Abstract
We have isolated mutations that block sporulation after formation of the polar septum in Bacillus subtilis. These mutations were mapped to the two genes of a new locus, spoIIS. Inactivation of the second gene, spoIISB, decreases sporulation efficiency by 4 orders of magnitude. Inactivation of the first gene, spoIISA, has no effect on sporulation but it fully restores sporulation of a spoIISB null mutant, indicating that SpoIISB is required only to counteract the negative effect of SpoIISA on sporulation. An internal promoter ensures the synthesis of an excess of SpoIISB over SpoIISA during exponential growth and sporulation. In the absence of SpoIISB, the sporulating cells show lethal damage of their envelope shortly after asymmetric septation, a defect that can be corrected by synthesizing SpoIISB only in the mother cell. However, forced synthesis of SpoIISA in exponentially growing cells or in the forespore leads to the same type of morphological damage and to cell death. In both cases protection against the killing effect of SpoIISA can be provided by simultaneous synthesis of SpoIISB. The spoIIS locus is unique to B. subtilis, and since it is completely dispensable for sporulation its physiological role remains elusive.
Nutrient deprivation of the gram-positive bacterium Bacillus subtilis triggers asymmetric cell division, a landmark event of the sporulation process. The smaller progeny cell, the forespore, becomes engulfed by the larger one, the mother cell, which transiently functions as a nurse cell before lysing to release the mature spore (26, 30). Although synthesized prior to septation and partitioned into both the forespore and the mother cell, the transcription factor ςF becomes active only in the forespore, therein initiating a genetic program that culminates in the formation of the dormant spore and launching by intercellular signaling the developmental program of the mother cell (17).
The specific release of ςF activity in the forespore is controlled by the complex interaction of three regulatory proteins, SpoIIAA, SpoIIAB, and SpoIIE, in conjunction with the formation of the sporulation septum (1, 9, 19, 22). Although the subject of intense investigation, the precise mechanisms that restrict ςF activity to the forespore are not yet fully understood (10, 15). In order to gain further insight into that important regulatory step, we have isolated mutations enhancing ςF activity. One previously identified class of ςF-activating mutations is characterized by the formation of abortive forespore compartments at both poles of the sporulating cell, the so-called disporic phenotype that is easily recognizable by phase contrast microscopy (26). The twofold increase of ςF activity, as monitored by ςF-dependent lacZ fusions carried by these mutants, is due to the activation of ςF in both forespore compartments (18). By screening for mutations enhancing ςF activity and accompanied by a nondisporic Spo− phenotype, we have identified a new locus in which some mutations block sporulation shortly after completion of the polar septum. Subsequent characterization of this locus indicated that it encodes two proteins, one of which prevents the lethal action of the other, with no direct bearing on the ςF activation cascade.
MATERIALS AND METHODS
Bacterial strains and techniques.
All B. subtilis strains were derivatives of the sporulation-proficient strain JH642 (trpC2 pheA1), with the exception of the xin15 L8460 strain obtained from D. Karamata. The reporter lacZ fusions to the spoIIR, spoIIQ, spoIID, and spoIIIG promoters have been described (20, 21, 29, 31), as have the gfp fusions to the spoIIQ and cotE promoters (20, 33). For monitoring sporulation efficiency, cells were grown at 37°C for about 40 h in Difco sporulation (DS) medium (28) and the number of spores was determined by their resistance to heat killing (10 min at 80°C). β-Galactosidase was assayed as previously described (29) and is expressed as nanomoles of 2-nitrophenyl-β-d-galactopyranoside hydrolyzed per minute per milligram of protein. Synthesis of green fluorescent protein from Aequorea victoria was monitored by fluorescence microscopy according to published protocols (33). UV mutagenesis of a wild-type strain containing a spoIIR-lacZ fusion at the amyE locus was performed according to standard protocols (8), and cells were directly plated on DS agar plates containing 400 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)/ml. Spontaneous Spo+ suppressor mutations obtained in a spoIISB null strain carrying an extra copy of spoIISA at amyE were identified on DS agar plates by the characteristic brown pigmentation of sporulating B. subtilis colonies. These mutations were mapped to spoIISA by their linkage with the tetracycline resistance marker in spoIISB or the chloramphenicol marker at amyE as assessed by DNA transformation. Antibiotic-resistance cassettes and conditions of selection for antibiotic resistance have been described (12).
Cloning the spoIIS locus.
Sporulation-deficient mutants exhibiting enhanced ςF activity were transformed with an integrative plasmid library (23), selecting for chloramphenicol resistance, and screened for sporulation recovery as judged by their pigmentation phenotype on DS agar plates. Prior to that step the spoIIR-lacZ fusion, associated in the mutants with a chloramphenicol resistance marker at amyE, was removed and replaced with a spectinomycin resistance marker (11). Rescuing plasmids could be recovered in Escherichia coli, and their insert could be characterized as previously described (21). Two plasmids were isolated that were able to correct the defect of the spoIIS mutants. In one plasmid the Sau3A insert did not extend further upstream than codon 7 in spoIISA, whereas the insert of the other plasmid contained 370 bp upstream of the spoIISA reading frame. The smaller insert contained 261 bp downstream of spoIISB and was followed by a Sau3A insert from another region of the B. subtilis chromosome, with a HindIII site located 41 bp after the junction between the two Sau3A fragments. Various subfragments from these two plasmids were cloned in integrative vectors that could recombine by single crossover at the spoIIS locus (24) or in vectors allowing ectopic integration by a double recombination event at amyE or thrC, occasionally after fusion to lacZ (11). Point mutations in spoIIS were mapped by using a series of overlapping integrative plasmids. They were cloned either by recovery in E. coli of a spontaneously excised plasmid (unable to rescue the original mutation) or by a chromosomal walk from a plasmid integrated in the vicinity of the mutation.
Manipulating the spoIIS locus.
A null mutation in spoIISA was created by replacing the 352-bp NdeI-HpaI fragment internal to spoIISA with a kanamycin resistance cassette. A null mutation in spoIISB was created by inserting a tetracycline resistance cassette in the DraI site located at codon 17 in spoIISB. A complete deletion of the spoIIS locus was constructed by cloning a kanamycin resistance cassette between PCR-amplified fragments bracketing an interval extending 48 bp upstream of spoIISA and 76 bp downstream of spoIISB. Because an intact spoIISA gene could not be cloned in E. coli without spoIISB, the spoIISA gene was introduced in two steps at the ectopic amyE site. First, a truncated spoIISA gene starting at codon 7 of spoIISA and extending to codon 17 of spoIISB was introduced at amyE with selection for spectinomycin resistance. Then, a complete spoIISA gene was reconstituted by recombination with a fragment containing 370 bp upstream of the spoIISA reading frame as well as 91 codons of spoIISA, exchanging the spectinomycin marker for a chloramphenicol resistance marker. A similar two-step strategy was followed to put the spoIISA gene under the control of foreign promoters by previously fusing a subfragment containing only 52 bp upstream of the spoIISA reading frame and 91 codons of spoIISA to the desired promoter-bearing fragment. The spoIID promoter (a 290-bp HindIII-PvuII fragment) and the spoIIQ promoter (a 566-bp SphI-Sau3A fragment) have been previously described (20, 29). A 1.5-kb fragment containing the xylA promoter and the xylR gene was a kind gift from F. Arigoni. The spoIISB gene was introduced at the ectopic amyE and thrC sites, either under the control of its own promoter as a 948-bp NaeI-HindIII fragment or under the control of foreign promoters as a 696-bp NdeI-HindIII fragment.
Ultrastructural studies.
Samples for electron microscopy were processed as described previously (3). Stained thin sections were examined and photographed on a JEOL-100 CX electron microscope. For quantification of morphological classes, at least 100 complete longitudinal sections were scored from random fields for each sample.
RESULTS
Identification of the spoIIS locus.
A B. subtilis strain carrying a spoIIR-lacZ transcriptional fusion was UV mutagenized to 98% killing, and cells were plated directly on DS agar plates containing the chromogenic β-galactosidase substrate X-Gal and incubated at 37°C. The spoIIR gene is expressed from a weak ςF-controlled promoter (14, 21), and colonies harboring a spoIIR-lacZ fusion become barely blue on such plates. The presence of a mutation leading to the disporic phenotype markedly increases the blue color after 2 days at 37°C, a feature that made that fusion ideal for our genetic screen. Forty-five colonies with a darker blue color were isolated from 150,000 colonies recovered after mutagenesis. Twelve of them were defective in sporulation, with sporulation efficiencies ranging from 10−2 to 10−7 of that observed with a wild-type strain. None of these mutants appeared to produce disporic cells as judged by phase contrast microscopy.
A mirror collection of mutant strains was constructed by substituting a spectinomycin resistance marker for the chloramphenicol resistance marker linked to the spoIIR-lacZ fusion at amyE in each of the 12 strains. The mutants were then sorted into complementation groups by transforming each original mutant with chromosomal DNAs prepared from the 11 other strains from the mirror set and selecting for spectinomycin resistance. Correction of the sporulation defect of the recipient strain could occur in a few cases by genetic congression (8), and the frequency of Spo+ transformants was interpreted as indicating the linkage of the spo mutations present in the donor and recipient strains. Closely linked mutations were expected to result in many fewer Spo+ transformants. The 12 spo mutations were found to define two linkage groups, with 10 mutations originally characterized by a similar darker intensity of coloration on X-Gal plates belonging to the same linkage group and 2 mutations with lesser activation of spoIIR-lacZ belonging to another group (data not shown).
A library of integrative plasmids containing sized Sau3A fragments from the B. subtilis genome (23) was screened for complementation of the sporulation defect of representative members of the two linkage groups. One plasmid was found to be able to restore sporulation to the 10 members of the larger linkage group. Sequence analysis of the plasmid insert identified the presence of the 5′ part of the spoIIIE gene. It is known that spoIIIE mutations prevent full partitioning of the chromosome into the forespore compartment (35) and lead to hyperactivation of ςF-dependent genes trapped in the forespore (21, 32, 36), although the basis for this phenomenon has not been elucidated. Since our genetic screen for enhanced ςF activity was carried out with a strain containing a ςF-dependent lacZ fusion inserted at amyE, a region of the chromosome trapped in the forespore, recovering spoIIIE mutations was not unexpected.
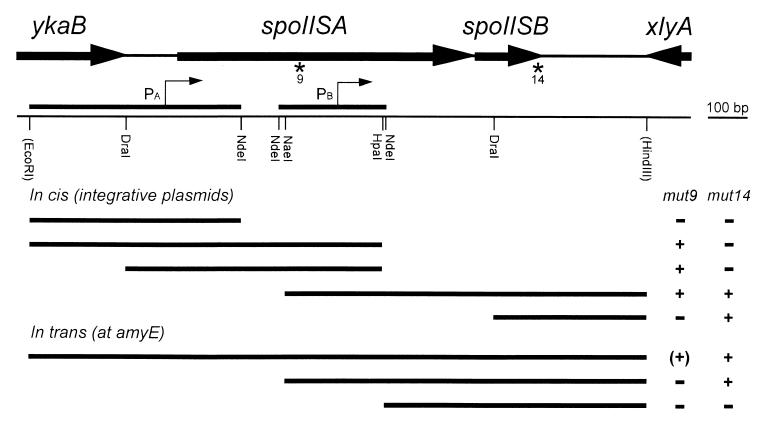
Two plasmids were isolated that were able to complement both mutants from the other linkage group. Restriction mapping showed that each plasmid contained at least two different inserts due to multiple ligation of Sau3A fragments into the vector. A 1,550-bp region overlapping the inserts present in the two plasmids was subcloned and sequenced, identifying the presence of two genes located between the ykaB and xlyA genes (Fig. 1). Sequencing data obtained in the frame of the B. subtilis genome project and kindly provided by K. Devine helped to solve a few ambiguities. Various subfragments of that region were cloned in an integrative vector and were checked for the ability to rescue the sporulation defect of the two mutants. From the results of these experiments, some of which are shown in Fig. 1, one mutation (mut9) was mapped in the upstream gene and the other mutation (mut14) in the downstream one. Since both mutations block sporulation at stage II (see below), this locus was called spoIIS and the two cistrons were called spoIISA and spoIISB.
FIG. 1.
Characterization of the spoIIS locus. The genetic organization of the spoIIS region is shown at the top, with partial or complete open reading frames displayed as thick arrows. Asterisks indicate the locations of the mut9 and mut14 mutations. In the simplified physical map, the bordering restriction sites originate from the plasmids used for cloning the region, either from the vector backbone (EcoRI) or from an additional genomic fragment present in the insert (HindIII). The two fragments fused to lacZ for monitoring promoter activity are shown with the presumed positions of the transcription starts (thin arrows). Thick bars in the bottom part of the figure indicate the extents of the DNA fragments that were used for complementation analysis of the two spoIIS mutants, with the results shown in the right-side columns. When these fragments were cloned in integrative plasmids, correction of the sporulation defect (indicated as +) was observed in a variable proportion of the recombinants, depending on the location of the mutation relative to the fragments. Conversely, introducing these DNA fragments at the ectopic amyE locus led to a homogeneous population of transformants. Partial restoration of sporulation of the mut9 strain is indicated by (+).
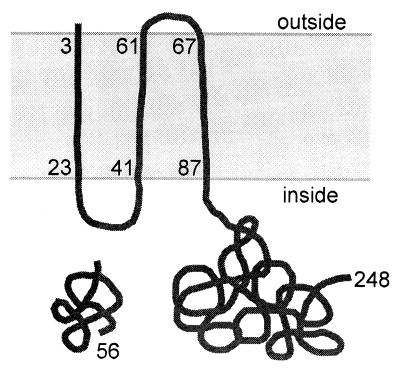
The spoIISA gene encodes a 248-residue protein containing three putative transmembrane domains (6), the last two-thirds of the protein being predicted to be located in the cytoplasm (Fig. 2). The spoIISB gene encodes a basic, hydrophilic, 56-residue protein. Neither protein bears any similarity to a protein of known function. The two mutations were cloned (see Materials and Methods) and sequenced. The mut9 mutation was found to be a missense mutation converting codon 103 of spoIISA from CTT (Leu) to TTT (Phe). The mut14 mutation was found to be a 2-bp deletion after codon 52 of spoIISB, leading to the replacement of the last four residues of SpoIISB by an unrelated stretch of 23 amino acids. The two mutations have similar effects on sporulation efficiency, which is decreased by about 4 orders of magnitude compared to that of the wild type (Table 1).
FIG. 2.
The two SpoIIS proteins. A schematic representation of the SpoIISA and SpoIISB proteins (248 and 56 residues, respectively) is shown with the coordinates of the three putative transmembrane domains of SpoIISA. The topological model for SpoIISA is based on the predictions of the TopPred II program (6) and includes the presence of six positively charged residues between the first two transmembrane segments.
TABLE 1.
Complementation studies with spoIIS mutants
| SpoIIS proteins encoded at:
|
Spores/mlc | |||||
|---|---|---|---|---|---|---|
| spoIISa | amyEb | |||||
| A | B | –d | – | 5.5 × 108 | ||
| – | B | – | – | 6.2 × 108 | ||
| – | – | – | – | 5.6 × 108 | ||
| A | B | A | – | 6.2 × 108 | ||
| Al103f | B | – | – | 1.9 × 104 | ||
| Al103f | – | – | – | 1.9 × 104 | ||
| Al103f | B | A | B | 2.9 × 107 | ||
| Al103f | B | A | – | 4.6 × 104 | ||
| Al103f | B | – | B | 2.7 × 104 | ||
| A | B∗ | – | – | 4.0 × 104 | ||
| A | – | – | – | 1.6 × 104 | ||
| A | – | A | B | 7.2 × 108 | ||
| A | – | – | B | 5.9 × 108 | ||
| A | – | – | (B) | 6.2 × 104 | ||
| A | – | A | – | 5.0 × 101 | ||
| Ar38q | – | A | – | 2.7 × 107 | ||
| Ar38q | – | – | – | 5.9 × 108 | ||
The null mutations in spoIISA, spoIISB, and the whole spoIIS locus are described in Materials and Methods. The abnormal B∗ protein is encoded by the spoIISB(mut14) mutant.
Although containing an intact spoIISB cistron, a DNA fragment extended only up to the NdeI site does not support SpoIISB synthesis, as indicated by (B), whereas SpoIISB is synthesized from a fragment extending up to the NaeI site.
Less than twofold variations were observed in several sporulation assays. A typical series of results is shown.
–, no SpoIISA (or SpoIISB) encoded.
Epistatic relationship between SpoIISA and SpoIISB.
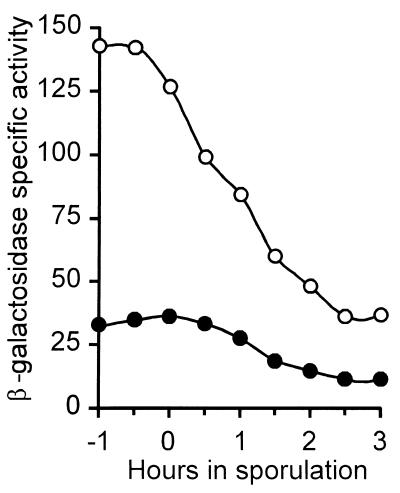
The SpoIISB translation start codon overlaps the spoIISA translation stop codon, a strong indication that the two genes constitute an operon. It was therefore unexpected that the mut14 mutation in spoIISB can be complemented in trans by a DNA fragment containing an intact spoIISB cistron but extending only up to the NaeI site located at codon 91 of spoIISA (Fig. 1). A shorter DNA fragment extending only up to the NdeI site located at codon 175 of spoIISA does not complement the mut14 mutation (Fig. 1), suggesting that a promoter allowing expression of spoIISB is present in the NaeI-NdeI interval. Indeed, this region of the chromosome is able to drive β-galactosidase synthesis when cloned upstream of a promoterless lacZ gene (Fig. 3). The spoIISB promoter is probably recognized by the major vegetative sigma factor ςA as suggested by the variations of its activity during exponential growth and sporulation (Fig. 3). These variations are not significantly altered by the absence of transcription factors involved in the transition to stationary phase, such as ςH, Spo0A, or AbrB (data not shown).
FIG. 3.
Expression of spoIIS-lacZ. The specific activity of β-galactosidase was monitored in a wild-type strain containing a transcriptional spoIIS-lacZ fusion, either from the Pa promoter (●) or from the Pb promoter (○), as defined in Fig. 1. Both fusions were inserted at the amyE locus. Bacteria were induced to sporulate by exhaustion in DS medium at 37°C, with the onset of sporulation defined as the time when cultures deviate from exponential growth.
A null mutation in spoIISB was constructed by inserting a tetracycline resistance cassette into the DraI site located at codon 17 of spoIISB. This mutation leads to the same sporulation defect as the original mut14 mutation and can be complemented by the same DNA fragments introduced at the amyE locus (Table 1). Therefore, the mut14 mutation is a loss-of-function mutation and the absence of the SpoIISB protein leads to a sporulation defect. It was then important to check that the Spo− phenotype of the mut9 mutant is not due to a cis-acting effect of the mut9 mutation on expression of spoIISB. This interpretation could be ruled out since the mut9 mutation is not complemented in trans by a DNA fragment carrying an intact spoIISB gene and its promoter (Fig. 1).
A null mutation in spoIISA was constructed by inserting a kanamycin resistance cassette between codons 54 and 172 of spoIISA. Although this insertion was anticipated to interfere with spoIISB expression, the resulting mutant is perfectly proficient at sporulation (Table 1). Moreover, replacement of the whole spoIIS locus by a kanamycin resistance cassette (see Materials and Methods) has no effect either on sporulation (Table 1), indicating that SpoIISB is dispensable for sporulation in the absence of SpoIISA and that SpoIISA itself does not play an essential role in the sporulation process. Therefore, the mut9 mutation is a gain-of-function mutation and the correlated sporulation defect is due to the presence of the altered SpoIISA protein. Indeed, disruption of the spoIISA gene carrying the mut9 mutation [spoIISA(mut9)], by integration of a plasmid containing a DNA fragment internal to the spoIISA reading frame, restores full sporulation (data not shown). The same integrative plasmid is also able to correct the sporulation defect of the spoIISB strain carrying the mut14 mutation, a further confirmation that SpoIISB is required for sporulation only if SpoIISA is present in the cell.
Altogether, these results show that SpoIISA prevents normal progression of the sporulation process and that SpoIISB neutralizes the action of SpoIISA whereas the SpoIISA(L103F) protein is resistant to SpoIISB. Indeed, the sporulation defect of the strain carrying the spoIISA(mut9) mutation is not aggravated by disruption of the spoIISB gene (Table 1). The mut9 mutation in spoIISA can be complemented in trans by a DNA fragment covering the whole spoIIS locus, albeit with only partial recovery of sporulation efficiency (about 5% of that of the wild type) as shown in Table 1. Since the SpoIISA(L103F) protein becomes partially sensitive to SpoIISB in the presence of wild-type SpoIISA, it is likely that SpoIISA acts as an oligomer. Strikingly, this partial complementation of the mut9 mutation requires the presence of two copies of the spoIISB cistron, indicating that the concentration of SpoIISB needed for efficiently antagonizing SpoIISA is higher than in cells containing two copies of wild-type spoIISA (Table 1). However, sporulation of the spoIISA(mut9) strain is not improved by the presence of two copies of the spoIISB gene (Table 1), presumably because SpoIISB can act only through wild-type SpoIISA.
A promoter driving expression of spoIISA (and consequently also of spoIISB) is located in the 123-bp ykaB-spoIISA interval, downstream of the DraI site (see Fig. 1). Otherwise, integration of a plasmid carrying a fragment of spoIISA extending up to that DraI site would correct the mut14 mutation in spoIISB by preventing transcription of spoIISA, which is not the case (Fig. 1). This promoter was further characterized by placing a promoterless lacZ gene under its control and following β-galactosidase synthesis during exponential growth and sporulation (Fig. 3). Although being about fourfold less active than the spoIISB promoter, the spoIISA promoter behaves similarly, sharing the same general features of a ςA-dependent promoter and the same independence regarding the transcription factors ςH, Spo0A, and AbrB (data not shown).
Sporulation phenotype of spoIIS mutants.
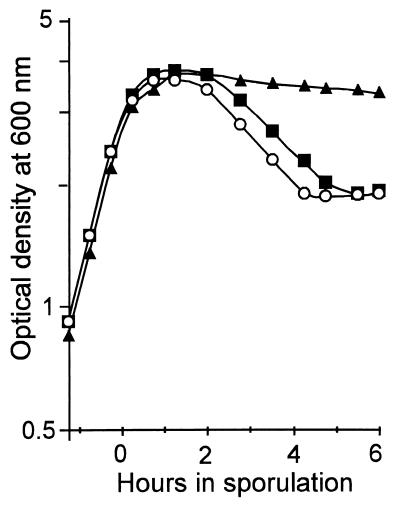
The strains carrying the spoIISA(mut9) mutation, the spoIISB(mut14) mutation, or the spoIISB null allele behave similarly when grown in sporulation medium (Fig. 4). They do not exhibit any obvious defect during exponential growth, and they reach stationary phase with the same optical density. However, about 2 h after the onset of sporulation the spoIIS mutants show a sudden drop in optical density, down to about 55% of the density of the wild type, a very unusual feature for a sporulation mutant.
FIG. 4.
Death of spoIIS mutants in stationary phase. The optical density of cells grown in DS medium at 37°C was monitored during exponential growth and sporulation. Strains contained either a wild-type spoIIS locus (▴), the spoIISA(mut9) mutation (■), or the spoIISB null mutation (○). The strain containing the spoIISB(mut14) mutation behaved exactly as the spoIISA(mut9) mutant and, for clarity, its results are not shown.
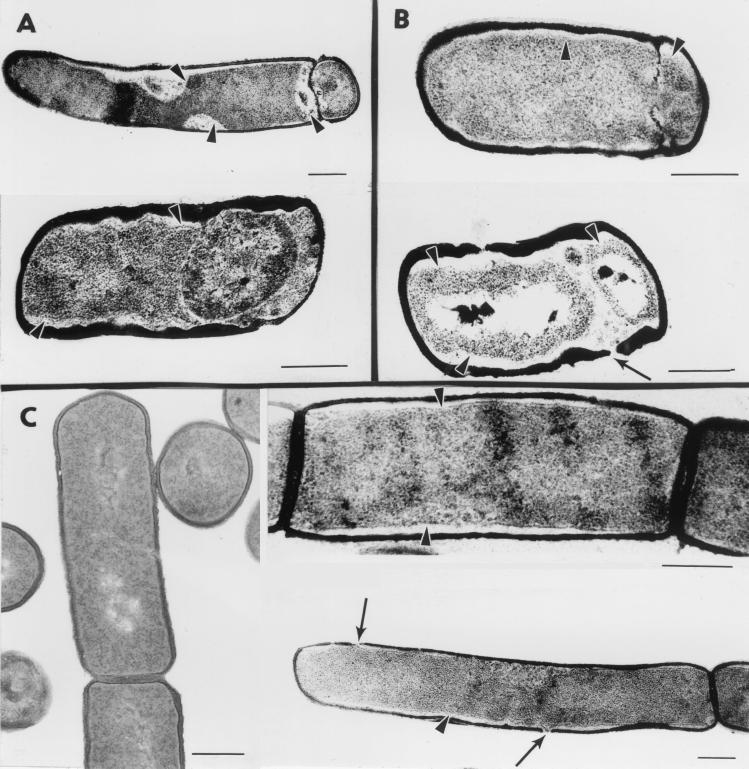
The morphological defects of the spoIISB null mutant were investigated by electron microscopy (Fig. 5A). When the cells were harvested 2 h after the onset of sporulation, about 50% had reached stage II and showed the presence of a polar septum. Thirty percent of the cells, with or without a polar septum, exhibited large plasmolysis zones where the cytoplasmic membrane had detached from the peptidoglycan layer (arrowheads in Fig. 5). These striking defects are not observed in wild-type cells grown in parallel and are reminiscent of the phenotype of spoIIAB mutants, which exhibit aberrantly high ςF activity (7). When the cells were harvested 4 h after the onset of sporulation, only 20% were blocked at stage II whereas most of the cells did not contain a septum, presumably as a consequence of selective lysis of post-stage II cells. In addition, a few cells appeared to have reached stage III and showed the presence of a free forespore (Fig. 5A, bottom). However, we favor the interpretation that these cells are actually stage II cells in which plasmolysis, combined with dissolution of the septal peptidoglycan, has allowed the forespore compartment to detach from the pole without being engulfed by the mother cell. Apparently, shortly after synthesis of the sporulation septum, the unleashed activity of SpoIISA in the absence of SpoIISB has dramatic consequences for the integrity of the cytoplasmic membrane and subsequently for cell viability, preventing any further development.
FIG. 5.
Morphological consequences of SpoIISA activity. (A) spoIISB cells were grown in DS medium and harvested 2 h (top) or 4 h (bottom) after the onset of sporulation. (B) Cells of a wild-type strain containing an extra copy of spoIISA under the control of the forespore-specific spoIIQ promoter were grown in DS medium and harvested 2 h (top) or 4 h (bottom) after the onset of sporulation. (C) Cells of a wild-type strain containing an extra copy of spoIISA under the control of the xylA promoter were grown in LB medium without xylose (left) or in the presence of 5 mM xylose (added at an optical density at 600 nm of 0.5) and harvested 1.5 h after xylose addition (right). Representative examples of cellular morphologies are shown. Arrowheads point to plasmolysis zones where the cytoplasmic membrane appears to be detached from the cell wall. Thin arrows indicate holes in the peptidoglycan layer. Bars represent 0.3 μm.
As a complement to these morphological studies, we determined the stage of blockage of the sporulation genetic pathway. Analysis of a few lacZ fusions indicated that the spoIISB null mutation slightly enhances the activity of ςF (about twofold), has little effect on the activity of the early mother cell sigma factor ςE, and completely blocks synthesis of the late forespore sigma factor ςG (data not shown). In addition, we checked that ςF and ςE activities are normally compartmentalized, as evidenced by cell-specific expression of selected green fluorescent protein fusions (data not shown).
Site of action of SpoIISA.
Since the absence of SpoIISB does not become apparent until cells enter sporulation, we wondered whether a spoIISB mutation would be corrected in cells synthesizing SpoIISB only during sporulation. Moreover, since spoIISB cells reach stage II after synthesis of the sporulation septum and activation of ςF and ςE, the spoIISB mutation could be rescued either in the forespore or in the mother cell by placing spoIISB under the control of a ςF- or a ςE-dependent promoter. The sporulation defect of the spoIISB null strain is fully corrected when SpoIISB is synthesized in the mother cell under the control of the spoIID promoter, whereas synthesis of SpoIISB in the forespore under the control of the spoIIQ promoter has no effect (Table 2). This result indicates that SpoIISA is acting mainly, if not exclusively, in the mother cell compartment of the sporulating cell. Interestingly, additional synthesis of SpoIISA in the mother cell from the spoIID promoter in a wild-type strain has no effect on sporulation (Table 2), suggesting the presence of an excess of SpoIISB molecules in the cell. However, it should be noted that the PspoIID-spoIISA hybrid gene has only a 20-fold negative effect on the sporulation of a strain lacking the whole spoIIS locus (Table 2) and might therefore not be fully functional.
TABLE 2.
Synthesis of the SpoIIS proteins after completion of the sporulation septum
| SpoIIS proteins synthesized in:
|
Spores/mld | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Growing cellsa | Foresporeb | Mother cellc | |||||||
| A | –e | – | – | – | – | 1.6 × 104 | |||
| A | – | – | B | – | – | 7.2 × 104 | |||
| A | – | – | – | – | B | 7.3 × 108 | |||
| A | B | – | – | – | – | 5.5 × 108 | |||
| A | B | – | – | A | – | 8.3 × 108 | |||
| A | B | A | – | – | – | 1.8 × 105 | |||
| A | B | A | B | – | – | 6.5 × 108 | |||
| – | – | – | – | A | – | 3.0 × 107 | |||
Depending on the genotype of the spoIIS locus.
From the spoIIQ promoter, either at amyE (spoIISA) or at thrC (spoIISB).
From the spoIID promoter, at the amyE locus.
Less than twofold variations were observed in several sporulation assays. A typical series of results is shown.
–, no SpoIISA (or SpoIISB) synthesized.
Since the spoIISA gene is transcribed during exponential growth, the SpoIISA protein is presumably present in the two cells generated by asymmetric septation. It was therefore intriguing that sporulation could be fully restored by synthesis of SpoIISB, the SpoIISA antagonist, exclusively in the mother cell. To check the possible immunity of the forespore to the action of SpoIISA, the spoIISA gene was placed under the control of the ςF-controlled spoIIQ promoter. The presence of the PspoIIQ-spoIISA hybrid gene decreases the sporulation of an otherwise wild-type strain by about 3 orders of magnitude, a defect that is fully corrected by intoducing at another chromosomal location the spoIISB gene under the control of the same spoIIQ promoter (Table 2). The morphological consequences of the activity of SpoIISA in the forespore were analyzed by electron microscopy (Fig. 5B). Stage II cells were present in proportions similar to those in the spoIISB strain grown in parallel. Plasmolysis zones were also observed in cells with complete or partially disrupted septa (Fig. 5B, top). Strikingly, plasmolysis was not confined to the forespore but also affected the mother cell and was sometimes associated with phenotypes as extreme as complete disappearance of the sporulation septum and local disruption of the cell wall (Fig. 5B, bottom). Thus, the SpoIISA protein is able to act in the forespore, when present in a sufficient amount, and from there to challenge the integrity and viability of the whole sporulating cell.
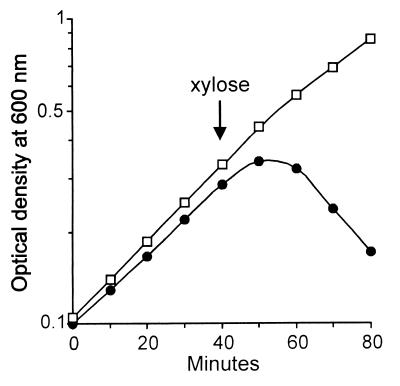
The apparent absence of phenotype of the spoIISB mutation during exponential growth suggested that growing cells are immune to the action of SpoIISA. Therefore, the spoIISA gene was placed under the control of the inducible xylA promoter. Addition of xylose to spoIIS+ cells grown in Luria-Bertani (LB) medium and containing the PxylA-spoIISA hybrid gene led to an almost immediate arrest of growth followed by an abrupt drop in optical density (Fig. 6), a phenomenon that could be countered by the presence in the strain of an additional copy of the spoIISB gene under the control of its own promoter. Electron microscopy analysis revealed the presence of large plasmolysis zones in about 70% of SpoIISA-challenged cells, especially along the main sides of the cells (Fig. 5C), as well as holes in the peptidoglycan layer (arrows in Fig. 5); none of these phenotypes were seen in cells grown in the absence of xylose. Therefore, the apparent immunity of exponentially growing cells to the absence of SpoIISB is not due to their intrinsic resistance to the action of SpoIISA but more likely reflects the existence of a threshold concentration below which SpoIISA does not significantly impair cell viability.
FIG. 6.
SpoIISA-induced death during exponential growth. Cells containing an extra copy of spoIISA at the amyE locus under the control of the xylA promoter were grown in LB medium at 37°C, and their optical density was monitored before and after addition of 5 mM xylose (arrow). Cells also contained a wild-type spoIIS locus with (□) or without (●) an additional copy of spoIISB at thrC.
In an effort to identify the molecular target of SpoIISA, we sought to isolate mutations extragenic to spoIISA that would restore sporulation to a spoIISB strain (see Materials and Methods). Despite the presence of an additional copy of spoIISA in the cells (which dramatically enhanced the sporulation defect due to the absence of SpoIISB), the only suppressor mutations that could be recovered were found to map in one of the spoIISA genes and to be dominant loss-of-function alleles (Table 1). One of these mutations was further characterized and found to convert codon 38 of spoIISA from CGG (Arg) to CAG (Gln) and to behave as a null mutation in spoIISA (Table 1). The dominance of such mutations provides additional evidence for SpoIISA acting as an oligomer, with the residual sporulation defect being exclusively due to the wild-type SpoIISA protein (Table 1).
DISCUSSION
Our results show that whenever its activity gets loose, SpoIISA induces striking abnormalities of the B. subtilis envelope and ultimately cell death. What makes SpoIISA a killer protein? Since it has structural features of an integral membrane protein, SpoIISA could act as a holin and allow some endolysin to gain access to the peptidoglycan (34). Local solubilization of the cell wall would obliterate its role as a protective barrier against osmotic pressure, leading to membrane disruption and consequently to the large plasmolysis zones observed by electron microscopy. It would also explain how SpoIISA toxic effects can spread to the mother cell in strains in which SpoIISA is synthesized exclusively in the forespore, since an endolysin would easily breach the thin septal cell wall separating the forespore from the mother cell. It is then intriguing that the spoIIS locus is located next to the PBSX prophage, immediately downstream of the xlyA gene encoding a phage muramidase (16). Yet, SpoIISA is not involved in PBSX-induced lysis since the presence of the xin15 mutation (that prevents induction of PBSX) does not suppress the sporulation defect of a spoIISB mutant nor the lethal effect of SpoIISA during exponential growth (data not shown).
However, SpoIISA does not show any similarity to known holins and is significantly larger than holins identified so far (34). It is therefore quite possible that the cytoplasmic membrane itself is the target of the toxic action of SpoIISA. For instance, SpoIISA could induce cell death directly by interfering with the respiration machinery whereas activation of autolysins and plasmolysis of the cytoplasmic membrane would be indirect consequences of the catastrophic failure of the dying cell. In this regard it should be noted that we have been unable to clone an intact spoIISA gene (without spoIISB) in E. coli, suggesting that SpoIISA is similarly toxic in E. coli (and that SpoIISB is similarly protective).
SpoIISB is the antidote neutralizing the killer protein SpoIISA. It is very likely that the two proteins interact directly and that the toxicity of SpoIISA (L103F) is due to the loss of that interaction. Several observations indicate that the relative levels of the two proteins are critical. For instance, the consequences of inducing SpoIISA synthesis during growth from the xylA promoter closely depend on the number of spoIISB genes in the cell. The presence of an internal promoter allowing sole transcription of spoIISB is a device that ensures an excess of SpoIISB molecules over SpoIISA, and it might be significant that this promoter is always at least threefold stronger than the promoter driving transcription of both spoIISA and spoIISB.
Complementation experiments strongly suggest that SpoIISA acts as an oligomer. On the one hand, the presence of wild-type SpoIISA makes SpoIISA(L103F) sensitive to SpoIISB provided that enough SpoIISB is supplied. On the other hand, SpoIISA(R38Q), which is apparently locked in an inactive conformation, prevents wild-type SpoIISA from releasing its activity in the absence of SpoIISB. In both cases the (partial) dominance of inactive SpoIISA is easily understood as a consequence of subunit mixing of a multimeric SpoIISA aggregate. It is then worth noting that such a molecular complex might be able to build a pore in the cytoplasmic membrane, a structural feature which could be the basis for the toxicity of SpoIISA.
Experiments in which SpoIISA was synthesized from the xylA or the spoIIQ promoters demonstrate that the vegetatively growing cell and the forespore are not immune to SpoIISA. Nevertheless, the absence of SpoIISB is cryptic during exponential growth when spoIISA is actively transcribed, and SpoIISA-induced death in stationary phase can be prevented by expressing spoIISB exclusively in the mother cell. Maybe some unidentified inhibitor restrains SpoIISA activity in growing cells and in the forespore of a spoIISB strain. Alternatively, SpoIISA might be intrinsically unstable and subjected to proteolysis but be stabilized in the mother cell. In both cases, synthesis of SpoIISA from a foreign promoter would override the mechanisms limiting its activity, with dramatic consequences for cell viability.
It is the deleterious action of SpoIISA on an essential function of the mother cell that prevents further morphological development and blocks the sporulation transcription program. The increase in ςF activity is probably the indirect consequence of the cells being stalled at stage II and the nonreplacement of ςF by ςG in the forespore. Deletion of the whole spoIIS locus has no effect on sporulation, and spoIIS is conspicuously absent from the genomes of all other sporulating gram-positive bacteria sequenced so far.
The genetic hierarchy between the two products of the spoIIS locus, one protein preventing the second one from hindering sporulation, has already been described for other pairs of proteins encoded by operons dispensable for sporulation. Such is the case for the antagonist of the starvation signaling pathway, the aspartate phosphatase RapA and its inhibitor, the imported peptide PhrA (25); for the repressor of some early sporulation genes, the DNA-binding protein Soj and its alternative partner, the chromosome partitioning protein Spo0J (5, 27); and for another repressor of early sporulation genes, the DNA-binding protein SinR and its inhibiting partner Sinl (2). Thus, a common gene organization of structurally and functionally unrelated sporulation regulatory circuits may be a general feature for B. subtilis genes encoding pairs of sporulation inhibitors and effectors.
Phenotypes formally similar to those of the spoIIS mutants have been reported for operons involved in bacterial cell death. Most of these operons are carried by plasmids and encode “addiction modules,” a device that kills the cells having lost the plasmid (13). A few others are present on bacterial chromosomes and code for “antidote-toxin” pairs, whose activation in response to environmental signals may have a selective advantage for a subpopulation (4). It is all the more intriguing that the spoIIS products appear to be unique to B. subtilis, providing no clue to their evolutionary origin and their physiological role. Elucidating the latter will require identification of natural conditions in which SpoIISA activity is released.
ACKNOWLEDGMENTS
We are grateful to Kevin Devine for providing sequence information prior to publication as well as the integrative plasmid library. We thank Fabrizio Arigoni for the xylose-controlled promoter, Dimitri Karamata for the xin15 strain, and Jozef Krištín for use of his electron microscope.
E. Adler was a postdoctoral fellow of the Fogarty Foundation and the Institut National de la Santé et de la Recherche Médicale. This work was supported by the CNRS (grant UPR 9073 to P.S.) and by the Slovak Academy of Sciences (grant 5025 to I.B.).
REFERENCES
- 1.Arigoni F, Guérout-Fleury A-M, Barák I, Stragier P. The SpoIIE phosphatase, the sporulation septum, and the establishment of forespore-specific transcription in Bacillus subtilis: a reassessment. Mol Microbiol. 1999;31:1407–1415. doi: 10.1046/j.1365-2958.1999.01282.x. [DOI] [PubMed] [Google Scholar]
- 2.Bai U, Mandic-Mulec I, Smith I. SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein-protein interaction. Genes Dev. 1993;7:139–148. doi: 10.1101/gad.7.1.139. [DOI] [PubMed] [Google Scholar]
- 3.Barák I, Youngman P. SpoIIE mutants of Bacillus subtilis comprise two distinct phenotypic classes consistent with a dual functional role for the SpoIIE protein. J Bacteriol. 1996;178:4984–4989. doi: 10.1128/jb.178.16.4984-4989.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop R E, Leskiw B K, Hodges R S, Kay C M, Weiner J H. The entericidin locus of Escherichia coli and its implications for programmed cell death. J Mol Biol. 1998;280:583–596. doi: 10.1006/jmbi.1998.1894. [DOI] [PubMed] [Google Scholar]
- 5.Cervin M A, Spiegelman G B, Raether B, Ohlsen K, Perego M, Hoch J A. A negative regulator linking chromosome segregation to developmental transcription in Bacillus subtilis. Mol Microbiol. 1998;29:85–95. doi: 10.1046/j.1365-2958.1998.00905.x. [DOI] [PubMed] [Google Scholar]
- 6.Claros M G, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Applic Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 7.Coppolecchia R, DeGrazia H, Moran C P., Jr Deletion of spoIIAB blocks endospore formation in Bacillus subtilis at an early stage. J Bacteriol. 1991;173:6678–6685. doi: 10.1128/jb.173.21.6678-6685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cutting S M, Vander Horn P B. Genetic analysis. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, United Kingdom: John Wiley & Sons Ltd.; 1990. pp. 27–74. [Google Scholar]
- 9.Duncan L, Alper S, Arigoni F, Losick R, Stragier P. Activation of cell-specific transcription by a serine phosphatase at the site of asymmetric division. Science. 1995;270:641–644. doi: 10.1126/science.270.5236.641. [DOI] [PubMed] [Google Scholar]
- 10.Frandsen N, Barák I, Karmazyn-Campelli C, Stragier P. Transient gene asymmetry during sporulation and establishment of cell specificity in Bacillus subtilis. Genes Dev. 1999;13:394–399. doi: 10.1101/gad.13.4.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guérout-Fleury A-M, Frandsen N, Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- 12.Guérout-Fleury A-M, Shazand K, Frandsen N, Stragier P. Antibiotic-resistance cassettes for Bacillus subtilis. Gene. 1995;167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- 13.Holc̆ik M, Iyer V N. Conditionally lethal genes associated with bacterial plasmids. Microbiology. 1997;143:3403–3416. doi: 10.1099/00221287-143-11-3403. [DOI] [PubMed] [Google Scholar]
- 14.Karow M L, Glaser P, Piggot P J. Identification of a gene, spoIIR, which links the activation of ςE to the transcriptional activity of ςF during sporulation in Bacillus subtilis. Proc Natl Acad Sci USA. 1995;92:2012–2016. doi: 10.1073/pnas.92.6.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King N, Dreesen O, Stragier P, Pogliano K, Losick R. Septation, dephosphorylation, and the activation of ςF during sporulation in Bacillus subtilis. Genes Dev. 1999;13:1156–1167. doi: 10.1101/gad.13.9.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krogh S, Jørgensen S T, Devine K M. Lysis genes of the Bacillus subtilis defective prophage PBSX. J Bacteriol. 1998;180:2110–2117. doi: 10.1128/jb.180.8.2110-2117.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levin P A, Losick R. Asymmetric division and cell fate during sporulation in Bacillus subtilis. In: Brun Y V, Shimkets L J, editors. Prokaryotic development. Washington, D.C.: American Society for Microbiology; 2000. pp. 167–189. [Google Scholar]
- 18.Lewis P J, Partridge S R, Errington J. ς factors, asymmetry, and the determination of cell fate in Bacillus subtilis. Proc Natl Acad Sci USA. 1994;91:3849–3853. doi: 10.1073/pnas.91.9.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis P J, Wu L J, Errington J. Establishment of prespore-specific gene expression in Bacillus subtilis: localization of SpoIIE phosphatase and initiation of compartment-specific proteolysis. J Bacteriol. 1998;180:3276–3284. doi: 10.1128/jb.180.13.3276-3284.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Londoño-Vallejo J-A, Fréhel C, Stragier P. spoIIQ, a forespore-expressed gene required for engulfment in Bacillus subtilis. Mol Microbiol. 1997;24:29–39. doi: 10.1046/j.1365-2958.1997.3181680.x. [DOI] [PubMed] [Google Scholar]
- 21.Londoño-Vallejo J-A, Stragier P. Cell-cell signaling pathway activating a developmental transcription factor in Bacillus subtilis. Genes Dev. 1995;9:503–508. doi: 10.1101/gad.9.4.503. [DOI] [PubMed] [Google Scholar]
- 22.Min K-T, Hilditch C M, Diederich B, Errington J, Yudkin M D. ςF, the first compartment specific transcription factor of Bacillus subtilis, is regulated by an anti-sigma factor which is also a protein kinase. Cell. 1993;74:735–742. doi: 10.1016/0092-8674(93)90520-z. [DOI] [PubMed] [Google Scholar]
- 23.O'Reilly M, Woodson K, Dowds B C A, Devine K M. The citruline biosynthetic operon, argC-F, and a ribose transport operon, rbs, from Bacillus subtilis are negatively regulated by Spo0A. Mol Microbiol. 1994;11:87–98. doi: 10.1111/j.1365-2958.1994.tb00292.x. [DOI] [PubMed] [Google Scholar]
- 24.Perego M. Integrational vectors for genetic manipulation in Bacillus subtilis. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C.: American Society for Microbiology; 1993. pp. 615–624. [Google Scholar]
- 25.Perego M, Hoch J A. Cell-cell communication regulates the effects of protein aspartate phosphatases on the phosphorelay controlling development in Bacillus subtilis. Proc Natl Acad Sci USA. 1996;93:1549–1553. doi: 10.1073/pnas.93.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piggot P J, Coote J G. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976;40:908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quisel J D, Grossman A D. Control of sporulation gene expression in Bacillus subtilis by the chromosome partitioning proteins Soj (ParA) and Spo0J (ParB) J Bacteriol. 2000;182:3446–3451. doi: 10.1128/jb.182.12.3446-3451.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaeffer P, Millet J, Aubert J-P. Catabolite repression of bacterial sporulation. Proc Natl Acad Sci USA. 1965;54:704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stragier P, Bonamy C, Karmazyn-Campelli C. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell. 1988;52:697–704. doi: 10.1016/0092-8674(88)90407-2. [DOI] [PubMed] [Google Scholar]
- 30.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 31.Sun D, Cabrera-Martinez R M, Setlow P. Control of transcription of the Bacillus subtilis spoIIIG gene, which codes for the forespore specific transcription factor ςG. J Bacteriol. 1991;173:2977–2984. doi: 10.1128/jb.173.9.2977-2984.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun D, Fajardo-Cavazos P, Sussman M D, Tovar-Rojo F, Cabrera-Martinez R M, Setlow P. Effect of chromosome location of Bacillus subtilis forespore genes on their spo gene dependence and transcription by EςF: identification of features of good EςF-dependent promoters. J Bacteriol. 1991;173:7867–7874. doi: 10.1128/jb.173.24.7867-7874.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Webb C D, Decatur A, Teleman A, Losick R. Use of green fluorescent protein for visualization of cell-specific gene expression and subcellular protein localization during sporulation in Bacillus subtilis. J Bacteriol. 1995;177:5906–5911. doi: 10.1128/jb.177.20.5906-5911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong I-N, Smith D L, Young R. Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol. 2000;54:799–825. doi: 10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- 35.Wu L J, Errington J. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science. 1994;264:572–575. doi: 10.1126/science.8160014. [DOI] [PubMed] [Google Scholar]
- 36.Wu L J, Errington J. Use of asymmetric cell division and spoIIIE mutants to probe chromosome orientation in Bacillus subtilis. Mol Microbiol. 1998;27:777–786. doi: 10.1046/j.1365-2958.1998.00724.x. [DOI] [PubMed] [Google Scholar]