Abstract
Necroptosis, a programmed form of necrotic cell death, plays critical regulatory roles in the progression and metastatic spread of cancers such as cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC). However, there are few articles systematically analyzing the necroptosis-related long non-coding RNAs (NRlncRNAs) correlated with CESC patients. Both RNA-sequencing and clinical data of CESC patients are downloaded from TCGA database in this study. Pearson correlation analysis, least absolute shrinkage, operator algorithm selection and Cox regression model are employed to screen and create a risk score model of eleven-NRlncRNAs (MIR100HG, LINC00996, SNHG30, LINC02688, HCG15, TUBA3FP, MIAT, DBH-AS1, ERICH6-AS1SCAT1, LINC01702) prognostic. Thereafter, a series of tests are carried out in sequence to evaluate the model for independent prognostic value. Gene set enrichment analytic paper, Gene Ontology analysis, Kyoto Encyclopedia of Genes and Genomes pathway enrichment analytic paper make it clear that immune-related signaling pathways are very rich in the high-risk subgroup. Additionally, the prognostic risk score model is correlated to immune cell infiltration, potential immune checkpoint, immune function, immune micro-environmental and m6A-related gene. Mutation frequency in mutated genes and survival probability trend are higher in the low-risk subgroup in most of test cases when compared to the high-risk subgroup. This study constructs a renewed prognostic model of eleven-NRlncRNAs, which may make some contribution to accurately predicting the prognosis and the immune response from CESC patients, and improve the recognition of CESC patients and optimize customized treatment regimens to some extent.
Subject terms: Cancer, Computational biology and bioinformatics, Genetics, Immunology, Molecular biology, Biomarkers
Introduction
Cervical cancer is the second most prevalent gynecological cancer1, one of leading causes of female death due to cancer in the world2. Cervical cancer is globally developing. More than 270,000 women worldwide die from cervical cancer each year3, especially in developing countries4,5. In the case of cervical cancer, the cervical squamous cell carcinoma and endocervical adenocarcinomas (CESC) accounts for about 10 to 15% of all tumor-related, female deaths6. The prognosis remains weak for patients at the terminal stage of CESC and most of CESC patients may have already gone into the middle or terminal stages when they get diagnosed initially, despite that the technology for diagnosis and treatment of CESC has developed a lot over the past 30 years. It is greatly significant to develop a novel and validated biomarker for CESC prognosis to guide clinical experience. With the deepening study of the cell death mechanisms, more and more novel cell-death modalities have been discovered and named. Necroptosis is a form of regulatory necrotic cell death adjusted by receptor-interacting protein [RIP] kinase 1(RIPK1), RIPK3 and activated by mixed lineage kinase domain-like pseudo kinase (MLKL), which was discovered and named in 2005 Degterev et al.7. Current studies have pointed out that necroptosis had the double roles in promoting and suppressing tumor and that a novel strategy to treat cancers might be – targeting necroptosis8. Park et al. reported that RIPK1 overexpression inhibited p53 pathway through the activation of NF-kappaB and that it had to do with a worse prognosis in glioblastoma9. A recent study showed that RIPK3 was a critical regulator in the malignant transformation of hepatocellular carcinoma (HCC) and that to regulate necroptosis may improve the chemosensitivity of HCC10. And by the same token, necroptosis has an important effect on cervical cancers. More tantalizingly, MLKL is a prognostic biomarker for the pancreatic cancer and the increased expression of MLKL may improve the overall survival rate11. There was a study which indicated that the expression status of RIPK3 acted as a critically influential factor for the release of IL-1α and the outcome of PolyIC-based immunotherapeutic approaches in cervical cancer cells12. These studies all showed that the key necroptotic factors which were mediated by escaping from necroptosis, presented one of the relevant molecular mechanisms of CESC progression and metastasis. There may be valuable implications for the treatment and prognostication of CESC patients in the future to induce necroptosis. Nevertheless, there is nearly no researches on the association between CESC and necroptosis in a wide and deep manner at present and the academia doesn’t figure out the clear role of necroptosis in the prognosis of CESC patients. Therefore, the team of this study needs to find out more novel necroptosis-related biomarkers for CESC patients to identify better treatments and predict prognostic effects in a better way.
Long non-coding RNAs (lncRNAs) are a subset of non-coding RNAs produced by the transcription of RNA polymerase II, which is 200-nucleotide longer in length13 and lacks the function of protein-coding14. A variety of lncRNAs expressions are abnormal in the tissues of cervical cancer, which is pointed out in some studies, and the expression level is significantly correlated with the invasion, apoptosis, proliferation and metastasis of cervical cancer cells in addition to the proliferation and metastasis of cervical cancer15. lncRNAs may help cervical cancer cells in combatting radiotherapy and chemotherapy, thus to promote the growth of cancer16. However, few systematically analytical papers focused on NRlncRNAs correlated to the prognosis of cervical cancer.
It is proven that high-throughput sequencing technology and bioinformatics approaches help explore valuable disease-specific biomarkers. This study investigated the molecular and signaling pathways recognizing NRlncRNAs so that the pavement can be provided for the research and development of a renewed risk-score model to predict the prognosis and select the immunotherapies of CESC patients. The first NRlncRNAs signature model with CESC was constructed, which may provide a new perspective to discover more accurate treatments for CESC patients.
Materials and methods
Raw data acquisition
Both mRNA and lncRNA sequencing data were selected from 303 CESC cases and 3 normal cervical tissue samples in The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov) and R (R 4.1.2, https://www.r-project.org/) was used to extract and standardize the data. A total of 104 necroptosis-related genes (NRGs) were retrieved from previous researches provided in the Appendix Table S1. The Pearson correlation coefficients were also calculated to evaluate the connection between lncRNAs and NRGs to identify lncRNAs related to NRGs. The absolute value of Pearson correlation coefficients greater than 0.4 (|R|> 0.4) and the p value less than 0.001 (p < 0.001) were used to filter the NRGs-related lncRNAs (NRGlncRNAs). Both LncRNA expression profile and relevant clinical data of CESC tissue samples originated from the GEO database (http://www.ncbi.nlm.nih.gov/geo). The TCGA samples, as validation datasets, were randomly divided into two equal numbers of groups and they were named Test1 and Test2, respectively. Since the TCGA database is publicly accessible, this study is exempted from the approval of the Ethics Committee. R is a software environment for statistical computing and graphics, which can be used for free. It compiles and runs on a wide variety of UNIX platforms, Windows and MacOS. R version 4.1.2; link: https://www.r-project.org/.
Construction and validation of a prognostic necroptosis-related lncrnas signature
Univariate Cox regression analysis is used to perform the screening of overall survival (OS) and identify the OS-related prognostic lncRNAs in the extracted genes (p < 0.05). Fifty-six genes associated with survival were selected for the further analysis. The least absolute shrinkage and selection operator (LASSO) are used to construct the Cox regression model using R software ‘glmnet’ package in order to precisely secure the range of the candidate genes and construct the prognostic model17,18. The risk gene was obtained through multivariate Cox regression analysis, while the risk score model was constructed to predict the survival time and the risk score coefficient (coefi) was included in the gene output of the model. According to the median risk score, these cases are categorized into two risk subgroups. The following is the calculation formula of the risk score model (X: coefficients, Y: gene expression level).
Predictive value of the LncRNA model
The principal component analysis (PCA) and t-SNE analysis were carried out to explore the data distribution for two risk subgroups and visualize the two groups using the “Rtsne” and “ggplot2” packages in R. The OS of the two risk subgroups were compared in the risk scores using the “survival” package. The “Time ROC” and “Clinical ROC” package were further used to evaluate the prediction accuracy and visualize the distribution of risk scores across clinical data. Univariate and multivariate Cox regression analyses were performed to evaluate the association between variables, while risk values and prognosis were used to test whether the risk model acts as an independent prognostic index in the treatment of CESC or not. A nomogram was used to predict the prognosis of CESC patients using the “rms” package. Calibration curves were plotted to compare and verify the survival probability rate of 1, 3 and 5-year nomogram prediction.
Construction of the mRNA-lncRNA co-expression network
The mRNA-lncRNA co-expression network was structured and visualized using Cytoscape software (software information is version 3.7.2 at http://www.cytoscape.org/) to additionally figure out the potential relationship and interaction between NRlncRNAs and their corresponding mRNAs.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA; http://www.broadinstitute.org/gsea) was conducted to explore the potential functional pathways or molecular mechanisms that contain the differentially-expressed NEGs between the high-risk and low-risk groups19. Min and max genomes of 15 and 500 genes were used to screen the NRGs sets.
Functional enrichment analysis
The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and the Gene Ontology (GO) analysis were conducted to identify the molecular functions (MF), cellular components (CC), biological process (BP), and NRlncRNAs-related signaling pathways.
Immune and M6A correlation analysis
A single-sample gene set enrichment analysis (ssGSEA) was performed to contrast the immune cell infiltration performance between the high-risk and low-risk groups and to discover the immune function20,21. Potential immune checkpoints were used to find out the association between immune-related genes and risk signatures. The immune microenvironment scores including stromal and immune scores were calculated using the “ESTIMATE” package, and the 'reshape2' and 'ggpubr' packages were used to visualized22. Spearman correlation analysis was conducted to investigate the connection between risk characteristics and M6A-related genes.
Grouping and survival analysis of TMB
The waterfall function within the "maftools" package was used to calculate the mutation frequency for every CESC sample and display the top 20 mutated genes in two high-risk and low-risk subgroups on a case-by-case basis. CESC samples were categorized into the high and low TMB groups based on the median value of tumor mutation burden (TMB). Kaplan–Meier analysis was carried out to compare the survival difference between the two TMB subgroups and the high-risk group and low-risk group.
Results
Construction of the prognostic model based on necroptosis-related lncRNAs for CESC
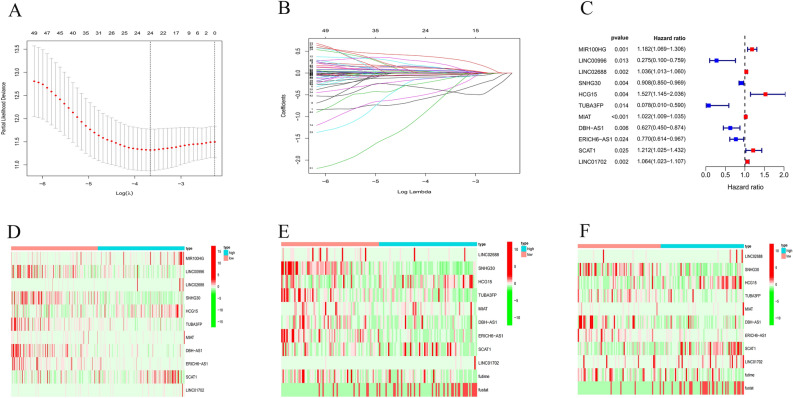
Univariate Cox regression analysis on OS was preliminarily carried out to identify 56 survival-related prognostic lncRNAs for the following analysis. Highly-correlated genes were screened out using the LASSO Cox regression model in order to prevent overfitting. The results showed that 24 out of 56 NRlncRNAs were beneficial to CESC prognostic model (Fig. 1A,B). And the Multivariate Cox regression analysis was adopted, and 11 NRlncRNAs including MIR100HG, LINC00996, LINC02688, SNHG30, HCG15, TUBA3FP, MIAT, DBH-AS1, ERICH6-AS1, SCAT1, LINC01702, were obtained eventually to develop an OS-related prognostic model (Fig. 1C–F).
Figure 1.
Identification of prognostic necroptosis-related lncRNAs in CESC patients. (A, B) Lasso Cox regression analysis showed that 24 of the 56 necroptosis-related lncRNAs were good candidates for construction prognostic model. (C–F) Forest plots and a heatmap (green: low expression level, red: high expression level) with two sets of validation datasets of the 11 necroptosis-related lncRNAs.
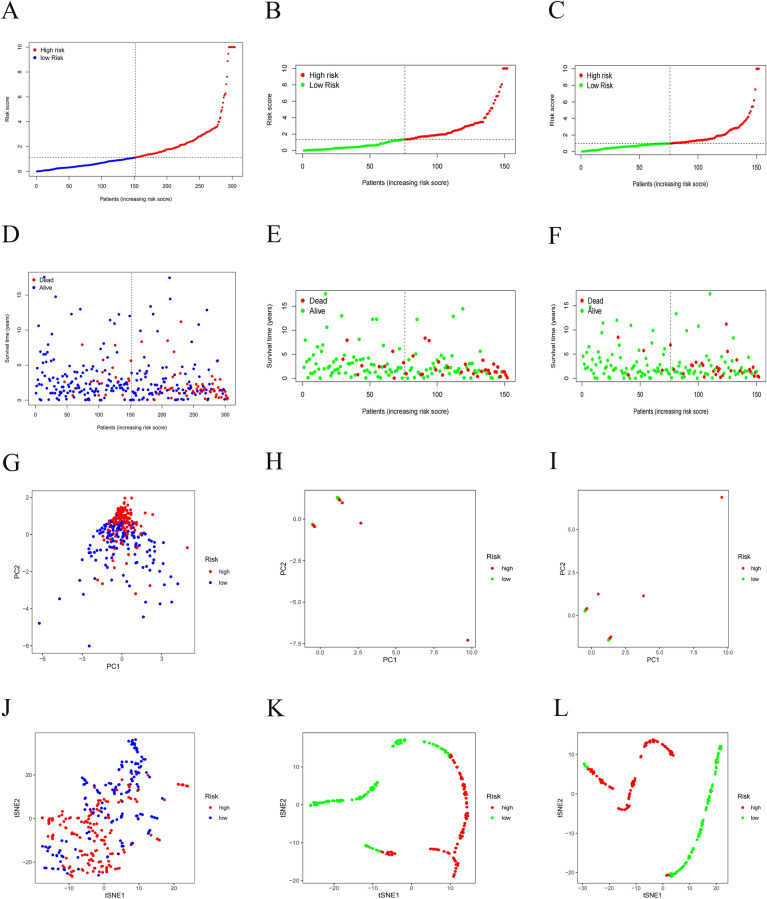
The median risk-score was used as a cut-off value to category CESC patients into low-risk and high-risk groups (Fig. 2A,D). PCA and t-SNE analyses confirmed that patients with CESC in the two risk subgroups were clearly distinguishable (Fig. 2G,J). At the same time, two sets of validation datasets were constructed to illustrate the generalization capacity of the models and the effect in other cohorts (Fig. 2B,C,E,F,H,I,K–L).
Figures 2.
Construction of the prognostic model based on necroptosis-related lncRNAs for CESC. (A–C) Risk score distribution with two sets of validation datasets, (D–F) survival status with two sets of validation datasets, (G–I) PCA plot, (J–L) t-SNE analysis with two sets of validation datasets of high and low-risk patients.
Associations between clinical characteristics and risk scores in CESC
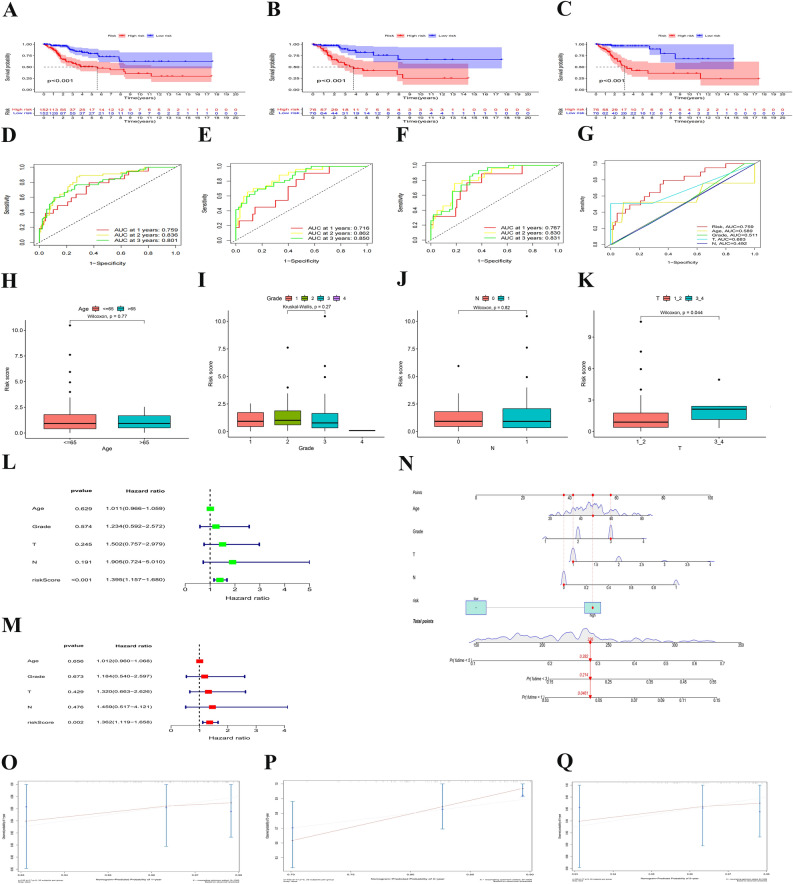
As shown in Fig. 3A–C, CESC patients in the low-risk group had a higher OS than that in the high-risk group. Through the time receiver operating characteristic (ROC) curve analysis, it was validated that the risk signature had moderate predictive accuracy at 1 (ROC = 0.759), 2 (ROC = 0.836) and 3 (ROC = 0.801) years (Fig. 3D). The remaining two datasets also supported the conclusion: The model had moderate prediction accuracy (Fig. 3E,F). At the same time, the Clinical ROC curve analysis proved that the signature had higher accuracy than other common clinicopathologic parameters (Fig. 3G). The prior data confirmed that the risk signature was a specific and sensitive indicator to predict the OS of CESC. The distribution of risk scores across clinical data was visualized (Fig. 3H–K).
Figure 3.
Associations between risk signature and clinicopathological factors. (A–C) Survival curve of CESC patients. (D–F) TimeROC curves with two sets of validation datasetsand and (G) Clinical ROC curves to forecast overall survival of patients. Visualization of risk scores in clinical data (H–K). Univariate (L) and multivariate Cox (M) regression of clinicopathological features in TCGE-CESC cohort. (N) Nomogram constructed to predit OS rates at 1, 3 and 5 years. (P–Q) The nomagram calibration curves were applied for investigation the deviation in nomagram-predicated and actual 1-, 3-, and 5-years survival probabilities.
The risk score was highly related to the OS of CESC patients according to multivariate and univariate Cox regression models, indicating that the risk score was an independent prognostic factor and that the above mentioned NRlncRNAs had independent prognostic benefits for CESC patients (Fig. 3L,M). Eventually, a nomogram was constructed to more accurately predict the survival outcome of CESC patients using the risk signature (Fig. 3N). Calibration curves were also used to verify the model's prediction accuracy (Fig. 3O–Q). The data showed that the 1, 3 and 5-year survival rate was in the high consistency with the actual survival rate according to the prediction of the constructed nomogram, which suggested a high prediction value.
Expression of the mRNA–lncRNA co-expression network
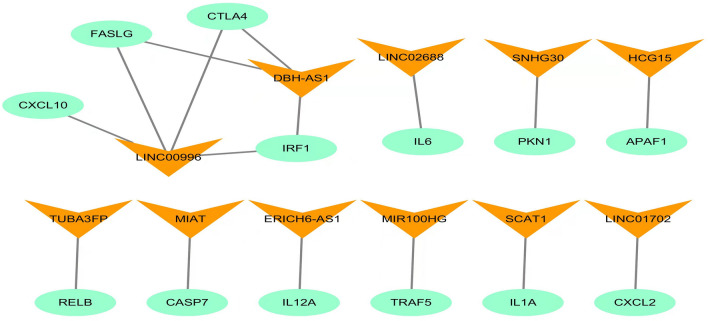
To investigate the potential effect of the 11 NRlncRNAs in CESC, Cytoscape was employed to construct the mRNA–lncRNA co-expression network. The lncRNA-mRNA co-expression network was involved in 16 mRNA–lncRNA pairs (Fig. 4). Of these, LncRNA LINC00996 was co-expressed with 4 NRGs (CXCL10, FASLG, CTLA4, IRF1), and LncRNA DBH − AS1 had the co-expression connection with 3 NRGs (FASLG, CTLA4, IRF1).
Figure 4.
Construction of the necroptosis-related lncRNA-mRNA co-expression network (A) Diagram of the necroptosis-related lncRNA-mRNA network.
Discovery of immunoregulatory pathways by GSEA
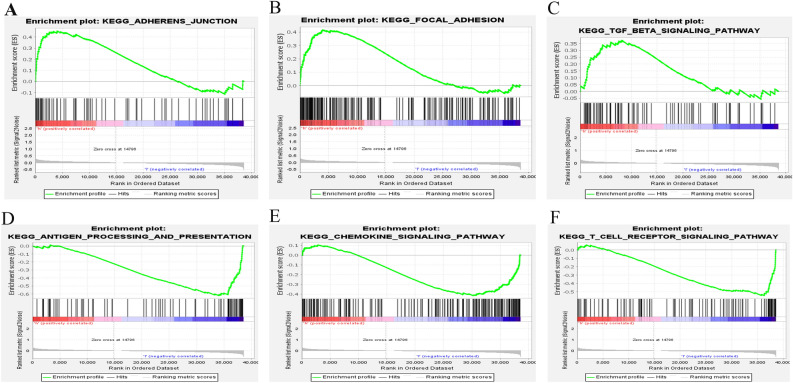
The GSEA was used to figure out the differences in the expressed NRGs between the two risk categories in the TCGA cohort to investigate the differential biological functions and the underlying signaling pathways associated with the NRlncRNAs. The findings revealed that “adherens junction”, “focal adhesion”, and “TGF beta signaling pathway” were all particularly active (Fig. 5A–C) in high-risk CESC patients. The results indicated that high risk was strongly associated with the occurrence, growth and metastasis of tumor. It was found that anti-cancer immunomodulatory pathways were distinctly up-regulated in the low-risk CESC patients such as antigen processing and presentation, chemokine signaling pathway and T cell receptor signaling pathway (Fig. 5D–F).
Figure 5.
Gene set enrichment analysis (GSEA) of high-risk group and low-risk group based on the necroptosis-related lncRNAs prognostic signature. (A–C) GSEA results show significant enrichment of adherens junction, focal adhesion, and TGF beta signaling pathway in the high-risk CESC patients. (D–F) GSEA results show significant enrichment of antigen processing and presentation, Chemokine signaling pathway and T cell receptor signaling pathway in the low-risk CESC patients.
Analysis of GO terms and KEGG pathway enrichment
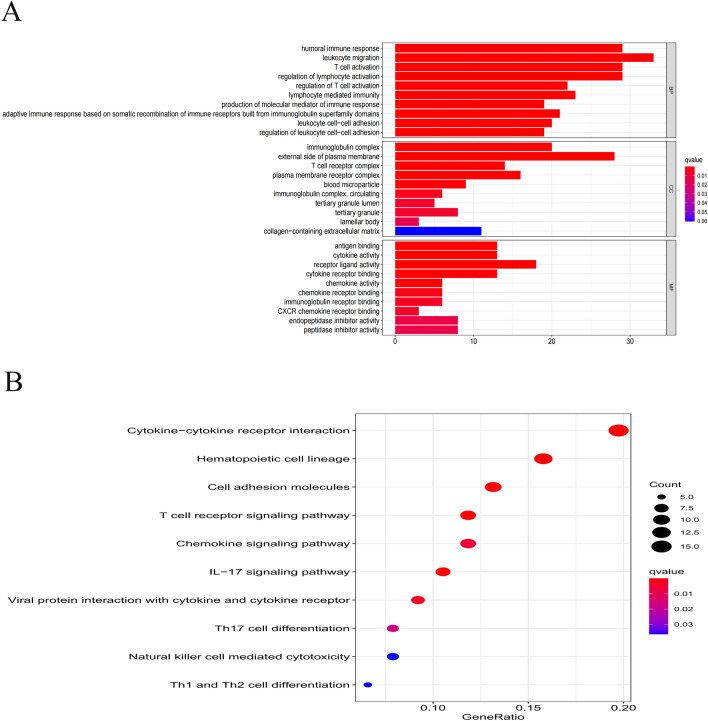
The GO analysis and KEGG pathway enrichment analyses were performed to progressively illuminate significant gene functions of the identified NRlncRNAs in CESC. The finding was that many classifications related to the immune processes showed significant enrichment changes by performing the GO enrichment analysis of differential expression levels of NEGs, especially in immune-related BP such as leukocyte migration, humoral immune response, T cell activation, regulation of lymphocyte activation and CC such as immunoglobulin complex, plasma membrane receptor complex and so on (p < 0.05; Fig. 6A). Further stated, KEGG pathway analyses demonstrated that the differentially-expressed genes were conspicuously enriched in cytokine -cytokine receptor interaction, hematopoietic cell lineage, cell adhesion molecules, T cell receptor signaling pathway, chemokine signaling pathway and so on (p < 0.05; Fig. 6B).
Figure 6.
Results of GO and KEGG analyses. (A) GO analysis showed differentially expressed genes between high- and low-risk groups were obviously enriched in immune-related biological processed, immune-related cell components. (B) KEGG analysis showed differentially expressed genes were enriched in immune-related pathway.
Associations with Immunity and M6A-related Genes
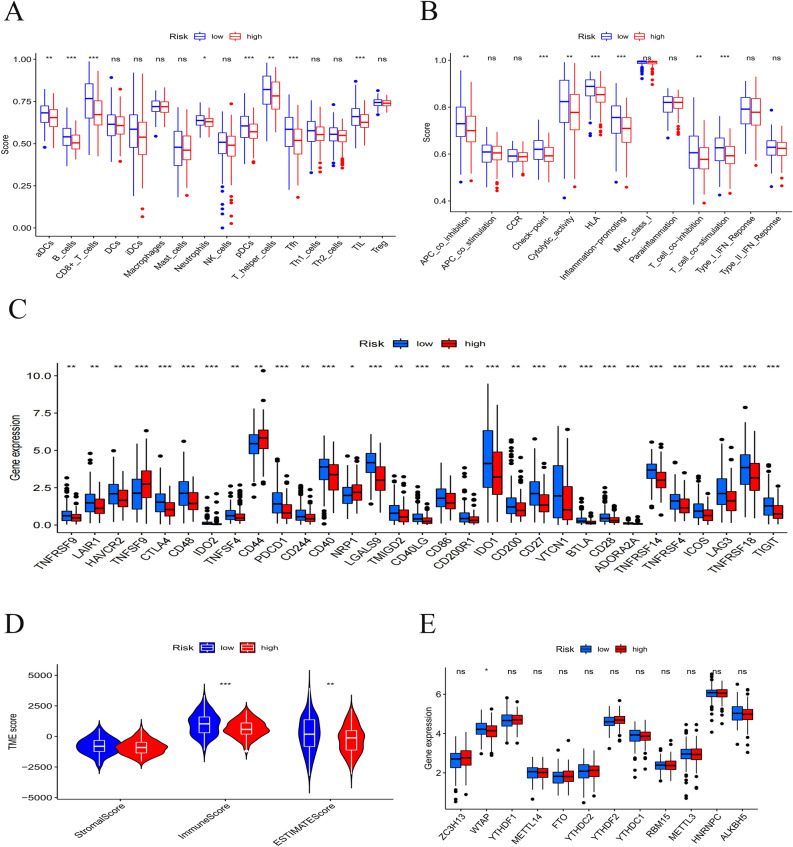
ssGSEA was employed to calculate diverse immune cell subpopulations, cellular functions, pathways and other factors to deeply figure out the relationship between prognostic risk score model and immunological state. As shown in Fig. 7A, the scores of most immune cells were largely different between the two risk subgroups. Not only the proportion of immune cell subpopulations, but also the level of immune-related functions in the high-risk subgroup was mostly lower than those in the low-risk subgroup (Fig. 7B). For immune checkpoints, all identified immune-related genes were expressed at a lower level in the high-risk subgroup, except TNFSF9, CD44 and NRP1 (Fig. 7C).
Figure 7.
Boxplots of scores of immune cells (A) and immune-associated functions (B) in risk subgroups. (C) Expression of immune checkpoints among two risk subgroups in CESC patients. Associations between risk signature immune microenvironment (D) and m6A-related genes (E).
Both immune micro-environments including stromal and immune scores and M6A-related genes played a critical role in regulating the proliferation and invasion of tumor cell. Through the existing cell infiltration data analysis, the ESTIMATE score model showed that the TME score of low-risk was higher than that in the high-risk group (Fig. 7D). For expressed levels of the M6A-related gene, WTAP were lower in the high-risk subgroup than those in the low-risk subgroup (Fig. 7E).
Tumor mutation burden (TMB) with survival outcomes
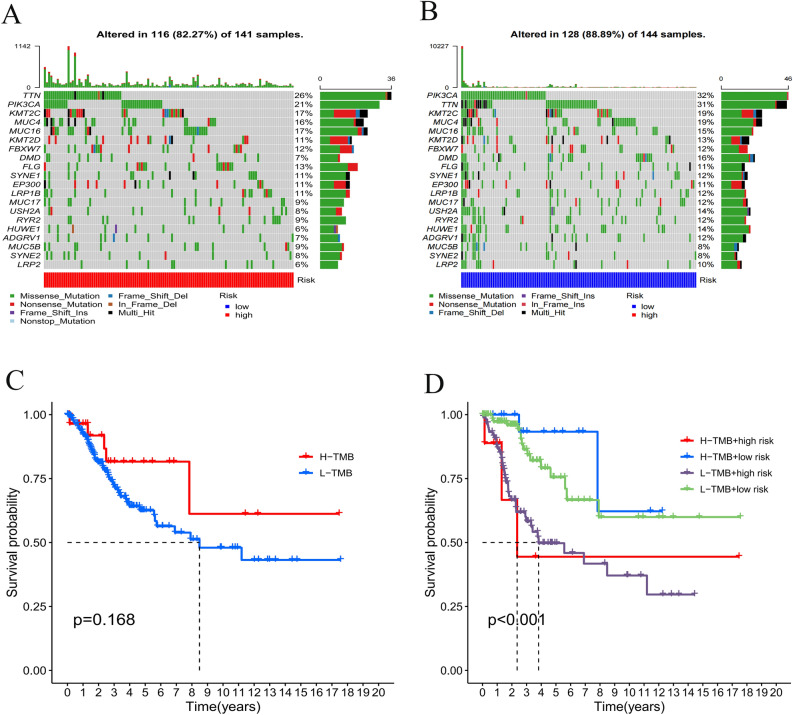
The maftools package was employed within R package maftools to display the mutation data findings. An exhaustive analysis on the gene mutation data of the CESC samples showed that the top 20 mutated genes were TTN, PIK3CA, KMT2C, MUC4, MUC16, KMT2D, FBXW7, DMD, FLG, SYNE1, EP300, LRP1B, MUC17, USH2A, RYR2, HUWE1, ADGRV1, MUC5B, SYNE2, LRP2. The results showed that the low-risk subgroup had a greater mutation frequency in the top 20 mutated genes than that in the high-risk group. (Fig. 8A,B). An examination of the Kaplan–Meier survival curves indicated a substantial difference between the two TMB subgroups (Fig. 8C), and the H-TMB + low risk group had the highest 2-year survival possibility trend in the four subgroups (Fig. 8D).
Figure 8.
Correlation of tumor mutation burden (TMB) with survival outcomes. (A) Waterfall chart of somatic mutations of necroptosis-related genes. The bar plot above showed the tumor mutation burden. The right number represented mutation frequency individually. (C) High TMB group was associated with higher survival probability. (D) The H-TMB + low risk group had the highest 2-year survival probability trend of the four subgroups.
Discussion
Cervical cancer is one of the leading causes of cancer deaths among women, and currently it is the fourth most common malignant tumor for torment women all over the world, with 604, 000 new diagnosed cases and approximately 342, 000 deaths reported in 2020 only23. CESC is considered the most common subtype of cervical cancer. The overall survival of CESC tends to be pessimistic due to the absence of early reliable and prognostic indicators, but some development has been made. The representatives are the screening technology for cervical cancer and prophylactic vaccination of HPV vaccine.
Necroptosis has advantages and disadvantages, and its specific effect depends on the cancer type and progression stage. Numerous studies showed that the downregulation of key necroptotic members such as CYLD24, RIPK325 and MLKL26 was common for some types of cancer, which might be one of the significant molecular mechanisms for tumor cells to escape necroptosis. Tumor necroptosis takes place in vivo and induces the immunosuppressive tumor microenvironment, playing a tumor-promoting role27. lncRNA is a key regulator in various biological processes of cells28. As an important regulatory molecule in tissues and cells of cervical cancer, lncRNA may become a molecular marker and therapeutic target for the diagnosis and prognosis of cervical cancer. Previous researches have widely developed LncRNAs differentially expression-based markers to predicte the prognosis of cervical cancer patients. A one-lncRNA (AC073320.1) signature could be used to predict lymph node metastasis (LNM) status and prognosis29. Another seven-lncRNA (HCG11, CASC15, HULC, LINC00173, LINC00189, LINC00905 and MIR22HG) signature has been proved to play a basic role in predicting the recurrence of CC and could be a potential prognostic biomarker30. Nevertheless, it remains unclear for what the role and the mechanism of necroptosis-related lncRNAs is in CC prognostic model.
This study screened out NRlncRNAs in CESC, and comprehensively analyzed the expression of NRlncRNAs in CESC and their relevance to a patient's prognosis, and successfully developed a unique prognostic risk model combining 11 NRlncRNAs(MIR100HG, LINC00996, LINC02688, SNHG30, HCG15, TUBA3FP, MIAT, DBH−AS1, ERICH6−AS1, SCAT1, LINC01702). Based on this prediction risk score model, the CESC patients were categorized into the high-risk and low-risk groups. After analyzing the differentially-expressed NRGs between the two risk subgroups, this study found that the prognostic model showed high accuracy in predicting the overall survival of CESC. Through gene set and functional enrichment analysis with the NRlncRNAs, immune-related BP and signaling pathways were observed to be dramatically different in the two risk subgroups. The mRNA-lncRNA co-expression network was built to investigate the potential relationship of the 11 NRlncRNAs in CESC. Finally, the levels of different immune cell infiltrations in CESC were investigated, at the same time, the immune functions detected, potential immune checkpoints detected and the expression level of M6A-related genes examined. It was the first NRlncRNA prognosis and immune response model for CESC patients, which may provide customized therapeutic strategies in treating CESC and offering clinicians the prognostic information earlier to improve clinical treatment effects. Based on the GSEA, it was found that the high-risk group exhibited significantly activated “adherence junction”, “focal adhesion”, and “TGF beta signaling pathway”. HPV induces changes in the expression levels of cell adhesion markers31, modulates composition and structure of keratinocyte junctions32 and perturbs the strength of cell–cell33, thus facilitating HIV embedded in cervical mucosa tissue34 and increasing the risk of cervical cancer. Focal adhesion kinase (FAK), a non-receptor cytoplasmic tyrosine kinase, plays a crucial role in cell adhesion, cell spreading and migration. Some previous studies proved that FAK overexpressed in different cancers and promoted the process of cancer invasion, migration and metastasis35–37, producing a low survival rate of cancer patients38,39. Meanwhile, it has been found that the significant over-expression of FAK in CESC cells was probably correlated to the carcinogenesis and the promotion of tumor metastasis, being a potential target for the treatment using anti-cancer drugs to inhibit the proliferation, migration and survival of cancer cell by inducing apoptosis40. In the early stage of tumor formation, TGF-β exerts a tumor suppressor effect in a large extension. In the late stage, however, it can change the extracellular matrix of tumor tissues, promote tumor invasion and metastasis by promoting neovascularization and weaken the killing effect of natural killer cells on tumor cells, and it can promote tumor grow by producing long-term immunosuppression to the host too41,42. TGF-β has a bidirectional regulator in different stages of cervical cancer. It is low-expressed in cervical tissues of patients with cervical intraepithelial neoplasia grade I–III although high-expressed in cervical cancer tissues43. In addition, TGF-β may be involved in the carcinogenesis and development of HPV infection-related cervical cancer by the interaction with the key pathogenic proteins (HPV16 E6, E7) in HPV infection44,45.
Additionally, it was also found that the risk signature was very rich in several immunoregulatory pathways such as antigen processing and presenting, chemokine signaling pathway and T cell receptor signaling pathway against cancers I. It can be reasonably hypothesized that NRlncRNAs signature was closely correlated with anti-tumor immunity in CESC.
In the context of necroptosis, a previous study discovered that specific molecules could be emitted to induce the adaptive immune response and stimulate antigen presenting cell (APC) to phagocytize tumor cell, and that signals released by necroptosis cells could be significant for surveillance against tumors46–48. Intra-tumoral necroptosis not only promoted tumor antigen loading through tumor antigen presenting cells based on RIPK1/ RIPK3 ectopic activation, but also enhanced DC-, and CD8 + T cell-mediated anti-tumor immunity function, which might provide a new idea for current T-cell-based immunotherapy49–51. These studies indicated that necroptosis was closely correlated with the anti-tumor immunity effect, implying that our hypothesis can be supported strongly.
ssGSEA was performed to further investigate the underlying relationship between necroptosis and immune cell infiltration in CESC. Nearly all infiltrated immune cells and immune-related functions of the high-risk group showed a considerable reduction in CESC tissues including B cells, CD8 + T cells, neutrophils pDCs and NK cells when compared with those in the low-risk group. It has been previously proved that the interaction of tumor-infiltrating immune cells with the microenvironment was effective in predicting malignant progression52 and that infiltrating lymphocytes were prognostic biomarkers53. Therefore, it is reasonable to conclude that the poor prognosis of CESC patients in the high-risk subgroup relates to the decrease in anti-tumor immunity degree.
In the early stage of cervical cancer, tumor can induce immune response. When human immunity is low, tumor can develop via immune escape54. However, there is still lack of effective treatments for patients diagnosed with advanced and recurrent cervical cancer, which may make an opportunity for the substitutional treatments including immune checkpoint inhibitor therapy. Programmed cell death 1 (PD-1) is a critically negative immunomodulatory factor, and its ligands consists of PD-L1 and PD-L255. PD-L1 expression can be detected in most cervical squamous cell carcinomas56. PD-L1 on the surface of tumor cells attenuates T cell-mediated cytotoxicity binding to PD-1 on the surface of activated T lymphocytes, which forms an immunosuppressive tumor microenvironment and promotes tumor cell proliferation and immune escape57. A completed phase Ib clinical trial (Keynote-028) demonstrated that the general response rate of the treatment with anti-PD-1 therapy (pembrolizumab) was 14.3% and that 91% of patients with the recurrent or metastatic cervical cancer had a response time of more than 6 months58. The positive expression of PD-L1 and PD-1 antibodies were observed to produce clinical outcomes and prognostic significance in cervical cancer59. Expression levels of nearly all immune checkpoint molecules were largely lower in high-risk group than those in the low-risk group, implying that the prognostic NRlncRNAs model could predict the expression of immune checkpoints in CESC, which might be beneficial to the efficacy of immune checkpoint blockade therapy in patients with CESC and offer a better option for anti-tumor immunity and immunotherapy. However, experimental and clinical data needs to be more for further validating the role of the necroptosis-related prognostic lncRNAs signature in predicting immunotherapy response from CESC patients.
The tumor microenvironment (TME) refers to a specific biological environment formed by tumor cells in conjunction with infiltrating immune cells, stromal cells, blood vessels, extracellular matrix and secretory factors in the tumor occurrence and development60,61. Immune cells of TME was observed to play an important role in tumor growth, invasion, and the stromal component is associated with tumor angiogenesis and extracellular matrix remodeling, thereby promoting tumor metastasis62,63. The results revealed that patients in the two risk subgroups had a higher immune score, which indicated a better response to immunotherapy. Therefore, TME might be used to predict the prognosis and immunotherapy efficacy in CESC patients. Wang et al.64 found that m6A levels in cervical cancer tissues were associated with tumor stage, size, differentiation, lymphatic infiltration and recurrence. NRlncRNAs signature of the study can not only effectively predict the expression levels of the m6A-related genes ZC3H13, FTO, YTHDF2, RBM15, WTAP, YTHDF1, METTL14, YTHDC2, YTHDC1, and ALKBH5 in CESC, but provide the academia with new idea and method to treat CESC.
TMB is a valuable biomarker to predict the clinical efficacy of immune checkpoint blockade (ICB) therapy for patients with many solid cancers65–69. Clinical trials and preclinical studies have shown that patients with tumors of higher somatic TMB (e. g. melanoma, breast cancer, non-small cell lung cancer) had longer life or better progression-free survival (PFS)70. This study investigated TMB of two risk subgroups. It was found that the lower group had a higher TMB and that there was a higher possibility for patients with high TMB to survive based on the findings of survival analysis. These results validated that TMB could be used to screen CESC patients for immunotherapy and predict the effect of immunotherapy and the prognostic status. Overall, TMB-based immunotherapy may be a promising oncology treatment for CESC patients.
Despite that there is a strong prediction value in the established risk model, there are some shortcomings in the current study. The significant shortcomings in this study lie in the lack of experimental validation of the identified targets. The mechanism by which necroptosis regulates the exact process of CESC remains unclear yet. It needs to be elucidated by further experimental studies.
Conclusions
In conclusion, this study constructs a novel necroptosis-related prognostic risk signature of eleven-lncRNAs which may lead to the accurate prediction of the prognosis and the immune response of CESC patients. It may be the first necroptosis-related lncRNA prognostic model for CESC. The findings may provide an insight into the specific role necroptosis-related lncRNAs plays in CESC, therefore promoting the prognosis improvement of CESC patients and optimizing the customized treatment regimens.
Supplementary Information
Acknowledgements
Thanks to everyone who contributed to this article.
Author contributions
Z.H.L. and J.N.Z. conceived and designed the study; Z.H.L. and X.H.S. searched the related articles; Z.H.L., X.H.S., J.N.Z., L.D.L. and S.J.Y. analyzed the data; Z.H.L., S.J.Y., L.D.L. and X.H.S. wrote the manuscript. J.D.Z. and J.L.W. supervised the whole process. All authors read and approved the final manuscript.
Data availability
The data for this study come from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). All the data in this paper support the results of this study. Details of R software: R is a free software environment for statistical computing and graphics. It compiles and runs on a wide variety of UNIX platforms, Windows and MacOS. R version 4.1.2; link: https://www.r-project.org/.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhiheng Lin, Jiani Zou, Junde Zhao and Jiuling Wang.
Contributor Information
Zhiheng Lin, Email: linzhiheng2020@163.com.
Jiuling Wang, Email: ayi-99@163.com.
Junde Zhao, Email: zhaojunde2020@163.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-20858-5.
References
- 1.Koh WJ, Abu-Rustum NR, Bean S, et al. Cervical cancer, Version 3.2019, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Canc. Netw. 2019;17(1):64–84. doi: 10.6004/jnccn.2019.0001. [DOI] [PubMed] [Google Scholar]
- 2.Small W, Jr, Bacon MA, Bajaj A, et al. Cervical cancer: A global health crisis. Cancer. 2017;123(13):2404–2412. doi: 10.1002/cncr.30667. [DOI] [PubMed] [Google Scholar]
- 3.Lahue BJ, Baginska E, Li SS, Parisi M. Health technology assessment on cervical cancer screening, 2000–2014. Int. J. Technol. Assess. Health Care. 2015;31(3):171–180. doi: 10.1017/S0266462315000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torre LA, Islami F, Siegel RL, Ward EM, Jemal A. Global cancer in women: Burden and trends. Cancer Epidemiol. Biomarkers Prev. 2017;26(4):444–457. doi: 10.1158/1055-9965.EPI-16-0858. [DOI] [PubMed] [Google Scholar]
- 5.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2020;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 6.Ojesina AI, Lichtenstein L, Freeman SS, et al. Landscape of genomic alterations in cervical carcinomas. Nature. 2014;506(7488):371–375. doi: 10.1038/nature12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Degterev A, Huang Z, Boyce M, et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat. Chem. Biol. 2005;1(2):112–119. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- 8.Yan J, Wan P, Choksi S, Liu ZG. Necroptosis and tumor progression. Trends Cancer. 2022;8(1):21–27. doi: 10.1016/j.trecan.2021.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park S, Hatanpaa KJ, Xie Y, et al. The receptor interacting protein 1 inhibits p53 induction through NF-kappaB activation and confers a worse prognosis in glioblastoma. Cancer Res. 2009;69(7):2809–2816. doi: 10.1158/0008-5472.CAN-08-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin X, Ma D, Tan YX, Wang HY, Cai Z. The role of necroptosis in cancer: A double-edged sword? Biochim. Biophys. Acta Rev. Cancer. 2019;1871(2):259–266. doi: 10.1016/j.bbcan.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Colbert LE, Fisher SB, Hardy CW, et al. Pronecrotic mixed lineage kinase domain-like protein expression is a prognostic biomarker in patients with early-stage resected pancreatic adenocarcinoma. Cancer. 2013;119(17):3148–3155. doi: 10.1002/cncr.28144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt SV, Seibert S, Walch-Rückheim B, et al. RIPK3 expression in cervical cancer cells is required for PolyIC-induced necroptosis, IL-1α release, and efficient paracrine dendritic cell activation. Oncotarget. 2015;6(11):8635–8647. doi: 10.18632/oncotarget.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zehendner CM, Valasarajan C, Werner A, et al. Long noncoding RNA TYKRIL plays a role in pulmonary hypertension via the p53-mediated regulation of PDGFRβ. Am. J. Respir. Crit. Care Med. 2020;202(10):1445–1457. doi: 10.1164/rccm.201910-2041OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rashid F, Shah A, Shan G. Long non-coding RNAs in the cytoplasm. Genom. Proteom. Bioinform. 2016;14(2):73–80. doi: 10.1016/j.gpb.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aalijahan H, Ghorbian S. Long non-coding RNAs and cervical cancer. Exp. Mol. Pathol. 2019;106:7–16. doi: 10.1016/j.yexmp.2018.11.010. [DOI] [PubMed] [Google Scholar]
- 16.Jing L, Yuan W, Ruofan D, Jinjin Y, Haifeng Q. HOTAIR enhanced aggressive biological behaviors and induced radio-resistance via inhibiting p21 in cervical cancer. Tumour Biol. 2015;36(5):3611–3619. doi: 10.1007/s13277-014-2998-2. [DOI] [PubMed] [Google Scholar]
- 17.Liu Z, Lu T, Li J, et al. Development and clinical validation of a novel six-gene signature for accurately predicting the recurrence risk of patients with stage II/III colorectal cancer. Cancer Cell Int. 2021;21(1):359. doi: 10.1186/s12935-021-02070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z, Weng S, Xu H, et al. Computational recognition and clinical verification of TGF-β-Derived miRNA signature with potential implications in prognosis and immunotherapy of intrahepatic cholangiocarcinoma. Front. Oncol. 2021;11:757919. doi: 10.3389/fonc.2021.757919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Liu Z, Li X, et al. Comprehensive molecular analyses of a six-gene signature for predicting late recurrence of hepatocellular carcinoma. Front. Oncol. 2021;11:732447. doi: 10.3389/fonc.2021.732447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Z, Liu L, Weng S, et al. Machine learning-based integration develops an immune-derived lncRNA signature for improving outcomes in colorectal cancer. Nat. Commun. 2022;13(1):816. doi: 10.1038/s41467-022-28421-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Z, Guo C, Dang Q, et al. Integrative analysis from multi-center studies identities a consensus machine learning-derived lncRNA signature for stage II/III colorectal cancer. EBioMedicine. 2022;75:103750. doi: 10.1016/j.ebiom.2021.103750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z, Guo C, Li J, et al. Somatic mutations in homologous recombination pathway predict favourable prognosis after immunotherapy across multiple cancer types. Clin. Transl. Med. 2021;11(12):e619. doi: 10.1002/ctm2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 24.Liu P, Xu B, Shen W, et al. Dysregulation of TNFα-induced necroptotic signaling in chronic lymphocytic leukemia: Suppression of CYLD gene by LEF1. Leukemia. 2012;26(6):1293–1300. doi: 10.1038/leu.2011.357. [DOI] [PubMed] [Google Scholar]
- 25.Koo GB, Morgan MJ, Lee DG, et al. Methylation-dependent loss of RIP3 expression in cancer represses programmed necrosis in response to chemotherapeutics. Cell Res. 2015;25(6):707–725. doi: 10.1038/cr.2015.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Guo J, Ding AP, et al. Association of mixed lineage kinase domain-like protein expression with prognosis in patients with colon cancer. Technol. Cancer Res. Treat. 2017;16(4):428–434. doi: 10.1177/1533034616655909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seifert L, Werba G, Tiwari S, et al. The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature. 2016;532(7598):245–249. doi: 10.1038/nature17403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen L, Dzakah EE, Shan G. Targetable long non-coding RNAs in cancer treatments. Cancer Lett. 2018;418:119–124. doi: 10.1016/j.canlet.2018.01.042. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Sun D, Song J, Yang N, Zhang Y. Integrated profiles analysis identified a coding-non-coding signature for predicting lymph node metastasis and prognosis in cervical cancer. Front. Cell Dev. Biol. 2021;8:631491. doi: 10.3389/fcell.2020.631491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Zhang X, Zhu H, et al. Identification of potential prognostic long non-coding RNA biomarkers for predicting recurrence in patients with cervical cancer. Cancer Manag. Res. 2020;12:719–730. doi: 10.2147/CMAR.S231796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Britto AMA, Goes LR, Sivro A, et al. HPV induces changes in innate immune and adhesion molecule markers in cervical mucosa with potential impact on HIV infection. Front. Immunol. 2020;11:2078. doi: 10.3389/fimmu.2020.02078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu D, Zhou J, Wang F, Shi H, Li Y, Li B. HPV-16 E6/E7 promotes cell migration and invasion in cervical cancer via regulating cadherin switch in vitro and in vivo. Arch. Gynecol. Obstet. 2015;292(6):1345–1354. doi: 10.1007/s00404-015-3787-x. [DOI] [PubMed] [Google Scholar]
- 33.Tugizov SM, Herrera R, Chin-Hong P, et al. HIV-associated disruption of mucosal epithelium facilitates paracellular penetration by human papillomavirus. Virology. 2013;446(1–2):378–388. doi: 10.1016/j.virol.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nazli A, Chan O, Dobson-Belaire WN, et al. Exposure to HIV-1 directly impairs mucosal epithelial barrier integrity allowing microbial translocation. PLoS Pathog. 2010;6(4):e1000852. doi: 10.1371/journal.ppat.1000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Judson PL, He X, Cance WG, Van Le L. Overexpression of focal adhesion kinase, a protein tyrosine kinase, in ovarian carcinoma. Cancer. 1999;86(8):1551–1556. doi: 10.1002/(sici)1097-0142(19991015)86:6<1551::aid-cncr23>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 36.Guo QS, Song Y, Hua KQ, Gao SJ. Involvement of FAK-ERK2 signaling pathway in CKAP2-induced proliferation and motility in cervical carcinoma cell lines. Sci. Rep. 2017;7(1):2117. doi: 10.1038/s41598-017-01832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halder J, Landen CN, Jr, Lutgendorf SK, et al. Focal adhesion kinase silencing augments docetaxel-mediated apoptosis in ovarian cancer cells. Clin. Cancer Res. 2005;11(24 Pt 1):8829–8836. doi: 10.1158/1078-0432.CCR-05-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cance WG, Harris JE, Iacocca MV, et al. Immunohistochemical analyses of focal adhesion kinase expression in benign and malignant human breast and colon tissues: Correlation with preinvasive and invasive phenotypes. Clin. Cancer Res. 2000;6(6):2417–2423. [PubMed] [Google Scholar]
- 39.Owens LV, Xu L, Craven RJ, et al. Overexpression of the focal adhesion kinase (p125FAK) in invasive human tumors. Cancer Res. 1995;55(13):2752–2755. [PubMed] [Google Scholar]
- 40.Hao H, Naomoto Y, Bao X, et al. Focal adhesion kinase as potential target for cancer therapy (Review) Oncol. Rep. 2009;22(5):973–979. doi: 10.3892/or_00000524. [DOI] [PubMed] [Google Scholar]
- 41.Principe DR, Doll JA, Bauer J, et al. TGF-β: Duality of function between tumor prevention and carcinogenesis. J. Natl. Cancer Inst. 2014;106(2):djt369. doi: 10.1093/jnci/djt369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ali A, Wang Z, Fu J, et al. Differential regulation of the REGγ-proteasome pathway by p53/TGF-β signalling and mutant p53 in cancer cells. Nat. Commun. 2013;4:2667. doi: 10.1038/ncomms3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu HS, Li YF, Chou CI, Yuan CC, Hung MW, Tsai LC. The concentration of serum transforming growth factor beta-1 (TGF-beta1) is decreased in cervical carcinoma patients. Cancer Invest. 2002;20(1):55–59. doi: 10.1081/cnv-120000366. [DOI] [PubMed] [Google Scholar]
- 44.Peralta-Zaragoza O, Bermúdez-Morales V, Gutiérrez-Xicotencatl L, Alcocer-González J, Recillas-Targa F, Madrid-Marina V. E6 and E7 oncoproteins from human papillomavirus type 16 induce activation of human transforming growth factor beta1 promoter throughout Sp1 recognition sequence. Viral Immunol. 2006;19(3):468–480. doi: 10.1089/vim.2006.19.468. [DOI] [PubMed] [Google Scholar]
- 45.Donalisio M, Cornaglia M, Landolfo S, Lembo D. TGF-beta1 and IL-4 downregulate human papillomavirus-16 oncogene expression but have differential effects on the malignant phenotype of cervical carcinoma cells. Virus Res. 2008;132(1–2):253–256. doi: 10.1016/j.virusres.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 46.Radogna F, Diederich M. Stress-induced cellular responses in immunogenic cell death: Implications for cancer immunotherapy. Biochem. Pharmacol. 2018;153:12–23. doi: 10.1016/j.bcp.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 47.Galluzzi L, Vitale I, Warren S, et al. Consensus guidelines for the definition, detection and interpretation of immunogenic cell death. J. Immunother. Cancer. 2020;8(1):e000337. doi: 10.1136/jitc-2019-000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fucikova J, Spisek R, Kroemer G, Galluzzi L. Calreticulin and cancer. Cell Res. 2021;31(1):5–16. doi: 10.1038/s41422-020-0383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yatim N, Jusforgues-Saklani H, Orozco S, et al. RIPK1 and NF-κB signaling in dying cells determines cross-priming of CD8+T cells. Science. 2015;350(6258):328–334. doi: 10.1126/science.aad0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Snyder AG, Hubbard NW, Messmer MN, et al. Intratumoral activation of the necroptotic pathway components RIPK1 and RIPK3 potentiates antitumor immunity. Sci. Immunol. 2019;4(36):eaaw2004. doi: 10.1126/sciimmunol.aaw2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park HH, Kim HR, Park SY, et al. RIPK3 activation induces TRIM28 derepression in cancer cells and enhances the anti-tumor microenvironment. Mol. Cancer. 2021;20(1):107. doi: 10.1186/s12943-021-01399-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liang Y, Lü W, Zhang X, Lü B. Tumor-infiltrating CD8+ and FOXP3+ lymphocytes before and after neoadjuvant chemotherapy in cervical cancer. Diagn. Pathol. 2018;13(1):93. doi: 10.1186/s13000-018-0770-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang S, Wan J, Chen M, Cai D, Xu J, Chen Q. Tumor-infiltrating CD8+ T cells driven by the immune checkpoint-associated gene IDO1 are associated with cervical cancer prognosis. Front. Oncol. 2021;11:720447. doi: 10.3389/fonc.2021.720447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pakish JB, Jazaeri AA. Immunotherapy in gynecologic cancers: Are we there yet? Curr. Treat. Options Oncol. 2017;18(10):59. doi: 10.1007/s11864-017-0504-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin Z, Xu Q, Miao D, Yu F. An inflammatory response-related gene signature can impact the immune status and predict the prognosis of hepatocellular carcinoma. Front. Oncol. 2021;11:644416. doi: 10.3389/fonc.2021.644416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mezache L, Paniccia B, Nyinawabera A, Nuovo GJ. Enhanced expression of PD L1 in cervical intraepithelial neoplasia and cervical cancers. Mod. Pathol. 2015;28(12):1594–1602. doi: 10.1038/modpathol.2015.108. [DOI] [PubMed] [Google Scholar]
- 57.Chitsike L, Duerksen-Hughes P. The potential of immune checkpoint blockade in cervical cancer: Can combinatorial regimens maximize response? A review of the literature. Curr. Treat. Options Oncol. 2020;21(12):95. doi: 10.1007/s11864-020-00790-4. [DOI] [PubMed] [Google Scholar]
- 58.Frenel JS, Le Tourneau C, O'Neil B, et al. Safety and efficacy of pembrolizumab in advanced, programmed death ligand 1-positive cervical cancer: Results from the phase Ib KEYNOTE-028 trial. J. Clin. Oncol. 2017;35(36):4035–4041. doi: 10.1200/JCO.2017.74.5471. [DOI] [PubMed] [Google Scholar]
- 59.Muñoz N, Bosch FX, de Sanjosé S, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 2003;348(6):518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 60.Vaupel P, Mayer A. Hypoxia in cancer: Significance and impact on clinical outcome. Cancer Metastasis Rev. 2007;26(2):225–239. doi: 10.1007/s10555-007-9055-1. [DOI] [PubMed] [Google Scholar]
- 61.Yang S, Gao H. Nanoparticles for modulating tumor microenvironment to improve drug delivery and tumor therapy. Pharmacol. Res. 2017;126:97–108. doi: 10.1016/j.phrs.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 62.Roma-Rodrigues C, Mendes R, Baptista PV, Fernandes AR. Targeting tumor microenvironment for cancer therapy. Int. J. Mol. Sci. 2019;20(4):840. doi: 10.3390/ijms20040840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bussard KM, Mutkus L, Stumpf K, Gomez-Manzano C, Marini FC. Tumor-associated stromal cells as key contributors to the tumor microenvironment. Breast Cancer Res. 2016;18(1):84. doi: 10.1186/s13058-016-0740-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang X, Li Z, Kong B, et al. Reduced m6A mRNA methylation is correlated with the progression of human cervical cancer. Oncotarget. 2017;8(58):98918–98930. doi: 10.18632/oncotarget.22041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carbone DP, Reck M, Paz-Ares L, et al. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N. Engl. J. Med. 2017;376(25):2415–2426. doi: 10.1056/NEJMoa1613493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hugo W, Zaretsky JM, Sun L, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165(1):35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 2014;371(23):2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Park SE, Park K, Lee E, et al. Clinical implication of tumor mutational burden in patients with HER2-positive refractory metastatic breast cancer. Oncoimmunology. 2018;7(8):e1466768. doi: 10.1080/2162402X.2018.1466768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goodman AM, Kato S, Bazhenova L, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol. Cancer Ther. 2017;16(11):2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data for this study come from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). All the data in this paper support the results of this study. Details of R software: R is a free software environment for statistical computing and graphics. It compiles and runs on a wide variety of UNIX platforms, Windows and MacOS. R version 4.1.2; link: https://www.r-project.org/.