Abstract
Objective
To investigate the agreement and reliability of estimating the volumes and normative percentiles (N%) of segmented brain regions among NeuroQuant (NQ), DeepBrain (DB), and FreeSurfer (FS) software programs, focusing on the comparison between NQ and DB.
Materials and Methods
Three-dimensional T1-weighted images of 145 participants (48 healthy participants, 50 patients with mild cognitive impairment, and 47 patients with Alzheimer’s disease) from a single medical center (SMC) dataset and 130 participants from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) dataset were included in this retrospective study. All images were analyzed with DB, NQ, and FS software to obtain volume estimates and N% of various segmented brain regions. We used Bland–Altman analysis, repeated measures ANOVA, reproducibility coefficient, effect size, and intraclass correlation coefficient (ICC) to evaluate inter-method agreement and reliability.
Results
Among the three software programs, the Bland–Altman plot showed a substantial bias, the ICC showed a broad range of reliability (0.004–0.97), and repeated-measures ANOVA revealed significant mean volume differences in all brain regions. Similarly, the volume differences of the three software programs had large effect sizes in most regions (0.73–5.51). The effect size was largest in the pallidum in both datasets and smallest in the thalamus and cerebral white matter in the SMC and ADNI datasets, respectively. N% of NQ and DB showed an unacceptably broad Bland–Altman limit of agreement in all brain regions and a very wide range of ICC values (-0.142–0.844) in most brain regions.
Conclusion
NQ and DB showed significant differences in the measured volume and N%, with limited agreement and reliability for most brain regions. Therefore, users should be aware of the lack of interchangeability between these software programs when they are applied in clinical practice.
Keywords: MR volumetry, Intermethod validation, Normative percentile, FreeSurfer, NeuroQuant, DeepBrain
INTRODUCTION
Alzheimer’s disease (AD), the most common neurodegenerative disease, is characterized by progressive amyloid deposition and neurofibrillary changes several years before the onset of symptoms [1]. Closely related to this histopathological process, AD brain atrophy precedes medial temporal lobe atrophy, followed by atrophy of other parts of the brain [1,2,3,4,5]. The volume, distribution, and proportion of brain atrophy significantly correlate with the degree of cognitive impairment [6,7]. Accordingly, previous studies on AD demonstrated that brain morphometry could be a radiological marker capable of distinguishing between normal patients and patients with dementia and predicting disease progression [8,9,10]. Therefore, three-dimensional T1-weighted imaging is currently recommended as an MRI protocol for neurodegenerative disorders such as AD [11].
To date, several brain morphometry software programs such as FreeSurfer (FS) [12], Advanced Normalization Tools (ANTs) [13], and FMRIB Software Library (FSL) [14] have been developed. Among these, FS provides the most diverse information, as it provides information on the measured volume, cortical thickness, and curvature of the cortical band [15]. However, the above-mentioned brain volumetry requires considerable time and complex processes to analyze and has been used mainly for research [9,10]. In fact, the evaluation of brain atrophy mainly relies on the visual assessment by radiologists, which has a poor interobserver agreement [16].
Currently, many commercial software programs for brain volume measurement are clinically available. Compared to research software, clinical software provides a simpler user interface and an intuitive result report that can be understood without special knowledge or a complex analytical process. It automatically analyzes the quantitative volume of gray matter (GM) and white matter (WM) in the brain, evaluates the degree of atrophy of a specific brain area, and provides statistical values, such as the normative percentiles (N%), using the healthy population data stored in the software [17,18,19,20,21]. Therefore, this software may help address the limitations of visual evaluations [17,18,22].
Currently, a clinical quantitative analysis software that applies deep learning technology called DeepBrain (DB) is being used with the approval of the Ministry of Food and Drug Safety (K-FDA) [21]. However, no comparative study has examined the inter-method agreement and reliability between DB and other commonly used commercial software, such as NeuroQuant (NQ), or research software, such as FS.
Therefore, this study aimed to investigate the agreement and reliability between two clinically available software programs, DB and NQ, in estimating the volumes and N% of segmented brain regions in patients with AD or mild cognitive impairment (MCI) and in healthy participants.
MATERIALS AND METHOD
This study had Institutional Review Board approval, which waived the need for written informed consent because of the retrospective design of this study (IRB No. HDT 2021-11-014-001).
Study Participants
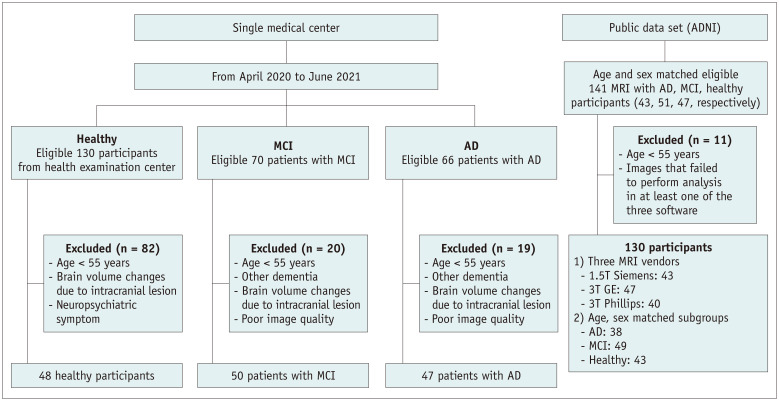
One hundred thirty-six consecutive patients with cognitive impairment (70 patients with MCI and 66 patients with AD) who visited a memory clinic and underwent brain 3T MRI from April 2020 to June 2021, and 130 healthy individuals who underwent brain MRI for medical check-ups at a health screening center during the same period were screened for this study. According to the clinical diagnosis and exclusion criteria, 50 patients with MCI, 47 patients with AD, and 48 normal elderly participants (NL) were finally included and referred to as the single medical center (SMC) dataset. To evaluate the generalizability of this study, we included additional 130 participants from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) dataset [23]. Further detailed study population information is summarized in the Supplementary Materials and Methods, Figure 1, Table 1, and Supplementary Table 1.
Fig. 1. Study design flow chart.
AD = Alzheimer’s disease, ADNI = Alzheimer’s Disease Neuroimaging Initiative, MCI = mild cognitive impairment
Table 1. Demographic Data of the Study Population.
| Data Source | SMC | ADNI | |||||
|---|---|---|---|---|---|---|---|
| Characteristics | NL | MCI | AD | NL | MCI | AD | |
| Number | 48 | 50 | 47 | 43 | 49 | 38 | |
| Age, years* | 60.75 ± 5.11 | 71.00 ± 9.04 | 77.85 ± 6.45 | 76.21 ± 5.77 | 76.53 ± 6.85 | 77.29 ± 8.93 | |
| Sex | |||||||
| Female | 22 | 29 | 39 | 24 | 24 | 17 | |
| Male | 26 | 21 | 8 | 19 | 25 | 21 | |
| MMSE score* | NA | 24.85 ± 1.23 | 16.83 ± 1.67 | 29.05 ± 1.24 | 27.08 ± 1.74 | 21.56 ± 4.85 | |
| CDR* | NA | 0.51 ± 0.02 | 1.25 ± 0.23 | 0.01 ± 0.08 | 0.50 ± 0.10 | 0.89 ± 0.38 | |
| Vendor | 3T Siemens | 3T Siemens | 3T Siemens | ||||
| 1.5T Siemens | 0 | 0 | 0 | 10 | 14 | 19 | |
| 3T Siemens | 48 | 50 | 47 | 0 | 0 | 0 | |
| 3T GE | 0 | 0 | 0 | 17 | 15 | 15 | |
| 3T Phillips | 0 | 0 | 0 | 11 | 14 | 15 | |
*The data are reported as the mean ± standard deviation. Otherwise, the data are number of patients. AD = Alzheimer’s disease, ADNI = Alzheimer’s Disease Neuroimaging Initiative, CDR = clinical dementia rating, MCI = mild cognitive impairment, MMSE = Mini-Mental State Examination, NA = not applicable, NL = normal elderly participants, SMC = single medical center
Image Acquisition
MR scans, which included routine brain MRI and additional T1-weighted volume images, were conducted with a 3T unit (Verio 3T, Siemens) using a 20-channel head coil in our SMC. All participants in this hospital, including the NL, MCI, and AD groups, were scanned to obtain T1-weighted volume images using the same sequence parameters. The specific MR imaging parameters of the T1-weighted volume images used for all participants, including the SMC and ADNI data, are presented in Supplementary Table 2.
Magnetic Resonance Volumetry
Sagittal T1-volume images of SMC and ADNI data were uploaded to the DB and NQ servers to perform automated quantification of the regional brain volume (Fig. 2). A detailed description of the analysis process for each software is provided in the Supplementary Materials and Methods.
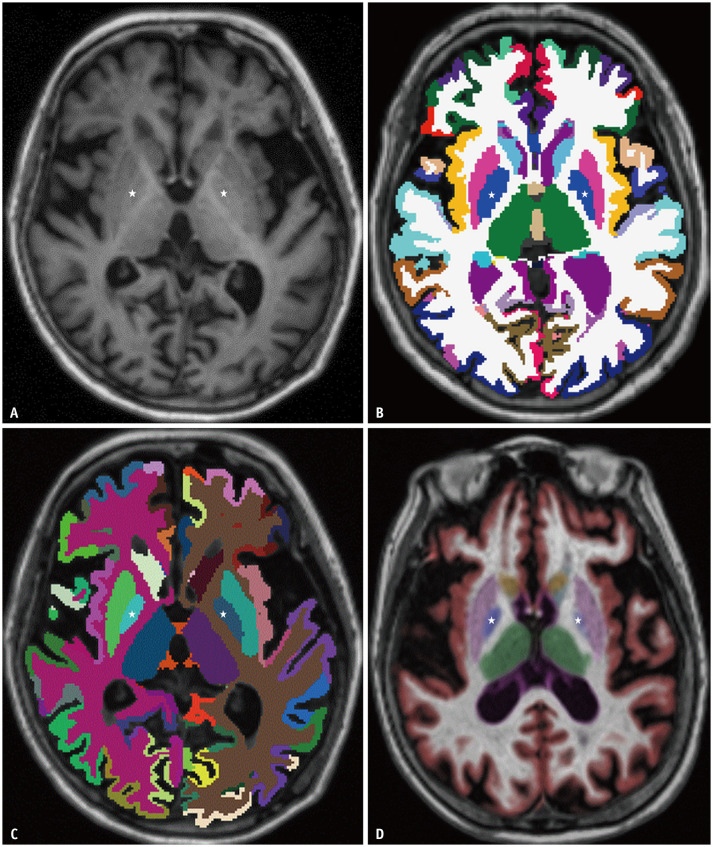
Fig. 2. Representative case of a 69-year-old female with Alzheimer’s disease.
A-D. Axial T1-weighted imaging (A); color overlays based on FreeSurfer (B), DeepBrain (C), and NeuroQuant (D). The overlaid area of the bilateral globus pallidus (marked with stars) is smaller with NeuroQuant (D) than with FreeSurfer (B) or DeepBrain (C).
Statistical Analysis
Bland–Altman analysis and reproducibility coefficient (RC) were used to analyze the inter-software agreements among NQ, DB, and FS in measuring the volumes [24]. The effect size (ES Cohen’s d) was used to evaluate the standardized mean difference for each software pair. The following guidelines were used to interpret ES values: small, 0.2; medium, 0.5; and large, 0.8 [25]. We then compared these three software volume datasets using repeated measures ANOVA and multiple comparisons for which adjusted p values were calculated by applying the Bonferroni correction method. Bland–Altman analysis was also used to compare the inter-software agreement between NQ and DB in obtaining N%.
Additionally, the reliability between the software programs was assessed using the intraclass correlation coefficient (ICC; two-way mixed model, single rater/measurement, absolute agreement) [26]. ICC values were interpreted as follows: poor, ICC < 0.5; moderate, 0.5 ≤ ICC < 0.75; good, 0.75–0.9; and excellent, ICC ≥ 0.9 [26].
Finally, the correlation between N% and the visual rating scales was analyzed using the Spearman correlation coefficient. Receiver operating characteristic analyses with area under the curve were performed to compute the discriminating power of N%.
Statistical analyses were performed using computer software packages (MedCalc version 20.014, MedCalc Software; SPSS, version 26 for Windows, IBM Corp.). In all analyses, p < 0.05 was considered to represent a significant difference.
RESULTS
Volume of Segmented Brain Regions
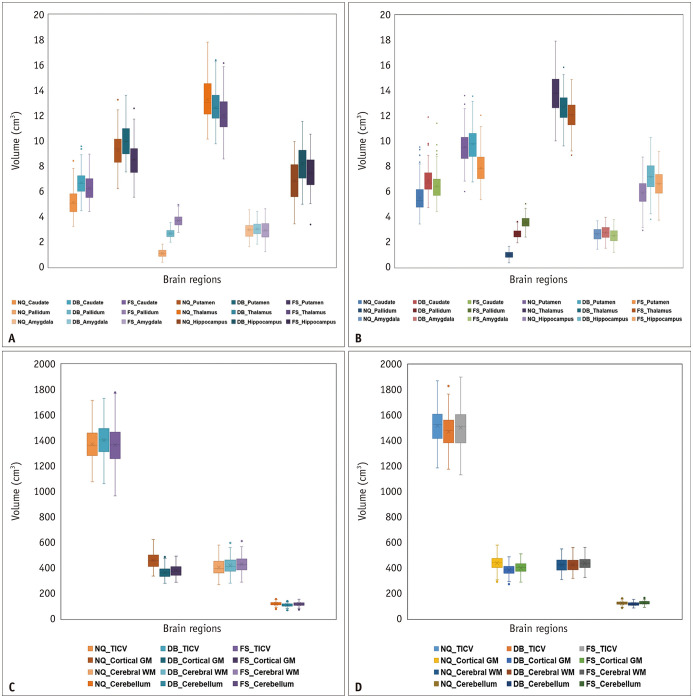
A graphical summary of all cortical and subcortical volumes of the brain regions is provided in Figure 3, and the specific numerical data are shown in Supplementary Table 3.
Fig. 3. Box-and-whisker plots illustrate differences in measured regional brain volume derived from NQ, DB, and FS in a SMC and ADNI data.
A-D. SMC (A) and ADNI (B) show smaller brain regions (caudate, pallidum, putamen, thalamus, amygdala, and hippocampus), and SMC (C) and ADNI (D) show the cortical gray matter, cerebral white matter, cerebellum, and total intracranial volume. The lines inside the boxes and the lower and upper boundary lines represent the median, 25th, and 75th percentile values, respectively, with whiskers extending from the median to the ± 1.5 × interquartile range; outliers beyond the whiskers are represented by points. ADNI = Alzheimer’s Disease Neuroimaging Initiative, DB = DeepBrain, FS = FreeSurfer, NQ = NeuroQuant, SMC = single medical center
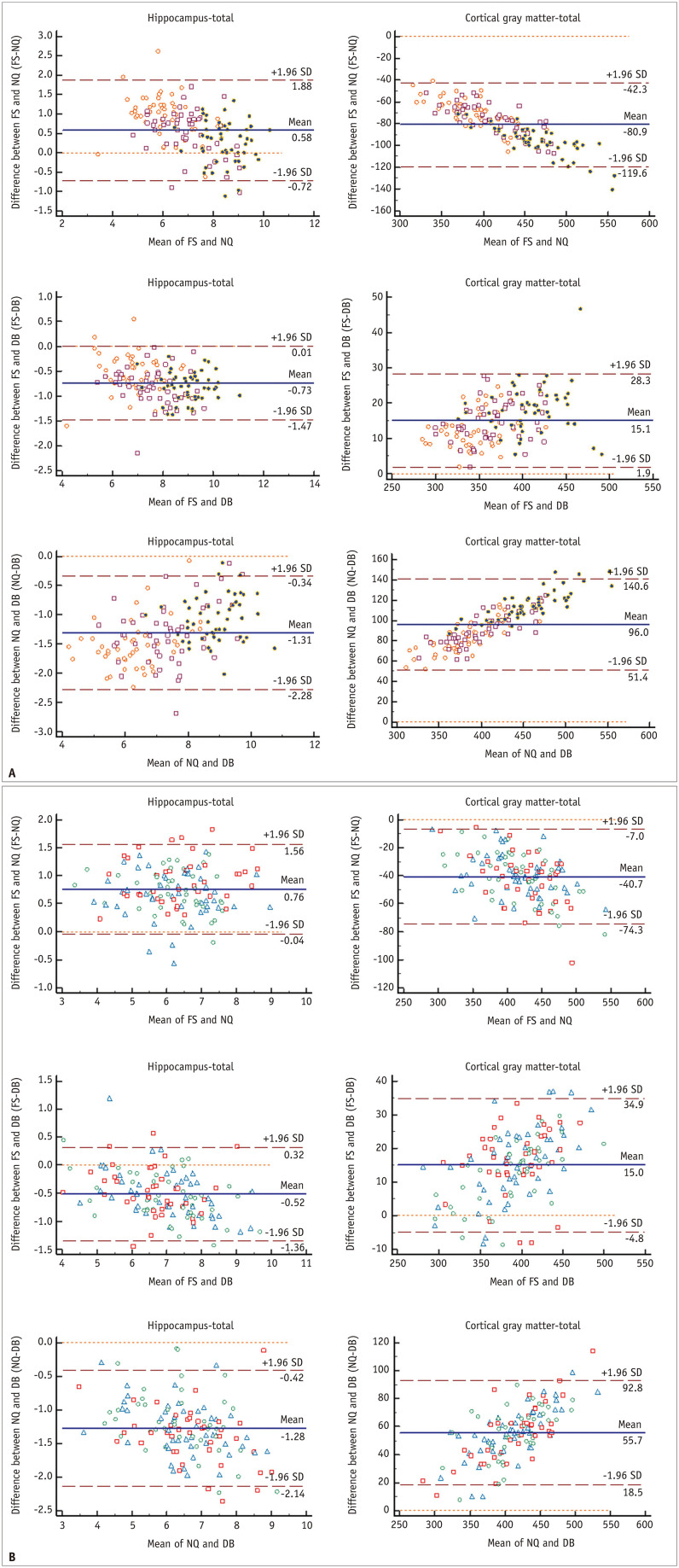
In the Bland–Altman analysis (Fig. 4, Supplementary Figs. 1-3), the mean difference between NQ and FS and those between DB and FS showed substantial bias in most brain regions except for total intracranial volume (TICV), when comparing NQ and FS. In particular, there was a significant bias in the pallidum among all the software comparisons. We found a tendency of NQ to overestimate the volume of large structures and underestimate the volume of small structures compared with FS in measuring cerebral cortical GM in both the SMC and ADNI datasets. In contrast, DB tends to underestimate the volume of large structures and overestimate the volume of small structures compared with FS in measuring the cerebral cortical GM in both the SMC and ADNI datasets and in measuring the amygdala in the SMC dataset.
Fig. 4. Bland–Altman plots for agreement between each software for regional brain volume.
A, B. Represent SMC and ADNI data, respectively. The units for both the x- and y-axes are cm3. There is a tendency for NQ to overestimate large volumes and underestimate small volumes compared with FS measurement of cerebral cortical GM in both SMC (A) and ADNI data (B). In contrast, DeepBrain slightly tends to underestimate large volumes and overestimate small volumes compared with FS measurement of the cerebral cortical GM in both A and B. The orange circle, brown square, and purple circle in A indicate the Alzheimer’s disease, mild cognitive impairment, and normal elderly subgroups, respectively. The blue triangle, red square, and green circle in B indicate the 1.5T Siemens, 3T GE, and 3T Phillips subgroups, respectively. The brown horizontal dashed lines delineate the 95% confidence intervals (the likelihood of individual measures to be within ± 1.96 SDs). The orange horizontal dashed line represents the equal (the difference between two software measurements is zero) line. The blue horizontal line indicates the difference between two software measurements. ADNI = Alzheimer’s Disease Neuroimaging Initiative, DB = DeepBrain, FS = FreeSurfer, GM = gray matter, NQ = NeuroQuant, SD = standard deviation, SMC = single medical center
Regarding the RC, DB gave volume estimates closer to those of FS than did NQ in most brain regions, including the cortical GM, caudate, pallidum, thalamus, amygdala, hippocampus, and cerebral WM; however, only two brain regions had a reversed relationship (NQ estimates instead of DB estimates were closer to those of FS), including the cerebellum and TICV (Table 2).
Table 2. RC of Measured Regional Brain Volume Obtained From Each Software in the Total Study Population.
| DB vs. FS | NQ vs. FS | DB vs. NQ | |||||
|---|---|---|---|---|---|---|---|
| Lt. Hemisphere | Rt. Hemisphere | Lt. Hemisphere | Rt. Hemisphere | Lt. Hemisphere | Rt. Hemisphere | ||
| Cortical GM | |||||||
| SMC | 16.63 | 16.08 | 79.13 | 83.40 | 94.30 | 98.10 | |
| ADNI | 16.38 | 19.46 | 42.91 | 43.76 | 55.75 | 59.63 | |
| Caudate | |||||||
| SMC | 0.48 | 0.76 | 1.39 | 1.17 | 1.59 | 1.63 | |
| ADNI | 0.66 | 0.59 | 1.12 | 1.21 | 1.50 | 1.52 | |
| Putamen | |||||||
| SMC | 1.77 | 1.29 | 1.12 | 0.89 | 0.97 | 0.91 | |
| ADNI | 2.25 | 1.63 | 2.08 | 1.64 | 1.00 | 0.92 | |
| Pallidum | |||||||
| SMC | 1.12 | 0.99 | 2.59 | 2.59 | 1.53 | 1.63 | |
| ADNI | 1.01 | 0.90 | 2.67 | 2.52 | 1.76 | 1.67 | |
| Thalamus | |||||||
| SMC | 1.20 | 0.51 | 1.47 | 1.42 | 0.85 | 1.36 | |
| ADNI | 1.17 | 0.70 | 1.99 | 1.90 | 1.41 | 1.58 | |
| Amygdala | |||||||
| SMC | 0.37 | 0.31 | 0.42 | 0.39 | 0.30 | 0.36 | |
| ADNI | 0.38 | 0.29 | 0.40 | 0.29 | 0.26 | 0.35 | |
| Hippocampus | |||||||
| SMC | 0.79 | 0.88 | 0.86 | 0.88 | 1.26 | 1.49 | |
| ADNI | 0.67 | 0.71 | 0.84 | 0.89 | 1.29 | 1.38 | |
| Cerebellum | |||||||
| SMC | 8.38 | 8.19 | 4.91 | 5.11 | 11.41 | 9.75 | |
| ADNI | 11.05 | 8.95 | 6.48 | 6.72 | 7.48 | 5.58 | |
| Total cortical GM | |||||||
| SMC | 32.20 | 162.35 | 192.30 | ||||
| ADNI | 35.45 | 86.39 | 115.17 | ||||
| Total cerebral WM | |||||||
| SMC | 28.29 | 55.48 | 35.49 | ||||
| ADNI | 37.40 | 59.22 | 54.67 | ||||
| TICV | |||||||
| SMC | 124.40 | 102.74 | 94.78 | ||||
| ADNI | 145.54 | 109.92 | 125.48 | ||||
ADNI = Alzheimer’s Disease Neuroimaging Initiative, DB = DeepBrain, FS = FreeSurfer, GM = gray matter, NQ = NeuroQuant, RC = reproducibility coefficient, SMC = single medical center, TICV = total intracranial volume, WM = white matter
According to the repeated measures ANOVA (Supplementary Table 3), there was a significant difference in volume values in all brain regions among the three software programs (DB, NQ, and FS; p < 0.05). Among these brain region volumes, the pallidum and cortical GM were areas with a particularly large mean volume difference between NQ and DB. As shown in Figure 3, the pallidum volumes in DB and FS were greater than those in NQ. The cortical GM volumes in DB and FS were smaller than those in NQ. The mean volume difference of the hippocampus between FS and NQ in the SMC data was slightly smaller than that between FS and DB, but this relationship was reversed in the ADNI data. All the other brain region volume comparisons, except for the pallidum, hippocampus, and cortical GM, are described in the Supplementary Materials and Methods.
Regarding ICC, the brain regions calculated by the three methods showed a wide range of concordance (0.004–0.97) (Table 3). In both datasets, the brain regions in which ICC values between DB and FS showed higher reliability than those between NQ and FS were the cortical GM, caudate, pallidum, thalamus, and cerebral WM. Conversely, the brain regions that showed higher ICC values between NQ and FS than between DB and FS were the cerebellum and TICV. Notably, the ICC values of the pallidum between each software showed the worst agreement (poor) among all the brain regions for the two datasets (DB vs. FS, 0.15–0.23; NQ vs. FS, 0.004–0.01; DB vs NQ, 0.01–0.04).
Table 3. ICC of Regional Brain Volume Measured by the Three Software Programs in the Total Study Population.
| DB vs. FS | NQ vs. FS | DB vs. NQ | |||||
|---|---|---|---|---|---|---|---|
| Lt. Hemisphere | Rt. Hemisphere | Lt. Hemisphere | Rt. Hemisphere | Lt. Hemisphere | Rt. Hemisphere | ||
| Cortical GM | |||||||
| SMC | 0.93 (0.03–0.98) | 0.93 (0.03–0.98) | 0.45 (-0.02–0.80) | 0.42 (-0.02–0.78) | 0.35 (-0.02–0.72) | 0.33 (-0.02–0.71) | |
| ADNI | 0.93 (0.28–0.98) | 0.90 (0.12–0.97) | 0.71 (-0.06–0.92) | 0.70 (-0.06–0.91) | 0.57 (-0.05–0.86) | 0.53 (-0.05–0.84) | |
| Caudate | |||||||
| SMC | 0.88 (0.66–0.94) | 0.74 (0.07–0.90) | 0.44 (-0.08–0.77) | 0.55 (-0.09–0.82) | 0.40 (-0.04–0.76) | 0.38 (-0.05–0.74) | |
| ADNI | 0.81 (0.41–0.92) | 0.86 (0.40–0.95) | 0.62 (-0.05–0.85) | 0.59 (-0.07–0.83) | 0.45 (-0.09–0.77) | 0.48 (-0.07–0.80) | |
| Putamen | |||||||
| SMC | 0.52(-0.05–0.83) | 0.65 (-0.07–0.89) | 0.72 (-0.02–0.90) | 0.78 (0.45–0.89) | 0.78 (0.04–0.92) | 0.77 (0.23–0.90) | |
| ADNI | 0.35(-0.04–0.72) | 0.50 (-0.04–0.82) | 0.36 (-0.09–0.70) | 0.46 (-0.09–0.76) | 0.72 (0.61–0.80) | 0.68 (0.57–0.76) | |
| Pallidum | |||||||
| SMC | 0.15(-0.04–0.46) | 0.18 (-0.04–0.50) | 0.01 (-0.01–0.07) | 0.01 (-0.01–0.05) | 0.04 (-0.03–0.17) | 0.04 (-0.02–0.17) | |
| ADNI | 0.21(-0.07–0.52) | 0.23 (-0.06–0.58) | 0.01 (-0.01–0.03) | 0.004 (-0.01–0.02) | 0.01 (-0.01–0.04) | 0.01 (-0.01–0.04) | |
| Thalamus | |||||||
| SMC | 0.74(-0.06–0.92) | 0.93 (0.90–0.95) | 0.65 (-0.06–0.87) | 0.67 (-0.07–0.89) | 0.85 (0.80–0.89) | 0.65 (-0.05–0.86) | |
| ADNI | 0.70 (0.02–0.88) | 0.83 (0.64–0.91) | 0.47 (-0.10–0.76) | 0.44 (-0.08–0.72) | 0.65 (0.37–0.80) | 0.48 (0.01–0.72) | |
| Amygdala | |||||||
| SMC | 0.83 (0.74–0.89) | 0.88 (0.83–0.92) | 0.82 (0.53–0.91) | 0.83 (0.68–0.90) | 0.87 (0.77–0.92) | 0.81 (0.12–0.93) | |
| ADNI | 0.77 (0.12–0.91) | 0.87 (0.77–0.92) | 0.77 (-0.02–0.93) | 0.86 (0.72–0.92) | 0.89 (0.84–0.92) | 0.79 (0.19–0.92) | |
| Hippocampus | |||||||
| SMC | 0.84 (0.05–0.95) | 0.81 (-0.05–0.95) | 0.82 (0.50–0.92) | 0.83 (0.37–0.93) | 0.72 (-0.05–0.92) | 0.63 (-0.06–0.88) | |
| ADNI | 0.86 (0.29–0.95) | 0.84 (0.28–0.94) | 0.75 (-0.03–0.91) | 0.69 (-0.02–0.88) | 0.60 (-0.07–0.87) | 0.55 (-0.08–0.84) | |
| Cerebellum | |||||||
| SMC | 0.81 (-0.05–0.95) | 0.84 (0.16–0.95) | 0.93 (0.67–0.97) | 0.93 (0.85–0.97) | 0.70 (-0.04–0.92) | 0.78 (-0.05–0.94) | |
| ADNI | 0.73 (-0.06–0.92) | 0.81 (-0.05–0.95) | 0.89 (0.65–0.95) | 0.88 (0.53–0.95) | 0.85 (0.05–0.96) | 0.91 (0.68–0.96) | |
| Total cortical GM | |||||||
| SMC | 0.93 (0.02–0.98) | 0.43 (-0.02–0.79) | 0.34 (-0.02–0.71) | ||||
| ADNI | 0.92 (0.16–0.98) | 0.70 (-0.06–0.91) | 0.54 (-0.05–0.85) | ||||
| Total cerebral WM | |||||||
| SMC | 0.97 (0.77–0.99) | 0.90 (0.06–0.97) | 0.96 (0.57–0.99) | ||||
| ADNI | 0.93 (0.80–0.96) | 0.79 (0.67–0.86) | 0.81 (0.74–0.86) | ||||
| TICV | |||||||
| SMC | 0.51 (-0.04–0.83) | 0.93 (0.91–0.95) | 0.50 (-0.02–0.83) | ||||
| ADNI | 0.86 (0.75–0.92) | 0.88 (0.83–0.91) | 0.82 (0.66–0.89) | ||||
Data are ICC (95% confidence interval). AD = Alzheimer’s disease, ADNI = Alzheimer’s Disease Neuroimaging Initiative, DB = DeepBrain, FS = FreeSurfer, GM = gray matter, ICC = intraclass correlation coefficient, MCI = mild cognitive impairment, NL = normal elderly participants, NQ = NeuroQuant, SMC = single medical center, TICV = total intracranial volume, WM = white matter
The comparison of the estimated volumes between the two datasets, the effect size of the measured volume, and all statistical results of the subgroup analysis are summarized in the Supplementary Materials and Methods, Supplementary Figures 1-3, and Supplementary Tables 4-16.
Normative Percentiles of Segmented Brain Regions
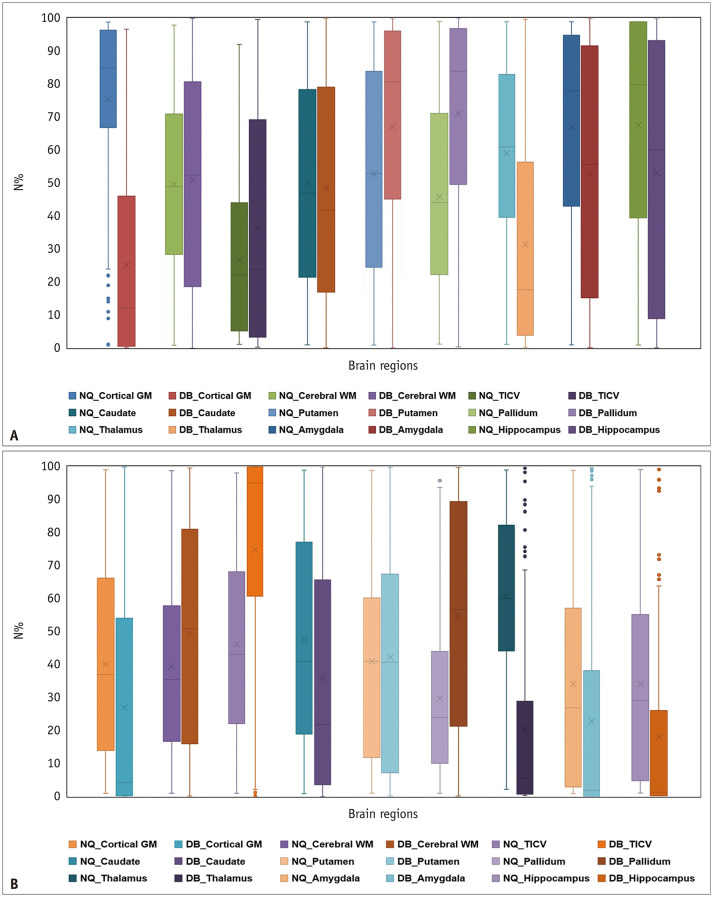
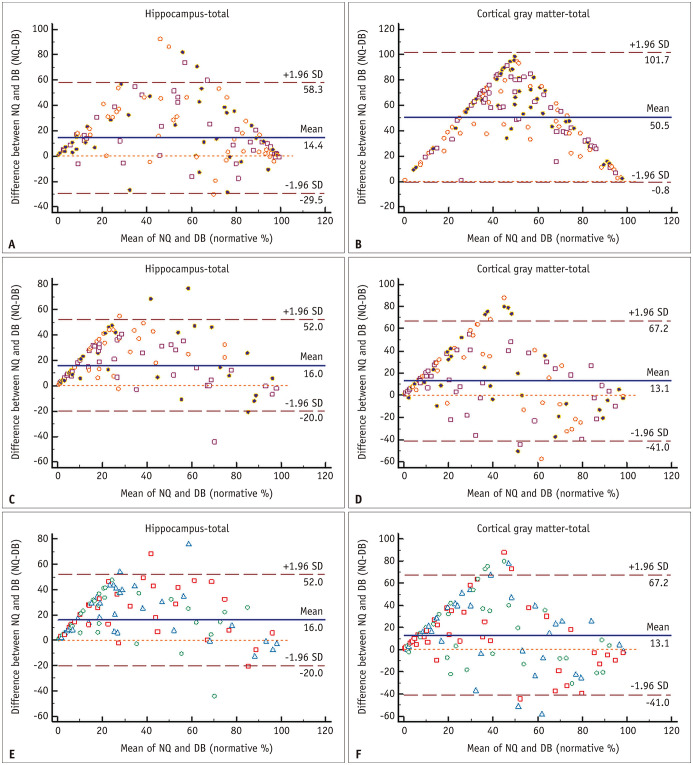
Table 4 and Figure 5 summarize the N% of each regional brain volume analyzed in NQ and DB. Table 5 and Figure 6 present the ICCs and Bland–Altman analysis results for N% of regional brain volumes. Unlike volume measurement, the Bland–Altman plots of N% were triangular or rhombus shaped with substantial bias (mean difference) and had an unacceptably broad limit of agreement for almost all brain regions. This means that the degree of agreement increased toward the smallest (near 0%) or largest (near 100%) N% values, but the degree of agreement was markedly decreased toward the median value. Furthermore, regarding the ICC of the N% between NQ and DB, it was revealed that there was poor to good agreement (ICC of -0.142–0.844) in almost all brain regions, except for some measurements of the caudate, hippocampus, cerebellar GM, and TICV.
Table 4. Comparison of Normative Percentiles of Regional Brain Volume Derived from NQ and DB in the Total Study Population.
| Lt. Hemisphere | Rt. Hemisphere | ||||
|---|---|---|---|---|---|
| NQ | DB | NQ | DB | ||
| Cortical GM | |||||
| SMC | 74.04 ± 26.54 | 23.40 ± 28.40 | 76.99 ± 24.62 | 27.85 ± 30.84 | |
| ADNI | 40.86 ± 30.00 | 27.74 ± 34.81 | 40.04 ± 30.47 | 27.18 ± 34.87 | |
| Caudate | |||||
| SMC | 49.32 ± 31.89 | 37.84 ± 39.73 | 50.85 ± 30.88 | 57.62 ± 34.11 | |
| ADNI | 47.71 ± 31.16 | 31.00 ± 33.42 | 47.76 ± 32.60 | 42.22 ± 36.58 | |
| Putamen | |||||
| SMC | 50.43 ± 31.21 | 65.98 ± 33.43 | 55.77 ± 30.47 | 68.16 ± 31.59 | |
| ADNI | 39.32 ± 29.93 | 45.02 ± 34.09 | 43.80 ± 30.43 | 39.67 ± 32.69 | |
| Pallidum | |||||
| SMC | 46.59 ± 29.53 | 65.44 ± 32.04 | 45.57 ± 28.47 | 74.47 ± 27.41 | |
| ADNI | 29.53 ± 24.12 | 57.78 ± 35.25 | 31.83 ± 24.57 | 49.68 ± 32.81 | |
| Thalamus | |||||
| SMC | 53.90 ± 28.17 | 24.81 ± 31.21 | 63.19 ± 26.32 | 43.50 ± 33.39 | |
| ADNI | 63.76 ± 24.11 | 20.15 ± 28.60 | 57.62 ± 26.72 | 23.85 ± 28.32 | |
| Amygdala | |||||
| SMC | 64.32 ± 32.31 | 50.03 ± 36.92 | 68.01 ± 31.14 | 54.91 ± 35.40 | |
| ADNI | 34.96 ± 32.86 | 24.82 ± 35.11 | 35.76 ± 31.20 | 24.74 ± 33.39 | |
| Hippocampus | |||||
| SMC | 65.30 ± 34.43 | 50.43 ± 38.96 | 69.19 ± 33.00 | 56.25 ± 37.57 | |
| ADNI | 87.98 ± 20.27 | 74.91 ± 30.73 | 91.77 ± 13.27 | 76.79 ± 26.56 | |
| Cerebellar GM | |||||
| SMC | 57.01 ± 27.52 | 40.07 ± 32.94 | 57.93 ± 27.01 | 35.64 ± 32.87 | |
| ADNI | 47.90 ± 30.35 | 54.00 ± 36.84 | 46.00 ± 30.45 | 48.75 ± 36.30 | |
| Cerebellar WM | |||||
| SMC | 73.56 ± 23.27 | 30.23 ± 31.29 | 76.37 ± 22.61 | 29.18 ± 30.32 | |
| ADNI | 59.93 ± 27.43 | 31.32 ± 32.21 | 61.44 ± 28.27 | 32.12 ± 32.36 | |
| Cerebral WM | |||||
| SMC | 48.03 ± 26.49 | 40.07 ± 32.94 | 51.45 ± 26.64 | 52.11 ± 32.82 | |
| ADNI | 38.54 ± 27.53 | 49.93 ± 34.47 | 39.94 ± 27.27 | 48.76 ± 34.82 | |
| Total cortical GM | |||||
| SMC | 75.64 ± 25.34 | 25.18 ± 29.74 | |||
| ADNI | 40.12 ± 30.22 | 27.07 ± 34.72 | |||
| Total cerebral WM | |||||
| SMC | 49.77 ± 26.22 | 51.08 ± 33.44 | |||
| ADNI | 39.18 ± 27.33 | 49.25 ± 34.18 | |||
| TICV | |||||
| SMC | 26.60 ± 23.84 | 36.41 ± 34.91 | |||
| ADNI | 45.92 ± 27.73 | 74.62 ± 34.52 | |||
Data are mean ± standard deviation. AD = Alzheimer’s disease, ADNI = Alzheimer’s Disease Neuroimaging Initiative, DB = DeepBrain, FS = FreeSurfer, GM = gray matter, MCI = mild cognitive impairment, NL = normal elderly participants, NQ = NeuroQuant, SMC = single medical center, TICV = total intracranial volume, WM = white matter
Fig. 5. Box-and-whisker plots showing differences in N% of regional brain volume derived from NQ and DB.
A, B. Represent N% of the single medical center and ADNI data, respectively. The lines inside the boxes and the lower and upper boundary lines represent the median, 25th, and 75th percentile values, respectively, with whiskers extending from the median to ± 1.5 × the interquartile range; outliers beyond the whiskers are represented by points. ADNI = Alzheimer’s Disease Neuroimaging Initiative, DB = DeepBrain, GM = gray matter, NQ = NeuroQuant, N% = normative percentiles, TICV = total intracranial volume, WM = white matter
Table 5. Intermethod Reliability of Normative Percentiles Presented by NQ and DB in the Total Study Population.
| NQ vs. DB | |||
|---|---|---|---|
| Lt. Hemisphere | Rt. Hemisphere | ||
| Cortical GM | |||
| SMC | 0.202 (-0.083–0.504) | 0.217 (-0.086–0.523) | |
| ADNI | 0.601 (0.403–0.730) | 0.599 (0.410–0.726) | |
| Caudate | |||
| SMC | 0.804 (0.542–0.881) | 0.784 (0.692–0.847) | |
| ADNI | 0.534 (0.277–0.696) | 0.692 (0.589–0.773) | |
| Putamen | |||
| SMC | 0.711 (0.352–0.850) | 0.749 (0.486–0.861) | |
| ADNI | 0.695 (0.591–0.776) | 0.619 (0.501–0.714) | |
| Pallidum | |||
| SMC | 0.245 (0.070–0.403) | 0.183 (-0.028–0.373) | |
| ADNI | -0.032 (-0.148–0.098) | -0.142(-0.291–0.019) | |
| Thalamus | |||
| SMC | 0.431 (-0.058–0.703) | 0.536 (0.152–0.734) | |
| ADNI | 0.163 (-0.082–0.409) | 0.325 (-0.091–0.620) | |
| Amygdala | |||
| SMC | 0.648 (0.432–0.774) | 0.670 (0.463–0.790) | |
| ADNI | 0.713 (0.569–0.806) | 0.672 (0.504–0.779) | |
| Hippocampus | |||
| SMC | 0.780 (0.470–0.889) | 0.716 (0.517–0.824) | |
| ADNI | 0.712 (0.409–0.843) | 0.629 (0.137–0.820) | |
| Cerebellar GM | |||
| SMC | 0.649 (0.246–0.816) | 0.602 (0.018–0.821) | |
| ADNI | 0.824 (0.746–0.878) | 0.844 (0.786–0.887) | |
| Cerebellar WM | |||
| SMC | 0.155 (-0.077–0.384) | 0.125 (-0.074–0.337) | |
| ADNI | 0.224 (-0.014–0.427) | 0.191 (-0.019–0.379) | |
| Total cortical GM | |||
| SMC | 0.207 (-0.083–0.512) | ||
| ADNI | 0.597 (0.402–0.726) | ||
| Total cerebral WM | |||
| SMC | 0.694 (0.599–0.769) | ||
| ADNI | 0.269 (0.107–0.418) | ||
| TICV | |||
| SMC | 0.817 (0.608–0.901) | ||
| ADNI | 0.450 (-0.035–0.712) | ||
Data are intraclass correlation coefficient (95% confidence interval). ADNI = the Alzheimer’s Disease Neuroimaging Initiative, DB = DeepBrain, GM = gray matter, NQ = NeuroQuant, SMC = single medical center, TICV = total intracranial volume, WM = white matter
Fig. 6. Bland-Altman plots for agreement of the normative percentile of the hippocampus (A, C, E) and cortical gray matter (B, D, F) between NQ and DB.
A-F. A and B represent SMC data, and C-F represent ADNI data. There is a tendency of a triangular or rhomboid shape on the Bland–Altman plot with unacceptably broad limits of agreement for all datasets. In A-D, the orange circle, brown square, and purple circle indicate the Alzheimer’s disease, MCI, and NL control subgroups, respectively, in both datasets. In E and F, the blue triangle, red square, and green circle indicate the 1.5T Siemens, 3T GE, and 3T Phillips subgroups, respectively. The brown horizontal dashed lines delineate the 95% confidence intervals (the likelihood of individual measures to be within ± 1.96 SDs). The orange horizontal dashed line represents the equal (the difference between two software measurements is zero) line. The blue horizontal line is the mean difference of two software measurements. ADNI = Alzheimer’s Disease Neuroimaging Initiative, DB = DeepBrain, MCI = mild cognitive impairment, NL = normal elderly participants, NQ = NeuroQuant, SD = standard deviation, SMC = single medical center
The comparison of the N% of the two datasets (SMC and ADNI), further subgroup analysis results, and clinical relevance of N% (correlation between N% and visual ratings and diagnostic performance of N% of two methods) are summarized in the Supplementary Materials and Methods, Supplementary Figures 4-7, and Supplementary Tables 17-24.
DISCUSSION
In this study, we compared two clinically available brain volumetry software programs, DB and NQ, in terms of their agreement and reliability in two different study populations consisting of different sites, vendors, magnetic field strengths, and participants with different cognitive functions. It was found that both NQ and DB had substantial bias according to the Bland–Altman analysis and broad (poor to excellent) inter-method reliability. In addition, the volume of most brain regions showed a significant difference between the values analyzed using the two methods. In particular, the cortical GM and pallidum were significantly different in terms of RC, effect size, and mean volume between the two software programs. Regarding the N% of the regional brain volume, there were significant differences in areas significantly related to cognitive functions, such as the hippocampus and cortical GM. Moreover, the difference between the two software programs in terms of the inter-method agreement of N% was even poorer.
Our study revealed that there were more brain regions, such as the cortical GM, pallidum, caudate, and thalamus, where the measured volumes of DB were closer to the values of FS than those of NQ on RC, mean volume, and ICC analysis, even though NQ and DB are commonly based on FS. We speculate that the main reason for this is the entirely different image analysis pipelines of the two methods. The NQ algorithm consists of the following steps: 1) quality check of the MR image sequence, 2) correction for gradient nonlinearity and B1 field inhomogeneity, 3) deletion of non-brain tissue using an active contour model, and 4) parcellation (segments anatomic structures) by nonlinear registration to the embedded probabilistic atlas, assignment of a neuroanatomic tag to each voxel, and repeated checking of each voxel to maximize the probability [17]. In particular, NQ uses a different probabilistic atlas than FS [27]. Previous studies have demonstrated that the anatomic atlas type affects volume measurement results; when different anatomic atlases are used for the hippocampus, the accuracy varies depending on the anatomic atlas used [28]. By contrast, DB is a trained deep convolutional neural network (CNN). The input for DB algorithm training was preprocessed (resampling, resizing, and intensity normalization) 3D-T1WI, and the corresponding output for training was the FS segmentation mask, which was corrected by anatomy experts [21]. Thus, unlike NQ, DB did not use a skull stripping algorithm or registration to the anatomic atlas, which could be a major reason for the discrepancy between NQ and DB in this study.
Moreover, in one recent study comparing the segmentation mask of FS with that of DB, the Dice similarity coefficients were 0.82 or greater, demonstrating relatively high inter-method reproducibility [21]. In another recent study, the inter-method agreement between segmentation masks of FS and those of the CNN, which comprised the modified AlexNet and was trained by the segmentation mask created in FS as a training output, was analyzed [15]. Compared with the ICCs between NQ and FS revealed in another previous study [27], the ICCs between CNN and FS were comparable in many brain regions; however, in the globus pallidus, the ICC between CNN and FS was significantly higher than that between NQ and FS [15,27]. Additionally, in this study, the ICC between FS and CNN for cortical thickness measurement showed ICCs comparable to the test-retest study results of FS [29] in many cortices [15]. Therefore, similar to the results of previous studies comparing FS and NQ [27,30], in our study, the cortical GM volume was significantly larger in NQ than in DB, and ICC in the pallidum was substantially higher between DB and FS than between NQ and FS, which can be explained in this context.
Overall, the measured volumes of the three software packages maintained a constant trend for both the SMC and ADNI datasets. However, there were only a few exceptional brain regions. For the measured volumes of the cortical GM, those of NQ were the largest, those of FS were the second largest, and those of DB were the smallest in both datasets. However, the difference in the measured volume of the cortical GM between NQ and FS was much smaller in the ADNI dataset than in the SMC dataset. The exact reason for this finding is difficult to explain. However, FS and NQ are softwares mainly based on the North American brain template. Therefore, we speculate that the difference between the two methods in the ADNI dataset, which is public data for North America, appears to be smaller than that in our SMC dataset. Another exceptional brain region was the TICV. The estimated TICV of DB was slightly larger than those of NQ and FS in the SMC data. However, in the ADNI data, those of DB were slightly smaller than those of the other software. The slight variation in the TICV of DB may be due to the different output masks used for the deep learning algorithm training. In contrast to the procedure used for other brain regions in the training process of the deep learning algorithm, manually segmented masks of the TICV were used as outputs for the training. Although manual segmentation is the gold standard ground truth, because TICV values between NQ and FS consistently show high ICC values regardless of the dataset, a comparative evaluation of TICV values between FS, DB, and manual segmentation is necessary in the future.
Among the brain regions, the pallidum showed the lowest (poor) reliability and the largest effect size among the three methods, especially between NQ and other software. Previous studies have revealed that the pallidum and adjacent WM are difficult to accurately distinguish and segment into two separate regions because they show similar signal intensities in T1-weighted images [27,31]. In addition, the pallidum volume is calculated by including the WM between the pallidum and putamen [27]. Another possible reason is that metal deposition as part of the aging or degeneration process, such as iron, calcium, and manganese deposition in the pallidum, may alter the T1 relaxation time [32], influencing the volume estimation of the software.
Almost all statistical tendencies of the total data were consistently maintained in the subgroup analysis. Because all statistical analyses were applied across different geographical regions, vendors, magnetic field strengths, and participants with different cognitive functions, the results of this study have higher generalizability than those of single-center studies. However, in the vendor subgroup of the ADNI data, some parts differed from the trend of the entire dataset. In the 3T GE subgroup using inversion recovery spoiled gradient-echo (IR-SPGR), TICV size was the largest in NQ and smallest in FS, unlike in other vendor subgroups. A study analyzing longitudinal brain volume changes using ADNI data reported that the volume differed by approximately 2.47% when magnetization-prepared rapid gradient-echo (MPRAGE) was changed to IR-SPGR within the same GE vendor [33]. This study suggested that the reason for this result is that there may be differences in tissue contrast and boundary delineation according to sequence changes [33], and our research results are likely to be understood in this context. In the 1.5T Siemens subgroup, cerebral WM was the largest in the NQ, unlike in other subgroup vendors. In a recent study using NQ, cerebral WM was larger at 1.2 mm slice thickness than at a 1 mm slice thickness [34]. In our study, there was no difference in slice thickness, but a difference in magnetic field strength; both factors are closely related to the matrix size. In fact, the matrix size in the 1.2 mm slice thickness protocol of the previous study and the 1.5T Siemens protocol (ADNI) of this study were the same at 192 × 192, which was smaller than the matrix size (256 × 256) in the 1 mm slice thickness protocol of the previous study [34] or other vendor subgroups in this study. Therefore, we hypothesize that NQ may tend to measure cerebral WM as larger than other brain regions when spatial resolution decreases.
The N% values analyzed for each software were significantly different. One reason for this, as explained above, is the significant difference in the measured volume of most brain regions analyzed in NQ and DB. Second, it is assumed that the reference values for the normal population built in each software were different. In the Bland–Altman analysis, almost all regional brain volumes except the cortical GM were almost randomly distributed with substantial bias (mean difference); in contrast, nearly all regional brain N% showed a triangular or rhomboid shape on the Bland–Altman plot with broad limits of agreement. Furthermore, the mean volume of the hippocampus itself was smaller in NQ than in DB, but the N% of the hippocampus was significantly larger in NQ than in DB for both datasets. Previous studies have shown that even if the same volume of data from normal participants are used, significantly different N% values are derived depending on the geographical region from which the reference values are derived [35,36]. Most of the reference population stored in the NQ server is from the United States, whereas most of it stored in the DB server is from South Korea; there is an essential difference in the geographic region where the reference population was obtained. Therefore, the results of this study are consistent with those of previous studies. Although there were significant differences in N% values between NQ and DB, they showed a similar discriminatory power to differentiate patients with NL, MCI, and AD, showing a high correlation with the visual atrophy rating scales.
This study has some limitations. First, the slice thickness of the MRI protocol used for our SMC dataset was kept constant at 1.2 mm during the study period, according to the MR protocol recommended by the NQ developer. In addition, all MRI protocols in the ADNI data had 1.2 mm slice thickness. If MRI is performed with a slice thickness of 1 mm or less, it is possible to obtain a slightly more accurate volume with a higher spatial resolution and a slightly higher ICC between NQ and FS [30]. However, in practice, a 1 mm slice thickness scan obeying the MR parameters recommended by the NQ vendor requires more time than a 1.2 mm slick thickness scan. This was also a clinical reason for choosing a 1.2 mm slice thickness, in addition to the recommendation of the NQ vendor in this study. It may be necessary to investigate inter-method reproducibility with a 1 mm slice thickness in a future study. Second, although this study tried to ensure the generalizability of the study results with ADNI data, we did not analyze other 1.5T MR machines except for the Siemens machine and recent faster scanning techniques, such as the new parallel imaging technique (Wave-CAIPI), compressed sensing, and deep-learning reconstruction. Third, FS is not the gold standard ground truth for brain volumetry, unlike manual segmentation. However, the accuracy and reliability of FS compared to manual segmentation performed by experts have been proven in several studies [37,38,39].
In conclusion, NQ, DB, and FS showed substantial biases when compared in terms of volume measurement of various brain regions. DB yielded results closer to those obtained with FS than NQ. Regarding the N% of the brain regions, the differences between NQ and DB were more remarkable. Users should be aware of the lack of interchangeability between these software programs when applying them in clinical practice, such as in the longitudinal follow-up for changes in patients with cognitive impairment and traumatic brain injury, to prevent confusion caused by the difference in software used.
Footnotes
Conflicts of Interest: Seung Hong Choi who is on the editorial board of the Korean Journal of Radiology was not involved in the editorial evaluation or decision to publish this article. All remaining authors have declared no conflicts of interest.
- Conceptualization: Sang Won Jo.
- Data curation: Sang Won Jo, Seun Ah Lee, Jae Ho Kim, Suk-Ki Chang, Yunji Lim, Yeong Seo Yoo.
- Formal analysis: Sang Won Jo, Seun Ah Lee, Huijin Song.
- Investigation: Sang Won Jo, Seun Ah Lee, Huijin Song, Jae Ho Kim, Suk-Ki Chang.
- Methodology: Sang Won Jo, Huijin Song.
- Project administration: Sang Won Jo.
- Resources: Seung Hong Choi, Chul-Ho Sohn.
- Software: Huijin Song, Seun Ah Lee, Sang Won Jo.
- Supervision: Seung Hong Choi, Chul-Ho Sohn.
- Visualization: Huijin Song, Seun Ah Lee, Sang Won Jo.
- Writing—original draft: Huijin Song, Seun Ah Lee.
- Writing—review & editing: Sang Won Jo.
Funding Statement: None
Availability of Data and Material
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
Supplement
The Supplement is available with this article at https://doi.org/10.3348/kjr.2022.0067.
References
- 1.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 2.Scahill RI, Schott JM, Stevens JM, Rossor MN, Fox NC. Mapping the evolution of regional atrophy in Alzheimer’s disease: unbiased analysis of fluid-registered serial MRI. Proc Natl Acad Sci U S A. 2002;99:4703–4707. doi: 10.1073/pnas.052587399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehéricy S, Baulac M, Chiras J, Piérot L, Martin N, Pillon B, et al. Amygdalohippocampal MR volume measurements in the early stages of Alzheimer disease. AJNR Am J Neuroradiol. 1994;15:929–937. [PMC free article] [PubMed] [Google Scholar]
- 4.Chan D, Fox NC, Scahill RI, Crum WR, Whitwell JL, Leschziner G, et al. Patterns of temporal lobe atrophy in semantic dementia and Alzheimer’s disease. Ann Neurol. 2001;49:433–442. [PubMed] [Google Scholar]
- 5.Killiany RJ, Hyman BT, Gomez-Isla T, Moss MB, Kikinis R, Jolesz F, et al. MRI measures of entorhinal cortex vs hippocampus in preclinical AD. Neurology. 2002;58:1188–1196. doi: 10.1212/wnl.58.8.1188. [DOI] [PubMed] [Google Scholar]
- 6.Fox NC, Scahill RI, Crum WR, Rossor MN. Correlation between rates of brain atrophy and cognitive decline in AD. Neurology. 1999;52:1687–1689. doi: 10.1212/wnl.52.8.1687. [DOI] [PubMed] [Google Scholar]
- 7.Cardenas VA, Chao LL, Studholme C, Yaffe K, Miller BL, Madison C, et al. Brain atrophy associated with baseline and longitudinal measures of cognition. Neurobiol Aging. 2011;32:572–580. doi: 10.1016/j.neurobiolaging.2009.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jack CR., Jr Alzheimer disease: new concepts on its neurobiology and the clinical role imaging will play. Radiology. 2012;263:344–361. doi: 10.1148/radiol.12110433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jack CR, Jr, Albert MS, Knopman DS, McKhann GM, Sperling RA, Carrillo MC, et al. Introduction to the recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:257–262. doi: 10.1016/j.jalz.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Jr, Kawas CH, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson KA, Fox NC, Sperling RA, Klunk WE. Brain imaging in Alzheimer disease. Cold Spring Harb Perspect Med. 2012;2:a006213. doi: 10.1101/cshperspect.a006213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischl B. FreeSurfer. Neuroimage. 2012;62:774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avants BB, Tustison NJ, Stauffer M, Song G, Wu B, Gee JC. The insight ToolKit image registration framework. Front Neuroinform. 2014;8:44. doi: 10.3389/fninf.2014.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. FSL. Neuroimage. 2012;62:782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 15.Rebsamen M, Suter Y, Wiest R, Reyes M, Rummel C. Brain morphometry estimation: from hours to seconds using deep learning. Front Neurol. 2020;11:244. doi: 10.3389/fneur.2020.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park M, Moon WJ. Structural MR imaging in the diagnosis of Alzheimer’s disease and other neurodegenerative dementia: current imaging approach and future perspectives. Korean J Radiol. 2016;17:827–845. doi: 10.3348/kjr.2016.17.6.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brewer JB, Magda S, Airriess C, Smith ME. Fully-automated quantification of regional brain volumes for improved detection of focal atrophy in Alzheimer disease. AJNR Am J Neuroradiol. 2009;30:578–580. doi: 10.3174/ajnr.A1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanpitukpongse TP, Mazurowski MA, Ikhena J, Petrella JR Alzheimer’s Disease Neuroimaging Initiative. Predictive utility of marketed volumetric software tools in subjects at risk for Alzheimer disease: do regions outside the hippocampus matter? AJNR Am J Neuroradiol. 2017;38:546–552. doi: 10.3174/ajnr.A5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Storelli L, Rocca MA, Pagani E, Van Hecke W, Horsfield MA, De Stefano N, et al. Measurement of whole-brain and gray matter atrophy in multiple sclerosis: assessment with MR imaging. Radiology. 2018;288:554–564. doi: 10.1148/radiol.2018172468. [DOI] [PubMed] [Google Scholar]
- 20.Lee JS, Kim C, Shin JH, Cho H, Shin DS, Kim N, et al. Machine learning-based individual assessment of cortical atrophy pattern in Alzheimer’s disease spectrum: development of the classifier and longitudinal evaluation. Sci Rep. 2018;8:4161. doi: 10.1038/s41598-018-22277-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suh CH, Shim WH, Kim SJ, Roh JH, Lee JH, Kim MJ, et al. Development and validation of a deep learning–based automatic brain segmentation and classification algorithm for Alzheimer disease using 3D T1-weighted volumetric images. AJNR Am J Neuroradiol. 2020;41:2227–2234. doi: 10.3174/ajnr.A6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang C, Beadnall HN, Hatton SN, Bader G, Tomic D, Silva DG, et al. Automated brain volumetrics in multiple sclerosis: a step closer to clinical application. J Neurol Neurosurg Psychiatry. 2016;87:754–757. doi: 10.1136/jnnp-2015-312304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, et al. Ways toward an early diagnosis in Alzheimer’s disease: the Alzheimer’s Disease Neuroimaging Initiative (ADNI) Alzheimers Dement. 2005;1:55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park JE, Han K, Sung YS, Chung MS, Koo HJ, Yoon HM, et al. Selection and reporting of statistical methods to assess reliability of a diagnostic test: conformity to recommended methods in a peer-reviewed journal. Korean J Radiol. 2017;18:888–897. doi: 10.3348/kjr.2017.18.6.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olejnik S, Algina J. Measures of effect size for comparative studies: applications, interpretations, and limitations. Contemp Educ Psychol. 2000;25:241–286. doi: 10.1006/ceps.2000.1040. [DOI] [PubMed] [Google Scholar]
- 26.Koo TK, Li MY. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med. 2016;15:155–163. doi: 10.1016/j.jcm.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ochs AL, Ross DE, Zannoni MD, Abildskov TJ, Bigler ED Alzheimer’s Disease Neuroimaging Initiative. Comparison of automated brain volume measures obtained with NeuroQuant and FreeSurfer. J Neuroimaging. 2015;25:721–727. doi: 10.1111/jon.12229. [DOI] [PubMed] [Google Scholar]
- 28.Nestor SM, Gibson E, Gao FQ, Kiss A, Black SE Alzheimer’s Disease Neuroimaging Initiative. A direct morphometric comparison of five labeling protocols for multi-atlas driven automatic segmentation of the hippocampus in Alzheimer’s disease. Neuroimage. 2013;66:50–70. doi: 10.1016/j.neuroimage.2012.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madan CR, Kensinger EA. Test-retest reliability of brain morphology estimates. Brain Inform. 2017;4:107–121. doi: 10.1007/s40708-016-0060-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yim Y, Lee JY, Oh SW, Chung MS, Park JE, Moon Y, et al. Comparison of automated brain volume measures by NeuroQuant vs. Freesurfer in patients with mild cognitive impairment: effect of slice thickness. Yonsei Med J. 2021;62:255–261. doi: 10.3349/ymj.2021.62.3.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 32.Kanda T, Nakai Y, Aoki S, Oba H, Toyoda K, Kitajima K, et al. Contribution of metals to brain MR signal intensity: review articles. Jpn J Radiol. 2016;34:258–266. doi: 10.1007/s11604-016-0532-8. [DOI] [PubMed] [Google Scholar]
- 33.Lee H, Nakamura K, Narayanan S, Brown RA, Arnold DL Alzheimer’s Disease Neuroimaging Initiative. Estimating and accounting for the effect of MRI scanner changes on longitudinal whole-brain volume change measurements. Neuroimage. 2019;184:555–565. doi: 10.1016/j.neuroimage.2018.09.062. [DOI] [PubMed] [Google Scholar]
- 34.Lee JY, Oh SW, Chung MS, Park JE, Moon Y, Jeon HJ, et al. Clinically available software for automatic brain volumetry: comparisons of volume measurements and validation of intermethod reliability. Korean J Radiol. 2021;22:405–414. doi: 10.3348/kjr.2020.0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finkelsztejn A, Fragoso YD, Bastos EA, Duarte JA, Varela JS, Houbrechts R, et al. Intercontinental validation of brain volume measurements using MSmetrix. Neuroradiol J. 2018;31:147–149. doi: 10.1177/1971400917721521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vinke EJ, Huizinga W, Bergtholdt M, Adams HH, Steketee RME, Papma JM, et al. Normative brain volumetry derived from different reference populations: impact on single-subject diagnostic assessment in dementia. Neurobiol Aging. 2019;84:9–16. doi: 10.1016/j.neurobiolaging.2019.07.008. [DOI] [PubMed] [Google Scholar]
- 37.Guenette JP, Stern RA, Tripodis Y, Chua AS, Schultz V, Sydnor VJ, et al. Automated versus manual segmentation of brain region volumes in former football players. Neuroimage Clin. 2018;18:888–896. doi: 10.1016/j.nicl.2018.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morey RA, Petty CM, Xu Y, Hayes JP, Wagner HR, 2nd, Lewis DV, et al. A comparison of automated segmentation and manual tracing for quantifying hippocampal and amygdala volumes. Neuroimage. 2009;45:855–866. doi: 10.1016/j.neuroimage.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tae WS, Kim SS, Lee KU, Nam EC, Kim KW. Validation of hippocampal volumes measured using a manual method and two automated methods (FreeSurfer and IBASPM) in chronic major depressive disorder. Neuroradiology. 2008;50:569–581. doi: 10.1007/s00234-008-0383-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.