Abstract
Vibrio anguillarum, which causes terminal hemorrhagic septicemia in fish, was previously shown to possess a LuxRI-type quorum-sensing system (vanRI) and to produce N-(3-oxodecanoyl)homoserine lactone (3-oxo-C10-HSL). However, a vanI null mutant still activated N-acylhomoserine lactone (AHL) biosensors, indicating the presence of an additional quorum-sensing circuit in V. anguillarum. In this study, we have characterized this second system. Using high-pressure liquid chromatography in conjunction with mass spectrometry and chemical analysis, we identified two additional AHLs as N-hexanoylhomoserine lactone (C6-HSL) and N-(3-hydroxyhexanoyl)homoserine lactone (3-hydroxy-C6-HSL). Quantification of each AHL present in stationary-phase V. anguillarum spent culture supernatants indicated that 3-oxo-C10-HSL, 3-hydroxy-C6-HSL, and C6-HSL are present at approximately 8.5, 9.5, and 0.3 nM, respectively. Furthermore, vanM, the gene responsible for the synthesis of these AHLs, was characterized and shown to be homologous to the luxL and luxM genes, which are required for the production of N-(3-hydroxybutanoyl)homoserine lactone in Vibrio harveyi. However, resequencing of the V. harveyi luxL/luxM junction revealed a sequencing error present in the published sequence, which when corrected resulted in a single open reading frame (termed luxM). Downstream of vanM, we identified a homologue of luxN (vanN) that encodes a hybrid sensor kinase which forms part of a phosphorelay cascade involved in the regulation of bioluminescence in V. harveyi. A mutation in vanM abolished the production of C6-HSL and 3-hydroxy-C6-HSL. In addition, production of 3-oxo-C10-HSL was abolished in the vanM mutant, suggesting that 3-hydroxy-C6-HSL and C6-HSL regulate the production of 3-oxo-C10-HSL via vanRI. However, a vanN mutant displayed a wild-type AHL profile. Neither mutation affected either the production of proteases or virulence in a fish infection model. These data indicate that V. anguillarum possesses a hierarchical quorum sensing system consisting of regulatory elements homologous to those found in both V. fischeri (the LuxRI homologues VanRI) and V. harveyi (the LuxMN homologues, VanMN).
Diverse gram-negative and gram-positive bacteria communicate intercellularly to regulate the transcription of multiple target genes in concert with cell density. This type of communication, termed quorum sensing, is mediated through the production of diffusible signal molecules, termed autoinducers or pheromones, which effectively enable a bacterium to monitor its own population density (for reviews, see references 13, 17, 21, and 44). Quorum sensing is now known to regulate diverse physiological processes such as bioluminescence, swarming, antibiotic biosynthesis, plasmid conjugal transfer, and the production of exoenzyme virulence determinants in human, animal, and plant pathogens. In gram-negative bacteria, the most intensively investigated autoinducer molecules are N-acylhomoserine lactones (AHLs) that vary in the length and saturation state of the N-acyl side chain (molecules with from 4 to 14 carbons have been characterized) and in the presence or absence of an acyl chain C-3 substituent (oxo- or hydroxy-).
The cell density-dependent regulation of bioluminescence in Vibrio (Photobacterium) fischeri (31, 32) is frequently used as the paradigm for quorum sensing. In this marine symbiont, as the bacterial cell population density increases, the level of the autoinducer, N-(3-oxohexanoyl)-l-homoserine lactone (3-oxo-C6-HSL) (14), accumulates until a critical threshold concentration is reached. 3-Oxo-C6-HSL then binds to the transcriptional activator protein, LuxR, and the resulting LuxR–3-oxo-C6-HSL complex triggers transcription of the luminescence (lux) operon, resulting in the emission of light.
In common with V. fischeri, the free-living marine bacterium Vibrio harveyi also regulates bioluminescence in a cell density-dependent manner at the level of transcription, involving the production and sensing of N-(3-hydroxybutanoyl)-l-homoserine lactone (3-hydroxy-C4-HSL [for a review, see reference 32]). However, the regulation of the V. harveyi system is very different and appears to be more complex than in V. fischeri. Based on genetic analyses, Freeman and Bassler (15, 16) have proposed a model for the regulation of bioluminescence in V. harveyi that involves two signaling systems and two autoinducer molecules. The first quorum-sensing system relies on 3-hydroxy-C4-HSL (9), the synthesis of which is directed by luxL and luxM. Interestingly, the gene products of luxL and luxM show no homology to the LuxI family of AHL synthases (4). Regulation of bioluminescence via the second quorum-sensing system utilizes another, as yet unidentified signal molecule (AI-2), which is chemically distinct from AHLs and is synthesized via LuxS (51). The sensors for 3-hydroxy-C4-HSL and AI-2, named LuxN and LuxQ, respectively, resemble proteins belonging to two-component signaling systems (4, 6) and possess both a histidine kinase and a response regulator domain. At low cell densities, in the absence of signal molecules, LuxN and LuxQ are suggested to work in parallel by relaying the phosphates from their response regulator domains to a shared phosphorelay protein, LuxU (16). The phosphate from LuxU is then transferred to the response regulator domain of the ς54 activator LuxO, which, when phosphorylated, represses bioluminescence by activating the expression of an unidentified repressor (5, 15, 28). In contrast, at high cell densities, the signal molecules accumulate and are believed to bind to their respective sensors (15, 16). It is thought that 3-hydroxy-C4-HSL may bind directly to LuxN, whereas AI-2 is postulated to bind to LuxQ via interaction with a putative periplasmic protein LuxP. Binding of the signals is suggested to switch the sensor kinase activities of LuxN and LuxQ into phosphatases, leading to the dephosphorylation of LuxO and derepression of the lux operon.
It is now known that V. fischeri also possesses multiple quorum-sensing circuits and produces at least three AHLs, which are involved in the regulation of bioluminescence (18, 26). While the production of both 3-oxo-C6-HSL and N-hexanoylhomoserine lactone (C6-HSL) is mediated via LuxI, an additional protein, AinS, is responsible for the synthesis of N-octanoylhomoserine lactone (C8-HSL) (18, 26). AinS is homologous to LuxM of V. harveyi but not to LuxI of V. fischeri, suggesting the existence of a new family of AHL synthases.
Vibrio anguillarum, a bacterial pathogen that causes hemorrhagic septicemia in marine fish, was previously shown to contain a LuxI and a LuxR homolog, termed VanI and VanR, respectively (36), and to produce N-(3-oxodecanoyl)homoserine lactone (3-oxo-C10-HSL), the synthesis of which is mediated by VanI. Although a null mutation in vanI abolished the production of 3-oxo-C10-HSL, this mutant was still capable of weakly activating AHL biosensors, suggesting that V. anguillarum possesses multiple quorum-sensing systems. In the present study, we describe the isolation and characterization of two additional AHLs, N-(3-hydroxyhexanoyl)homoserine lactone (3-hydroxy-C6-HSL) and C6-HSL. The protein responsible for the synthesis of these AHLs, VanM, is shown to share homology with both LuxLM and AinS from V. harveyi and V. fischeri, respectively. In addition, we characterized a second gene, vanN, the product of which is homologous to LuxN from V. harveyi. A null mutation of vanM abolished the production of both 3-hydroxy-C6-HSL and C6-HSL. Surprisingly, however, the production of 3-oxo-C10-HSL was also downregulated, suggesting that the vanM quorum-sensing system is involved in regulating the vanIR locus responsible for the synthesis of 3-oxo-C10-HSL.
MATERIALS AND METHODS
Strains, phage, plasmids, and media.
Bacterial strains and plasmids are described in Table 1. V. anguillarum NB10 (serotype O1) is a clinical isolate from the Gulf of Bothnia outside Umeå Marine Center, Norrbyn, Sweden (39). Escherichia coli JM109 (45) was used as the bacterial background for the purification of AHLs. The wild-type V. harveyi strain BB120 (7) was used for resequencing the junction between luxL and luxM. E. coli SY327 (33) was used for transformation after subcloning fragments into either the pNQ705-1 or pDM4 vector. All plasmids to be conjugated into V. anguillarum were transformed into E. coli S17-1 (49), which was used as the donor strain. Plasmid transfers from E. coli to V. anguillarum were done as previously described (35). E. coli XL1-Blue (Stratagene) was used for bacteriophage lambda infections and for routine cloning. The medium routinely used for E. coli was Luria broth, which contains Bacto Tryptone (10 g/liter), Bacto Yeast Extract (5 g/liter), and sodium chloride (10 g/liter). For V. anguillarum, Trypticase soy medium (BBL) was used for routine growth. For purification and identification of AHLs, V. anguillarum was grown at 20°C with shaking in M9 medium (45) supplemented with 2% (wt/vol) NaCl. For purification of AHLs produced via recombinant VanM, E. coli JM109(pBSVanMN) and E. coli JM109(pDKVanM) were grown at 30°C with shaking in M9 medium containing ampicillin (100 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype and/or relevant markers | References or source |
|---|---|---|
| E. coli strains | ||
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB) | 46 |
| SY327 | Δ(lac pro) argE(Am) rif malA recA56 | 34 |
| S17-1 | thi pro hsdR hsdM+ recA RP4-2-Tc::Mu-Km::Tn7 | 50 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 Δ(lac-pro) [F′ proAB lacIqlacZΔM15 Tn10(Tetr)] | Stratagene |
| C. violaceum CV026 | Kmr; strain ATCC 31532 defective in C6-HSL production due to a mini-Tn5 insertion in the cviI gene | 28, 30, 58 |
| V. harveyi BB120 | Wild type | 7 |
| V. anguillarum strains | ||
| NB10 | Wild type, serotype O1 | 40 |
| DM21 | In-frame deletion of vanI | 37 |
| DM27 | In-frame deletion of vanM | This study |
| DM34 | Cmr; polar mutation in vanN | This study |
| Plasmids | ||
| pBluescript | Apr; ColE1 origin | Stratagene |
| pBSVanN-320 | Apr; pBluescript containing a 320-bp PCR fragment from vanN (bp 1706–1990) | This study |
| pBSVanN10-1 | Apr; pBluescript containing a cloned fragment with the putative vanMN operon to bp 4061 | This study |
| pBSVanN10-5 | Apr; pBluescript containing a cloned fragment with vanN from bp 3731 to 4099 | This study |
| pBSVanMN | Apr; pBluescript containing subcloned fragments that give a complete putative vanMN operon | This study |
| pSup202P | Cmr, Tcr; pSup202 derivative (RP4 Mob+) with a polylinker cloned into PstI within the Apr gene | 35 |
| pVanM-2 | Cmr, Tcr; pSup202P derivative containing the vanM gene and its promoter (bp 108–1627) | This study |
| pDM4 | Cmr; suicide vector with an R6K origin and the sacBR genes from Bacillus subtilis | 36 |
| pDMVanM1 | Cmr; pDM4 derivative containing vanM bp 108–324 fused in frame to bp 1309–1518 | This study |
| pNQ705-1 | Cmr; suicide vector that contains an R6K origin | 31 |
| pNQVanN2 | Cmr; pNQ705-1 derivative containing vanN bp 1548–1787 | This study |
| pGEM-T Easy | Apr; ColE1 origin | Promega |
| pDKVanM | Apr; pGEM-T Easy derivative containing vanM | This study |
| pSB401 | Tcr; contains a fusion of luxRI′::luxCDABE in pACYC184 | 58 |
| pSB1075 | Apr; contains a fusion of lasRI′::luxCDABE in pUC18 | 59 |
Plasmid pVanM-2 is a pSup202P (34) derivative which contains the vanM gene. A fragment containing vanM and its promoter (bp 108 to 1627 [Fig. 1]) was obtained by PCR. A restriction enzyme site, either NheI or BglII, was added to the 5′ end of each PCR primer to aid cloning of the PCR product. This fragment was ligated into the SpeI and BglII sites of pSup202P.
FIG. 1.
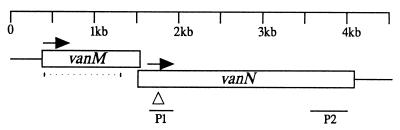
Genetic map of the vanMN locus of V. anguillarum. The vanN gene overlaps the vanM gene by 8 bp. The horizontal arrows indicate the direction of transcription. The open arrowhead indicates where pNQVanN2 was inserted into vanN to make strain DM34. The dotted line indicates the region of vanM that was deleted in frame to make strain DM27. The line labeled P1 indicates the region of vanN that was amplified using the degenerate primers complementary to ainS and luxN. This P1 region of vanN was used as the probe for first screening of the gene library from which pBSVanN10-1 was isolated. The line labeled P2 indicates the region of vanN that was used as a probe in the second screening of the gene library from which pBSVanN10-5 was isolated.
AHL reporter assays.
AHLs were detected using a Tn5-generated Chromobacterium violaceum mutant termed CV026 which responds to a range of AHLs (with from C4 to C8 acyl side chains) by inducing the synthesis of the purple pigment violacein (27, 29, 57). For this assay, AHL-producing strains are cross-streaked vertically against a horizontal streak of CV026 on Trypticase soy agar plates. Alternatively, spent culture supernatants, solvent extracts, or high-pressure liquid chromatography (HPLC) fractions (see below) were analyzed for the presence of short- and long-chain AHLs using the direct and reverse CV026 seeded agar plate assays, respectively, as described previously (29, 36). For the more sensitive detection of long-chain AHLs (with C10 to C14 acyl side chains), a bioluminescent E. coli lux-based AHL biosensor termed E. coli JM109(pSB1075), which contains an intact lasR gene and the lasI promoter from Pseudomonas aeruginosa fused to luxCDABE from Photorhabdus luminescens, was used (58). Thin-layer chromatography (TLC) was also used to separate AHLs and overlaid with soft top agar seeded with the appropriate biosensor as described by McClean et al. (29). For analysis of short-chain AHLs, reverse-phase aluminum-backed RP18 F254S TLC plates (20 by 20 cm; Merck) and a mobile phase of 60% (vol/vol) methanol in water were used. Long-chain AHLs were analyzed on aluminum-backed Silica Gel 60 F254 normal-phase TLC plates (20 by 20 cm; Merck) using a 45%-55% (vol/vol) hexane-acetone mix as the mobile phase.
Isolation, purification, and chemical characterization of AHLs.
AHLs were purified and characterized as described by Bainton et al. (3) and Cámara et al. (8). Essentially, spent supernatants (4 liters) from stationary-phase cultures of V. anguillarum NB10, E. coli JM109(pBSVanMN), or E. coli JM109(pDKVanM) were extracted with dichloromethane, and solvent extracts were separated by semipreparative reverse-phase HPLC as previously described (36). Fractions were tested for the presence of AHLs using the AHL biosensor C. violaceum CV026 or E. coli(pSB401) in conjunction with TLC as described before (29). Following preparative HPLC, the active subfractions were analyzed by HPLC-mass spectrometry (LC-MS) as described previously (2). The spectra obtained were compared with those for synthetic AHL standards subjected to the same LC-MS conditions.
Synthesis of AHLs.
The AHLs 3-oxo-C10-HSL, 3-hydroxy-C6-HSL, and C6-HSL were synthesized, purified, and characterized as described previously (8, 10). Each compound was subjected to MS, proton nuclear magnetic resonance spectroscopy, and infrared spectroscopy. For 3-hydroxy-C6-HSL and C6-HSL, spectroscopic data are provided in references 8 and 36 for 3-oxo-C10-HSL.
Quantification of AHL production.
To determine the relative levels of 3-hydroxy-C6-HSL, C6-HSL, and 3-oxo-C10-HSL in V. anguillarum wild type and vanM (DM27)- and vanI (DM21)-negative mutants, 100 ml of each stationary-phase culture supernatant was extracted with dichloromethane as described above, evaporated to dryness, and resuspended in 100 μl of acetonitrile. Each sample was subjected to LC-MS, and the concentration was determined by comparison with a calibration curve constructed for molecular ion abundance, using each of the appropriate AHL synthetic standards.
Cloning of the vanMN DNA locus.
To identify genes homologous to luxLMN from V. harveyi (4) and to ainSR from V. fischeri (18), protein comparisons were performed and two degenerate inosine-containing oligonucleotides were designed to complement both the ainR and the luxN DNA sequences and to contain a restriction endonuclease site, either SacI or SpeI, at each 5′ end. These two primers were used in PCR to generate a 320-bp fragment from the chromosome of V. anguillarum (Fig. 1). This 320-bp fragment was cloned into the SpeI and SacI sites of pBluescript to give pBSVanN-320. The DNA sequence of this fragment was similar to that of the partial ainR gene of V. fischeri (18) and the luxN gene of V. harveyi (4). The 320-bp fragment was used as a probe to screen a Lambda Zap II (Stratagene)-based gene bank of V. anguillarum (34) as previously described (35). From a positive plaque, pBluescript containing the chromosomal fragment was excised from the bacteriophage DNA as described previously (34). This plasmid, pBSVanN10-1, contained a 7-kb chromosomal insert with the entire vanM gene and all but the last 39 bp of the vanN gene. Thus, a second screening was done as above, using a 500-bp KpnI fragment from the 3′ end of the vanN gene as the probe (Fig. 1). The excised plasmid, pBSVanN10-5, contained a 6.5-kb chromosomal insert with only 368 bp of the 3′ end of vanN. The overlap was sufficient to complete the sequencing of both the vanN and vanM genes. From the sequence information, pBSVanMN, a pBluescript derivative that contained the entire putative vanMN operon, was designed. To create pBSVanMN, pBSVanN10-1, which was missing only the last 39 bp of this putative operon, was used. To make the chromosomal fragment insert smaller, pBSVanN10-1 was digested with BamHI and MluI to remove approximately 1,800 bp upstream of vanM. The vector fragment containing vanM was gel purified, and the ends were filled in using Klenow enzyme and then ligated together. To create a complete vanN gene on the now smaller chromosomal fragment insert, approximately 500 bp was removed from the 3′ end of the fragment insert using the internal KpnI site, which is about 500 bp from the 3′ end of vanN, and the KpnI site in the vector polylinker. The vector ends were then dephosphorylated using calf intestinal phosphatase, and a PCR fragment containing the vanN sequence from the internal KpnI site to 320 bp downstream of vanN was ligated to the KpnI-cut pBSVanN10-1. This region was sequenced to ensure that it was identical to the wild-type sequence.
Construction of pDKVanM.
The vanM gene, from 10 bp upstream of the start codon through the stop codon, was amplified from V. anguillarum NB10 chromosomal DNA by PCR and then cloned into the plasmid pGEM-T Easy (Promega) by T/A cloning as described in the manufacturer's instructions. A clone (pDKVanM) was selected in which the vanM gene was under the control of the plasmid-borne lacZ promoter.
DNA techniques and sequencing.
Oligonucleotides for primers were synthesized using a model 394 Applied Biosystems DNA/RNA synthesizer. Unless otherwise stated, all conditions for DNA techniques and enzymatic reactions were done as described by Sambrook et al. (45) or as suggested by the manufacturers. Double-strand DNA sequencing was performed either by the dideoxy-chain termination method with T7 DNA polymerase (Pharmacia Biotech) or by automated sequencing using an Applied Biosystems 373A sequencer. Using pBSVanN10-1 and pBSVanN10-5 plasmid DNA, both strands of a 4,514-bp fragment containing vanM and vanN were sequenced by primer walking in two directions.
PCR conditions.
PCRs were performed as previously described (30). For resequencing of the junction between the V. harveyi luxL and luxM genes, two primers were designed from the DNA sequence (GenBank accession number L13940) to amplify bp 587 to 1035. PCR began with template denaturation for 5 min at 95°C before the addition of Taq DNA polymerase. After enzyme addition, the PCR was continued for 29 cycles of 2 min at 55°C, 1 min at 72°C, and 1 min at 95°C, followed by one cycle of 55°C for 2 min and 72°C for 10 min.
Construction of the vanM and vanN mutations.
The vanN null mutant DM34 was made by the integration of a suicide plasmid, pNQVanN2, a derivative of pNQ705-1 (30), into the vanN gene as described previously (34). Using SacI and SpeI, a 239-bp PCR fragment (residues 1548 to 1787 [Fig. 1]) complementary to vanN was cloned into the SacI and SpeI sites of pNQ705-1, creating pNQVanN2. The entire pNQVanN2 was inserted 28 bp downstream of the vanN start codon. Since the PCR fragment cloned onto pNQVanN2 did not contain the vanN promoter or start codon, this plasmid insertion created a null mutation of vanN. The small piece of the vanN gene carried on pNQVanN2 used to target the insertion of the suicide plasmid allows for only the first 89 amino acids of VanN to be translated. The insertion of the plasmid was checked by PCR analysis using a previously described primer (34) complementary to a region on the plasmid just outside the polylinker region and a primer complementary to a chromosomal DNA region just outside the PCR fragment cloned into pNQVanN2. The PCR fragments obtained from the mutant were analyzed by restriction mapping to ensure that the PCR fragments obtained were from the correct region of the chromosome. Stability of the insertion mutation was tested by growth for 30 generations in the absence of chloramphenicol. Of 100 colonies tested, no loss in chloramphenicol resistance was seen.
The vanM mutant DM27 contains an in-frame deletion created by allelic exchange as described previously (35). To create the new deletion allele, overlap PCR was performed joining vanM residues 108 to 324 to residues 1309 to 1518. By using NheI and BglII, this PCR fragment was cloned into the SpeI and BglII sites of pDM4, creating pDMVanM1. After allelic exchange using pDMVanM1, the start codon was fused to the last 72 amino acid codons of vanM (Fig. 2). To confirm that an in-frame deletion was made in DM27, this region from the chromosome was PCR amplified and cloned. Several clones were then sequenced to ensure that the deletion was made and that no errors were inserted within the new allele.
FIG. 2.
Protein sequence alignments. (A) The corrected V. harveyi LuxM* protein sequence, the V. fischeri AinS protein sequence, and the V. anguillarum VanM protein sequence were aligned and compared for similarities. The LuxL-LuxM fused protein will be called LuxM (Bassler, personal communication), but for the sake of discussion it is called LuxM* in this report. (B) The V. harveyi LuxN protein sequence, the V. fischeri AinR partial protein sequence, and the V. anguillarum VanN protein sequence were aligned and compared for similarities. Asterisks indicate amino acids that are identical in the V. harveyi and V. anguillarum protein sequences; plus signs indicate amino acids that are identical in all aligned protein sequences. The amino acids within boxes represent the various conserved motifs important for the function of hybrid sensor kinases (10).
Computer analysis.
Database searches were done using the sequence analysis software of the Genetics Computer Group, Inc. (University of Wisconsin) (12).
Fish infections.
Rainbow trout (Oncorhynchus mykiss) with an approximate weight of 10 to 15 g were infected with V. anguillarum either by intraperitoneal injections or by immersion of the fish in seawater containing V. anguillarum as previously described (35). The immersion and intraperitoneal infections were done at least two times. Five fish were infected for each bacterial dilution used. The 50% lethal doses (LD50s) were calculated as described by Reed and Muench (43). The LD50s recorded are an averaged number of all infections for each strain. To aid comparative analysis between strains, the standard deviation of the wild-type LD50 was calculated for both routes of infection. LD50 values were collected from previous studies in our lab and used in determining a standard deviation for the wild-type strain. For infection by immersion, 37 LD50 values were used to give a standard deviation of 3.9 × 103 bacteria per ml of seawater. For the intraperitoneal route, 35 LD50 values were used, giving a standard deviation of 29 bacterial cells.
Nucleotide sequence accession number.
The vanM and vanN DNA sequence has been submitted to GenBank with accession number AF288163. The sequence from V. harveyi with the amended sequence for luxLM sequence in V. harveyi has also been submitted; its accession number is AF286004.
RESULTS
Cloning and sequencing of the vanMN locus.
Using AHL bioassays, we have previously detected AHLs in bacterial culture supernatants from both V. anguillarum NB10 and DM21 (36), a vanI mutant which does not produce 3-oxo-C10-HSL. This result suggests that V. anguillarum most likely contains a second quorum-sensing system which may be responsible for this activity. Since V. anguillarum is related to V. harveyi and V. fischeri and since both V. harveyi and V. fischeri contain luxLMN quorum-sensing systems (4, 18), we rationalized that V. anguillarum may also have a luxLMN-type circuit. To determine whether V. anguillarum contains a second quorum-sensing system, degenerate PCR primers were designed from the luxN and ainR genes, which code for AHL sensor proteins in V. harveyi (4) and V. fischeri (18), respectively. A DNA fragment highly homologous to both luxN and ainR was amplified from the V. anguillarum chromosome and used as a probe to screen a V. anguillarum chromosomal gene bank. Positive clones were sequenced revealing the presence of two open reading frames (ORFs), which we termed vanM and vanN. Further analysis showed that the last 8 bp of vanM overlapped with the start of vanN, suggesting that these genes may share the same promoter. Figure 1 shows a genetic map of this DNA locus.
The predicted amino acid sequences for VanM and VanN were compared with the their homologues in both V. harveyi and V. fischeri. The amino terminus of VanM is 58% identical to LuxL, and the carboxy terminus of VanM is 66% identical to LuxM (4). Over the whole protein, VanM is 33% identical to AinS (Fig. 2A) (18). Figure 2B shows that VanN is 76% identical to the entire LuxN protein (4) and 38% identical to the partial sequence of AinR that is available (18). Like LuxN (4, 16), VanN is also homologous to numerous members of the two-component family of adaptive response regulators (for a review, see reference 40), with the highest similarity to hybrid sensor kinases that contain both sensor histidine kinase and response regulator domains (40). This group includes proteins such as BvgS from Bordetella pertussis (1), LemA (GacS) from Pseudomonas syringae pv. syringae (22), and RpfC from Xanthomonas campestris (54), all of which regulate virulence gene expression, as well as RcsC, BarA, and ArcB from E. coli, which regulate capsule production, gene expression in response to changes in osmolarity, and gene expression in response to oxygen levels, respectively (23, 38, 50).
The V. harveyi luxL and luxM genes are within a single ORF.
The sequence homology analyses and the functional data (see below) presented in this report show that VanM is the third member of a new family of AHL synthases. However, we were intrigued as to why a single protein is sufficient for AHL synthesis in V. anguillarum and V. fischeri whereas two proteins, LuxL and LuxM, are apparently required in V. harveyi. Given the similarity of VanM to LuxL and LuxM and the fact that the combined molecular mass of LuxL and LuxM (42.5 kDa) is approximately equal to that of AinS (43.5 kDa) and VanM (44.1 kDa), we wondered whether there is a sequencing error in the junction between luxL and luxM that leads to a frameshift which results in two ORFs instead of one. To test this hypothesis, PCR was used to amplify the junction between the luxL and luxM genes from V. harveyi BB120 (7). PCR products from three independent reactions were sequenced, and all showed an extra base pair at the 3′ end of luxL compared to the previously published sequence (GenBank accession number L13940). The addition of this base pair alters the predicted polypeptide sequences of LuxL and LuxM, fusing them into one 44-kDa protein which will be called LuxM (B. Bassler, personal communication). However, for ease of discussion, we will call the 44-kDa protein LuxM* in this report. VanM is 64% identical to LuxM*, indicating that VanM and LuxM∗ are more similar to each other than to AinS (Fig. 2A).
Identification of AHLs synthesized by the vanMN and vanM DNA loci in E. coli JM109.
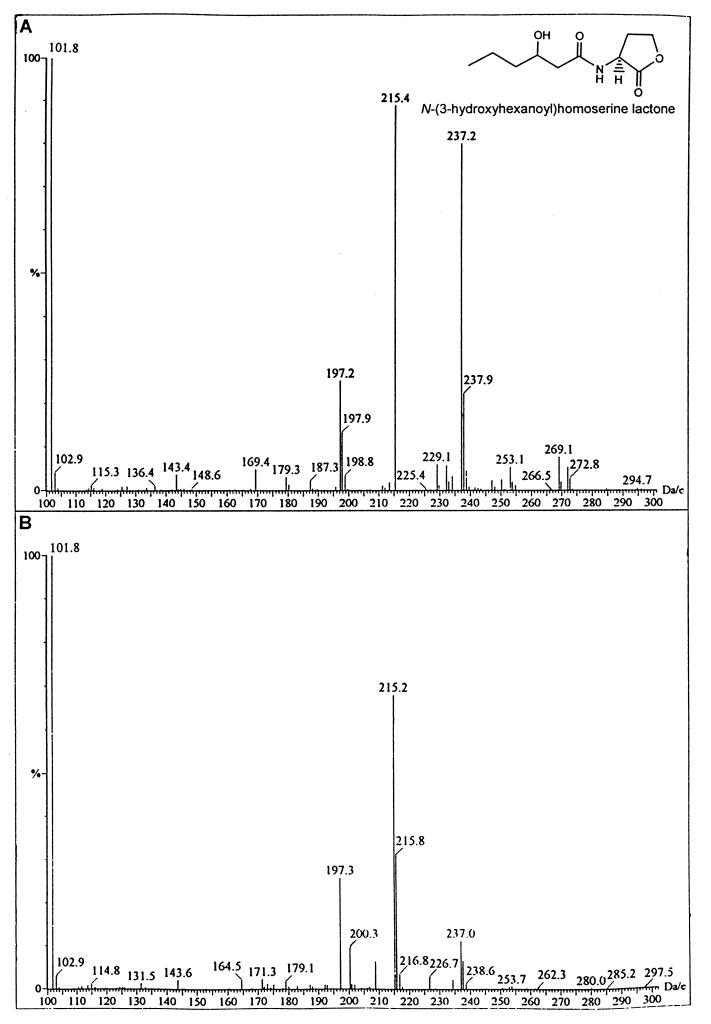
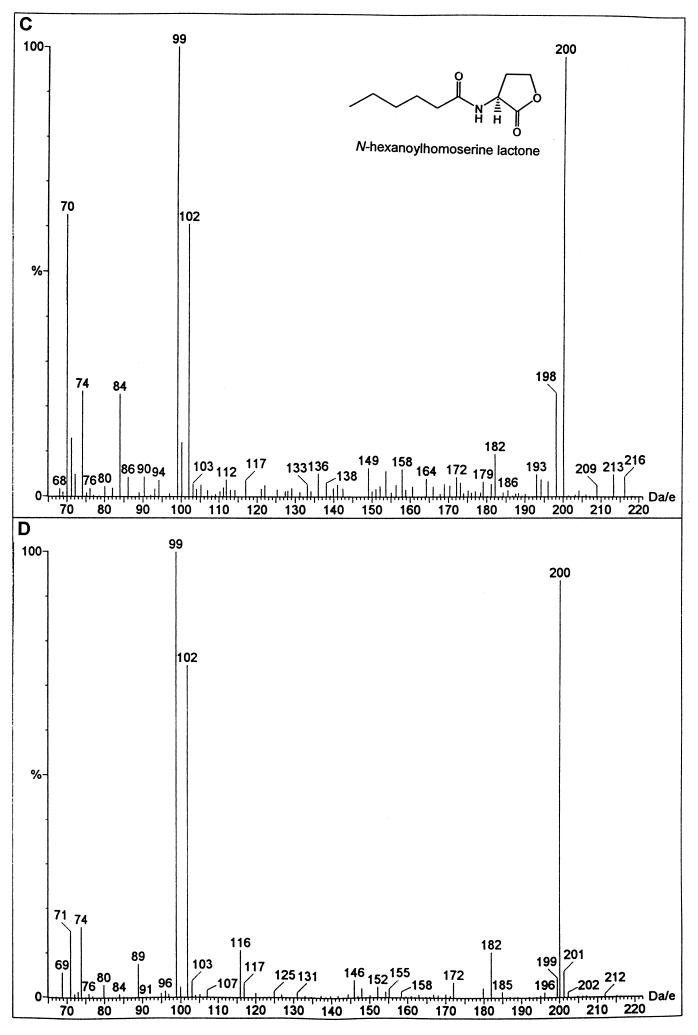
To determine what AHLs, if any, are synthesized via this DNA locus, the vanMN genes were subcloned into pBluescript and introduced into E. coli JM109. Large-scale solvent extractions were performed on spent supernatant from stationary-phase E. coli JM109(pBSVanMN) cultures grown in M9 culture medium. The concentrated crude extract was fractionated by HPLC using a linear acetonitrile-in-water gradient. The resulting fractions were assayed for AHL activity using the CV026 biosensor in conjunction with TLC. Two active fractions (fractions 2 and 3) were then analyzed by LC-MS. The LC-MS spectrum for the molecule present in fraction 2 revealed a molecular ion of 215 and a fragmentation product of 102 (which corresponds to the homoserine lactone moiety). This suggests that the AHL present is 3-hydroxy-C6-HSL (Fig. 3). Furthermore, we observed a peak at 197 arising from the expected loss of a molecule of water which is characteristic of 3-hydroxy AHL derivatives (48). 3-Hydroxy-C6-HSL was synthesized as described previously (10) and shown to possess chromatographic and spectral properties identical to those of the natural compound. The presence of C6-HSL in fraction 3 was deduced from the presence of a molecular ion of 200 (M + 1) together with fragmentation ions of 102 and 99, which correspond to the homoserine lactone moiety and the hexanoyl side chain, respectively (Fig. 3). To confirm that vanM alone was responsible for the production of 3-hydroxy-C6-HSL and C6-HSL, the gene was amplified from the V. anguillarum NB10 chromosome by PCR and cloned into pGEM-T to create pDKVanM. Cell-free culture supernatant extracts were subjected to TLC and overlaid with either CV026 or E. coli(pSB401). In both cases, two spots were obtained which migrated with the same Rf values as the synthetic 3-hydroxy-C6-HSL and C6-HSL standards (data not shown).
FIG. 3.
Mass spectra (LC-MS) of two compounds purified from the spent culture supernatant of E. coli JM109(pBSVanMN) (B and D) are indistinguishable from those of synthetic 3-hydroxy-C6-HSL (A) and C6-HSL (C), respectively.
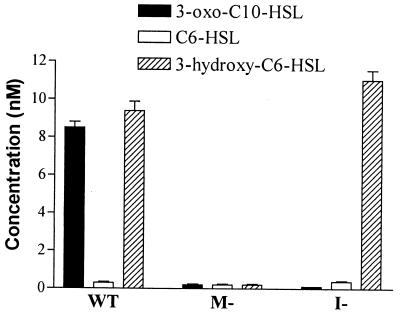
To determine the relative levels of the three AHLs produced by V. anguillarum NB10, cell-free culture supernatants were extracted with solvent and subjected to LC-MS. The concentration of each AHL present was determined by reference to a calibration curve constructed for each synthetic standard. Figure 4 reveals that 3-oxo-C10-HSL and 3-hydroxy-C6-HSL are the major AHLs and are present at approximately 8.5 and 9.5 nM, respectively. C6-HSL is a minor component present at around 0.3 nM.
FIG. 4.
Quantification of AHLs produced by V. anguillarum wild type (WT) and vanM (DM27) and vanI (DM21) mutants (M- and I-) in stationary-phase supernatant determined by LC-MS. Each sample was subjected to LC-MS, and the concentration was determined by comparison with a calibration curve constructed for molecular ion abundance using each of the corresponding AHL synthetic standards.
VanM is required for synthesis of C6-HSL and 3-hydroxy-C6-HSL in V. anguillarum.
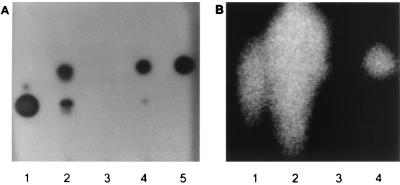
To determine whether vanM and vanN are required for AHL synthesis, isogenic mutations that resulted in the inactivation of either of these genes in V. anguillarum were constructed. The vanM mutant DM27 contained an in-frame deletion removing codons 2 through 336 of VanM (Fig. 1). Analysis of spent cell-free culture supernatants, using the CV026/TLC bioassays and LC-MS (Fig. 4 and 5A), revealed that DM27 did not produce either 3-hydroxy-C6-HSL or C6-HSL. Interestingly, further analysis using the E. coli(pSB1075) lux-based sensor showed that the production of 3-oxo-C10-HSL, which is driven by VanI, had also been downregulated (Fig. 5B). Using LC-MS, no 3-oxo-C10-HSL could be detected in the vanM mutant (Fig. 4). These results demonstrate that VanM directs the synthesis of C6-HSL and 3-hydroxy-C6-HSL in V. anguillarum and that these AHLs are required for the production of 3-oxo-C10-HSL. To confirm that the lack of all three AHLs was due to the loss of vanM, pVanM-2, containing the wild-type vanM gene, was introduced into the vanM isogenic mutant DM27. TLC analysis showed that the production of C6-HSL, 3-hydroxy-C6-HSL, and 3-oxo-C10-HSL was restored in the transcomplemented mutant (Fig. 5). Furthermore, growth of DM27 in medium previously conditioned by growth with the vanI mutant DM21 (which does not produce 3-oxo-C10-HSL but still produces C6-HSL and 3-hydroxy-C6-HSL [Fig. 4]) restored production of 3-oxo-C10-HSL in the vanM mutant (data not shown).
FIG. 5.
TLC analyses of the V. anguillarum vanM mutant (DM27) showing the loss of 3-hydroxy-C6-HSL, C6-HSL (A), and 3-oxo-C10-HSL (B) and the restoration of AHL synthesis in the vanM mutant (DM27) complemented with plasmid-borne copies of vanM [DM27(pVanM-2)]. For this assay, stationary-phase, cell-free supernatants together with synthetic standards were analyzed by TLC in conjunction with the AHL biosensor, C. violaceum CV026 (A) or E. coli JM109(pSB1075) (B), for the detection of short or long-chain AHLs, respectively. (A) C6-HSL (lane 1), wild type (lane 2), DM27 (lane 3), DM27(pVanM-2) (lane 4), and 3-hydroxy-C6-HSL (lane 5); (B) 3-oxo-C10-HSL (lane 1), wild type (lane 2), DM27 (lane 3), and DM27(pVanM-2) (lane 4).
To assess the role of vanN in AHL production, a vanN isogenic null mutant was constructed. A suicide plasmid, pNQVanN2, was inserted 28 bp downstream of the vanN start codon, resulting in mutant DM34 (Fig. 1). Using CV026 and E. coli(pSB1075) bioassays, cell-free supernatants from DM34 displayed wild-type levels of all three AHLs, suggesting that VanN does not play a role in the regulation of AHL production (data not shown).
Virulence analysis.
Since VanN is homologous to several two-component regulatory proteins involved in controlling virulence in other bacteria, the LD50s for the vanM and vanN mutants were compared to that of the wild type in a fish infection model. Both the immersion and intraperitoneal infection routes were used. For the immersion route, the LD50s were 4 × 103, 3 × 104, and 4 × 103 bacteria per ml of seawater for the wild type, the vanM mutant (DM27), and the vanN mutant (DM34), respectively. A sevenfold difference in virulence was seen for the vanM mutant; however, the standard deviation of the wild-type LD50 was determined to be ±3.9 × 103, indicating that this sevenfold difference may not be significant. For the intraperitoneal route, the LD50s were similar: 34 (standard deviation of ±29), 74, and 70 bacteria for the wild type, the vanM mutant (DM27), and the vanN mutant (DM34), respectively. These data suggest that the virulence of V. anguillarum is likely not affected by the vanM and vanN mutations.
Protease production.
The metalloprotease, EmpA, of V. anguillarum is 69% identical to Hap, the V. cholerae hemagglutinin protease (33). The hap gene is regulated via HapR, a homologue of the LuxR protein of V. harveyi (25). Therefore, since LuxR is a component of the same quorum sensing circuitry as LuxM* and LuxN, we reasoned that empA expression in V. anguillarum may be affected in the vanM and vanN mutants. However, on skimmed milk agar plates, the zones of clearance were similar for both mutants and the wild type.
DISCUSSION
In recent years, diverse gram-negative bacteria have been shown to produce AHLs that stimulate LuxI/LuxR-type regulatory circuits. Some of these bacteria possess multiple LuxI/LuxR-type regulatory circuits and produce multiple AHLs, which are involved in the regulation of multiple target genes (for a review, see reference 13). Despite the low similarity (∼30%) among members of the LuxI family, these homologues appear to have similar enzymatic activities. LuxI homologues utilize S-adenosylmethionine and an acyl-acyl carrier protein (acyl-ACP) in the synthesis of AHLs (19, 37, 41, 47, 55), although some have also been shown to use the appropriately charged acyl-coenzyme A (acyl-CoA) as an alternative source of the acyl side chain (24, 41). However, the molecular basis by which different LuxI homologues are able to select the appropriate acyl chain from ACP or CoA derivatives to produce the desired AHL(s) is not known. Recently, a second family of AHL synthases, which bears no homology to LuxI, has been described (18). To date, members of this new family have been identified in V. harveyi (LuxM*) (4) and V. fischeri (AinS) (18), where they direct the synthesis of 3-hydroxy-C4-HSL and C8-HSL, respectively. Hanzelka et al. (20) have shown that AinS utilizes substrates and has enzyme kinetics similar to those for LuxI. However, in addition to octanoyl-ACP, AinS also utilized octanoyl-CoA as the acyl group donor. Consequently, Hanzelka et al. (20) speculated that possession of two different types of AHL synthases is beneficial to V. fischeri, as it may permit the production of AHLs and quorum sensing even when the prevailing environmental conditions are not optimal for AHL synthesis via LuxI.
The results presented in this study add V. anguillarum to the list of gram-negative bacteria that produce multiple AHLs and possess multiple quorum-sensing regulatory circuits. Specifically, we have identified two additional AHLs that are produced by V. anguillarum. Moreover, we have characterized the gene responsible for their synthesis, vanM. VanM is homologous to AinS of V. fischeri and LuxM* of V. harveyi. Thus, like V. fischeri, V. anguillarum contains an AHL synthase from the LuxI (36) and the LuxM*/AinS families.
Protein sequence comparisons revealed that VanM and LuxM* are 64% identical, whereas AinS is only 35% identical to LuxM*. Thus, AinS appears to be significantly different from VanM and LuxM*. Accordingly, AinS could represent a different subset of synthases within this new family. Even though LuxM* and VanM are closely related, they direct the synthesis of different AHLs. However, both make 3-hydroxylated AHLs, in contrast to the AHL synthesized via AinS, which lacks a C-3 acyl side chain substituent.
VanN belongs to a family of hybrid sensor kinases that contain two or more signaling domains within a single polypeptide (for a review, see reference 40). Like LuxN, VanN contains nine possible membrane-spanning regions at its amino terminus, suggesting that VanN is a membrane-bound protein. This region (Fig. 2B) may contain the signal input domain that responds to the AHLs produced by VanM. Attempts to complement a V. harveyi luxN mutant with vanN have not been successful. However, spent culture supernatants from V. anguillarum NB10 do not activate either the wild-type V. harveyi or a luxQ-negative mutant (unpublished data), suggesting that vanN is unlikely to complement a luxN mutation unless vanM or exogenous 3-hydroxy-C6-HSL is also provided. As shown in Fig. 2, VanN appears to contain a centrally located sensor kinase domain required for autophosphorylation, nucleotide binding, and autokinase activity (reviewed in reference 39). A typical response regulator domain is found at the carboxy terminus that contains the characteristic asparagine, which receives a phosphoryl group from a histidine residue in a sensor kinase domain. A Lys-109 is also present, which may play a role in triggering a conformational change in the protein upon phosphorylation of the asparagine residue (reviewed in reference 39). In V. harveyi, the phosphoryl group of LuxN is passed to an additional phosphotransfer protein, LuxU, and then to the ς54 activator LuxO (15, 16). Once phosphorylated, LuxO, together with ς54, activates the expression of an unidentified repressor of bioluminescence (5, 15, 28). Thus, V. anguillarum may contain homologues of the V. harveyi proteins LuxU and LuxO. Furthermore, V. anguillarum may possess a gene with homology to luxR from V. harveyi. Jobling and Holmes (25) have shown, by Southern blot analysis, that the hapR gene from V. cholerae, also a homologue of the V. harveyi luxR gene, cross-hybridizes to chromosomal DNA from V. anguillarum.
In addition to the VanMN system identified in this study, V. anguillarum may also possess homologues of LuxS, LuxP, and LuxQ from V. harveyi (6, 51). These proteins are part of a second quorum-sensing system that works in parallel to the LuxLMN circuit (15, 16) by transferring a phosphate group onto the same phosphorelay protein, LuxU. The presence of this second circuit may provide an explanation as to why protease production and/or virulence were not affected in the vanM and vanN mutants. In V. harveyi, both sensory channels need to be inactivated for loss of bioluminescence; if only one channel is inactivated, the second channel will compensate, ensuring that the bacterium still produces light (4, 6). In our studies, we have made mutations in only one of two possible sensory channels. If V. anguillarum does indeed contain a second sensory channel as does V. harveyi, then we would not expect a loss of function in the mutants that we have made. Therefore, we cannot rule out that quorum sensing does not regulate virulence genes or protease production until we have determined whether a second sensory channel is present. Moreover, Denkin and Nelson (11) have shown that the expression of the metalloprotease gene, empA, is ninefold higher in broth containing fish gastrointestinal mucus than in conventional broth. Consequently, metalloprotease production may be affected differently in vanM and vanN mutants grown in the presence of fish gastrointestinal mucus.
The AHL profiles for the vanM and vanN mutants, however, may conflict with the presence of a putative dual-sensory-channel model in V. anguillarum. A mutation in vanM not only abolished the production of C6-HSL and 3-hydroxy-C6-HSL but also affected 3-oxo-C10-HSL production, suggesting that the VanMN system, in some way, regulates VanIR. In contrast, the vanN mutant showed wild-type AHL production. However, in a dual-pathway model, a mutation in vanM should not affect the production of 3-oxo-C10-HSL. There are several possibilities to explain how a vanM mutation may affect the production of 3-oxo-C10-HSL. First, the two-sensory-channel model may not exist in V. anguillarum, and quorum sensing may not regulate virulence genes or protease production. If this is true, then the vanN mutant should also be negative for AHL production, as there would be no second compensatory system as in V. harveyi. Second, the V. anguillarum VanR protein, a homolog of LuxR from V. fischeri (36), may bind 3-hydroxy-C6-HSL and/or C6-HSL as well as 3-oxo-C10-HSL to become an active transcriptional activator of vanI, which directs the synthesis of 3-oxo-C10-HSL. However, for the LuxR homologues studied to date, the cognate AHL seems to be required for maximal activation of gene expression (42, 46). Alternatively, multiple AHL signals may be needed for activation of VanR. Finally, perhaps there is an additional, unidentified protein or signal molecule in this system that allows the VanMN circuit to function independently of a second sensory channel.
Thus far, the only clear function of either of the two quorum-sensing circuits in V. anguillarum is that the VanMN quorum-sensing circuit regulates the production of 3-oxo-C10-HSL via the VanRI quorum-sensing circuit, which could potentially regulate the expression of target genes. Additional phenotypic analyses have not yet revealed any further role for the AHLs produced by V. anguillarum. However, given the potential complexity of the V. anguillarum quorum-sensing systems, the mutant strains that we are using may not be sufficient to permit the complete elucidation of the role of quorum sensing in this organism. Characterization of any additional genes in the VanMN circuit should help to identify AHL-regulated genes in V. anguillarum. One possible role of 3-oxo-C10-HSL may be as an effector of genes found in other bacteria besides V. anguillarum. Interestingly, 3-oxo-C10-HSL has been shown to antagonize protease activities of Aeromonas hydrophila (53) and Aeromonas salmonicida (52). These proteases are known to contribute to virulence and to be regulated by quorum sensing. Since these organisms are all fish pathogens, Swift et al. (53) have suggested that a possible role for 3-oxo-C10-HSL may be to antagonize the quorum-sensing dependent virulence of A. hydrophila and A. salmonicida, which in turn may provide V. anguillarum with a competitive edge during fish infection. Another possibility is that the long-acyl-chain AHL, 3-oxo-C10-HSL, has an effect on eukaryotic cell behavior. Previously it has been shown that 3-oxo-C12-HSL, produced by the human pathogen P. aeruginosa, has immunomodulatory and vasodilator activities in various animal models processes (for a review, see reference 56). Furthermore, activity seems to be dependent on the presence of a long acyl chain since 3-oxo-C6-HSL was inactive. These studies imply that 3-oxo-C12-HSL plays a role not only in regulating P. aeruginosa virulence gene expression but also in the orchestration of eukaryotic cells to maximize the provision of nutrients via the bloodstream while downregulating host defense mechanisms. Since V. anguillarum produces a long-acyl-chain AHL, it is possible that 3-oxo-C10-HSL may regulate eukaryotic cell behavior and modulate the disease process within fish.
ACKNOWLEDGMENTS
We thank Karen McGee for making many of the mutants, Mavis Daykin for HPLC analysis, Cath Otori for LC-MS analysis, and Ram Chhabra and Chris Harty for the synthesis of AHLs.
This work was supported by grants from the Swedish Council for Forestry and Agricultural Research, the Swedish Research Council for Engineering Sciences, and the Carl Tryggers Foundation, Sweden (to D.L.M.), and by grants and a studentship from the Biotechnology and Biological Sciences Research Council, United Kingdom (to A.H., M.C., and P.W.), which are gratefully acknowledged.
REFERENCES
- 1.Arico B, Miller J F, Roy C, Stibitz S, Monack D, Falkow S, Gross R, Rappuoli R. Sequences required for expression of Bordetella pertussis virulence factors share homology with prokaryotic signal transduction proteins. Proc Natl Acad Sci USA. 1989;86:6671–6675. doi: 10.1073/pnas.86.17.6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson S, Throup J P, Stewart G S A B, Williams P. A hierarchical quorum-sensing system in Yersinia pseudotuberculosis is involved in the regulation of motility and clumping. Mol Microbiol. 1999;33:1267–1277. doi: 10.1046/j.1365-2958.1999.01578.x. [DOI] [PubMed] [Google Scholar]
- 3.Bainton N J, Stead P, Chhabra S R, Bycroft B W, Salmond G P C, Stewart G S A B, Williams P. N-(3-Oxohexanoyl)-l-homoserine lactone regulates carbapenem antibiotic production in Erwinia carotovora. Biochem J. 1992;288:997–1004. doi: 10.1042/bj2880997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassler B L, Wright M, Showalter R E, Silverman M R. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol. 1993;9:773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- 5.Bassler B L, Wright M, Silverman M R. Sequence and function of LuxO, a negative regulator of luminescence in Vibrio harveyi. Mol Microbiol: 1994;12:403–412. doi: 10.1111/j.1365-2958.1994.tb01029.x. [DOI] [PubMed] [Google Scholar]
- 6.Bassler B L, Wright M, Silverman M R. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol Microbiol. 1994;13:273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- 7.Bassler B, Greenberg E P, Stevens A M. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J Bacteriol. 1997;179:4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cámara M, Daykin M, Chhabra S R. Detection, purification and synthesis of N-acyl homoserine lactone quorum sensing molecules. Methods Microb Bacter Pathog. 1998;27:319–330. [Google Scholar]
- 9.Cao J-G, Meighen E A. Purification and structural identification of an autoinducer for the luminescence system of V. harveyi. J Biol Chem. 1989;264:21670–21676. [PubMed] [Google Scholar]
- 10.Chhabra S R, Stead P, Bainton N J, salmond G P C, Stewart G S A B, Williams P, Bycroft B W. Autoregulation of carbapenem biosynthesis in Erwinia carotovora by analogues of N-(3-oxohexanoyl)-l-homoserine lactone. J Antibiot. 1993;46:441–449. doi: 10.7164/antibiotics.46.441. [DOI] [PubMed] [Google Scholar]
- 11.Denkin S M, Nelson D R. Induction of protease activity in Vibrio anguillarum by gastrointestinal mucus. Appl Environ Microbiol. 1999;65:3555–3560. doi: 10.1128/aem.65.8.3555-3560.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunny G M, Winans S C. Cell-cell signaling in bacteria. Washington, D.C.: American Society for Microbiology; 1999. [Google Scholar]
- 14.Eberhard A, Burlingame A L, Kenyon G L, Nealson K H, Oppenheimer N J. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry. 1981;20:2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- 15.Freeman J A, Bassier B L. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibiro harveyi. Mol Microbiol. 1999;31:665–677. doi: 10.1046/j.1365-2958.1999.01208.x. [DOI] [PubMed] [Google Scholar]
- 16.Freeman J A, Bassler B L. Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J Bacteriol. 1999;181:899–906. doi: 10.1128/jb.181.3.899-906.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuqua C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 18.Gilson L, Kuo A, Dunlap P V. AinS and a new family of autoinducer synthesis proteins. J Bacteriol. 1995;177:6946–6951. doi: 10.1128/jb.177.23.6946-6951.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanzelka B L, Greenberg E P. Quorum sensing in Vibrio fischeri: evidence that S-adenosylmethionine is the amino acid substrate for the autoinducer synthesis. J Bacteriol. 1996;178:5291–5294. doi: 10.1128/jb.178.17.5291-5294.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanzelka B L, Parsek M R, Val D L, Dunlap P V, Cronan J E, Jr, Greenberg E P. Acylhomoserine lactone synthase activity of the Vibrio fischeri AinS protein. J Bacteriol. 1999;181:5766–5770. doi: 10.1128/jb.181.18.5766-5770.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardman A M, Stewart G S A B, Williams P. Quorum sensing and the cell-cell communication dependent regulation of gene expression in pathogenic and non-pathogenic bacteria. Antonie Leeuwenhoek J Microbiol. 1998;74:199–210. doi: 10.1023/a:1001178702503. [DOI] [PubMed] [Google Scholar]
- 22.Hrabak E M, Willis D K. The lemA gene required for pathogenicity of Pseudomonas syringae pv. syringae on bean is a member of a family of two-component regulators. J Bacteriol. 1992;174:3011–3020. doi: 10.1128/jb.174.9.3011-3020.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iuchi S, Matsuda Z, Fujiwara T, Lin E C. The arcB gene of Escherichia coli encodes a sensor-regulator protein for anaerobic repression of the arc modulon. Mol Microbiol. 1990;4:715–727. doi: 10.1111/j.1365-2958.1990.tb00642.x. [DOI] [PubMed] [Google Scholar]
- 24.Jiang Y, Cámara M, Chhabra S R, Hardie K H, Bycroft B W, Lazdunski A, Salmond G P, Stewart G S A B, Williams P. In vitro biosynthesis of the Pseudomonas aeruginosa quorum sensing signal molecule N-butanoyl-l-homoscrine lactone. Mol Microbiol. 1998;28:193–203. doi: 10.1046/j.1365-2958.1998.00789.x. [DOI] [PubMed] [Google Scholar]
- 25.Jobling M G, Holmes R K. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol Microbiol. 1997;26:1023–1034. doi: 10.1046/j.1365-2958.1997.6402011.x. [DOI] [PubMed] [Google Scholar]
- 26.Kuo A N, Blough V, Dunlap P V. Multiple N-acyl-l-homoserine lactone autoinducers of luminescence in the marine symbiotic bacterium Vibrio fischeri. J Bacteriol. 1994;176:7558–7565. doi: 10.1128/jb.176.24.7558-7565.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Latifi A, Winson M K, Foglino M, Bycroft B W, Stewart G S A B, Lazdunski A, Williams P. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol Microbiol. 1995;17:333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- 28.Lilly B N, Bassler B L. Regulation of quorum sensing in Vibrio harveyi by LuxO and Sigma 54. Mol Microbiol. 2000;36:940–954. doi: 10.1046/j.1365-2958.2000.01913.x. [DOI] [PubMed] [Google Scholar]
- 29.McClean K H, Winson M K, Fish L, Taylor A, Chhabra S R, Camara M, Daykin M, Lamb J H, Swift S, Bycroft B W, Stewart G S A B, Williams P. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition of the detection of N-acylhomoserine lactones. Microbiol. 1997;143:3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 30.McGee K, Hörstedt P, Milton D L. Identification and characterization of additional flagellin genes from Vibrio anguillarum. J Bacteriol. 1996;178:5188–5198. doi: 10.1128/jb.178.17.5188-5198.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meighen E A. Molecular biology of bacterial bioluminescence. Microbiol Rev. 1991;55:123–142. doi: 10.1128/mr.55.1.123-142.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meighen E A. Genetics of bacterial bioluminescence. Annu Rev Genet. 1994;28:117–139. doi: 10.1146/annurev.ge.28.120194.001001. [DOI] [PubMed] [Google Scholar]
- 33.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milton D L, Norqvist A, Wolf-Watz H. Cloning of a metalloprotease gene involved in the virulence mechanism of Vibrio anguillarum. J Bacteriol. 1992;174:7235–7244. doi: 10.1128/jb.174.22.7235-7244.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milton D L, O'Toole R, Hörstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. J Bacteriol. 1996;178:1310–1319. doi: 10.1128/jb.178.5.1310-1319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milton D L, Hardman A, Camara M, Chhabra S R, Bycroft B W, Stewart G S A B, Williams P. Quorum sensing in Vibrio anguillarum: characterization of the vanI/vanR locus and identification of the autoinducer N-(3-oxodecanoyl)-l-homoserine lactone. J Bacteriol. 1997;179:3004–3012. doi: 10.1128/jb.179.9.3004-3012.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moré M I, Finger L D, Stryker J L, Fuqua C, Eberhard A, Winans S C. Enzymatic synthesis of a quorum-sensing autoinducer through use of defined substrates. Science. 1996;272:1655–1658. doi: 10.1126/science.272.5268.1655. [DOI] [PubMed] [Google Scholar]
- 38.Nagasawa S, Tokishita S, Aiba H, Mizuno T. A novel sensor-regulator protein that belongs to the homologous family of signal transduction proteins involved in adaptive responses in Escherichia coli. Mol Microbiol. 1992;6:799–807. doi: 10.1111/j.1365-2958.1992.tb01530.x. [DOI] [PubMed] [Google Scholar]
- 39.Norqvist A, Hagström Å, Wolf-Watz H. Protection of rainbow trout against vibriosis and furunculosis by the use of attenuated strains of Vibrio anguillarum. Appl Environ Microbiol. 1989;55:1400–1405. doi: 10.1128/aem.55.6.1400-1405.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 41.Parsek M R, Val D L, Hanzelka B L, Cronan J E, Jr, Greenberg E P. Acyl homoserine lactone quorum-sensing signal generation. Proc Natl Acad Sci USA. 1999;96:4360–4365. doi: 10.1073/pnas.96.8.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Passador L, Tucker K D, Guertin K R, Journet M P, Kende A S, Iglewski B H. Functional analysis of the Pseudomonas aeruginosa autoinducer PAI. J Bacteriol. 1996;178:5995–6000. doi: 10.1128/jb.178.20.5995-6000.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reed L J, Muench J. A simple method of estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 44.Salmond G P C, Bycroft B W, Stewart G S A B, Williams P. The bacterial ‘enigma’: cracking the code of cell-cell communication. Mol Microbiol. 1995;16:615–624. doi: 10.1111/j.1365-2958.1995.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 46.Schaefer A L, Hanzelka B L, Eberhard A, Greenberg E P. Quorum sensing in Vibrio fischeri: probing autoinducer-LuxR interactions with autoinducer analogs. J Bacteriol. 1996;178:2897–2901. doi: 10.1128/jb.178.10.2897-2901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaefer A L, Val D L, Hanzelka B L, Cronan J E, Jr, Greenberg E P. Generation of cell-to cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc Natl Acad Sci USA. 1996;93:9505–9509. doi: 10.1073/pnas.93.18.9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shaw P D, Ping G, Daly S L, Chung C, Cronan Jr J E, Rhinehart K L, Farrand S K. Detecting and characterizing N-acylhomoserine lactone signal molecules by thin-layer chromotagraphy. Proc Nat Acad Sci USA. 1997;94:6036–6041. doi: 10.1073/pnas.94.12.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:787–796. [Google Scholar]
- 50.Stout V, Gottesman S. RcsB and RcsC: A two-component regulator of capsule synthesis in Escherichia coli. J Bacteriol. 1990;172:659–669. doi: 10.1128/jb.172.2.659-669.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Surette M G, Miller M B, Bassler B L. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci USA. 1999;96:1639–1644. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swift S, Karlyshev A V, Durant E L, Winson M K, Chhabra S R, Williams P, Macintyre S, Stewart G S A B. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologues AhyRI and AsaRI and their cognate signal molecules. J Bacteriol. 1997;179:5271–5281. doi: 10.1128/jb.179.17.5271-5281.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swift S, Lynch M J, Fish L, Kirke D F, Tomás J M, Stewart G S A B, Williams P. Quorum sensing-dependent regulation and blockade of exoprotease production in Aeromonas hydrophila. Infect Immun. 1999;67:5192–5199. doi: 10.1128/iai.67.10.5192-5199.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang J L, Liu Y N, Barber C E, Dow J M, Wootton J C, Daniels M J. Genetic and molecular analysis of a cluster of rpf genes involved in positive regulation of synthesis of extracellular enzymes and polysaccharide in Xanthomonas campestris pathovar campestris. Mol Gen Genet. 1991;226:409–417. doi: 10.1007/BF00260653. [DOI] [PubMed] [Google Scholar]
- 55.Val D L, Cronan J E., Jr In vivo evidence that S-adenosylmethionine and fatty acid synthesis intermediates are the substrates for the LuxI family of autoinducer substrates. J Bacteriol. 1998;180:2644–2651. doi: 10.1128/jb.180.10.2644-2651.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williams P, Camara M, Hardman A, Swift S, Milton D, Hope V J, Winzer K, Middleton B, Pritchard D I, Bycroft B W. Quorum sensing and the population dependent control of virulence. Philos Trans R Soc Lond Sect B. 2000;355:1–14. doi: 10.1098/rstb.2000.0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Winson M K, Camara M, Latifi A, Foglino M, Chhabra S R, Daykin M, Bally M, Chapon V, Salmond G P C, Bycroft B W, Lazdunski A, Stewart G S A B, Williams P. Multiple N-acyl-l-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:9427–9431. doi: 10.1073/pnas.92.20.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Winson M K, Swift S, Fish L, Throup J P, Jorgensen F, Chhabra S R, Bycroft B W, Williams P, Stewart G S A B. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acylhomoserine lactone-mediated quorum sensing. FEMS Microbiol Lett. 1998;163:185–192. doi: 10.1111/j.1574-6968.1998.tb13044.x. [DOI] [PubMed] [Google Scholar]