Abstract
The role of proteases in pathogenesis is well established for several microorganisms but has not been described for Yersinia enterocolitica. Previously, we identified a gene, hreP, which showed significant similarity to proteases in a screen for chromosomal genes of Y. enterocolitica that were exclusively expressed during an infection of mice. We cloned this gene by chromosome capture and subsequently determined its nucleotide sequence. Like inv, the gene encoding the invasin protein of Y. enterocolitica, hreP is located in a cluster of flagellum biosynthesis and chemotaxis genes. The genomic organization of this chromosomal region is different in Escherichia coli, Salmonella, and Yersinia pestis than in Y. enterocolitica. Analysis of the distribution of hreP between different Yersinia isolates and the relatively low G+C content of the gene suggests acquisition by horizontal gene transfer. Sequence analysis also revealed that HreP belongs to a family of eukaryotic subtilisin/kexin-like proteases. Together with the calcium-dependent protease PrcA of Anabaena variabilis, HreP forms a new subfamily of bacterial subtilisin/kexin-like proteases which might have originated from a common eukaryotic ancestor. Like other proteases of this family, HreP is expressed with an N-terminal prosequence. Expression of an HreP-His6 tag fusion protein in E. coli revealed that HreP undergoes autocatalytic processing at a consensus cleavage site of subtilisin/kexin-like proteases, thereby releasing the proprotein.
The genus Yersinia consists of several species, three of which are considered to be pathogens for mammals. Yersinia pestis is the etiologic agent of plague, while Yersinia pseudotuberculosis and Yersinia enterocolitica primarily cause gastrointestinal syndroms. Yersinia enterocolitica is a pathogen for humans and can cause a variety of syndromes, including acute enteritis, mesenteric lymphadenitis, and enterocolitis (15). All three species have a tropism for lymphoid tissues and contain a 70-kb virulence plasmid which encodes a system that delivers antiphagocytic effector proteins into the cytosol of eukaryotic cells (16).
In contrast to the virulence plasmid, relatively little is known about chromosomally borne virulence genes of Y. enterocolitica. In addition to the genes inv and ail, which encode proteins that enable the bacterium to adhere to and invade eukaryotic cells (29, 39), genes for the acquisition of iron and the synthesis of the O antigen component of lipopolysaccharide and an enterotoxin gene have been described as chromosomally borne virulence-associated genes (1, 13, 27, 52, 53, 59). By signature-tagged transposon mutagenesis, chromosomal genes of Y. enterocolitica were identified that were previously not described as required for in vivo survival. They include genes for the synthesis of outer-membrane components, stress response, and nutrient acquisition (17). One transposon insertion was localized in pspC, and the corresponding strain was severely attenuated for virulence (17).
Recently, in vivo expression technology (IVET) was established as a powerful tool to select for and identify genes that are expressed during an infection (35, 36). To overcome the discrepancy between the availability of information about chromosomally and virulence plasmid-borne virulence genes, IVET was used with Y. enterocolitica in a mouse model of infection (58). This study led to the identification of 45 different chromosomal loci, designated hre for host responsive elements, that are expressed early during an infection but not under standard laboratory conditions. The identified hre loci were grouped according to their predicted function or property and comprise genes important for stress response, iron starvation response, or cell envelope maintenance. Another group contains hre loci with noncategorized functions, including those with unknown function or no similarity to genes in the database (58). The same IVET pool was also used to identify genes expressed during later stages of infection (21). These were designated sif for systemic infection factor. Comparison of sif and hre genes suggests that different sets of genes are active during different stages of infection.
After the identification of hre genes, it was important to further characterize these genes and their products and to elucidate their role for pathogenesis. Three of four different hre mutant strains tested showed reduced virulence in the mouse model of infection, indicating their importance for pathogenesis. One of these, hreP (previously referred to as hre-22), showed similarity to the protease PrcA of Anabaena variabilis (5, 37, 58). The importance of HreP for the pathogenesis of Y. enterocolitica is clearly reflected in the reduction of virulence of an hreP mutant strain by both a 50% lethal dose and an in vivo survival assay (58). While it is possible that this protease serves a housekeeping function during infection, proteases are well established as bacterial virulence factors (33, 54). There are several examples of bacterial proteases that contribute to pathogenesis by interfering with host tissues or proteins and by inactivating key proteins important in host defense (33).
To characterize the in vivo-expressed protease encoded by hreP, the gene and its flanking regions were cloned and sequenced. The genomic organization, together with the G+C content of hreP and the distribution among different Yersinia species, suggests that hreP was acquired by horizontal gene transfer. This is especially interesting, as HreP has significant similarity to eukaryotic subtilisin/kexin-like proprotein convertases. In addition, we could show that purified HreP undergoes autocatalytic processing of its amino-terminal prosequence.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The Y. enterocolitica strain GY4J7, a derivative of the previously described strain JB580v (30), is the original cat fusion strain isolated in a screen for in vivo-expressed genes (58). The pGY2-based plasmid pGY49, which has oriR6K and is mobilizable, was maintained in Escherichia coli S17-1λpir (31, 57). For the expression of recombinant His6-tagged proteins, plasmid pET-24(+) (Novagen) and the T7 RNA polymerase expressing E. coli strain ER2566 were used. All strains were grown in Luria-Bertani broth or agar plates at 26°C (Y. enterocolitica) or 37°C (E. coli) unless otherwise mentioned. Antibiotics were used in the following concentrations where appropriate: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 25 μg/ml (for E. coli) or 12.5 μg/ml (for Y. enterocolitica); and nalidixic acid, 20 μg/ml.
Cloning, subcloning, and site-directed mutagenesis.
For the cloning of the hreP gene by chromosome capture, chromosomal DNA of Y. enterocolitica strain GY4J7 was digested with EcoRI, ligated after the restriction enzyme was heat inactivated, and subsequently electroporated into E. coli S17-1λpir. Because EcoRI does not cut within pGY2 (integrated on the chromosome), this should release a fragment containing pGY2 along with flanking chromosomal sequences. Analysis of the transformants revealed a plasmid containing a fragment of captured chromosomal DNA of approximately 4.3 kb that was named pGY49. The insert in this fragment was subcloned as a ClaI fragment in pWSK29 (55), resulting in plasmid pGY50, and subsequently sequenced. For the recombinant expression of the hreP gene as a His6 tag fusion protein, PCR amplification with pGY50 as template and primers GH-P8 (5′-GGAATTCTATTAAAGGGAAATTAAAATG-3′) and GH-P4-3 (5′-CCGCTCGAGTTTATGGCACCCTACCATTTC-3′) was performed (EcoRI and XhoI linkers, respectively, are underlined). The 1,684-nucleotide (nt) PCR product, which contains the entire hreP coding region plus an additional 18 nt upstream of the start codon, was digested with EcoRI and XhoI and ligated into the EcoRI- and XhoI-restricted vector pET-24(+), resulting in plasmid pET-P8/P4-3. The correct sequence of the fragment was confirmed by sequencing. Site-directed mutagenesis was performed by recombination PCR as previously described (56, 61), resulting in plasmid pET-P8/P4-3 S471-A. Confirmation of the introduced nucleotide exchanges was done by sequence analysis.
The propeptide of hreP was expressed in E. coli ER2566 as a fusion protein with intein-chitin binding protein (CBP) using the IMPACT T7 system (New England Biolabs). For this purpose, a 552-nt PCR product was generated, using primers GH-P1 (5′-GGAATTCCATATGAAACGATTTGACGTTACTTAT-3′) (the NdeI linker is underlined) and GH-P2 (5′-TTTGTGTTCTTTGTGATAAAT-3′) and pGY50 as template. The PCR product was treated with T4 DNA polymerase, phosphorylated with polynucleotide kinase, digested with NdeI, and ligated with NdeI/SmaI-digested pTYB2 to result in the expression plasmid pTYB-P1/P2.
Southern analysis.
Chromosomal DNA was purified from Yersinia, Salmonella, and E. coli strains as previously described (38) and digested with HindIII. The restricted DNA was separated by electrophoresis on a 0.8% agarose gel and subsequently transferred to a nitrocellulose membrane by the method of Southern (50). An internal fragment of the hreP gene corresponding to nt 594 to 1097 of the hreP coding region was generated by PCR amplification and used as the probe. Labeling of the probe, hybridization, and detection were done with the enhanced chemiluminescence (ECL) Southern blotting system as described by the manufacturer (Amersham Pharmacia Biotech).
Sequence analysis.
DNA sequence analysis was carried out using the Applied Biosystems DNA sequencing system and the BigDye terminator cycle sequencing kit according to the manufacturer's instructions. Sequencing reactions were processed by the Washington University Protein and Nucleic Acid Chemistry Laboratory. Sequence alignments were generated with the Genetics Computer Group program Bestfit, and database searches were conducted with the Blast program.
For the amino-terminal sequence analysis of processing products, proteins were blotted on polyvinylidene difluoride membrane, stained with 0.025% (wt/vol) Coomassie brilliant blue R-250 in 40% (vol/vol) methanol and destained in 50% (vol/vol) methanol. The appropriate area of the membrane was isolated, and the N-terminal sequence of the bound protein was determined by automated Edman degradation by standard procedures by the Washington University Protein and Nucleic Acid Chemistry Laboratory.
Protein expression and affinity purification.
E. coli ER2566 containing plasmid pET-P8/P4-3 or pET-P8/P4-3 S471-A was grown to an optical density at 600 nm of 0.6 at 37°C. After addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.33 mM, cells were incubated with shaking at 25°C for 5 h to induce expression of the HreP-His6 tag fusion protein. Cells were pelleted, resuspended in lysis buffer (50 mM Tris-Cl [pH 8.0], 500 mM NaCl, 10 mM imidazole, 10% glycerol, 0.1% Triton X-100), and sonicated to break open the cells. After centrifugation, the supernatant was applied to a Ni-nitriloacetic acid agarose column (Qiagen) that was subsequently washed with washing buffer (50 mM Tris-Cl [pH 8.0], 500 mM NaCl, 20 mM imidazole, 10% glycerol, 0.1% Triton X-100) to remove nonspecifically bound proteins. The His6 tag fusion protein was eluted with elution buffer (50 mM Tris-Cl [pH 8.0], 500 mM NaCl, 250 mM imidazole, 10% glycerol, 0.1% Triton X-100), and five 1.5-ml fractions were collected. The purification was monitored by separating samples of each fraction by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and staining with Coomassie brilliant blue.
For the expression of the propeptide, E. coli ER2566 containing pTYB-P1/P2 was induced as described for the His6 tag fusion proteins. This resulted in the expression of a 79-kDa propeptide-intein-CBP fusion protein. Cells were pelleted, resuspended in lysis buffer without imidazole, and sonicated. Soluble proteins were applied to a chitin column for affinity purification. After being washed with washing buffer without imidazole, the column was flushed with cleavage buffer (50 mM Tris-Cl [pH 8.0], 500 mM NaCl, 10% glycerol, 30 mM dithiothreitol [DTT]) and incubated overnight for DTT-induced intein-mediated cleavage of the fusion protein. The next day, the propeptide was eluted from the column with cleavage buffer without DTT, leaving the fusion partner (intein-CBP) bound to the column. Fractions were collected and monitored by separating samples by SDS-PAGE and staining with Coomassie brilliant blue. This protein was then used to produce polyclonal antiserum in New Zealand White rabbits by standard procedures (Cocalico Biologicals, Inc., Reamstown, Pa.).
Western blot analysis.
Proteins were separated by SDS-PAGE and transferred to nitrocellulose for immunoblot analysis (26). Antipropeptide polyclonal antiserum was used at a 1:750 dilution. Anti-His monoclonal antibody was used at a 1:2,000 dilution as recommended by the manufacturer (Qiagen). Binding was detected by incubation with horseradish peroxidase-conjugated secondary antibody and with a chemiluminescent substrate (ECL; Amersham).
Protease activity assay.
The standard assay for protease activity was performed at 37°C in 50 mM Tris [pH 8.0] and 0.5 mM CaCl2, with various amounts of affinity-purified HreP, depending on the preparation. BAPNA (Nα-benzoyl-dl-arginine-p-nitroanilide) was dissolved in dimethyl sulfoxide and used as a substrate at a final concentration of 1 mM. The absorbance at 405 nm was measured to detect the appearance of 4-nitroanilide.
When azocasein was used as a substrate, 100 μl of azocasein (5 mg/ml in 50 mM Tris [pH 8.0], 0.5 mM CaCl2, 0.04% NaN3) was incubated with HreP in a total volume of 200 μl. The reaction was terminated by adding 400 μl of 10% (wt/vol) trichloroacetic acid and incubating the mixture on ice for 30 min to precipitate proteins. Activity was assayed as an increase in trichloroacetic acid-soluble azopeptides by adding 700 μl of NaOH (525 mM) to supernatants and measuring the absorbance at 442 nm.
For the peptide-based cleavage assay, 1 mM peptide (NH2-Ile-Tyr-His-Lys-Glu-His-Lys-Thr-Ile-His-Pro-Asn-COOH; Anaspec, San Jose, Calif.) was incubated in 50 mM Tris (pH 8.0) and 0.5 mM CaCl2, with various amounts of protease in a total volume of 50 μl. The reactions were terminated by the addition of 50 μl of 0.1% trifluoroacetic acid (TFA) and the mixtures were analyzed by reversed-phase high-performance liquid chromatography (HPLC) on a C18 column. Cleavage was determined by resolving the reactions with a 20-min gradient from 0 to 60% acetonitrile in 0.1% TFA, followed by a 1-min gradient from 60 to 90% acetonitrile in 0.1% TFA. The absorbance was determined at 215 nm.
Nucleotide sequence accession number.
The complete sequence of hreP and surrounding DNA can be found in GenBank (accession number AF354753).
RESULTS
Sequence analysis of hreP.
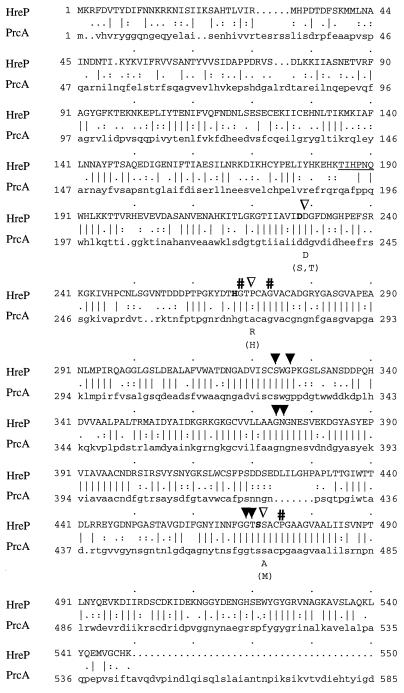
The nucleotide sequence of a 4.3-kb DNA fragment from Y. enterocolitica JB580v corresponding to the complete gene of hreP and its surrounding regions was determined after plasmid rescue from the original cat fusion strain GY4J7 (58). The hreP open reading frame (ORF) comprises 1,650 nt, encoding a hypothetical protein 550 amino acids (aa) in length with a calculated molecular weight of 60,013. A putative Shine-Dalgarno sequence (AAGGGA) was located 7 nt upstream of the initiation codon. The amino acid sequence of the hypothetical protein was compared to entries in databases with the BLAST program (2). The highest similarity was to the cyanobacterial calcium-stimulated protease PrcA of A. variabilis (5, 37) (Fig. 1). However, significant similarity was found to several subtilases (subtilisin-like serine proteases) belonging to the family of eukaryotic subtilisin/kexin-like proprotein convertases (Table 1). These proteases are often referred to as convertases because they are synthesized in a proform, where the amino-terminal proprotein serves as an intramolecular chaperone that is autocatalytically cleaved off after folding, thereby releasing the mature enzyme (6, 18).
FIG. 1.
Amino acid alignment of Y. enterocolitica HreP and A. variabilis PrcA. Identical residues are indicated by vertical lines, and similar residues are indicated by periods or colons (Bestfit; Genetics Computer Group). The amino acids that make up the typical catalytic triad of serine proteases are highlighted by boldface letters. Solid arrowheads, amino acids that are predicted to be located in the putative substrate binding region; open arrowheads, positions of amino acids typical for the kexin subfamily of subtilases; #, amino acids predicted to be located in internal helices. Those amino acids that are characteristic for kexin proteases or serine proteases (in brackets) are indicated under the alignment. The amino acids that were determined by N-terminal sequencing of the 42-kDa polypeptide are underlined and correspond to the amino-terminal end of the mature HreP protease.
TABLE 1.
HreP shows significant similarity to various subtilisin/kexin-like proteases at the amino acid levela
| % identity on amino acid level to HreP (E value) |
Similar protein |
|---|---|
| 52 (e−134) | Calcium-dependent protease PrcA of A. variabilis |
| 28 (2e−23) | Kex2 protease of Candida albicans |
| 28 (1e−22) | Hypothetical protein of Caenorhabditis elegans with similarity to subtilases |
| 27 (3e−22) | Subtilisin-like proprotein convertase PC6; human |
| 28 (5e−22) | Furin 2 of Drosophila melanogaster |
| 27 (5e−22) | Prohormone convertase PC5; human |
| 28 (7e−21) | Kex1 protease of Kluyveromyces lactis |
| 28 (1e−20) | Proprotein convertase subtilisin/kexin type 7; Mus musculus |
The alignment was performed using full-length HreP. The similarity was usually to the mature protease, although the similarity extended into the C-terminal part of the propeptide in some cases. The identity of HreP to all subtilisin/kexin-like proteases besides PrcA is between 27 and 28%, and only some of the proteases that came up in a BLAST search are shown here.
Serine proteases are characterized by their active-site residues, Asp, His, and Ser. In HreP, Asp228, His267, and Ser471 (Fig. 1) are predicted to be active-site residues by sequence comparison to PrcA and other proteases. Other conserved amino acids in all subtilisin-like proteases include those that are located in the putative substrate binding region and in internal helices (46). The corresponding amino acids in HreP that make up the putative substrate binding site are Ser324, Gly326, Gly374, Gly469, and Thr470, whereas Gly268, Gly273, and Pro475 are predicted to be located in internal helices. In addition to residues Ser324 and Gly326, Asn375 is conserved in HreP (Fig. 1). These three amino acids are predicted to make up the oxyanion hole of serine proteases that stabilizes the oxyanion in the transition state during catalysis (32, 46). Additionally, there are conserved residues characteristic only for members of the kexin subfamily of subtilases (8, 43). In HreP, Asp229, immediately following the active site Asp, is conserved as in other kexin proteases. In other subfamilies of subtilases there is usually a Ser or Thr at this position. Two other positions that are different between the kexin subfamily and other subtilases correspond to Pro270 and Ser472 of HreP; both of these positions are different in HreP from either typical subtilases or the kexin subfamily (Fig. 1). However, Ser472 is conserved between HreP and PrcA. Additionally, PrcA and HreP do not seem to have a typical presequence at the amino terminus, which is rather unusual for subtilisin/kexin-like proteases. From these data we suggest that HreP and PrcA may be representatives of a new subfamily of bacterial subtilisin/kexin-like proteases.
Genomic organization of the hreP locus.
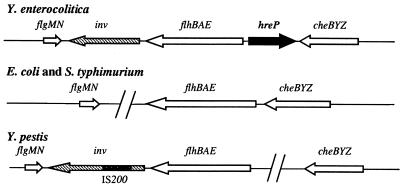
Sequence analysis of the neighboring regions of hreP revealed that hreP is located in a cluster of genes related to flagellum biosynthesis and chemotaxis. Upstream of hreP the 5′ end of the divergently transcribed flhB gene of Y. enterocolitica was identified. Downstream of hreP, the cheZ, cheY, and cheB genes of Y. enterocolitica were identified by sequence identity to the corresponding genes of E. coli (identities of 67% [cheZ], 81% [cheY], and 74% [cheB], respectively). As flhBAE and cheZYB are transcribed divergently and convergently, respectively, from hreP, a polar effect of the previously described Y. enterocolitica hreP mutant that was tested for pathogenicity in the mouse model of infection (58) is unlikely.
The genomic organization of the flhBAE-cheBYZ region is arranged differently in Y. enterocolitica from that in E. coli and Salmonella (Fig. 2) (10; http://genome.wustl.edu/gsc/bacterial/salmonella.shtml). First, there is no additional ORF between the cheZ and flhB loci in E. coli or Salmonella enterica serovar Typhimurium. Second, the inv gene is located downstream of the flhBAE operon in Y. enterocolitica, followed by the flgMN genes (19). E. coli and S. enterica serovar Typhimurium have no inv gene downstream of the flhBAE operon, and flgMN is located in a different region of the chromosome. Interestingly, the genomic organization of this region is again different in Y. pestis (30; http://www.sanger.ac.uk/Projects/Y_pestis). There is no ORF with similarity to hreP in the available Y. pestis genome, neither downstream of cheZ nor upstream of flhB, and the cheBYZ operon is located elsewhere on the chromosome in comparison to flhBAE and flgMN. Furthermore, the inv gene of Y. pestis is disrupted by an IS200-like element and hence is not functional (47).
FIG. 2.
Genomic organization of the chromosomal region surrounding the flhBAE operons of Y. enterocolitica, Y. pestis, E. coli, and Salmonella. Each operon and/or ORF is indicated by an arrow, whereas large gaps between genes are indicated by two slashes.
The G+C content of hreP (42.5 mol% G+C) is relatively low compared to the rest of the genome of Y. enterocolitica (about 53 mol% G+C in conserved genes). This is also true for the inv gene (45 mol% G+C) that interrupts the flagellum locus, and another chromosomal, virulence-associated gene, ail (43 mol% G+C). In addition, the virulence-plasmid-borne genes of Y. enterocolitica that have homologues in other enteropathogenic bacteria, often on pathogenicity islands, have a G+C content much lower than that of the Y. enterocolitica chromosome (34 to 45 mol% G+C) and are suggested to be recently acquired horizontally. This suggests that hreP might also have been acquired by a mechanism involving horizontal gene transfer and chromosomal rearrangement. The corresponding region of the chromosome seems to be a hot spot for genome rearrangement and the integration of virulence factors in Y. enterocolitica, since two genes, inv and hreP, have probably integrated independently into this region.
Analysis of the distribution of hreP in different Yersinia isolates.
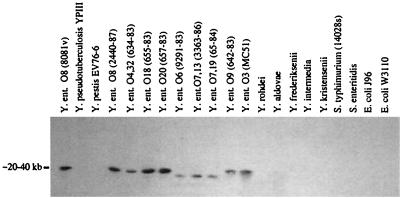
We were interested in the distribution of hreP in different pathogenic and environmental Yersinia isolates as well as in E. coli and Salmonella species, because the genomic organization of the hreP region of the chromosome of Y. enterocolitica and the G+C content of the gene suggested the acquisition of hreP by horizontal gene transfer. The chromosomal DNA of these species was isolated and analyzed by Southern hybridization with hreP as the probe under medium-stringency conditions (Fig. 3). All isolates of Y. enterocolitica tested gave a positive signal for hreP, although the serotypes that are generally considered to be environmental isolates (O6, O7,13, and O7,19, respectively) showed a different hybridization pattern from that of the pathogenic strains. The other pathogenic Yersinia species tested, Y. pestis and Y. pseudotuberculosis, did not react with the hreP probe; this is consistent with results from the search of the Y. pestis genome for hreP-related sequences. Likewise, DNA from environmental isolates of Yersinia (Yersinia rohdei, Yersinia aldovae, Yersinia frederiksenii, Yersinia intermedia, and Yersinia kristensenii), S. enterica serovar Typhimurium, Salmonella enterica serovar Enteritidis, the uropathogenic E. coli strain J96, and the wild-type E. coli strain W3110 did not hybridize with the hreP probe. In some cases a very weak signal could be detected with Y. kristensenii DNA under low-stringency conditions (data not shown).
FIG. 3.
Southern analysis of chromosomal digests of various Yersinia, Salmonella, and E. coli isolates. Chromosomal DNA was isolated and prepared for Southern hybridization as described in Materials and Methods. A PCR product corresponding to an internal fragment of hreP (nt 594 to 1097) was used as the probe.
Expression of HreP in E. coli.
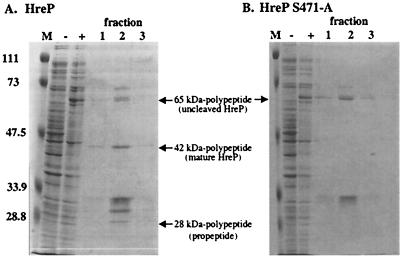
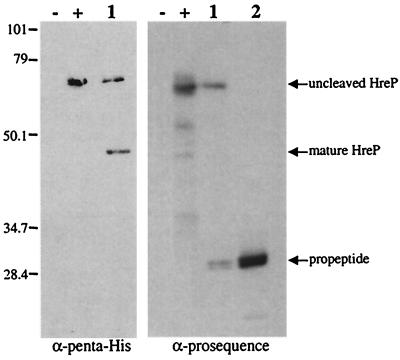
To determine the proteolytic properties of HreP, we expressed the protein with a carboxy-terminal His6 tag from plasmid pET-P8/P4-3 in E. coli and affinity purified it via a Ni-nitriloacetic acid agarose column. Induction of expression with IPTG in E. coli led to the synthesis of a polypeptide with an apparent molecular mass of 65 kDa that could be detected in whole-cell lysates of induced but not uninduced E. coli cells carrying the expression plasmid. After affinity purification was done, we detected several other polypeptides in addition to the 65-kDa polypeptide on Coomassie-stained SDS-polyacrylamide gels, which we first assumed to be contaminants (Fig. 4A). However, closer examination of the molecular masses suggested that in analogy to the processing of PrcA of A. variabilis, two of these polypeptides with apparent molecular masses of 42 and 28 kDa, respectively, might be processing products of HreP. To test this hypothesis, we performed Western blot analysis using either penta-His antibody that recognizes the carboxy-terminal His6 tag of the fusion protein or rabbit antiserum raised against aa 1 to 184 of HreP, corresponding to the presumed amino-terminal prosequence (Fig. 5). As expected, the penta-His antibody recognizes the full-length HreP as well as the 42-kDa polypeptide, which shows that this polypeptide derives from the carboxy-terminal end of HreP. The antiserum raised against aa 1 to 184 recognizes the full-length HreP as well as the 28-kDa polypeptide. Additionally, the protein that was used to immunize the rabbits ran at the same size as the potential 28-kDa processing product, suggesting that processing occurs close to the presumed site (Fig. 5). These data identify the 42- and the 28-kDa polypeptides as processing products of the full-length HreP. This is not surprising, since sequence analysis suggested that HreP belongs to a class of proprotein convertases which are expressed with an amino-terminal prosequence that is usually autocatalytically cleaved off after correct folding of the mature enzyme.
FIG. 4.
Expression of HreP and HreP S471-A. E. coli ER2566 carrying plasmid pET-P8/P4-3 (A) or pET-P8/P4-3 S471-A (B) was either not induced (−) or induced (+) by the addition of IPTG. Aliquots of 30 μl of culture were mixed with SDS sample buffer, boiled, and stored until analysis by SDS-PAGE. After affinity purification of the induced proteins, fractions were collected and 30-μl aliquots of fractions 1 to 3 were separated on SDS–10% PAGE gels. M, molecular mass standards (in kilodaltons).
FIG. 5.
Western blot analysis of HreP processing products. Proteins were prepared as described in the legend for Fig. 4. −, uninduced cells ER2566 (pET-P8/P4-3); +, IPTG-induced cells ER2566 (pET-P8/P4-3); 1, purified HreP; 2; purified HreP proprotein (aa 1 to 184); M, molecular mass standard (in kilodaltons).
Autocatalytic processing of HreP.
To determine if the observed cleavage is due to autocatalytic processing or to processing by another unidentified protease in the preparation, we constructed a mutant form of HreP in which the potential catalytic Ser471 is replaced by Ala. This mutant protease should no longer be able to autocatalytically cleave off its prosequence but should still be a substrate for another processing protease. Induction of expression of HreP S471-A led to the synthesis of a polypeptide with the same molecular mass as uncleaved wild-type HreP (Fig. 4B). After affinity purification, the bands previously identified as processing products could no longer be detected, suggesting that the observed processing is not the result of a cleavage mediated by another protease in the preparation. In addition, this result suggests that HreP S471-A is no longer able to undergo autocatalytic processing and that S471 is part of the catalytic triad.
Determination of the processing site of HreP.
For multiple reasons, we were interested in determining the site where proprotein processing of HreP occurs. First, this should provide further evidence that the 42-kDa polypeptide detected on SDS-polyacrylamide gels is the amino-terminally truncated version of HreP. Second, we can positively identify the amino terminus of the mature enzyme. Third, we should be able to identify a preferred cleavage site which could provide clues to the potential substrate specificity of the protease. For this purpose, the affinity-purified HreP preparation was transferred to a polyvinylidene difluoride membrane, the area of the membrane corresponding to the 42-kDa polypeptide was cut out, and the sequence of the first 6 aa was determined by automated Edman degradation. The analysis identified the amino acids Thr-Ile-His-Pro-Asn-Gln, corresponding to aa 185 to 190 of HreP. Therefore, cleavage occurs between Lys184 and Thr185 of the HreP amino acid sequence, leaving Thr185 as the amino-terminal residue of the mature HreP protease. This further demonstrated that the 42-kDa polypeptide is, as expected, the processed, mature form of HreP. Mammalian proprotein convertases cleave precursor polypeptides at specific sites with the consensus motif (R/K)-(X)n-(K/R)*, where n = 0, 2, 4, or 6, X is any amino acid except Cys, and the asterisk indicates the cleavage site (41, 44, 45, 51). The cleavage site at which autocatalytic processing occurs in HreP (K-E-H-K*; aa 181 to 184 of the HreP coding sequence) is in accordance with this consensus sequence.
Protease activity assays.
As HreP is capable of autocatalytically processing its amino terminus, we also examined its ability to cleave other substrates. Casein was chosen as a substrate to assay protease activity because it has an almost random three-dimensional structure and a wide range of both hydrophobic and hydrophilic sites incorporating most possible types of peptide bonds (4). However, we were not able to detect any specific hydrolysis of azocasein even after prolonged incubation was carried out with different concentrations of affinity-purified HreP at different temperatures (20 to 37°C) (data not shown). As PrcA of A. variabilis as well as other proprotein convertases are calcium dependent, we tested the possibility that trans activity of HreP is dependent on calcium. Again, we could not detect any specific hydrolysis of azocasein in the presence of 0.05 to 1 mM CaCl2. As PrcA efficiently cleaves BAPNA (5), we tested this substrate for hydrolysis by HreP, but again there was no detectable activity. Another substrate that we tested was the synthetic peptide NH2-Ile-Tyr-His-Lys-Glu-His-Lys-Thr-Ile-His-Pro-Asn-COOH, corresponding to amino acids 178 to 190 of HreP and spanning the previously identified cleavage site used for autocatalytic processing. However, we were not able to detect any specific cleavage of the peptide substrate after reversed-phase HPLC analysis. Although HreP is able to autocatalytically cleave off its prosequence, it does not appear to be active in trans against the substrates under the conditions tested.
DISCUSSION
The hre genes of Y. enterocolitica are potential virulence factors, as their expression is specifically induced during an infection of mice, but not under standard laboratory conditions. The hreP gene was previously shown to be important for virulence, as an hreP mutant strain showed a 33-fold-higher 50% lethal dose and a decreased rate of survival in Peyer's patches and mesenteric lymph nodes of infected mice after 3 and 5 days postinfection (58). The role of a protease as a virulence factor is novel for Y. enterocolitica, which prompted us to examine this hre gene in more detail.
Sequencing of the whole hreP gene and its flanking regions revealed an ORF encoding a hypothetical protein with a molecular mass of approximately 60 kDa. Interestingly, the gene showed significant similarity to a class of eukaryotic proteases that belong to the family of subtilisin/kexin-like proprotein convertases. Only one prokaryotic protease, PrcA of A. variabilis, showed significant similarity. Both HreP and PrcA share typically conserved residues that have been identified by sequence alignments and by a comparison of the crystal structures of several subtilases (5, 32, 46). One of three residues that are specific for the kexin subfamily of subtilases is conserved in HreP as well as in PrcA. At the other two residues specific for the kexin subfamily, HreP and PrcA differ not only from the kexin subfamily but also from typical subtilases. We conclude that these two bacterial proteases are members of a new subfamily of subtilisin/kexin-like proteases that might have evolved from the same protease ancestor as its eukaryotic relatives. This hypothesis is especially intriguing, as the genomic organization of this region and the relatively low G+C content of hreP make an acquisition by horizontal gene transfer likely.
Horizontal gene transfer has been evoked recently especially in the context of so-called pathogenicity islands (23, 25). However, hreP cannot be considered a pathogenicity island, as it does not have most of the characteristic features. Although hreP is present in Y. enterocolitica, it is absent from the genome of other related species and has a G+C content different from the rest of the genome. It is also much smaller than a typical pathogenicity island. In addition, hreP is not associated with a tRNA locus and does not seem to carry other cryptic genes such as phage attachment sites, phage integrase genes, or plasmid origins of replication. The acquisition of genes is a widespread phenomenon in the bacterial world. In Salmonella, for example, at least three pathogenicity islands and several smaller so-called pathogenicity islets, structurally comparable to hreP, have been described (9, 22). Interestingly, some Salmonella genes that were identified by an IVET screen show an atypical base composition and may also have been acquired horizontally (14, 28). The inv gene of Y. enterocolitica also could be a horizontally acquired virulence factor, as it is located in a cluster of flagellar genes that have a different G+C content (19).
The origin of hreP remains unknown; besides PrcA of A. variabilis, the highest similarities of HreP are to eukaryotic proteases. No other protease of prokaryotic origin shows the same characteristic features; thus, it is tempting to speculate that hreP has been acquired from a eukaryotic ancestor. Horizontal gene transfer from eukaryotes to prokaryotes has been postulated, although these events are difficult to prove (48). For example, YopH and YpkA, two effector proteins of the virulence plasmid-encoded type III secretion system of Yersinia, also show similarities to eukaryotic proteins. The C terminus of YopH contains a domain that is homologous to catalytic domains of eukaryotic protein tyrosine phosphatases (11, 24), and YpkA is a protein kinase with extensive homology to eukaryotic Ser/Thr protein kinases (20).
By expressing HreP in E. coli we were able to show that HreP undergoes an autocatalytic cleavage, thereby releasing its prosequence. This was shown by determining the amino terminus of the released mature protease and by blocking processing by exchanging the catalytic Ser471 with Ala by site-directed mutagenesis. The determined cleavage site consists of a typical consensus motif of subtilisin/kexin-like proteases (41, 44, 45, 51). Although HreP shows the highest similarity to PrcA of A. variabilis, PrcA is autocatalytically processed after an Arg residue, in contrast to processing after Lys by HreP. Furthermore, the PrcA cleavage site does not appear to have a consensus sequence (R-E-F-R-Q-R*) typical for subtilisin/kexin-like proteases (5). This suggests that HreP and PrcA cleave different substrates and may also have different functions. Eukaryotic proprotein convertases cleave a wide variety of proproteins in the secretory pathway. Targets of this type of protease include peptide hormones, neuropeptides, and growth factors. These proteases also process many other proteins into their biologically active forms, thereby fulfilling an important regulatory role. Furin, a mammalian proprotein convertase with a broad range of different substrates, has been shown not only to process cellular proproteins but also to activate viral envelope glycoproteins and bacterial exotoxins like Shiga toxin, diphtheria toxin, and Pseudomonas exotoxin A (41).
Protease activity of HreP could be shown as autocatalytic processing in cis. However, we were not able to show trans activity against various substrates under different conditions. There are several possible explanations why HreP protease activity has not been detected in trans. First, the assay conditions used might not reflect conditions under which the protease is active, although they are commonly used for proteases of this class. Second, HreP might not be stable under the conditions used. We detected decreased amounts of HreP but not of HreP S471-A after prolonged storage at 4°C, although we never saw degradation of HreP during an assay, as determined by SDS-PAGE. Third, HreP activity in trans might be very specific for a certain substrate that needs to be identified.
Another possibility for why we could not detect trans activity arises from the fact that we were not able to further purify the mature HreP protease from the prosequence, even by gel filtration (G. Heusipp and V. L. Miller, unpublished results). The inhibitory effect of prosequences has been extensively studied. The prosequence can remain associated with the cleaved protease and inhibit its activity by steric occlusion of the active site until a further activation step occurs (7, 34, 49, 60). Mammalian proprotein convertases are autocatalytically processed in the endoplasmic reticulum. Under these conditions, the protease is not activated, as the prosegment remains associated with the enzyme until it is transported through the trans-Golgi network to its final cellular destination. During this process, a change in H+ and/or Ca2+ concentrations leads to a second cleavage event that releases the prosegment and activates the enzyme (3, 12, 40, 42). The association of the prosequence with the mature enzyme after autocatalytic cleavage serves as a mechanism for the spatial and temporal regulation of the proteolytic activity. A similar mechanism can also be proposed for HreP, in which the enzyme has to be activated by a second event under conditions yet to be identified. Further analysis of this potential activation in Y. enterocolitica has been limited by the fact that in vitro conditions that lead to expression of HreP have not been identified (58). Future studies will focus on the regulation of hreP to shed further light on the signals that lead to its expression and also to investigate the activation of the enzyme.
ACKNOWLEDGMENTS
We thank Ulf Sommer for help with HPLC analysis; Amy Strohmeier Gort, Peter Dube and Kristin Nelson for experimental help; and Silke Kügler for critical reading of the manuscript.
This work was supported by National Institutes of Health research grant AI42736 to V.L.M. G.H. was supported by a Forschungsstipendium of the DFG (He3079/1–1).
REFERENCES
- 1.Al-Hendy A, Toivanen P, Skurnik M. Expression cloning of Yersinia enterocolitica O:3 rfb gene cluster in Escherichia coli K12. Microb Pathog. 1991a;10:47–59. doi: 10.1016/0882-4010(91)90065-i. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson E D, VanSlyke J K, Thulin C D, Jean F, Thomas G. Activation of the furin endoprotease is a multiple-step process: requirements for acidification and internal propeptide cleavage. EMBO J. 1997;16:1508–1518. doi: 10.1093/emboj/16.7.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews A T, editor. Peptidases, proteinases and their inhibitors. Weinheim, Germany: Verlag Chemie; 1981. [Google Scholar]
- 5.Baier K, Nicklisch S, Lockau W. Evidence for propeptide-assisted folding of the calcium-dependent protease of the cyanobacterium Anabaena. Eur J Biochem. 1996;241:750–755. doi: 10.1111/j.1432-1033.1996.00750.x. [DOI] [PubMed] [Google Scholar]
- 6.Baker D, Shiau K, Agard D A. The role of pro regions in protein folding. Curr Opin Cell Biol. 1993;5:966–970. doi: 10.1016/0955-0674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 7.Baker D, Silen J L, Agard D A. Protease pro region required for folding is a potent inhibitor of the mature enzyme. Proteins. 1992;12:339–344. doi: 10.1002/prot.340120406. [DOI] [PubMed] [Google Scholar]
- 8.Barr P J. Mammalian subtilisins: the long-sought dibasic processing endoproteases. Cell. 1991;66:1–3. doi: 10.1016/0092-8674(91)90129-m. [DOI] [PubMed] [Google Scholar]
- 9.Blanc-Potard A-B, Groisman E A. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 1997;16:5376–5385. doi: 10.1093/emboj/16.17.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blattner F R, Plunkett G I, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G W, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao J. The complete genome sequence of Escherichia coli K12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 11.Bliska J B. Crystal structure of the Yersinia tyrosine phosphatase. Trends Microbiol. 1995;3:125–127. doi: 10.1016/S0966-842X(00)88898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boudreault A, Gauthier D, Lazure C. Proprotein convertase PC1/3-related peptides are potent slow tight-binding inhibitors of murine PC1/3 and Hfurin. J Biol Chem. 1998;273:31574–31580. doi: 10.1074/jbc.273.47.31574. [DOI] [PubMed] [Google Scholar]
- 13.Carniel E, Mercereau-Puijalon O, Bonnefoy S. The gene coding for the 190,000-dalton iron-regulated protein of Yersinia species is present only in the highly pathogenic strains. Infect Immun. 1989;57:1211–1217. doi: 10.1128/iai.57.4.1211-1217.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conner C P, Heithoff D M, Julio S M, Sinsheimer R L, Mahan M J. Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc Natl Acad Sci USA. 1998;95:4641–4645. doi: 10.1073/pnas.95.8.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornelis G, Laroche Y, Balligand G, Sory M-P, Wauters G. Y. enterocolitica, a primary model for bacterial invasiveness. Rev Infect Dis. 1987;9:64–87. doi: 10.1093/clinids/9.1.64. [DOI] [PubMed] [Google Scholar]
- 16.Cornelis G R, Boland A, Boyd A P, Geuijen C, Iriarte M, Neyt C, Sory M-P, Stainier I. The virulence plasmid of Yersinia, an antihost genome. Microbiol Mol Biol Rev. 1998;62:1315–1352. doi: 10.1128/mmbr.62.4.1315-1352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darwin A J, Miller V L. Identification of Yersinia enterocolitica genes affecting survival in an animal host using signature-tagged transposon mutagenesis. Mol Microbiol. 1999;32:51–62. doi: 10.1046/j.1365-2958.1999.01324.x. [DOI] [PubMed] [Google Scholar]
- 18.Eder J, Fersht A R. Pro-sequence-assisted protein folding. Mol Microbiol. 1995;16:609–614. doi: 10.1111/j.1365-2958.1995.tb02423.x. [DOI] [PubMed] [Google Scholar]
- 19.Fauconnier A, Allaoui A, Campos A, Van Elsen A, Cornelis G, Bollen A. Flagellar flhA, flhB, and flhE genes, organized in an operon, cluster upstream of the inv locus in Yersinia enterocolitica. Microbiology. 1997;143:3461–3471. doi: 10.1099/00221287-143-11-3461. [DOI] [PubMed] [Google Scholar]
- 20.Galyov E E, Håkansson S, Forsberg Å, Wolf-Watz H. A secreted protein kinase of Yersinia pseudotuberculosis is an indispensable virulence determinant. Nature. 1993;361:730–732. doi: 10.1038/361730a0. [DOI] [PubMed] [Google Scholar]
- 21.Gort A S, Miller V L. Identification and characterization of Yersinia enterocolitica genes induced during systemic infection. Infect Immun. 2000;68:6633–6642. doi: 10.1128/iai.68.12.6633-6642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groisman E A, Ochman H. How Salmonella became a pathogen. Trends Microbiol. 1997;5:343–349. doi: 10.1016/S0966-842X(97)01099-8. [DOI] [PubMed] [Google Scholar]
- 23.Groisman E A, Ochman H. Pathogenicity islands: bacterial evolution in quantum leaps. Cell. 1996;87:791–794. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 24.Guan K, Dixon J E. Protein tyrosine phosphatase activity of an essential virulence determinant in Yersinia. Science. 1990;249:553–556. doi: 10.1126/science.2166336. [DOI] [PubMed] [Google Scholar]
- 25.Hacker J, Bluhm-Oehler G, Mühldorfer I, Tschäpe H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 26.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 27.Heesemann J, Hanke K, Vocke T, Saken E, Rakin A, Stojiljkovic I, Berner R. Virulence of Yersinia enterocolitica is closely associated with siderophore production, expression of an iron-repressible outer membrane polypeptide of 65,000 Da and pesticin sensitivity. Mol Microbiol. 1993;8:397–408. doi: 10.1111/j.1365-2958.1993.tb01583.x. [DOI] [PubMed] [Google Scholar]
- 28.Heithoff D M, Conner C P, Hanna P C, Julio S M, Hentschel U, Mahan M J. Bacterial infection as assessed by in vivo gene expression. Proc Natl Acad Sci USA. 1997;94:934–939. doi: 10.1073/pnas.94.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isberg R R, Falkow S. A single genetic locus encoded by Yersinia pseudotuberculosis permits invasion of cultured animal cells by Escherichia coli K-12. Nature. 1985;317:262–264. doi: 10.1038/317262a0. [DOI] [PubMed] [Google Scholar]
- 30.Kinder S A, Badger J L, Bryant G O, Pepe J C, Miller V L. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O:8 and construction of a transformable R−/+ mutant. Gene. 1993;136:271–275. doi: 10.1016/0378-1119(93)90478-l. [DOI] [PubMed] [Google Scholar]
- 31.Kolter R, Inuzuka M, Helinski D R. Transcomplementation-dependent replication of a low molecular weight origin fragment from plasmid RK6. Cell. 1978;15:1199–1208. doi: 10.1016/0092-8674(78)90046-6. [DOI] [PubMed] [Google Scholar]
- 32.Kraut J. Serine proteases: structure and mechanism of catalysis. Annu Rev Biochem. 1977;46:331–358. doi: 10.1146/annurev.bi.46.070177.001555. [DOI] [PubMed] [Google Scholar]
- 33.Lantz M S. Are bacterial proteases important virulence factors? J Periodontal Res. 1977;32:126–132. doi: 10.1111/j.1600-0765.1997.tb01393.x. [DOI] [PubMed] [Google Scholar]
- 34.Li Y, Hu Z, Jordan F, Inouye M. Functional analysis of the propeptide of subtilisin E as an intramolecular chaperone for protein folding. J Biol Chem. 1995;270:25127–25132. doi: 10.1074/jbc.270.42.25127. [DOI] [PubMed] [Google Scholar]
- 35.Mahan M J, Slauch J M, Mekalanos J J. Selection of bacterial virulence genes that are specifically induced in host tissues. Science. 1993;259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- 36.Mahan M J, Tobias J W, Slauch J M, Hanna P C, Collier R J, Mekalanos J J. Antibiotic-based selection for bacterial genes that are specifically induced during infection of a host. Proc Natl Acad Sci USA. 1995;92:669–673. doi: 10.1073/pnas.92.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maldener I, Lockau W, Cai Y, Wolk C P. Calcium-dependent protease of the cyanobacterium Anabaena: molecular cloning and expression of the gene in Escherichia coli, sequencing and site-directed mutagenesis. Mol Gen Genet. 1991;225:113–120. doi: 10.1007/BF00282649. [DOI] [PubMed] [Google Scholar]
- 38.Mekalanos J J. Duplication and amplification of toxin genes in Vibrio cholerae. Cell. 1983;35:253–263. doi: 10.1016/0092-8674(83)90228-3. [DOI] [PubMed] [Google Scholar]
- 39.Miller V L, Falkow S. Evidence for two genetic loci from Yersinia enterocolitica that can promote invasion of epithelial cells. Infect Immun. 1988;56:1242–1248. doi: 10.1128/iai.56.5.1242-1248.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molloy S S, Thomas L, VanSlyke J K, Stenberg P E, Thomas G. Intracellular trafficking and activation of the furin proprotein convertase: localization to the TGN and recycling from the cell surface. EMBO J. 1994;13:18–33. doi: 10.1002/j.1460-2075.1994.tb06231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakayama K. Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem J. 1997;327:625–635. doi: 10.1042/bj3270625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Powner D, Davey J. Activation of the kexin from Schizosaccharomyces pombe requires internal cleavage of initially cleaved prosequence. Mol Cell Biol. 1998;18:400–408. doi: 10.1128/mcb.18.1.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rawlings N D, Barrett A J. Families of serine peptidases. Methods Enzymol. 1994;244:19–61. doi: 10.1016/0076-6879(94)44004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seidah N G, Chrétien M. Eukaryotic protein processing: endoproteolysis of precursor proteins. Curr Opin Biotechnol. 1997;8:602–607. doi: 10.1016/s0958-1669(97)80036-5. [DOI] [PubMed] [Google Scholar]
- 45.Seidah N G, Day R, Marcinkiewicz M, Chrétien M. Precursor convertases: an evolutionary ancient, cell-specific, combinatorial mechanism yielding diverse bioactive peptides and proteins. Ann N Y Acad Sci. 1998;839:9–24. doi: 10.1111/j.1749-6632.1998.tb10727.x. [DOI] [PubMed] [Google Scholar]
- 46.Siezen R J, de Vos W M, Leunissen J A M, Dijkstra B W. Homology modelling and protein engineering strategy of subtilases, the family of subtilisin-like serine proteinases. Protein Eng. 1991;4:719–737. doi: 10.1093/protein/4.7.719. [DOI] [PubMed] [Google Scholar]
- 47.Simonet M, Riot B, Fortineau N, Berche P. Invasin production by Yersinia pestis is abolished by insertion of an IS200-like element within the inv gene. Infect Immun. 1996;64:375–379. doi: 10.1128/iai.64.1.375-379.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith M W, Feng D-F, Doolittle R F. Evolution by acquisition: the case for horizontal gene transfer. Trends Biochem Sci. 1992;17:489–493. doi: 10.1016/0968-0004(92)90335-7. [DOI] [PubMed] [Google Scholar]
- 49.Sohl J L, Shiau A K, Rader S D, Wilk B J, Agard D A. Inhibition of α-lytic protease by pro region C-terminal steric occlusion of the active site. Biochemistry. 1997;36:3894–3902. doi: 10.1021/bi962341o. [DOI] [PubMed] [Google Scholar]
- 50.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 51.Steiner D F. The proprotein convertases. Curr Opin Chem Biol. 1998;2:31–39. doi: 10.1016/s1367-5931(98)80033-1. [DOI] [PubMed] [Google Scholar]
- 52.Stojiljkovic I, Hantke K. Hemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in gram-negative bacteria. EMBO J. 1992;11:4359–4367. doi: 10.1002/j.1460-2075.1992.tb05535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stojiljkovic I, Hantke K. Transport of haemin across the cytoplasmic membrane through a haemin-specific periplasmic binding-protein-dependent transport system in Yersinia enterocolitica. Mol Microbiol. 1994;13:719–732. doi: 10.1111/j.1365-2958.1994.tb00465.x. [DOI] [PubMed] [Google Scholar]
- 54.Travis J, Potempa J, Maeda H. Are bacterial proteinases pathogenic factors? Trends Microbiol. 1995;3:405–407. doi: 10.1016/s0966-842x(00)88988-x. [DOI] [PubMed] [Google Scholar]
- 55.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 56.Yao Z, Jones D H, Grose C. Site-directed mutagenesis of herpesvirus glycoprotein phosphorylation sites by recombination polymerase chain reaction. PCR Methods Appl. 1992;1:205–207. doi: 10.1101/gr.1.3.205. [DOI] [PubMed] [Google Scholar]
- 57.Young G M, Amid D, Miller V L. A bifunctional urease enhances survival of pathogenic Yersinia enterocolitica and Morganella morganii at low pH. J Bacteriol. 1996;178:6487–6495. doi: 10.1128/jb.178.22.6487-6495.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Young G M, Miller V L. Identification of novel chromosomal loci affecting Yersinia enterocolitica pathogenesis. Mol Microbiol. 1997;25:319–328. doi: 10.1046/j.1365-2958.1997.4661829.x. [DOI] [PubMed] [Google Scholar]
- 59.Zhang L, Radziejewshka-Lebrecht J, Krajewska-Pietrasik D, Toivanen P, Skurnik M. Molecular and chemical characterization of lipopolysaccharide O-antigen and its role in the virulence of Yersinia enterocolitica serotype O:8. Mol Microbiol. 1997;23:63–76. doi: 10.1046/j.1365-2958.1997.1871558.x. [DOI] [PubMed] [Google Scholar]
- 60.Zhong M, Munzer J S, Basak A, Benjannet S, Mowla S J, Decroly E, Chrétien M, Seidah N G. The prosegments of furin and PC7 as potent inhibitors of proprotein convertases. J Biol Chem. 1999;274:33913–33920. doi: 10.1074/jbc.274.48.33913. [DOI] [PubMed] [Google Scholar]
- 61.Ziebuhr J, Heusipp G, Siddell S G. Biosynthesis, purification, and characterization of the human coronavirus 229E 3C-like proteinase. J Virol. 1997;71:3992–3997. doi: 10.1128/jvi.71.5.3992-3997.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]