and defective erythropoiesis [39, 40]. On the other hand, deregulation of inflammatory signaling plays a central role in the pathogenesis of MDS [34]. In fact, MDS patients present higher levels of pro-inflammatory cytokines in serum [41]. In this regard, splicing factor mutations and Tet2 deficiency have been associated with increased inflammatory cytokine production [42, 43]. Cytokine multiplex assay reveals that Zrsr2m/mTet2−/− mice with more aggressive MDS phenotype display higher levels of pro-inflammatory cytokines. This suggests a possible role of inflammation favoring MDS.
Since mutations in ZRSR2 alter RNA splicing and HSC are the cell-of-origin of MDS, we performed an alternative splicing analysis in LSK cells. Unexpectedly, this analysis showed that exon skipping but not intron retention was the category most affected in Zrsr2 mutant mice. This contrasts with most of the literature associating ZRSR2 to intron retention of U12-type introns [4–8]. However, our data are in agreement with one study reporting that ZRSR2 mutations in humans cause more exon skipping than intron retention [44]. One of the most affected pathways by mis-splicing in Zrsr2m/mTet2−/− LSK cells was MAPK signaling. Among the nine genes that were found to be aberrantly spliced in this pathway, we confirmed Dusp1, Tgfbr2, and Fgf11. Interestingly, we found that these MAPK-related targets converged into MAPK14, an essential kinase for definitive erythropoiesis in mice [45]. In addition, MAPK14 has a key role in regulating pro-inflammatory cytokine production via MK2 and MSK1/2. Aberrant mRNA splicing in this pathway, thus, may deregulate MAPK14 function and explain, in combination with other transcriptional changes, the level of pro-inflammatory cytokines found in Zrsr2m/mTet2−/− mice. This finding coincides with previous work showing MAPK14 as a down-regulated target downstream of ZRSR2 mutation [6].
Collectively, these data provide a unique insight into the mechanisms by which Zrsr2 and Tet2 mutations affect MDS pathogenesis. Our results indicate that concomitant mutations in Zrsr2 and Tet2 can initiate MDS in mice. We identify alterations in HSC function and myelo-erythroid differentiation as well as transcriptional and splicing changes in relevant biological pathways (MAPK family, Fanconi Anemia pathway) that may contribute to MDS tumorigenesis. Given these findings, MAPK modulators, anti-inflammatory agents, and drugs aiming to restore normal myelo-erythroid differentiation may be tested in our model. This is particularly important given the unmet clinical need to find new curative approaches beyond HSPC transplantation. Since co-occurrence of ZRSR2 and TET2 mutations have also been observed in BPDCN, our model could also be exploited to explore the biology of dendritic leukemia. Overall, this study expands on mouse models based on recurrent mutations in splicing and epigenetic regulators and offers an opportunity to elucidate further the mechanisms governing tumorigenesis in minor spliceosome-mutated MDS.
Supplementary information
Acknowledgements
The authors would like to thank Dr. Francisco Vera and Almudena Vidal for their assistance with histology and Dr. Leonor Senent for the help in cytomorphology evaluation. We thank the Cytomics Unit, Instituto de Investigación Sanitaria La Fe (IIS La Fe) for the technical support. This investigation was supported by the grant SAF2017-82171-R to ASP from the Spanish Ministry of Economy, Industry, and Competitiveness. CGR is founded by JAP Valencia AECC predoctoral fellowships. CMV is sponsored by a predoctoral fellowship from Generalitat Valenciana (ACIF/2018/255). AL is funded by Generalitat Valenciana (APOSTD/2021/212). AGA was funded by grant RTI2018-093548-B-I00 from the Spanish Ministry of Science and Innovation. ASP is supported by the Ramón y Cajal Program (RYC2015-17534).
Author contributions
ASP designed the study; CGR and ASP designed experiments; CGR and CMV performed experiments; AGA, RFG and EP generated the mouse lines; CGR, CMV, LC, JS, and ASP analyzed data; AL, JC, JS and AGA performed critical reading of the manuscript; CGR and ASP wrote the manuscript; ASP supervised the study.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41375-022-01674-2.
References
- 1.Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–27. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen H, Zheng X, Luecke S, Green MR. The U2AF35-related protein Urp contacts the 3’ splice site to promote U12-type intron splicing and the second step of U2-type intron splicing. Genes Dev. 2010;24:2389–94. doi: 10.1101/gad.1974810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–9. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 4.Madan V, Kanojia D, Li J, Okamoto R, Sato-Otsubo A, Kohlmann A, et al. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat Commun. 2015;6:6042. doi: 10.1038/ncomms7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madan V, Li J, Zhou S, Teoh WW, Han L, Meggendorfer M, et al. Distinct and convergent consequences of splice factor mutations in myelodysplastic syndromes. Am J Hematol. 2020;95:133–43. doi: 10.1002/ajh.25673. [DOI] [PubMed] [Google Scholar]
- 6.Madan V, Cao Z, Teoh WW, Dakle P, Han L, Shyamsunder P, et al. ZRSR1 cooperates with ZRSR2 in regulating splicing of U12-type introns in murine hematopoietic cells. Haematologica. 2021;107:680–9. doi: 10.3324/haematol.2020.260562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inoue D, Polaski JT, Taylor J, Castel P, Chen S, Kobayashi S, et al. Minor intron retention drives clonal hematopoietic disorders and diverse cancer predisposition. Nat Genet. 2021;53:707–18. doi: 10.1038/s41588-021-00828-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Togami K, Chung SS, Madan V, Booth CAG, Kenyon CM, Cabal-Hierro L, et al. Sex-biased ZRSR2 mutations in myeloid malignancies impair plasmacytoid dendritic cell activation and apoptosis. Cancer Discov. 2022;12:522–41. doi: 10.1158/2159-8290.CD-20-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28:241–7. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Awada H, Nagata Y, Goyal A, Asad MF, Patel B, Hirsch CM, et al. Invariant phenotype and molecular association of biallelic TET2 mutant myeloid neoplasia. Blood Adv. 2019;3:339–49. doi: 10.1182/bloodadvances.2018024216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;377:111–21. doi: 10.1056/NEJMoa1701719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016;30:733–50. doi: 10.1101/gad.276568.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, et al. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. PNAS. 2011;108:14566–71. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Cai X, Cai C-L, Wang J, Zhang W, Petersen BE, et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509–18. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quivoron C, Couronné L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Tara S, Isshiki Y, Nakajima-Takagi Y, Oshima M, Aoyama K, Tanaka T, et al. Bcor insufficiency promotes initiation and progression of myelodysplastic syndrome. Blood. 2018;132:2470–83. doi: 10.1182/blood-2018-01-827964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Obeng EA, Chappell RJ, Seiler M, Chen MC, Campagna DR, Schmidt PJ, et al. Physiologic expression of Sf3b1(K700E) causes impaired erythropoiesis, aberrant splicing, and sensitivity to therapeutic spliceosome modulation. Cancer Cell. 2016;30:404–17. doi: 10.1016/j.ccell.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gómez-Redondo I, Pericuesta E, Navarrete-Lopez P, Ramos-Ibeas P, Planells B, Fonseca-Balvís N, et al. Zrsr2 and functional U12-dependent spliceosome are necessary for follicular development. iScience. 2022;25:103860. doi: 10.1016/j.isci.2022.103860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martínez‐valiente C, Garcia‐ruiz C, Rosón B, Liquori A, González‐romero E, Fernández‐gonzález R, et al. Aberrant alternative splicing in u2af1/tet2 double mutant mice contributes to major hematological phenotypes. Int J Mol Sci. 2021;22:6963. doi: 10.3390/ijms22136963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen S, Park JW, Lu Z, Lin L, Henry MD, Wu YN, et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. PNAS. 2014;111:E5593–601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–13. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–21. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 29.Pietras EM, Reynaud D, Kang YA, Carlin D, Calero-Nieto FJ, Leavitt AD, et al. Functionally distinct subsets of lineage-biased multipotent progenitors control blood production in normal and regenerative conditions. Cell Stem Cell. 2015;17:35–46. doi: 10.1016/j.stem.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oguro H, Ding L, Morrison SJ. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 2013;13:102–16. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pronk CJH, Rossi DJ, Månsson R, Attema JL, Norddahl GL, Chan CKF, et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–42. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Zhou T, Kinney MC, Scott LM, Zinkel SS, Rebel VI. Revisiting the case for genetically engineered mouse models in human myelodysplastic syndrome research. Blood. 2015;126:1057–68. doi: 10.1182/blood-2015-01-624239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grover A, Mancini E, Moore S, Mead AJ, Atkinson D, Rasmussen KD, et al. Erythropoietin guides multipotent hematopoietic progenitor cells toward an erythroid fate. J Exp Med. 2014;211:181–8. doi: 10.1084/jem.20131189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sallman DA, List A. The central role of inflammatory signaling in the pathogenesis of myelodysplastic syndromes. Blood. 2019;133:1039–48. doi: 10.1182/blood-2018-10-844654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fleischman RA, Stockton SS, Cogle CR. Refractory macrocytic anemias in patients with clonal hematopoietic disorders and isolated mutations of the spliceosome gene ZRSR2. Leuk Res. 2017;61:104–7. doi: 10.1016/j.leukres.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 36.Muto T, Sashida G, Oshima M, Wendt GR, Mochizuki-Kashio M, Nagata Y, et al. Concurrent loss of Ezh2 and Tet2 cooperates in the pathogenesis of myelodysplastic disorders. J Exp Med. 2013;210:2627–39. doi: 10.1084/jem.20131144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abdel-Wahab O, Gao J, Adli M, Dey A, Trimarchi T, Chung YR, et al. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J Exp Med. 2013;210:2641–59. doi: 10.1084/jem.20131141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bravo GM, Lee E, Merchan B, Kantarjian HM, García-Manero G. Integrating genetics and epigenetics in myelodysplastic syndromes: advances in pathogenesis and disease evolution. Br J Haematol. 2014;166:646–59. doi: 10.1111/bjh.12957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galili N, Qasim SA, Raza A. Defective ribosome biogenesis in myelodysplastic syndromes. Haematologica. 2009;94:1336–8. doi: 10.3324/haematol.2009.012021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khajuria RK, Munschauer M, Ulirsch JC, Fiorini C, Ludwig LS, McFarland SK, et al. Ribosome levels selectively regulate translation and lineage commitment in human hematopoiesis. Cell. 2018;173:90–103. doi: 10.1016/j.cell.2018.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verhoef GEG, De Schouwer P, Ceuppens JL, Van Damme J, Goossens W, Boogaerts MA. Measurement of serum cytokine levels in patients with myelodysplastic syndromes. Leukemia. 1992;6:1268–72. [PubMed] [Google Scholar]
- 42.Pollyea DA, Harris C, Rabe JL, Hedin BR, De Arras L, Katz S, et al. Myelodysplastic syndrome-associated spliceosome gene mutations enhance innate immune signaling. Haematologica. 2019;104:e388–92. doi: 10.3324/haematol.2018.214155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cull AH, Snetsinger B, Buckstein R, Wells RA, Rauh MJ. Tet2 restrains inflammatory gene expression in macrophages. Exp Hematol. 2017;55:56–70. doi: 10.1016/j.exphem.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 44.Pellagatti A, Armstrong RN, Steeples V, Sharma E, Repapi E, Singh S, et al. Impact of spliceosome mutations on RNA splicing in myelodysplasia: dysregulated genes/pathways and clinical associations. Blood. 2018;132:1225–40. doi: 10.1182/blood-2018-04-843771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamura K, Sudo T, Senftleben U, Dadak AM, Johnson R, Karin M. Requirement for p38α in erythropoietin expression: a role for stress kinases in erythropoiesis. Cell. 2000;102:221–31. doi: 10.1016/s0092-8674(00)00027-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.