Abstract
Two genes encoding Na+-ATPases from Debaryomyces hansenii were cloned and sequenced. The genes, designated ENA1 from D. hansenii (DhENA1) and DhENA2, exhibited high homology with the corresponding genes from Schwanniomyces occidentalis. DhENA1 was expressed in the presence of high Na+ concentrations, while the expression of DhENA2 also required high pH. A mutant of Saccharomyces cerevisiae lacking the Na+ efflux systems and sensitive to Na+, when transformed with DhENA1 or DhENA2, recovered Na+ tolerance and also the ability to extrude Na+.
Two strategies for adapting to the presence of high salt have-been identified in different organisms. The best-known mechanism is to exclude sodium from the cytoplasm and to accumulate high concentrations of compatible solutes to avoid water loss (excluder organisms), while some organisms use and accumulate high Na+ concentrations without becoming intoxicated (includer organisms) (26).
Saccharomyces cerevisiae is an excluder yeast which keeps cytoplasmic sodium at low levels by extruding the cation out of the cell or driving it into the vacuole. Nha1 and Ena1-4 mediate sodium efflux processes. Nha1 works as a Na+ (K+)/H+ antiporter (4, 23), and Ena proteins are P-type ATPases (5, 9, 11, 28). Deletion of the corresponding genes renders the cells sensitive to sodium, specially in the case of ENA1 (11). In addition, Nhx1 drives sodium into the vacuole in exchange with H+ (17). Schwanniomyces occidentalis and Schizosaccharomyces pombe are also excluder yeasts. While two ENA genes have been identified in S. occidentalis (3), in the case of the fission yeast, which is more sensitive to external sodium, only a Na+/H+ antiporter (Sod2) has been found (10, 13). Finally, in Zygosaccharomyces rouxii, an osmotolerant yeast, there is evidence of the existence of both, two antiporters (ZSOD2 and ZSOD22) (12, 19) and one ATPase (ZrENA) (unpublished data) (GenBank TM/EMBL accession number D78567).
Debaryomyces hansenii can be isolated from salty environments (14, 32). More than 30 years ago, it was reported that among several marine yeasts, D. hansenii was the least affected by high concentrations of NaCl (21, 27). D. hansenii accumulates high amounts of Na+, and in this yeast, Na+ is not more toxic than K+ (18, 24, 30). More recently it was shown that Na+ improves the performance of D. hansenii under different stress conditions (1). Therefore, we have proposed that D. hansenii behaves as a Na+ includer yeast and that it may be considered a halophilic yeast, in particular at low K+ concentrations. In any case, a role for glycerol, as a compatible solute, must be reserved (2, 20), since a glycerol/Na+ symporter with homeosmotic function in this yeast has been described (15, 16). In addition, biochemical work has shown the existence of sodium efflux processes that may be involved in sodium tolerance (for a review, see reference 26). There is no specific molecular information on the processes involved in such good performance in the presence of salt and in sodium fluxes in D. hansenii. This information would be essential in the understanding of the molecular basis of halotolerance and halophilism in yeasts.
Effect of pH on the Na+ efflux in D. hansenii.
The existence of Na+ efflux processes in D. hansenii has been previously reported (22, 24, 30). The mechanisms described may involve Na+-ATPases or Na+/H+ antiporters. While antiporters can be strongly affected by pH, the activity of Na+-ATPases is constant over a wide range of pH. As a first approach to investigate the nature of the system(s) involved in Na+ extrusion, we measured the effect of pH on this process. D. hansenii (strain PYCC 2968) was grown in YNB medium (Difco), and cells were harvested at the early exponential growth phase. Cells were then loaded with 200 mM NaCl over 3 h, resulting in an increase in Na+ content from 77 to 290 nmol/mg (dry weight) of cells. Cells were resuspended in assay buffer: Tris-citrate (10 mM Tris brought to pH 3.5 with citric acid), MES [10 mM MES (2-morpholinoethanesulfonic acid) brought to pH 5.5 with Ca(OH)2], or TAPS {10 mM TAPS [N-Tris(hydroxymethyl)methyl-3-aminopropanesulfonic acid] brought to pH 7.5 with Ca(OH)2}. Tris buffer was supplemented with CaCl2 (1 mM), and all buffers also contained 0.1 mM MgCl2 and 2% (wt/vol) glucose. The efflux process was studied as described previously (24), and results indicate that it was not significantly affected by pH in the range from 3.5 to 7.5 (data not shown). This was an indication that under the conditions tested, a Na+/H+ antiport was not the main mechanism involved. This idea is supported by the observation that D. hansenii can grow at high pH (7.8) and high Na+ concentration (1.5 M) (1). Under these conditions, the activity of a Na+ ATPase should be required in order to extrude the cation in the absence of favorable Na+ or pH gradients.
Cloning and sequence analysis of ENA genes in D. hansenii.
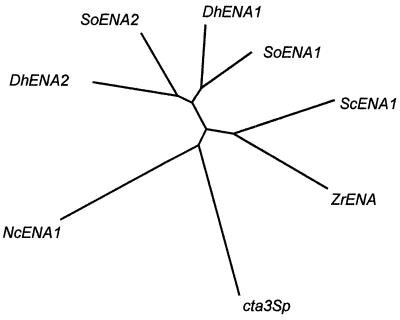
Taking into account the previous result and the similarity of the kinetics of Na+ extrusion in D. hansenii and in S. cerevisiae (24), we investigated the existence of ENA-type genes in D. hansenii by a PCR approach. Manipulation of nucleic acids was performed by standard protocols (29) or by following the manufacturer's instructions. We used degenerated primers designed from conserved sequences of Na+- and Ca2+-ATPase genes (6). Using two of these primers corresponding to the conserved amino acids, CSDK (as the sense primer) and DDNFASSI (as the antisense primer), we amplified from genomic DNA of D. hansenii two fragments of 1.0 and 1.2 kb. In order to obtain the complete sequence of the cDNAs, we extended the sequences up to the 5′ end by reverse transcription-PCR using a 5′3′RACE kit (Boehringer) and to the 3′ end by reverse transcription-PCR using first an anchored Not-dT18 commercial primer for reverse transcription and then an antisense anchor primer and a sense specific primer for the PCR amplification of each cDNA. Full-length cDNAs of 3,309 and 3,219 bp, respectively, were obtained. Analysis of their translated sequences showed 75 and 73% identity with SoEna1 and SoEna2, respectively. Figure 1 shows the phylogenetic relationships between the different fungal Na+-ATPases. It is interesting that cta3Sp, a putative calcium transporter, appears to be closely related to the Ena proteins.
FIG. 1.
Phylogenetic tree of Na+- and Ca2+-ATPases from fungi. Alignment of the sequences was performed with the CLUSTALX program (31) using the core sequences starting at the beginning of the f and ending at the end of the j conserved regions (28). Accession numbers are as follows: ScENA1, X67136; ZrENA, D78567; cta3Sp, P22189; NcENA1, AJ243520; DhENA2, AF263248; SoENA2, AF030861; DhENA1, AF247561; SoENA1, AF030860.
On the basis of sequence homologies, the corresponding genes were designated ENA1 of D. hansenii (DhENA1) and DhENA2. Southern blot analyses revealed that the probes prepared for DhENA1and DhENA2 were specific and hybridized only with their own genes and with total DNA from D. hansenii.
Regulation of transcription of DhENA1 and DhENA2.
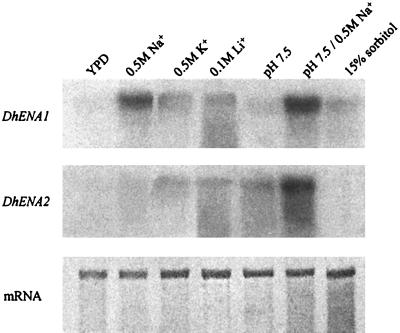
In order to perform Northern blot analyses, cells were grown in YPD medium (1% [wt/vol] yeast extract, 2% [wt/vol] peptone, and 2% [wt/vol] glucose) (pH 5.8) at 28°C and harvested at the early exponential phase. Total RNA was prepared from D. hansenii cells incubated for 2 h under different conditions (Fig. 2) which were chosen by taking into account previous data. The concentration of 0.5 M NaCl improves the performance of D. hansenii (1). KCl and sorbitol were used at concentrations equivalent to that of NaCl. The selected concentration of LiCl was lower (100 mM) because Li+ is toxic for D. hansenii (24). Finally, a high pH was selected because it regulates the expression of ENA genes in other yeasts (3). Total RNA purification and hybridizations were performed as described previously (3).
FIG. 2.
Northern blot analyses of DhENA1 and DhENA2 transcripts in D. hansenii cells grown under the conditions indicated at the top of the figure. Total RNA was fractionated, transferred to a nylon membrane, and probed. Filters were stripped and stained with methylene blue as a loading control (bottom).
Northern blot analyses showed that DhENA1 and DhENA2 were differentially expressed. While the expression of DhENA1 was mainly dependent on the presence of high Na+ concentrations, high expression of DhENA2 required high Na+ concentrations together with high pH. This is not surprising, since these two factors are present in natural habitats of D. hansenii, such as marine water. K+ and Li+ triggered poor expression of both genes. When the cells were incubated with 15% (wt/vol) sorbitol, the expression of the genes was also poor, even at pH 7.5 (data not shown). This observation eliminated the hypothesis that osmotic factors were involved in the regulation of expression. Similar results were obtained when the cells were grown under the same conditions, instead of having been incubated for 2 h (not shown).
Functional expression of DhENA1 and DhENA2 in S. cerevisiae.
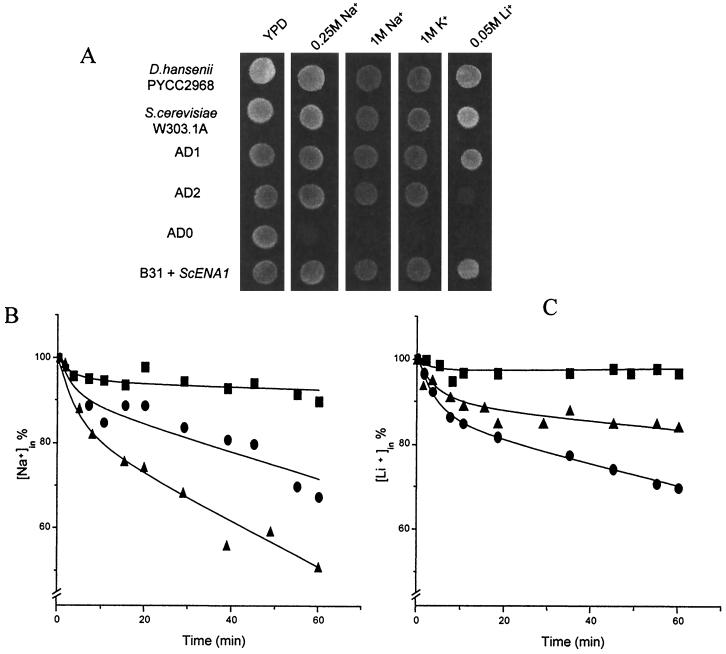
DhENA1 and DhENA2 were inserted in pYPGE15 after the PGK1 promoter (8). A strain of S. cerevisiae (B31) (4) lacking all Na+ efflux systems was transformed with the plasmid without any insert (strain AD0) or with the plasmid containing the DhENA1 or DhENA2 gene. Growth of several transformants was assessed in plates with 0.5 M NaCl, in which the performance of strains carrying DhENA1 and DhENA2 genes was similar. From these, two were selected to study heterologous expression in more detail. The two transformants were named AD1, carrying DhENA1, and AD2, carrying DhENA2. Salt tolerance was studied in these strains. Growth experiments were carried out in solid medium (YPD) with Na+, K+, or Li+ (Fig. 3A). For comparison, growth of S. cerevisiae (wild type and AD0 strain) and of D. hansenii is also shown. Strain AD0 did not grow in the presence of the cations tested. The strains carrying any of the genes were able to grow in plates with 1 M NaCl after 60 h of incubation. It is worth mentioning that Na+ tolerance in both strains was at the same level as in the wild strain of S. cerevisiae but never reached the level in D. hansenii, which grew well in the presence of 2 M NaCl (data not shown). The tolerance to Li+ was only slightly increased by DhENA2 but was clearly improved by DhENA1, suggesting a selective role of both genes in the process of tolerance to cations. As in the case of Na+, the presence of any of the ENA genes increased the capacity to grow at high K+ concentrations and with the same level of tolerance as the wild strain of S. cerevisiae. In order to compare the efficiency of Ena1 from D. hansenii and from S. cerevisiae, strain B31 was also transformed with ScENA1 under the control of the same promoter. Tolerance tests showed that this strain behaved like AD1 (Fig. 3A).
FIG. 3.
Alkali cation tolerance and efflux in strains of S. cerevisiae transformed with DhENA1 and DhENA2. (A) Tolerance to alkali cations was tested in solid YPD in the S. cerevisiae B31 null mutant (ena 1-4− nha1−) transformed with DhENA1 (AD1) or with DhENA2 (AD2). The same strain containing the plasmid without any insert (AD0) or with ScENA1 and a D. hansenii strain were used as controls. Plates were inoculated with 10-μl drops of cultures at the middle of the exponential phase, and growth was scored after 60 h. The efflux of Na+ (B) or Li+ (C) was evaluated in AD1 (●), AD2 (▴), and AD0 (■). For determination of Na+ and Li+ efflux rates, cells were grown in YNB medium and loaded with the cations by incubation in the same medium plus 200 mM NaCl or 100 mM LiCl for 3 h. Cells were harvested and resuspended in the assay buffer (pH 5.5) supplemented with 50 mM KCl to trigger the efflux process. Samples were taken at regular intervals, filtered, and treated as previously described (24, 25). The experiments were repeated at least three times. The standard errors were 20% lower than the corresponding mean values.
In addition, we performed growth experiments in liquid media containing NaCl or LiCl. The results confirmed that expression of DhENA1 or DhENA2 improved growth in the presence of either or both cations. The doubling time of strains AD1 and AD2 in the presence of 0.5 M NaCl were 3.4 and 4.3 h, respectively, significantly lower than the value obtained with AD0 (6.9 h). In the presence of Li+, growth was significantly improved in the case of AD1 strain and only very slightly in AD2 (the doubling times of strains AD0, AD1, and AD2 in the presence of 0.05 M LiCl were 8.5, 4.1, and 7.0 h, respectively). All these results suggest that the increase in Na+ and Li+ tolerance could be due to an increase in the process of Na+ and Li+ efflux in cells carrying the DhENA genes. In order to confirm this hypothesis, we studied Na+ and Li+ efflux in Na+-loaded (Na+in, 205 to 221 nmol/mg [dry weight] of cells) or Li+-loaded (Li+in, 52 to 59 nmol/mg [dry weight] of cells) cells of the three strains. The results indicate that the proteins were located at the plasma membrane (Fig. 3B and C). Interestingly, the efflux of Li+ in strain AD2 was very poor, a result that fits with the fact that DhENA2 improved Li+ tolerance only to a small extent. It is worth mentioning that Na+ efflux was clearly higher in the strain expressing DhENA2 than in the strain expressing DhENA1, although this fact was not linked to a higher tolerance to Na+. It has been reported that a certain amount of the protein could be located in some intracellular organelles, increasing tolerance without important changes in the efflux process (5). It is worth noting that the different ENA genes previously described for other yeasts confer Na+ and Li+ tolerance, although Wieland et al. (33) have proposed a specific role in Li+ tolerance in the case of ScENA2 (also named PMR2B). In our case, DhENA1 and DhENA2 conferred Na+ and K+ tolerance, but while DhENA1 improved Li+ tolerance, the effect of DhENA2 in this process was almost negligible. Two results indicate that this is the case: very little complementation was conferred by DhENA2 when Li+ was the toxic cation in the growth medium, and no important Li+ efflux was detected in S. cerevisiae cells carrying DhENA2.
We have cloned, sequenced, and expressed two genes coding for sodium efflux systems in D. hansenii. To our knowledge, this is the first report on genes specifically involved in salt tolerance in a halophilic yeast. The existence of several Na+ transporter genes has been previously described for the osmotolerant yeast Z. rouxii. This yeast is considered to be highly sugar tolerant, but different strains exhibit dramatically different responses to NaCl (reference 7 and references therein). Four different results support the idea that DhENA1 and DhENA2 are P-type ATPases involved in Na+ extrusion: (i) homology with sequences of previously reported ENA genes from S. cerevisiae, S. occidentalis, or Z. rouxii; (ii) complementation of salt tolerance of a mutant of S. cerevisiae lacking the Na+ efflux systems; (iii) complementation of the sodium efflux defect in the same mutant; and (iv) the observation that the extrusion of sodium is pH independent in D. hansenii.
A question remains: what is the physiological relevance of a Na+ efflux system in a yeast which is not normally intoxicated by the cation? We propose that Ena proteins play an important role in maintaining balanced levels of intracellular cations, ensuring the ionic homeostasis of the cell. Therefore, these pumps would transport alkali cations out of the cells, not because they are specifically toxic for D. hansenii but because excessive Na+ (or K+) accumulation would cause osmotic problems.
Our research on the mechanisms involved in salt tolerance in D. hansenii will continue. We are looking for other genes involved in this process. The heterologous expression of DhENA genes in a sensitive mutant of S. cerevisiae lacking the Na+ extrusion systems recovered salt tolerance in this mutant up to the level of the wild strain but still far from the tolerance level of D. hansenii. Salt tolerance is a complex challenge for which nature has developed a number of strategies. Some have been identified in a physiological perspective. We expect to identify other genes and find clues to establish the relative importance of each mechanism in overall salt tolerance.
Acknowledgments
We gratefully acknowledge A. Rodríguez-Navarro and B. Garciadeblas for stimulating discussion of the manuscript and technical assistance.
This research was partially supported by grants BIO4-CT97-2210 (Commission of the European Communities) and PB98-1036 (M.E.C.) to J.R. C.P. was the recipient of grant PRAXIS XXI BD/9089/96.
REFERENCES
- 1.Almagro A, Prista C, Castro S, Quintas C, Madeira-Lopes A, Ramos J, Loureiro-Dias M C. Effects of salts on Debaryomyces hansenii and Saccharomyces cerevisiae under stress conditions. Int J Food Microbiol. 2000;56:191–197. doi: 10.1016/s0168-1605(00)00220-8. [DOI] [PubMed] [Google Scholar]
- 2.André L, Nilsson A, Adler L. The role of glycerol in osmotolerance of the yeast Debaryomyces hansenii. J Gen Microbiol. 1988;134:669–677. [Google Scholar]
- 3.Bañuelos M A, Rodríguez-Navarro A. P-type ATPases mediate sodium and potassium effluxes in Schwanniomyces occidentalis. J Biol Chem. 1998;273:1640–1646. doi: 10.1074/jbc.273.3.1640. [DOI] [PubMed] [Google Scholar]
- 4.Bañuelos M A, Sychrová H, Bleykasten-Grosshans C, Souciet J L, Potier S. The Nha1 antiporter of Saccharomyces cerevisiae mediates sodium and potassium efflux. Microbiology. 1998;144:2749–2758. doi: 10.1099/00221287-144-10-2749. [DOI] [PubMed] [Google Scholar]
- 5.Benito B, Quintero F J, Rodríguez-Navarro A. Overexpression of the sodium ATPase of Saccharomyces cerevisiae: conditions for phosphorylation from ATP and Pi. Biochim Biophys Acta. 1997;1328:214–225. doi: 10.1016/s0005-2736(97)00098-9. [DOI] [PubMed] [Google Scholar]
- 6.Benito B, Garciadeblas B, Rodríguez-Navarro A. Molecular cloning of the calcium and sodium ATPases in Neurospora crassa. Mol Microbiol. 2000;35:1079–1088. doi: 10.1046/j.1365-2958.2000.01776.x. [DOI] [PubMed] [Google Scholar]
- 7.Blomberg A, Adler L. Tolerance of fungi to NaCl. In: Jenings D H, editor. Stress tolerance of fungi. New York, N.Y: Marcel Dekker Inc.; 1993. pp. 209–232. [Google Scholar]
- 8.Brunelli J P, Pall M L. A series of yeast shuttle vectors for expression of cDNAs and other DNA sequences. Yeast. 1993;9:1299–1308. doi: 10.1002/yea.320091203. [DOI] [PubMed] [Google Scholar]
- 9.Garciadeblas B, Rubio F, Quintero F J, Bañuelos M A, Haro R, Rodríguez-Navarro A. Differential expression of two genes encoding isoforms of the ATPase involved in sodium efflux in Saccharomyces cerevisiae. Mol Gen Genet. 1993;236:363–368. doi: 10.1007/BF00277134. [DOI] [PubMed] [Google Scholar]
- 10.Hahnenberg K, Jia Z P, Young P C. Functional expression of the Schizosaccharomyces pombe Na+/H+ gene, sod2, in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5031–5036. doi: 10.1073/pnas.93.10.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haro R, Garciadeblas B, Rodríguez-Navarro A. A novel P-type ATPase from yeast involved in sodium transport. FEBS Lett. 1991;291:189–191. doi: 10.1016/0014-5793(91)81280-l. [DOI] [PubMed] [Google Scholar]
- 12.Iwaki T, Higashida Y, Tsuji H, Tamai Y, Watanabe Y. Characterization of a second gene (ZSOD22) of Na+/H+ antiporter from salt-tolerant yeast Zygosaccharomyces rouxii and functional expression of ZSOD2 and ZSOD22 in Saccharomyces cerevisiae. Yeast. 1998;14:1167–1174. doi: 10.1002/(SICI)1097-0061(19980930)14:13<1167::AID-YEA318>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 13.Jia Z P, McCullogh N, Martel R, Hemmingsen S, Young P G. Gene amplification at a locus encoding a putative Na+/H+ antiporter confers sodium and lithium tolerance in fission yeast. EMBO J. 1992;11:1631–1640. doi: 10.1002/j.1460-2075.1992.tb05209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kreger van Rij N J W, editor. The yeasts: a taxonomic study. 3rd ed. Amsterdam, The Netherlands: Elsevier Science Publishers; 1984. pp. 135–145. [Google Scholar]
- 15.Lages F, Silva-Graça M, Lucas C. Active glycerol uptake is a mechanism underlying halotolerance in yeasts: a study of 42 species. Microbiology. 1999;145:2577–2585. doi: 10.1099/00221287-145-9-2577. [DOI] [PubMed] [Google Scholar]
- 16.Lucas C, da Costa M, van Uden N. Osmoregulatory active sodium-glycerol co-transport in the halotolerant yeast Debaryomyces hansenii. Yeast. 1990;6:187–191. [Google Scholar]
- 17.Nass R, Cunningham K W, Rao R. Intracellular sequestration of sodium by a novel Na+/H+ exchanger in yeast is enhaced by mutations in the plasma membrane H+-ATPase. J Biol Chem. 1997;272:26145–26152. doi: 10.1074/jbc.272.42.26145. [DOI] [PubMed] [Google Scholar]
- 18.Neves M L, Oliveira R P, Lucas C. Metabolic flux response to salt-induced stress in the halotolerant yeast Debaryomyces hansenii. Microbiology. 1997;143:1133–1139. doi: 10.1099/00221287-143-4-1133. [DOI] [PubMed] [Google Scholar]
- 19.Nishi T, Yagi T. Efflux of sodium ions by a Na+/H+ antiporter during salt stress in the salt tolerant yeast Zygosaccharomyces rouxii. J Gen Appl Microbiol. 1995;41:87–97. [Google Scholar]
- 20.Nobre M F, da Costa M S. The accumulation of polyols by the yeast Debaryomyces hansenii in response to water stress. Can J Microbiol. 1985;31:1061–1064. [Google Scholar]
- 21.Norkrans B. Studies on marine-occurring yeasts: growth related to pH, NaCl concentration and temperature. Arch Microbiol. 1966;54:374–392. [Google Scholar]
- 22.Norkrans B, Kylin A. Regulation of the potassium to sodium ratio and of the osmotic potential in relation to salt tolerance in yeasts. J Bacteriol. 1969;100:836–845. doi: 10.1128/jb.100.2.836-845.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prior C, Potier S, Souciet J L, Sychrová H. Characterization of the NHA1 gene encoding a Na+/H+-antiporter of the yeast Saccharomyces cerevisiae. FEBS Lett. 1996;387:89–93. doi: 10.1016/0014-5793(96)00470-x. [DOI] [PubMed] [Google Scholar]
- 24.Prista C, Almagro A, Loureiro-Dias M C, Ramos J. Physiological basis for the high salt tolerance of Debaryomyces hansenii. Appl Environ Microbiol. 1997;63:4005–4009. doi: 10.1128/aem.63.10.4005-4009.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramos J, Alijo R, Haro R, Rodríguez-Navarro A. TRK2 is not a low-affinity potassium transporter in Saccharomyces cerevisiae. J Bacteriol. 1994;176:249–252. doi: 10.1128/jb.176.1.249-252.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramos J. Contrasting salt tolerance mechanisms in Saccharomyces cerevisiae and Debaryomyces hansenii. In: Pandalai S G, editor. Recent research developments in microbiology. Trivandrum, India: Research Signpost Publishers; 1999. pp. 377–390. [Google Scholar]
- 27.Ross S S, Morris E O. Effect of sodium chloride on the growth of certain yeasts of marine origin. J Sci Food Agric. 1962;13:377–390. [Google Scholar]
- 28.Rudolph H K, Antebi A, Fink G R, Buckley C M, Dorman T E, LeVitre J A, Davidow L S, Mao J, Moir D T. The yeast secretory pathway is perturbed by mutations in PMR1, a member of Ca2+ ATPase family. Cell. 1989;58:133–145. doi: 10.1016/0092-8674(89)90410-8. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Thomé-Ortiz P E, Peña A, Ramírez J. Monovalent cation fluxes and physiological changes of Debaryomyces hansenii grown at high concentrations of KCl and NaCl. Yeast. 1998;14:1355–1371. [PubMed] [Google Scholar]
- 31.Thompson J D. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tokuoka K. Sugar-and salt-tolerant yeasts. J Appl Bacteriol. 1993;74:101–110. [Google Scholar]
- 33.Wieland J, Nitsche A M, Strayle J, Steiner H, Rudolph H K. The PMR2 gene cluster encodes functionally distinct isoforms of a putative Na+ pump in the yeast plasma membrane. EMBO J. 1995;14:3870–3882. doi: 10.1002/j.1460-2075.1995.tb00059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]