Key Points
A deep-scale proteomic and phosphoproteomic database of AML was produced and can be explored interactively (www.leylab.org/amlproteome).
Posttranscriptionally regulated proteins were identified globally and associated with specific AML-initiating events.
Visual Abstract
Abstract
We have developed a deep-scale proteome and phosphoproteome database from 44 representative acute myeloid leukemia (AML) patients from the LAML TCGA dataset and 6 healthy bone marrow–derived controls. After confirming data quality, we orthogonally validated several previously undescribed features of AML revealed by the proteomic data. We identified examples of posttranscriptionally regulated proteins both globally (ie, in all AML samples) and also in patients with recurrent AML driver mutations. For example, samples with IDH1/2 mutations displayed elevated levels of the 2-oxoglutarate–dependent histone demethylases KDM4A/B/C, despite no changes in messenger RNA levels for these genes; we confirmed this finding in vitro. In samples with NPMc mutations, we identified several nuclear importins with posttranscriptionally increased protein abundance and showed that they interact with NPMc but not wild-type NPM1. We identified 2 cell surface proteins (CD180 and MRC1/CD206) expressed on AML blasts of many patients (but not healthy CD34+ stem/progenitor cells) that could represent novel targets for immunologic therapies and confirmed these targets via flow cytometry. Finally, we detected nearly 30 000 phosphosites in these samples; globally, AML samples were associated with the abnormal phosphorylation of specific residues in PTPN11, STAT3, AKT1, and PRKCD. FLT3-TKD samples were associated with increased phosphorylation of activating tyrosines on the cytoplasmic Src-family tyrosine kinases FGR and HCK and related signaling proteins. PML-RARA–initiated AML samples displayed a unique phosphorylation signature, and TP53-mutant samples showed abundant phosphorylation of serine-183 on TP53 itself. This publicly available database will serve as a foundation for further investigations of protein dysregulation in AML pathogenesis.
Introduction
Proteins, despite being the primary effectors of cellular processes, are often studied only indirectly through transcriptomic analysis. However, it has been repeatedly shown that the relationship between messenger RNA (mRNA) expression and protein expression is only approximate in many cancers.1-8 Furthermore, the phosphoproteome provides a unique, global look at active signaling pathways not visible with the transcriptome. In acute myeloid leukemia (AML), the genome and transcriptome have been extensively characterized9-11; limited studies of the proteome and phosphoproteome have yielded promising insights.12-20 Here, we present a deep-scale study of the proteomes and phosphoproteomes of 44 primary AML bone marrow samples representing a wide range of AML across the spectrum of cytogenetic risk, common mutations, and driver fusions, validate several unique findings revealed by these data, and provide an interactive platform for exploration of these databases.
Methods
Sample collection and preparation
Bone marrow samples were collected at presentation from adult patients with de novo AML on an Institutional Review Board–approved banking protocol (#201011766) that provided all 200 samples for the LAML TCGA study. From these 200, 70 still had ≥8 cryovials available for analysis. From these 70, we chose 55 samples representing major cytogenetic and mutational landscapes; 44 yielded adequate, high-quality material for study. Bone marrow buffy coat cells were immediately cryopreserved without further manipulation, as described previously.9 Cryovials were thawed in the presence of the cell-permeable, irreversible serine protease inhibitor diisopropyl fluorophosphate (DFP) to inactivate the myeloid serine proteases. Healthy control bone marrow cells from 3 independent donors were depleted of cells with terminal differentiation markers to enrich for progenitors and precursors (“lineage-depleted,” using the Miltenyi Biotec reagent 130-092-211). Bone marrow cells from 3 different healthy donors were used to enrich for CD34+ stem/progenitor cells (“CD34-selected,” using the Miltenyi Biotec reagent 130-100-453) Both lineage depletion and CD34 selection were performed using an autoMACS separator per manufacturer instructions.
Proteomic and phosphoproteomic methods
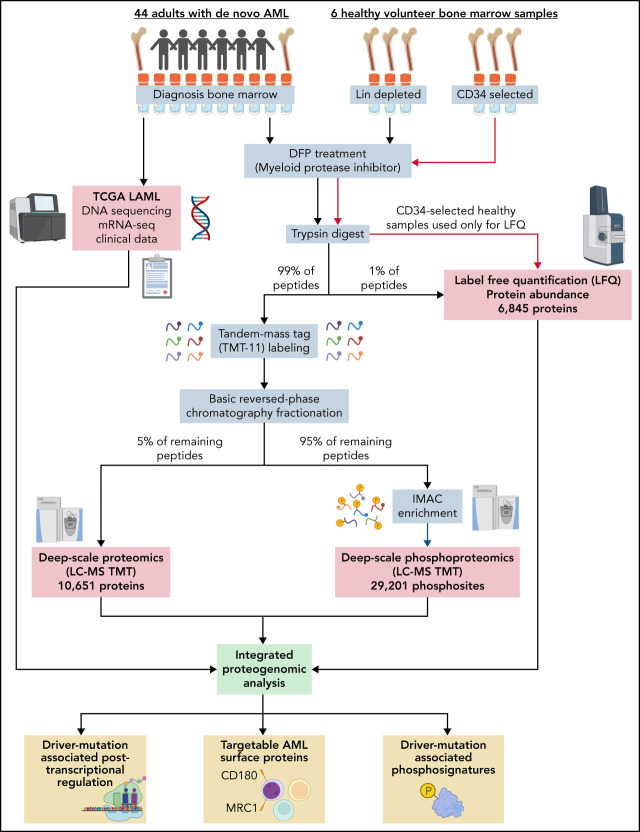
An overview of the workflow is provided in the visual abstract. Peptides and phosphopeptides for deep-scale proteomic analysis were prepared as previously described.21 Peptides and phosphopeptides from individual samples were labeled with tandem mass tag (TMT) reagents (TMT-11; supplemental Table 1). Before labeling, an aliquot was removed for label-free quantification. Labeled peptides and phosphopeptides were combined into TMT-11 plexes (9 samples plus 2 reference pools). Plexes were fractionated offline using basic reversed-phase high-performance liquid chromatography; 24 concatenated fractions from each plex were prepared as previously described.21 An aliquot (5%) of each concatenated fraction and fraction A was analyzed using ultraperformance liquid chromatography Orbitrap mass spectrometry. The remainder was combined into 12 fractions for phosphopeptide enrichment.
Labeled peptides were analyzed using high-resolution liquid chromatography-tandem mass spectrometry. Unlabeled, unfractionated peptides from individual samples were analyzed using trapped ion mobility time-of-flight mass spectrometry22 using ultraperformance liquid chromatography timsTOF Pro (Bruker Daltonics). The mass spectrometer was operated in parallel accumulation–serial fragmentation mode.22 The chromatographic instrument parameters and setup are in supplemental Methods (available on the Blood Web site). Protein and phosphopeptide identification and quantification algorithms are in supplemental Methods.
Unsupervised clustering
Unsupervised clustering of protein/phosphosite abundance was performed using the unweighted pair group method with arithmetic mean.23 Similarity scores between samples were Pearson correlations across all proteins/phosphosites detected in all samples.
K562 nucleofection
K562 cells were nucleofected using the Lonza 4D Nucleofector X-unit as per manufacturer instructions using Amaxa Kit (Catalog #V4XC-2024). pcDNA3-Flag-IDH1 and pcDNA3-Flag-IDH1-R132H were gifts from Yue Xiong (Addgene #62906 and #62907).24 Cells were harvested 48 hours after nucleofection.
Western blotting
Western blots were performed using the Jess western blotting system and total protein normalization module and analyzed using Compass software (ProteinSimple). Antibodies are in supplemental Methods.
TurboID and immunofluorescence
TurboID was performed as described previously,25 with details in supplemental Methods.
Results
A deep-scale proteome recapitulates many well-recognized features of AML
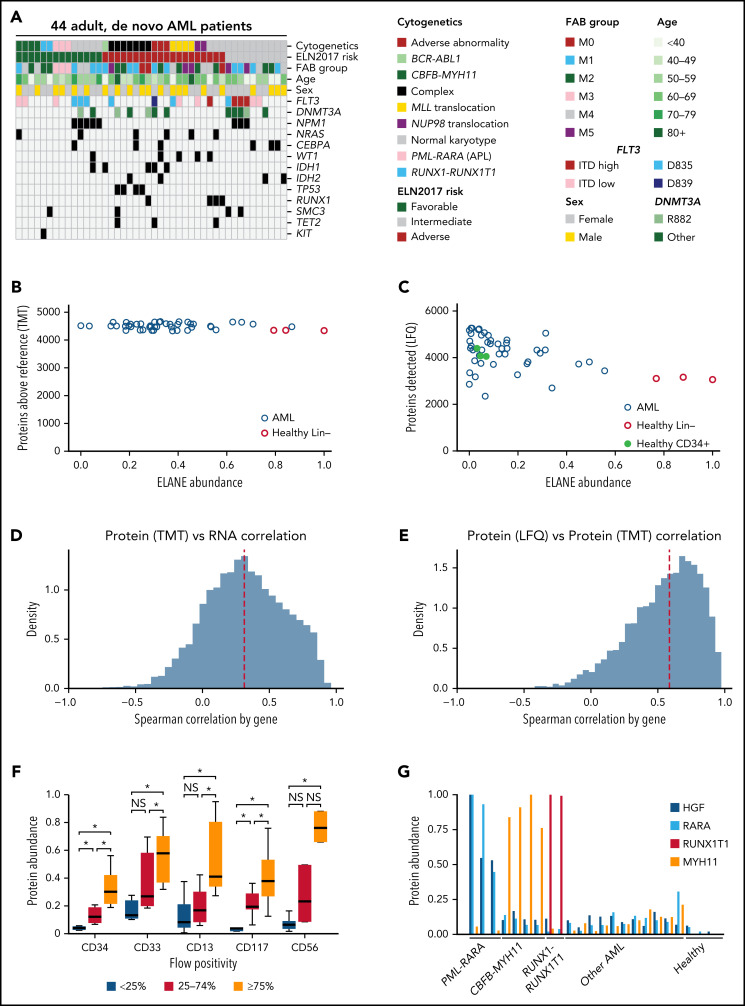
Using the LAML TCGA sample set,9 we obtained protein extracts from 44 fully characterized AML bone marrow samples, representing a wide range of AML subtypes across the spectrum of cytogenetic risk, common mutations, and driver fusions (Figure 1A; supplemental Table 2).
Figure 1.
Characteristics of the AML patients in the study and data quality. (A) Relevant clinical parameters and common genetic findings in TCGA patients selected for this study. (B) Number of proteins detected at above-average abundance (greater than reference pool) for each patient or healthy donor sample compared with the normalized protein abundance of the neutrophil elastase (ELANE) serine protease in each sample (defined by TMT). Similar distributions were noted for the other abundant myeloid serine proteases (cathepsin G [CTSG] and proteinase 3 [PRTN3]). Note that high levels of ELANE are not correlated with reduced numbers of detected proteins, suggesting that tryptic peptides are not being cleaved by endogenous proteases during sample preparation. (C) Number of proteins detected for each sample compared with the normalized protein abundance of ELANE using label-free quantification (LFQ). (D-E) Distribution of gene-wise Spearman correlation between proteomic (TMT) and bulk RNA sequencing data (D) and between LFQ and TMT platforms (E). For both panels D and E, only genes quantifiable by both technologies in at least 20% of AML samples were included in the analysis. Dashed red lines represent median values. (F) Measured protein abundance (TMT) for each of the identified cell surface proteins in patients grouped by the percentage of cells from their presentation marrow sample that displayed the same respective marker using clinical flow cytometry. Protein abundance is calculated based on summed reporter ion intensity, which was normalized and median-centered across TMT plexes to a reference sample. Each protein expression value is then scaled to have a maximum value across all measured samples of 1, and a minimum value of 0, for this display. *P < .05 by 1-sided Mann-Whitney U test between groups. (G) Measured protein abundance (TMT) for each of 4 proteins known to be expressed in only 1 AML subtype. Shown are 3 AMLs with PML-RARA fusions, 4 with CBFB-MYH11 fusions, and 2 with RUNX1-RUNX1T1 fusions; 11 other representative AML samples and 3 healthy adult donor bone marrow samples are shown. HGF and RARA are overexpressed only in PML-RARA–initiated AML, MYH11 is overexpressed only in CBFB-MYH11 initiated AML, and RUNX1T1 is overexpressed only in RUNX-RUNX1T1 AML. ITD, internal tandem duplication; NS, not significant.
Because AML cells contain abundant, highly active serine proteases (ELANE, CTSG, PRTN3, and neutrophil serine protease 4/PRSS57), sample preparation was optimized to avoid proteolysis (which could affect the quantitation of tryptic peptides if they were cleaved by another proteinase). Cryovials of patient bone marrow samples collected at presentation were thawed in the presence of the cell-permeable, covalent serine-protease inhibitor DFP and processed for mass spectrometry with a standard cocktail of protease inhibitors. First, the deep-scale and phosphoproteomic workflow was validated against standards from a previously published workflow21 (supplemental Figure 1). Then, both LFQ and TMT deep-scale proteomics were performed on 44 patient samples, as well as 3 lineage-depleted bone marrow samples from healthy adult donors. LFQ was also performed on 3 CD34-selected bone marrow samples from healthy adult donors.
Although the TMT and LFQ mass spectrometry platforms both measure protein abundance, they were used in tandem for this study because of their complementary strengths. TMT has higher sensitivity than LFQ; we detected 10 651 proteins in the TMT dataset and 6845 proteins in the LFQ dataset (supplemental Tables 3 and 4). TMT more accurately defines the relative abundance of an individual protein across a set of patient samples due to the use of simultaneous loading of multiple samples and comparison with a common reference pool comprised of all samples; however, only relative (ie, compared with other samples), not absolute, protein abundance measurements are obtained. One major advantage of LFQ is its ability to determine the absolute abundance of proteins, allowing for direct comparisons of abundance among different proteins. LFQ also has a wider dynamic range, allowing for the detection of large differences among samples, and it requires less total protein. For example, we were able to obtain LFQ measurements from highly enriched CD34+ healthy control bone marrows; this was not possible using TMT due to the rarity of these cells in healthy bone marrow samples (<1% of cells). In this study, we have provided information from both datasets whenever possible, which provides orthogonal support for many biological conclusions.
Importantly, because of the use of the cell-permeable DFP proteinase inhibitor, we did not find evidence for protein degradation caused by the endogenous myeloid serine proteases, with only a modest relationship between ELANE abundance and the detection of proteins in the TMT dataset (R2 = −0.18); in each sample, between 4328 and 4664 unique proteins were detected at above-average abundance (Figure 1B). A slightly stronger relationship was seen in the LFQ dataset between ELANE abundance and total proteins detected (Figure 1C, R2 = −0.46), with healthy lineage-depleted marrows containing the most ELANE; all samples had 2347 to 5265 detected proteins. Similar trends were seen for CTSG, PRTN3, and neutrophil serine protease 4 (supplemental Figure 2). Using previously measured mRNA abundance for these samples from the TCGA study, we found a median Spearman correlation between protein and RNA abundance of 0.31 for TMT data (Figure 1D) and 0.23 for LFQ data (supplemental Figure 3A), similar to previously published large-scale proteomics studies,7 which have also demonstrated that RNA serves as only an approximation of protein abundance. Reflecting the quality of these data, the correlation between protein abundance measurements across platforms was higher, with a median Spearman correlation of 0.59 (Figure 1E; supplemental Table 5), consistent with other recent large-scale proteomics studies.7
One thousand, four hundred and seventy-four proteins showed anticorrelation between protein and mRNA abundance (Spearman correlation < 0 across AML samples using TMT protein abundance). These proteins included TP53, RAD51B, RAD52, NRAS, HRAS, EGFR, and ERBB3/HER3. In pathway analyses using the Kyoto Encyclopedia of Genes and Genomes,26 these 1474 anticorrelated proteins were significantly enriched for the spliceosome (q < 10−21, including SRSF2, SF3B1, and U2AF1), the ribosome (q < 10−4), the oxidative phosphorylation pathway (q < 10−5), and RNA polymerase (q < 10−3). Conversely, 1198 proteins showed a Spearman correlation between protein and mRNA abundance of >0.7. These included CEBPA, GATA1, GATA2, RB1, JAK2, FGR, HCK, SYK, PRKCA/B/C, and 108 of the 247 detected cell differentiation markers27 (q < 10−15, including CD34, KIT, and FLT3).
We then examined the ability of these data to recapitulate known features of AML. First, we examined the correlation with clinical flow cytometry of common AML surface proteins (Figure 1F [TMT]; supplemental Figure 3B [LFQ]); we found that protein abundance measurements were significantly correlated with the fraction of cells in each sample bearing cell surface proteins CD34, CD33, CD13, CD117, and CD56. Next, we looked at proteins known to be overexpressed in AML samples driven by common fusion events (PML-RARA, CBFB-MYH11, and RUNX1-RUNX1T1).9 For each subtype, overexpression of the unique, expected proteins was detected in relevant samples but not in other AMLs or healthy controls (Figure 1G [TMT]; supplemental Figure 3C [LFQ]). We also found the mean quantitative ratio of β-globin (HBB) to α-globin (HBA2) was ∼1.2, with minimal variance, consistent with the nearly equal abundance of these proteins in hemoglobin A in red blood cell precursors in bone marrow28 (supplemental Figure 3D, using LFQ). We next evaluated sex-based differences in AML patient samples, looking for differential protein expression between the 23 female and 21 male patients; 4 significantly different proteins (DDX3Y, EIF1AY, RPS4Y1, and ZFY) were identified in the TMT data, all of which are Y-linked proteins widely expressed in males (supplemental Figure 3E). Two of these (DDX3Y and RPS4Y1) were also detected in the LFQ data and were present only in male patients (DDX3Y was detected at a low level in 1 of 23 female patients). Taken together, these analyses confirm the quality and reproducibility of the proteomic dataset, which reflects many relevant features of AML biology.
Inclusion of healthy control samples
As noted above, the LFQ dataset includes both lineage-depleted and CD34-selected healthy bone marrow control samples, whereas the TMT dataset includes only lineage-depleted healthy bone marrow controls (due to the rarity of CD34+ cells in healthy bone marrow, we could not purify enough protein from single donors for TMT determinations). Lineage depletion removes the most mature hematopoietic cells, leaving a heterogeneous mixture of progenitors and precursors, whereas CD34 selection enriches directly for stem and progenitor cells. Both of these populations represent important comparators for the AML samples because AML originates from hematopoietic stem/progenitor populations.
Global analysis of the AML proteomic landscape
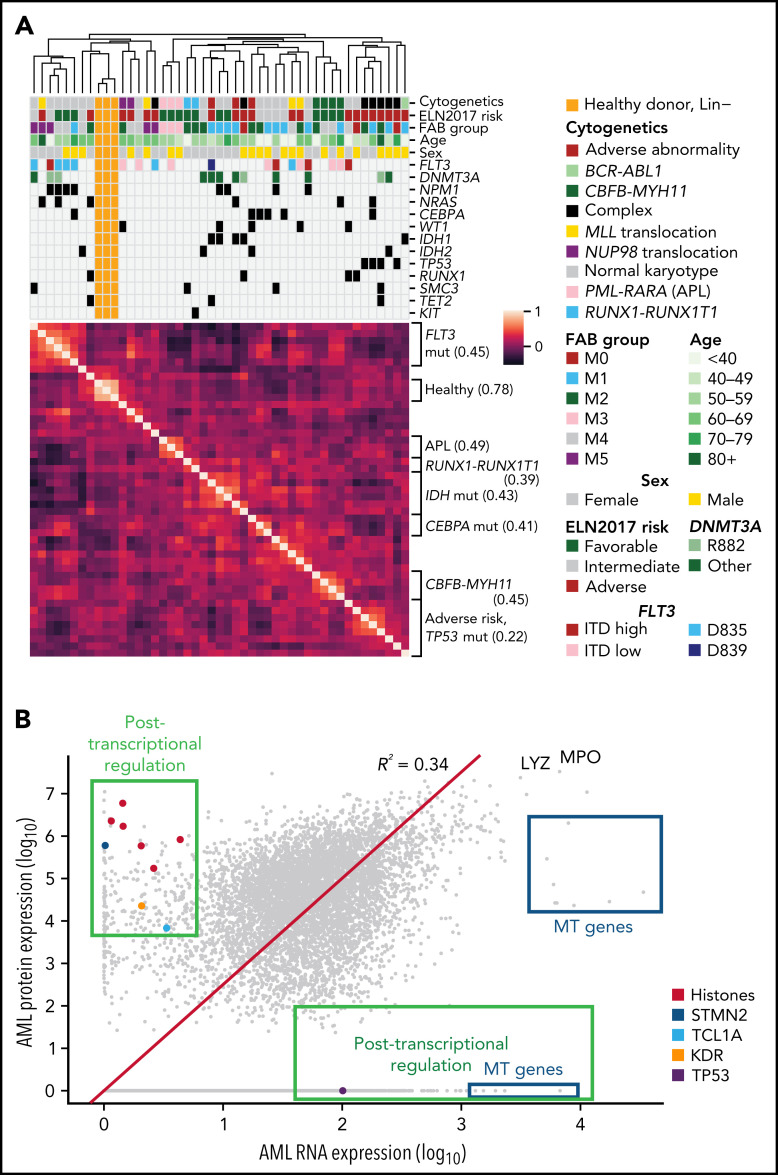
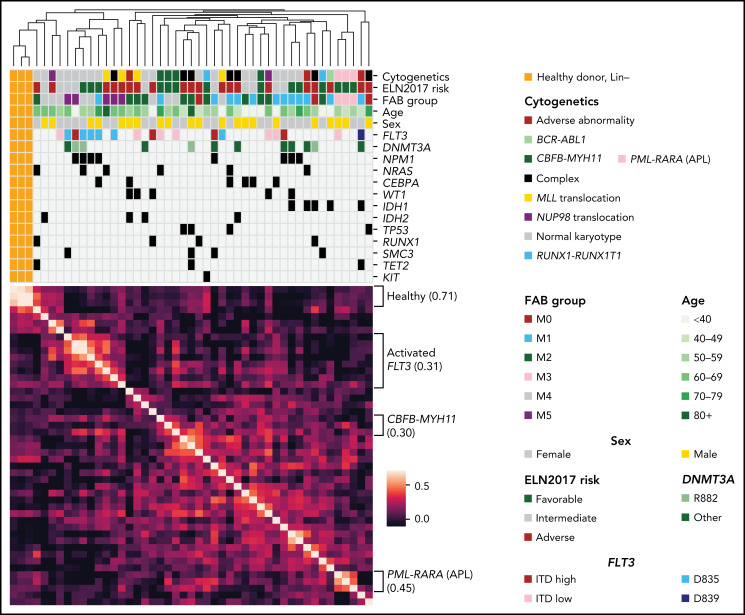
We next performed unsupervised hierarchical clustering of samples based on protein abundance measured on the TMT platform. We found that samples organized primarily by important clinical covariates, including cytogenetics and/or common mutations, suggesting that these supervised proteomic signatures (Figure 2A). Clustering with LFQ data recapitulated some (but not all) of these features (supplemental Figure 4), emphasizing the importance of the deep-scale TMT dataset, which more accurately captures interpatient, relative protein abundance. The first 2 principal components separated most AML patients from healthy donor samples in both TMT and LFQ datasets (supplemental Figures 5A-B), whereas higher-dimensional analysis using t-Distributed Stochastic Neighbor Embedding29 recapitulated the groups seen using hierarchical clustering, highlighting their robustness across algorithms (supplemental Figures 5C-D and 6). Clustering without lineage-associated proteins (as previously published30) did not markedly change the results (supplemental Figure 7).
Figure 2.
Protein expression levels are often correlated with clinical features and reveal evidence for posttranscriptional regulation. (A) Unsupervised clustering of proteomic profiles, revealing distinct clusters of samples, many of which correlate with known molecular covariates, including cytogenetic alterations, FAB subgroups, and recurrent mutations. The heatmap shows a Pearson correlation of protein expression levels among all patients using TMT proteomic measurements. Clustering was based on the unweighted pair group method with arithmetic mean algorithm, with similarity scores as shown in the heatmap. Brackets on the side of the heatmap indicate subgroups with shared clinical or molecular features; the value in parentheses indicates the mean Pearson correlation among members of that subgroup. (B) Mean log10 expression values of protein and RNA abundance for 7916 proteins measured in the bone marrow samples of 44 de novo, primary AML patients at time of diagnosis. RNA expression was quantified using transcripts per million from RNA sequencing after log10 transformation. Protein expression was measured using LFQ tandem mass spectrometry with normalized precursor ion intensities representing protein abundance after log10 transformation. The red line shows a line of best fit using linear regression with no intercept. Proteins displaying evidence for posttranscriptional regulation (high protein expression with low RNA expression or vice versa) are boxed in green and include the labeled histones H1-3, H1-4, H1-5, H2AC21, H3C1, and H3C15, as well as STMN2, the AKT co-activator/oncogene TCL1A, the protein tyrosine kinase receptor KDR, and the key tumor suppressor TP53. High protein, low RNA green box includes proteins with at least half of the maximal protein expression detected (log10 scale) and at most 15% of the maximal RNA expression. High RNA, low protein green box includes proteins with <25% of the maximal protein expression and at least median RNA expression. All proteins in the blue boxes are mitochondrially (MT) encoded and have lower-than-expected protein expression values as predicted by RNA expression. LYZ and MPO are known highly abundant proteins in myeloid cells. mut, mutant.
Comparing global protein vs RNA expression in all 44 AML samples using the LFQ data (which allows measurement of absolute protein abundance), we saw numerous examples of posttranscriptional regulation, where RNA abundance did not predict protein abundance (eg, high RNA and low protein or vice versa, Figure 2B). We noted that several histone proteins, STMN2, the AKT coactivator/oncogene TCL1A, and the protein tyrosine kinase receptor KDR, all displayed markedly increased protein abundance out of proportion to RNA expression. Conversely, TP53 protein was undetectable in LFQ and minimally detectable in TMT data, despite significant mRNA expression equivalent to most detectable proteins (previous in-depth analysis of TP53 protein abundance in AML used reverse-phase protein arrays, which may be more sensitive31). Still, the small amount of TP53 detected suggests that posttranscriptional mechanisms may influence TP53 protein abundance. Other mass spectrometry–based proteomics studies have detected TP53 in only some tumor types, suggesting this may reflect differences in tumor biology; a recent compendium of 2002 human cancers of 14 different types32 detected TP53 in 1142 of 2002 patient samples (705 of 805 TP53-mutant patients), and a recent study of intrahepatic cholangiocarcinoma33 (with 39 of 262 patients harboring a TP53 mutation) did not detect TP53 among 10 529 detected proteins. Further, multiple families of proteins were abundant in AML samples compared with healthy control marrow, without corresponding changes in mRNA abundance; examples include the H/ACA box small nucleolar ribonucleoprotein core complex (which is involved in both telomere maintenance and pseudouridylation of mRNA34) as well as the THO complex (which is involved in the formation and export of messenger ribonucleoparticles35) (supplemental Figure 8).
IDH1/2 mutations are associated with increased abundance of KDM4A/B/C histone demethylases
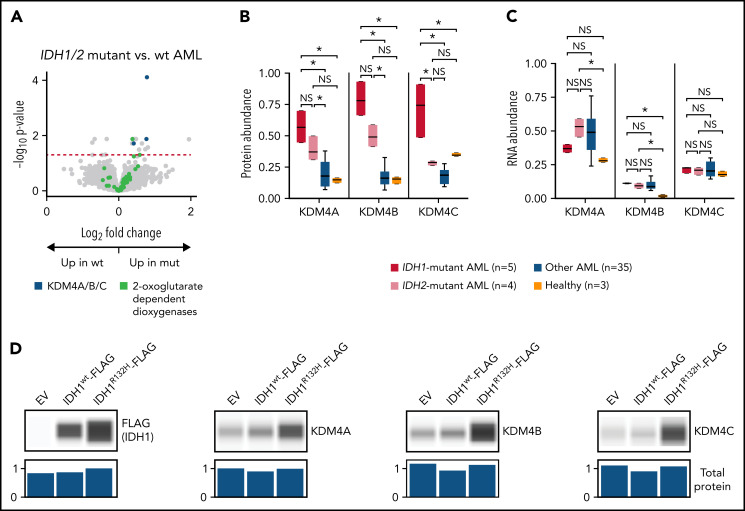
Given the protein abundance signature for IDH mutations in unsupervised clustering, we looked for differentially abundant proteins in patients with IDH1 (n = 5) or IDH2 (n = 4) mutations compared with other AML samples (n = 35). We found 17 differentially abundant proteins after multiple-hypothesis correction (13 with increased abundance in IDH-mutant patients) and noted that the 2-oxoglutarate–dependent H3K9/27/36 histone demethylases KDM4A, KDM4B, and KDM4C36 were all affected similarly (Figure 3A). Given the known dysregulation of 2-oxoglutarate metabolism caused by IDH1/2 mutations,37 we looked for evidence of dysregulation for all 2-oxoglutarate–dependent dioxygenases; there was not a general effect (Figure 3A). The trend of increased abundance generally held for both IDH1- and IDH2-mutant samples in the TMT dataset, although the effect was strongest for IDH1 (Figure 3B). Although detection of these proteins was limited in the less sensitive LFQ dataset, a similar trend was seen, with KDM4B abundance significantly higher (P < .05 by Mann-Whitney U test) in IDH1- and IDH2-mutant samples than IDH1/2 wild-type (wt) samples (KDM4A was detected in LFQ in only 3 of 44 samples, 2 of which were IDH1/2-mutant samples; KDM4C was not detected using LFQ). Strikingly, no significant change was seen in mRNA abundance, suggesting that the protein abundance of KDM4A/B/C is controlled at a posttranscriptional level (Figure 3C).
Figure 3.
AML samples with IDH1/2 mutations are associated with increased abundance of KDM4A/B/C histone demethylases. (A) Volcano plot showing protein abundance in IDH1/IDH2-mutated vs wt AML samples. P values are calculated using the t test and corrected for multiple-hypothesis testing with the Benjamini-Hochberg method. Dashed red line shows P = .05. IDH1 and IDH2 mutations in AML cause dysregulation of 2-oxoglutarate metabolism, and KDM4A/B/C are some of the known 2-oxoglutarate–dependent dioxygenases. (B-C) Normalized abundance of KDM4A/B/C classified by IDH1/2 mutation status in TMT data (B) and bulk RNA sequencing (C). *P < .05 by t test between groups. (D) K562 cells were transfected in vitro with pcDNA3-EV, pcDNA3-FLAG-IDH1wt, or pcDNA3-FLAG-IDH1R132H plasmids and cultured for 2 days prior to cell lysis. Western blots show 1 of 3 representative biologic replicates using antibodies specific for the indicated proteins. Normalized total protein values as a loading control were calculated using the protein normalization module on the Jess western blotting system. EV, empty vector; mut, mutant; NS, not significant.
To determine whether the IDH1 mutation alone was sufficient to cause this effect, we transfected the erythroleukemia cell line K562 with plasmids containing either an empty vector, a wt IDH1-FLAG construct, or an IDH1R132H-FLAG construct representing the most common IDH1 mutation in AML. We found that transient expression of the IDH1R132H protein (but not wt IDH1) for 48 hours caused increased abundance of KDM4A, KDM4B, and KDM4C as detected by western blotting (Figure 3D). Taken together, these results suggest that the IDH1/2 mutations affect the protein abundance of the oncogenic KDM4 family36 through posttranscriptional mechanisms.
Mutant NPMc protein is associated with increased abundance of (and physical interaction with) several nuclear importins
We next identified differential protein abundance in samples with mutant NPM1. The NPMc mutation was present in 8 cases in this dataset and leads to aberrant cytoplasmic mislocalization of NPM1.38 We found 11 differentially abundant proteins between NPMc-mutant and NPM1wt samples after multiple-hypothesis testing correction; 8 showed increased abundance in NPMc-mutant samples. Of these, 2 belonged to the nuclear importin family:39 KPNA4 and KPNB1 (Figure 4A). Across the entire family of nuclear importins (KPNA1-6 and KPNB1), we noticed a general trend of increased protein abundance in NPMc-mutant AML and an even more pronounced increase when compared with healthy, lineage-depleted bone marrow (Figure 4B). A similar (but not statistically significant) trend was seen in the LFQ dataset. No similar trend was seen in mRNA, where transcript abundance was similar between NPMc AML, NPM1wt AML, and healthy CD34+ cells; for KPNA2 and KPNA4, there was actually decreased mRNA abundance in the AML samples compared with healthy controls (Figure 4C).
Figure 4.
AML samples with the NPMc mutation are associated with increased abundance of several nuclear importins, and NPMc interacts directly with several family members. (A) Volcano plot showing protein abundance in NPM1-mutated vs wt AML samples. P values are calculated using the t test and corrected for multiple-hypothesis testing with Benjamini-Hochberg method. Dashed red line shows P = .05. (B-C) Normalized abundance of the nuclear importins organized by NPM1 mutation status in TMT data (B) and bulk RNA sequencing data (C). *P < .05 by t test between groups. (D) TurboID vectors were created with no fused complementary DNA (cDNA) (“TurboID only”) or fused at either the N or C terminus of wt NPM1 or mutant NPMc (T-NPM1 and T-NPMc indicate N-terminal fusions, whereas NPM1-T and NPMc-T indicate C-terminal fusions). Each vector was stably transduced into primary mouse hematopoietic stem/progenitor cells and, after 4 days, cultured in the presence of biotin for 4 hours. Biotin-labeled proteins were then enriched with streptavidin beads and stringently washed, and tryptic peptides were released from the beads and identified by mass spectrometry. Z-scores are calculated based on spectral counts across 10 TurboID-only biological replicates and 3 biological replicates for each of the other indicated vectors. The 30 interacting proteins with the greatest fold change for the NPM1-TurboID constructs are shown; all display significant differences (t test, multiple-hypothesis correction by Benjamini-Hochberg method with P < .05) from samples expressing TurboID only and NPMc-TurboID. Similarly, 30 proteins with the greatest fold change were selected for the NPMc-TurboID vector, and all displayed significant differences from both wt NPM1 and TurboID only. KPNA3 and KPNA4, 2 members of the nuclear importin family, are highlighted in green. (E) Spectral counts detected in the TurboID experiments for each of the displayed nuclear importins are normalized for display between 0 and 1. *P < .05 by t test between groups. N- and C-terminal TurboID constructs are analyzed together for the NPM1 and NPMc vectors. (F) Western blotting of protein abundance for the indicated proteins in cell lysates created from the stably transduced mouse bone marrow cells prior to streptavidin pulldown. Normalized total protein for each lane is shown as a loading control as determined on the Jess western blotting system. In this short-term expression system, the abundance of the nuclear importins is not increased, suggesting that the detection of interactions with NPMc is not due to an increase in total importin protein abundance. One representative example from 3 biologic replicates is shown. mut, mutant; NS, not significant.
To identify a potential mechanism for this posttranscriptional regulation, we performed a screen for physical interactions specific for NPMc in primary murine hematopoietic cells using the TurboID system for proximity tagging of tightly associated proteins with biotin.25 We stably transduced primary mouse hematopoietic stem/progenitor cells with viral constructs expressing the TurboID cDNA alone or fused in frame to either the N or C terminus of wt NPM1 or mutant NPMc. An internal ribosome entry site–green fluorescent protein cassette was downstream from the expressed cDNA in all vectors. We confirmed that the constructs displayed nucleolar localization for wt NPM1 fusions vs nuclear and cytoplasmic localization for NPMc fusions (supplemental Figure 9). After 4 days, the transduced, GFP+ cells were cultured with biotin for 4 hours. Biotin-labeled proteins were enriched with streptavidin bead pulldowns, and tryptic peptides released from the beads were identified by mass spectrometry. This system showed reproducible results across technical replicates and across both N- and C-terminal fusions; the nuclear importins KPNA3 and KPNA4 were among the top 30 biotinylated proteins from NPMc compared with NPM1wt TurboID fusions, indicating physical proximity with mutant NPMc (Figure 4D; supplemental Table 6). Focusing specifically on the nuclear importins, we found that mutant NPMc TurboID constructs showed significantly increased interactions with KPNA1, KPNA3, KPNA4, KPNA6, and KPNB1 compared with the wt NPM1 TurboID vectors (Figure 4E). The total abundance of these proteins was minimally altered after this brief overexpression (Figure 4F), suggesting that increased labeling was due to physical proximity, not increased protein abundance.
Analysis of protein abundance signatures for recurrently mutated AML-associated genes
We also defined differentially expressed proteins for all samples with recurrently mutated genes (n ≥ 3) in this dataset and identified many additional examples of posttranscriptional regulation associated with AML-specific mutations (supplemental Figures 10-14; see leylab.org/amlproteome for an interactive interface).
CD180 and MRC1/CD206 are expressed on AML cells but not normal CD34 cells
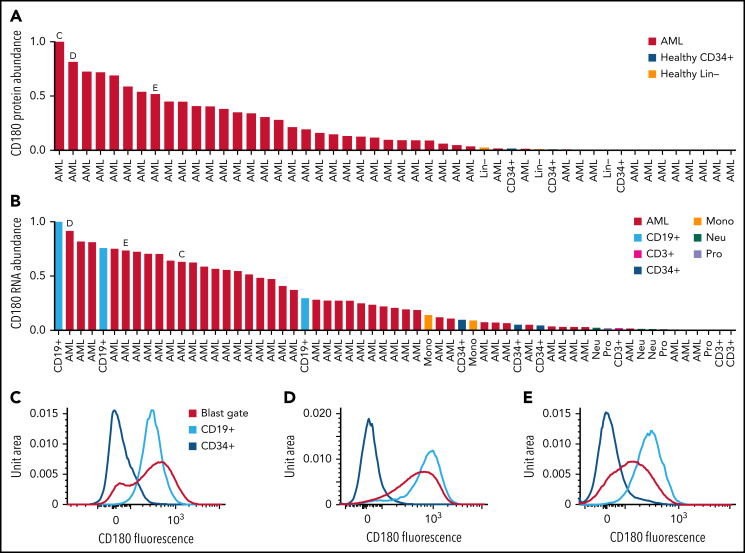
Because AML-specific proteins on the cell surface could serve as targets for immunologic therapies (eg, antibody-drug conjugates, bispecific T-cell engagers, and/or chimeric antigen receptor T cells), we searched the proteomic database for evidence of these proteins. We first compiled a permissive list of 4092 probable surface proteins by combining lists from the Human Protein Atlas40 and the in silico human surfaceome.41 We identified 27 proteins from the list with a median protein expression difference between AML and healthy lineage-depleted bone marrow of >2 standard deviations above the median expression difference in the TMT dataset. This list included the folate receptor β (FOLR2), previously identified as an AML-specific target for chimeric antigen receptor T cells.42 We further filtered for proteins with minimal or no expression on healthy CD34+ cells in the LFQ dataset and that did not show significant expression in other tissue types in the public Human Protein Atlas database.43 This analysis nominated CD180 and MRC1/CD206 as candidate proteins.
CD180 is a Toll-like receptor expressed primarily on B cells and involved in activation signaling44; it has been identified as a possible target for treatment of B-cell non-Hodgkin lymphoma.45 We found that CD180 is expressed highly in many AML samples but not in healthy CD34+ enriched stem/progenitor cells or in lineage-depleted healthy bone marrow (Figure 5A; supplemental Figure 15A). Corresponding RNA sequencing shows that CD180 is primarily expressed in some AML samples and CD19+ B cells and, to a lesser extent, in monocytes/macrophages, consistent with previous reports46 (Figure 5B; supplemental Figure 15B). Similarly, the Human Protein Atlas single-cell transcriptomics database43 showed expression primarily in B cells and macrophages/monocytes, with minimal expression elsewhere. Furthermore, flow cytometry of selected patient samples and healthy bone marrow confirmed expression of CD180 on AML blasts, and CD19+ B cells, with no detectable expression on healthy CD34+ stem/progenitor cells (Figure 5C-E).
Figure 5.
CD180 and MRC1 are highly expressed on AML blasts from some patients but not on CD34 stem/progenitor cells. (A) Normalized protein abundance of CD180 in LFQ data for AML patient bone marrow samples, lineage-depleted bone marrow from healthy donors (Healthy Lin−), and CD34-selected bone marrow from healthy donors (Healthy CD34+). (B) Normalized RNA abundance of CD180 in AML patient samples and the indicated cell types purified from healthy donor bone marrow samples. In both panels A and B, the letters C, D, and E indicate patient samples selected for flow cytometry as shown in the indicated panels. (C-E) Flow cytometry results with staining for CD180 on both AML cells in the blast gate and CD19+ B cells (cells that normally express CD180, as a positive control) in the indicated patient samples. Staining of CD34+ cells from healthy donor marrow is also shown. (F) Normalized protein abundance of MRC1 in LFQ data for AML patient bone marrow samples, lineage-depleted bone marrow cells from healthy donors (Healthy Lin−), and CD34-enriched bone marrow cells from healthy donors (Healthy CD34+). (G) Normalized RNA abundance of MRC1 in AML patient samples, and the indicated cell types from healthy donor bone marrow, are shown. In both panels F and G, the letters H, I, and J indicate patient samples selected for flow cytometry and are shown in the indicated panels. (H-J) Flow cytometry results with staining for MRC on AML cells in the blast gate in the indicated patient samples. Staining of CD34+ cells and monocytes (cells that normally express MRC1, as a positive control) is shown from healthy donor marrow.
MRC1/CD206 is a mannose receptor primarily expressed on the surface of M2 immunosuppressive macrophages47 and tumor-associated macrophages (TAMs), which are thought to promote an immunosuppressive and pro-tumorigenic microenvironment.48,49 We found that MRC1/CD206 protein is highly expressed in a subset of AML samples, but not in CD34+ healthy bone marrow cells (Figure 5F; supplemental Figure 15C). RNA expression corroborates high levels of expression in many AML samples with lower expression in monocytes, and minimal to no expression in healthy CD34+ cells (Figure 5G; supplemental Figure 15D). The Human Protein Atlas single-cell transcriptomics database43 showed expression primarily on macrophages and hepatic stellate cells. Flow cytometry of selected patient samples confirmed expression of MRC1/CD206 was indeed on high on AML blasts and healthy monocytes, but not on healthy CD34+ cells (Figure 5H-J). Taken together, these data suggest that CD180 and MRC1/CD206 may be candidates for targeting AML, with potentially tolerable “on-target, off-cancer” toxicity.
The phosphoproteomic landscape of AML cells
Using a previously validated protocol21 for deep-scale phosphoproteomics, we measured global phosphopeptides in the 44 AML samples and 3 lineage-depleted healthy bone marrow controls. We detected 29 201 unique phosphosites on 5407 unique proteins in the dataset. Unsupervised clustering of patients based on phosphoproteomic profiles revealed clear segregation between AML and healthy controls, as well as groups corresponding to activating FLT3 mutations, PML-RARA fusions (acute promyelocytic leukemia [APL]), and CBFB-MYH11 fusions (Figure 6; supplemental Table 7; supplemental Figure 16).
Figure 6.
Phosphoproteomic analysis of AML samples reveals associations with some initiating events and with FLT3 mutations. Unsupervised clustering of phosphoproteomic data from 44 AML samples and 3 healthy control bone marrow samples using the unweighted pair group method with arithmetic mean algorithm. The heatmap shows Pearson correlation of total phosphosite abundance between each patient. Clinical correlates are noted as in Figure 1.
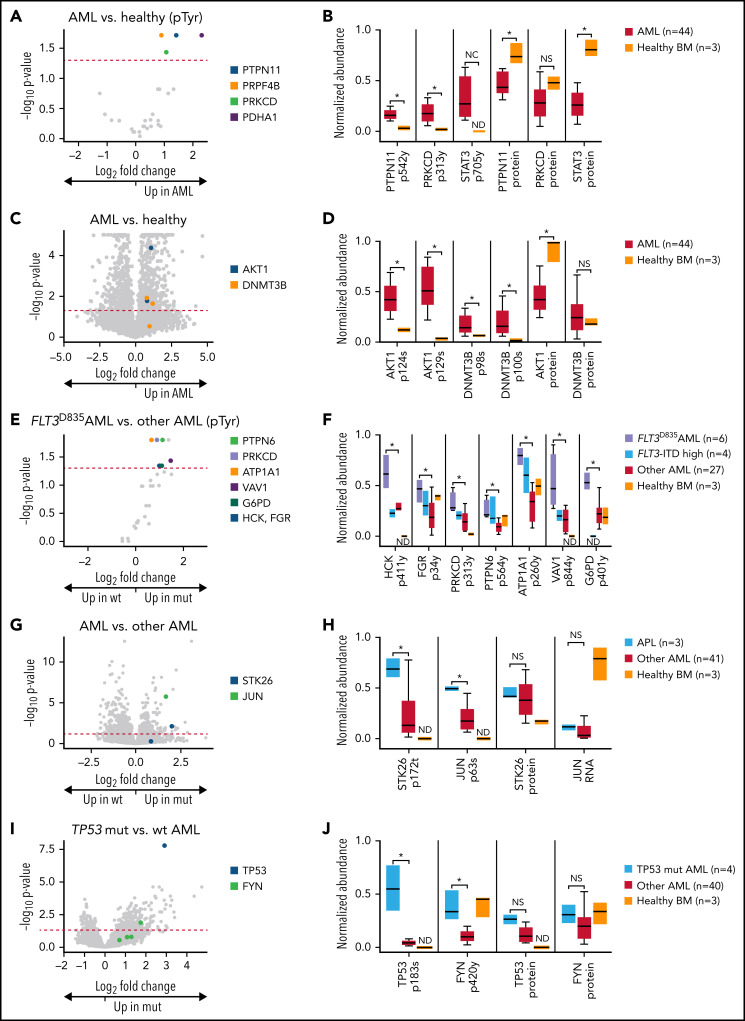
We next identified specific phosphosites driving these signatures. In a global analysis of AML samples vs healthy control samples, we detected significantly increased tyrosine phosphorylation of 4 sites (PTPN11 tyrosine-542, protein kinase C δ (PRKCD) tyrosine-313, PRPF4B tyrosine-849, and PDHA1 tyrosine-242; Figure 7A. Note that immobilized metal affinity chromatography enriches all phosphorylated peptides and is not tyrosine specific). For PTPN11/SHP-2, a recurrently mutated gene in AML50 (Figure 7B), the increased phosphorylation of the activating site tyrosine-54251 is not significantly driven by PTPN11 mutations; in this dataset, only 2 patients had PTPN11 mutations, and neither displayed aberrant PTPN11 tyrosine-542 phosphorylation. The AML samples also displayed increased phosphorylation of the activating site tyrosine-313 on PRKCD.52 STAT3 tyrosine-705, a site essential for its transcriptional activity53 and previously shown to be phosphorylated in many AML patients,54 was also found to be phosphorylated in AML samples and was undetectable in healthy bone marrow samples (Figure 7B). None of PTPN11, PRKCD, or STAT3 proteins had increased abundance in AML samples; in fact, both PTPN11 and STAT3 had significantly decreased abundance in AML compared with healthy, lineage-depleted bone marrow cells (Figure 7B). Taken together, these results confirm the association between increased STAT3 signaling and AML, strengthen evidence for PRKCD activation,55 and suggest widespread signaling via PTPN11 in AML cells.
Figure 7.
Phosphoproteomic analyses of AML samples associated with specific mutations. (A) Volcano plot showing phosphorylated tyrosine sites in AML samples vs lineage-depleted bone marrow from healthy donors. P values are calculated using the Mann-Whitney U test and corrected for multiple-hypothesis testing with Benjamini-Hochberg method. Dashed red line shows P = .05. (B) Normalized abundance of selected tyrosine phosphosites with differences between AML and healthy samples, including the activating site tyrosine-546 on the phosphatase PTPN11/SHP2, the activating site tyrosine-313 on PRKCD, and the activating site tyrosine-705 on STAT3. Total protein abundance for all 3 proteins is shown as well, indicating that increased phosphorylation of these sites is not due to changes in overall protein abundance in AML samples. *P < .05 represent significantly different sites after multiple hypothesis correction as calculated in panel A. All sites were normalized to between 0 and 1 for display. (C) Volcano plot showing all phosphorylated sites in AML samples vs lineage-depleted bone marrow from healthy donors. P values are calculated using the t test and corrected for multiple-hypothesis testing with Benjamini-Hochberg method. Dashed red line shows P = .05. (D) Normalized abundance of selected phosphosites with differences between AML and healthy samples, including the serine-124 in the linker domain involved in optimal activation of AKT1 and multiple sites on DNMT3B of unknown function. Total protein abundance of AKT1 and DNMT3B are shown as well. *P < .05 after multiple-hypothesis correction as in panel C. (E) Volcano plot showing tyrosine phosphorylation sites comparing between FLT3D835-mutant AML samples and FLT3 wt AML samples as determined by 1-sided Mann-Whitney U test with multiple-hypothesis correction by Benjamin-Hochberg method. (F) Normalized abundance for each of the indicated phosphosites in the indicated patient groups. *P < .05 represents significantly different tyrosine phosphorylation sites between FLT3D835-mutant AML samples and FLT3 wt AML samples as determined in panel E. ND indicates a phosphosite was not detected in that group. Only FLT3-ITD samples with high variant allele frequency are shown. (G) Volcano plot showing differentially phosphorylated sites in samples initiated by PML-RARA (APL) vs other AML. P values are calculated using the t test and corrected for multiple-hypothesis testing with Benjamini-Hochberg method. (H) Normalized abundance of phosphorylated threonine-172 in the activation loop site on the kinase STK26 in APL vs other AML and healthy bone marrow. Total STK26 protein abundance is also shown. Normalized abundance of phosphorylation of the known activating site serine-63 on the transcription factor JUN in APL vs other AML and healthy bone marrow. Normalized abundance of JUN RNA is shown, since total JUN protein was below the limits of detection in this dataset. (I) Volcano plot showing differentially phosphorylated sites in TP53-mutant vs wt AML. P values are calculated using the t test and corrected for multiple-hypothesis testing with Benjamini-Hochberg method. (J) Abundance of the degradation related site serine-183 on TP53 and the activating site tyrosine-420 on FYN. NC, not calculable due to site being not detected in healthy samples; BM, bone marrow; ND, not detected; NS, not significant.
We next evaluated all phosphorylation sites and found 970 with significant differences (P < .05, Benjamini-Hochberg method56 for multiple-hypothesis correction) between AML samples and healthy controls (Figure 7C). These included 2 activating sites (serine-12457 and serine-12958) on the oncogenic serine/threonine protein kinase AKT1 and multiple sites on DNMT3B (1 of the 2 de novo methyltransferases expressed in AML cells; the other is DNMT3A, one of the most frequently mutated genes in AML9-11,59; Figure 7D), among others.
We also looked for evidence of recurrent phosphorylation patterns in AML. Using unsupervised clustering, we found 5 groups containing at least 10 phosphoproteins and where the average Pearson correlation among all phosphosites in the group across all samples was >0.8. One group was enriched for chromatin-organization proteins, 1 for histone modifiers, 2 for RNA processing/splicing proteins, and 1 for cytoskeletal proteins.
FLT3-TKD mutations are associated with activation of the SRC-family tyrosine kinases FGR and HCK
As expected, we observed a strong phospho-signature associated with activating tyrosine kinase domain (TKD) mutations in the receptor tyrosine kinase FLT3, as evidenced by the grouping of FLT3-TKD samples in unsupervised clustering (Figure 6). We sought to characterize downstream pathways of FLT3-TKD signaling by identifying phosphorylated tyrosines in samples with the common D835 mutation (6 of 44 patients in this dataset). Nine tyrosines had increased phosphorylation in FLT3-TKD samples compared with FLT3 wt samples (Figure 7E). These include the activating site tyrosine-411 on HCK,60,61 and tyrosine-34 on FGR, both of which are SRC-family cytoplasmic tyrosine kinases62 (Figure 7F). We also identified the activating site tyrosine-313 on PRKCD,52 the activating site tyrosine-56463 on the tyrosine-protein phosphatase PTPN6 (which plays a known role in deactivation of Src signaling),64 the highly conserved tyrosine-844 site on the ρ/Rac guanine nucleotide exchange factor VAV1 (downstream from HCK and FGR in neutrophil activation),65,66 and the tyrosine-401 site on G6PD (which increases activity when phosphorylated by SRC-family kinases).67 Finally, we identified increased phosphorylation of tyrosine-260 on ATP1A1, which regulates SRC-family kinase activity.68 Taken together, these data suggest that the SRC-family kinases HCK and FGR, and the downstream kinase PRKCD, are activated in AML cells containing FLT3-TKD mutations. Many of these phosphosites demonstrated similar trends in the 4 samples containing FLT3-ITD mutations in dominant subclones.
AMLs initiated by PML-RARA display a phosphoproteomic signature
In APL samples initiated by PML-RARA, we identified 490 phosphosites that were significantly different from other AML samples (Figure 7G). These included the threonine-172 site on the serine/threonine kinase STK26/MST4, just adjacent to the activation loop,69 and the critical activating site serine-63 on the oncogenic transcription factor JUN70; no significant increase in STK26 protein levels or JUN RNA (total JUN protein not detected) was detected in APL samples (Figure 7H).
TP53-mutated AML displays abundant phosphorylation of TP53 and activated FYN
In TP53-mutated AML samples, there were 344 phosphosites that were significantly different from other AMLs (Figure 7I). These included phosphorylation of the serine-183 site on TP53 itself (which marks TP53 for degradation71) and the activating site tyrosine-420 on the FYN tyrosine kinase72; no significant changes in TP53 or FYN protein levels were detected (Figure 7J).
Discussion
This study presents a deep-scale proteomic and phosphoproteomic database of AML samples as a resource for the AML community. Several vignettes highlight the value and novelty of this dataset, including the identification of posttranscriptionally regulated protein abundance, the identification of cell surface markers for immunologic targeting of AML, and phosphorylation changes in signaling pathways of relevance for AML pathogenesis. An intuitive, interactive database is available at leylab.org/amlproteome.
This study adds to the strong body of evidence of that mRNA and protein abundance in AML cells are relatively limited (median Spearman correlation only 0.31). We found considerable evidence of posttranscriptional regulation of protein abundance in AML, consistent with that described for other cancers.1-8,14 One striking example is the H/ACA box small nucleolar ribonucleoprotein core complex, consisting of DKC1, NHP2, NOP10, and GAR1 (supplemental Figure 8A-B).73 This complex has 2 main roles: it is a component of the telomerase complex (in combination with TERT, which does not show increased abundance in AML samples in these data), and it is important for RNA pseudouridylation.73 Recent work has shown the importance of pseudouridylation for maintenance of hematopoietic stem cells,74 suggesting a possible role in AML pathogenesis.
In IDH1- and IDH2-mutated AML, neomorphic enzyme activity and R-2-hydroxyglutarate (R-2HG) production lead to epigenetic changes and dysfunction of the 2-oxoglutarate–dependent TET enzymes.75 However, R-2HG inhibits a variety of 2-oxoglutarate–dependent enzymes; in fact, prior work demonstrated KDM4A/B/C to be particularly sensitive to R-2HG inhibition.76,77 Indeed, we found increased protein abundance of the KDM4A/B/C family of H3K9/27/36 demethylases in IDH-mutant AML despite no change in RNA abundance; we also show that transient expression of IDH1R132H in K562 cells recapitulates this phenotype, suggesting a direct link between this mutation and increased abundance of this protein family. This link is further suggested by the previous observation that IDH1-mutant gliomas show significantly increased H3K9 trimethylation,78 and expression of mutant IDH1 leads to increases in H3K9, H3K27, and H3K36 trimethylation.79 Additional experiments will be required to determine whether the increased abundance of the KDM4A/B/C proteins represents a “futile” adaptive mechanism in the face of strong R-2HG inhibition, or whether alternative mechanisms affect the abundance of these proteins in IDH-mutant AML samples.
In NPMc-mutant AML, a key feature of pathogenesis is the cytoplasmic mislocalization of NPM1 through the formation of NPMc/NPM1wt heterodimers.38,80 The loss of functional NPM1 from the nucleolus is thought to be relevant for AML pathogenesis; recently, however, it has been suggested that NPMc may also exhibit novel “gain-of-function” activity in the cytoplasm.81 Here, we show that NPMc-mutant AML samples have increased abundance of several nuclear importins (caused by a posttranscriptional mechanism), perhaps related to the fact that NPMc (but not wt NPM) interacts directly with several of these importins, which may stabilize them. The functional relevance of this novel “gain-of-function” property of NPMc is not yet clear.
In this study, we also performed an unbiased evaluation of the AML phosphoproteome. We have highlighted several findings relevant to signaling in AML samples globally or in the context of specific AML-initiating events. For example, we identified a signature for signaling downstream of activating FLT3-TKD mutations, which are correlated with activation of the SRC-family kinases HCK and FGR, and also a highly active PRKCD. Of note, the clinically effective, but “dirty,” kinase inhibitor midostaurin (which improves overall survival when added to induction chemotherapy in patients with FLT3 mutations82) was initially developed as an inhibitor of protein kinase C83 and exhibits strong activity against the SRC-family kinases.84 These data suggest that midostaurin's “off-target” inhibition of these kinases may contribute to its clinical activity in patients with FLT3 mutations by blocking activated signaling pathways downstream from FLT3. Other SRC-family kinases, including LYN85 and SRC,86 have previously been implicated in signaling downstream of FLT3-ITD mutations. In this dataset, samples with FLT3-ITD mutations did not have a unique phospho-signature. However, when we limited the analysis to the 4 AML samples with the highest FLT3-ITD allelic ratios, many of the phosphopeptides identified with FLT3-TKD-mutant samples demonstrated similar trends. Targeted phosphoproteomic studies of FLT3-ITD samples have demonstrated increased power to identify phosphosignatures.19,55,87-89
In summary, we have generated a publicly available, deep-scale proteomic and phosphoproteomic database of AML that provides a missing data layer for a representative part of the LAML TCGA dataset, and we provide an interactive interface for easy access to the data. We identified previously known and novel dysregulated proteins in AML samples and validated several of these findings orthogonally. This publicly available dataset will serve as a valuable resource for the AML research community.
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgments
The expert technical assistance of Alan Davis, James Malone, and Rose Connors is gratefully acknowledged. The proteomic experiments were performed at the Washington University (WU) Proteomics Shared Resource (R.R.T., director). The visual abstract was created with Biorender.com.
This work was supported by National Institutes of Health (NIH) grants T32 HL007088 (M.H.K.; National Heart, Lung and Blood Institute [NHLBI]), CA211782 (C.A.M.; National Cancer Institute [NCI]), CA101937 (T.J.L.; NCI), and CA197561 (T.J.L.; NCI) and the Barnes Jewish Hospital Foundation (T.J.L.). The Proteomics Shared Resource is supported in part by the Washington University Institute of Clinical and Translational Sciences (National Center for Advancing Translational Sciences [NCATS] grant UL1 TR000448), the Mass Spectrometry Research Resource (National Institute of General Medical Sciences grants P41 GM103422 and R24GM136766), and the Siteman Comprehensive Cancer Center Support Grant (NCI grant P30 CA091842).
Footnotes
All datasets are available at www.leylab.org/amlproteome and as supplemental tables. Mass spectrometry machine files and metadata are in the MassIVE database (MassIVE IDs: MSV00089012 [TMT], MSV000089029 [LFQ], MSV000089028 [TurboID], and MSV000089035 [PDX validation]).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: M.H.K., R.B.D., Y.L., Z.X., N.M.H., D.R.G., and C.A.M. performed research; M.H.K., Z.X., S.M.R., and T.J.L. analyzed data; Q.Z., R.S., P.E.-G., Y.M., and R.R.T. optimized and performed mass spectrometry and data processing; P.W., J.E.P., C.A.M., D.C.L., J.F.D., M.J.W., R.R.T., and T.J.L. designed the study; and M.H.K. and T.J.L. wrote the manuscript.
Conflict-of-interest disclosure: J.F.D. has an equity ownership position in Magenta Therapeutics and WUGEN and receives research funding from Amphivena Therapeutics, NeoImmune Tech, Macrogenics, Incyte, Bioline Rx, and WUGEN. None of that funding was used for this study. The remaining authors declare no competing financial interests.
Correspondence: Timothy J. Ley, Washington University School of Medicine, 660 S. Euclid Ave, Box 8007, St Louis, MO 63110; e-mail: timley@wustl.edu.
REFERENCES
- 1.Clark DJ, Dhanasekaran SM, Petralia F, et al. ; Clinical Proteomic Tumor Analysis Consortium . Integrated proteogenomic characterization of clear cell renal cell carcinoma [published correction appears in Cell. 2019;179(4):964–983.e31]. Cell. 2020;180(1): 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dou Y, Kawaler EA, Cui Zhou D, et al. ; Clinical Proteomic Tumor Analysis Consortium . Proteogenomic characterization of endometrial carcinoma. Cell. 2020;180(4):729-748.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gillette MA, Satpathy S, Cao S, et al. ; Clinical Proteomic Tumor Analysis Consortium . Proteogenomic characterization reveals therapeutic vulnerabilities in lung adenocarcinoma. Cell. 2020;182(1): 200-225.e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang C, Chen L, Savage SR, et al. ; Clinical Proteomic Tumor Analysis Consortium . Proteogenomic insights into the biology and treatment of HPV-negative head and neck squamous cell carcinoma. Cancer Cell. 2021;39(3):361-379.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krug K, Jaehnig EJ, Satpathy S, et al. ; Clinical Proteomic Tumor Analysis Consortium . Proteogenomic landscape of breast cancer tumorigenesis and targeted therapy. Cell. 2020;183(5):1436-1456.e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petralia F, Tignor N, Reva B, et al. Integrated proteogenomic characterization across major histological types of pediatric brain cancer. Cell. 2020;183(7): 1962-1985.e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasaikar S, Huang C, Wang X, et al. ; Clinical Proteomic Tumor Analysis Consortium . Proteogenomic analysis of human colon cancer reveals new therapeutic opportunities. Cell. 2019;177(4):1035-1049.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang LB, Karpova A, Gritsenko MA, et al. ; Clinical Proteomic Tumor Analysis Consortium . Proteogenomic and metabolomic characterization of human glioblastoma. Cancer Cell. 2021;39(4): 509-528.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ley TJ, Miller C, Ding L, et al. ; Cancer Genome Atlas Research Network . Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tyner JW, Tognon CE, Bottomly D, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562(7728):526-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23):2209-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoff FW, Hu CW, Qutub AA, et al. Proteomic profiling of acute promyelocytic leukemia identifies two protein signatures associated with relapse. Proteomics Clin Appl. 2019;13(4):e1800133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kornblau SM, Qutub A, Yao H, et al. Proteomic profiling identifies distinct protein patterns in acute myelogenous leukemia CD34+CD38- stem-like cells. PLoS One. 2013;8(10):e78453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoff FW, Van Dijk AD, Qiu Y, et al. Clinical relevance of proteomic profiling in de novo pediatric acute myeloid leukemia: a Children’s Oncology Group study [published online ahead of print 13 January 2022]. Haematologica. 2022. doi: 10.3324/haematol.2021.279672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Dijk AD, de Bont ESJM, Kornblau SM. Targeted therapy in acute myeloid leukemia: current status and new insights from a proteomic perspective. Expert Rev Proteomics. 2020;17(1):1-10. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez-Valladares M, Wangen R, Aasebø E, et al. Proteomic studies of primary acute myeloid leukemia cells derived from patients before and during disease-stabilizing treatment based on all-trans retinoic acid and valproic acid. Cancers (Basel). 2021;13(9):2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alanazi B, Munje CR, Rastogi N, et al. Integrated nuclear proteomics and transcriptomics identifies S100A4 as a therapeutic target in acute myeloid leukemia. Leukemia. 2020;34(2):427-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aasebø E, Berven FS, Bartaula-Brevik S, et al. Proteome and phosphoproteome changes associated with prognosis in acute myeloid leukemia. Cancers (Basel). 2020; 12(3):709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joshi SK, Nechiporuk T, Bottomly D, et al. The AML microenvironment catalyzes a stepwise evolution to gilteritinib resistance. Cancer Cell. 2021;39(7):999-1014.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu CW, Qiu Y, Ligeralde A, et al. A quantitative analysis of heterogeneities and hallmarks in acute myelogenous leukaemia. Nat Biomed Eng. 2019;3(11): 889-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mertins P, Tang LC, Krug K, et al. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nat Protoc. 2018;13(7):1632-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meier F, Brunner AD, Koch S, et al. Online Parallel Accumulation-Serial Fragmentation (PASEF) with a novel trapped Ion mobility mass spectrometer. Mol Cell Proteomics. 2018;17(12):2534-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sokal RR, Michener CD, Kansas U. A Statistical Method for Evaluating Systematic Relationships. Lawrence, KS: University of Kansas; 1958. [Google Scholar]
- 24.Zhao S, Lin Y, Xu W, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324(5924):261-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Branon TC, Bosch JA, Sanchez AD, et al. Efficient proximity labeling in living cells and organisms with TurboID [published correction appears in Nat Biotechnol. 2020;38(1):108]. Nat Biotechnol. 2018;36(9):880-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clark G, Stockinger H, Balderas R, et al. Nomenclature of CD molecules from the Tenth Human Leucocyte Differentiation Antigen Workshop. Clin Transl Immunology. 2016;5(1):e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lukin JA, Kontaxis G, Simplaceanu V, Yuan Y, Bax A, Ho C. Quaternary structure of hemoglobin in solution. Proc Natl Acad Sci USA. 2003;100(2):517-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van der Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9(86):2579-2605. [Google Scholar]
- 30.Abbas HA, Mohanty V, Wang R, et al. Decoupling lineage-associated genes in acute myeloid leukemia reveals inflammatory and metabolic signatures associated with outcomes. Front Oncol. 2021;11:705627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quintás-Cardama A, Hu C, Qutub A, et al. p53 pathway dysfunction is highly prevalent in acute myeloid leukemia independent of TP53 mutational status. Leukemia. 2017; 31(6):1296-1305. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Chen F, Chandrashekar DS, Varambally S, Creighton CJ. Proteogenomic characterization of 2002 human cancers reveals pan-cancer molecular subtypes and associated pathways. Nat Commun. 2022;13(1):2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong L, Lu D, Chen R, et al. Proteogenomic characterization identifies clinically relevant subgroups of intrahepatic cholangiocarcinoma. Cancer Cell. 2022; 40(1):70-87.e15. [DOI] [PubMed] [Google Scholar]
- 34.McCann KL, Kavari SL, Burkholder AB, Phillips BT, Hall TMT. H/ACA snoRNA levels are regulated during stem cell differentiation. Nucleic Acids Res. 2020;48(15):8686-8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pühringer T, Hohmann U, Fin L, et al. Structure of the human core transcription-export complex reveals a hub for multivalent interactions. eLife. 2020;9:e61503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee DH, Kim GW, Jeon YH, Yoo J, Lee SW, Kwon SH. Advances in histone demethylase KDM4 as cancer therapeutic targets. FASEB J. 2020;34(3):3461-3484. [DOI] [PubMed] [Google Scholar]
- 37.Losman JA, Koivunen P, Kaelin WG Jr. 2-Oxoglutarate-dependent dioxygenases in cancer. Nat Rev Cancer. 2020;20(12): 710-726. [DOI] [PubMed] [Google Scholar]
- 38.Falini B, Brunetti L, Sportoletti P, Martelli MP. NPM1-mutated acute myeloid leukemia: from bench to bedside. Blood. 2020; 136(15):1707-1721. [DOI] [PubMed] [Google Scholar]
- 39.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282(8):5101-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thul PJ, Åkesson L, Wiking M, et al. A subcellular map of the human proteome. Science. 2017;356(6340):eaal3321. [DOI] [PubMed] [Google Scholar]
- 41.Bausch-Fluck D, Goldmann U, Müller S, et al. The in silico human surfaceome. Proc Natl Acad Sci USA. 2018;115(46):E10988-E10997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lynn RC, Poussin M, Kalota A, et al. Targeting of folate receptor β on acute myeloid leukemia blasts with chimeric antigen receptor-expressing T cells. Blood. 2015;125(22):3466-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alam O. A single-cell-type transcriptomics map of human tissues. Nat Genet. 2021; 53(9):1275. [DOI] [PubMed] [Google Scholar]
- 44.Miura Y, Shimazu R, Miyake K, et al. RP105 is associated with MD-1 and transmits an activation signal in human B cells. Blood. 1998;92(8):2815-2822. [PubMed] [Google Scholar]
- 45.Polson AG, Calemine-Fenaux J, Chan P, et al. Antibody-drug conjugates for the treatment of non-Hodgkin’s lymphoma: target and linker-drug selection [published correction appears in Cancer Res. 2010;70(3):1275]. Cancer Res. 2009;69(6):2358-2364. [DOI] [PubMed] [Google Scholar]
- 46.Divanovic S, Trompette A, Atabani SF, et al. Negative regulation of Toll-like receptor 4 signaling by the Toll-like receptor homolog RP105. Nat Immunol. 2005;6(6):571-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roca H, Varsos ZS, Sud S, Craig MJ, Ying C, Pienta KJ. CCL2 and interleukin-6 promote survival of human CD11b+ peripheral blood mononuclear cells and induce M2-type macrophage polarization. J Biol Chem. 2009;284(49):34342-34354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaynes JM, Sable R, Ronzetti M, et al. Mannose receptor (CD206) activation in tumor-associated macrophages enhances adaptive and innate antitumor immune responses. Sci Transl Med. 2020;12(530):eaax6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lepland A, Asciutto EK, Malfanti A, et al. Targeting pro-tumoral macrophages in early primary and metastatic breast tumors with the CD206-binding mUNO peptide. Mol Pharm. 2020;17(7):2518-2531. [DOI] [PubMed] [Google Scholar]
- 50.Alfayez M, Issa GC, Patel KP, et al. The Clinical impact of PTPN11 mutations in adults with acute myeloid leukemia. Leukemia. 2021;35(3):691-700. [DOI] [PubMed] [Google Scholar]
- 51.Araki T, Nawa H, Neel BG. Tyrosyl phosphorylation of Shp2 is required for normal ERK activation in response to some, but not all, growth factors. J Biol Chem. 2003;278(43):41677-41684. [DOI] [PubMed] [Google Scholar]
- 52.Hall KJ, Jones ML, Poole AW. Coincident regulation of PKCdelta in human platelets by phosphorylation of Tyr311 and Tyr565 and phospholipase C signalling. Biochem J. 2007;406(3):501-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang G, Yan H, Ye S, Tong C, Ying QL. STAT3 phosphorylation at tyrosine 705 and serine 727 differentially regulates mouse ESC fates. Stem Cells. 2014;32(5): 1149-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Redell MS, Ruiz MJ, Alonzo TA, Gerbing RB, Tweardy DJ. Stat3 signaling in acute myeloid leukemia: ligand-dependent and -independent activation and induction of apoptosis by a novel small-molecule Stat3 inhibitor. Blood. 2011;117(21):5701-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Casado P, Wilkes EH, Miraki-Moud F, et al. Proteomic and genomic integration identifies kinase and differentiation determinants of kinase inhibitor sensitivity in leukemia cells. Leukemia. 2018;32(8):1818-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57(1):289-300. [Google Scholar]
- 57.Guo H, Gao M, Lu Y, et al. Coordinate phosphorylation of multiple residues on single AKT1 and AKT2 molecules. Oncogene. 2014;33(26):3463-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ponce DP, Maturana JL, Cabello P, et al. Phosphorylation of AKT/PKB by CK2 is necessary for the AKT-dependent up-regulation of β-catenin transcriptional activity. J Cell Physiol. 2011;226(7): 1953-1959. [DOI] [PubMed] [Google Scholar]
- 59.Ley TJ, Ding L, Walter MJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pecquet C, Nyga R, Penard-Lacronique V, et al. The Src tyrosine kinase Hck is required for Tel-Abl- but not for Tel-Jak2-induced cell transformation. Oncogene. 2007;26(11):1577-1585. [DOI] [PubMed] [Google Scholar]
- 61.Bureau JF, Cassonnet P, Grange L, et al. The SRC-family tyrosine kinase HCK shapes the landscape of SKAP2 interactome. Oncotarget. 2018;9(17):13102-13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Amata I, Maffei M, Pons M. Phosphorylation of unique domains of Src family kinases. Front Genet. 2014;5:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Z, Shen K, Lu W, Cole PA. The role of C-terminal tyrosine phosphorylation in the regulation of SHP-1 explored via expressed protein ligation. J Biol Chem. 2003;278(7):4668-4674. [DOI] [PubMed] [Google Scholar]
- 64.Abram CL, Roberge GL, Pao LI, Neel BG, Lowell CA. Distinct roles for neutrophils and dendritic cells in inflammation and autoimmunity in motheaten mice. Immunity. 2013;38(3):489-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lazer G, Pe’er L, Farago M, Machida K, Mayer BJ, Katzav S. Tyrosine residues at the carboxyl terminus of Vav1 play an important role in regulation of its biological activity. J Biol Chem. 2010;285(30):23075-23085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fumagalli L, Zhang H, Baruzzi A, Lowell CA, Berton G. The Src family kinases Hck and Fgr regulate neutrophil responses to N-formyl-methionyl-leucyl-phenylalanine. J Immunol. 2007;178(6):3874-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mattè A, Lupo F, Tibaldi E, et al. Fyn specifically Regulates the activity of red cell glucose-6-phosphate-dehydrogenase. Redox Biol. 2020;36:101639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Banerjee M, Cui X, Li Z, et al. Na/K-ATPase Y260 phosphorylation-mediated Src regulation in control of aerobic glycolysis and tumor growth. Sci Rep. 2018;8(1):12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Record CJ, Chaikuad A, Rellos P, et al. Structural comparison of human mammalian ste20-like kinases. PLoS One. 2010;5(8):e11905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smeal T, Binetruy B, Mercola DA, Birrer M, Karin M. Oncogenic and transcriptional cooperation with Ha-Ras requires phosphorylation of c-Jun on serines 63 and 73. Nature. 1991;354(6353):494-496. [DOI] [PubMed] [Google Scholar]
- 71.Gully CP, Velazquez-Torres G, Shin JH, et al. Aurora B kinase phosphorylates and instigates degradation of p53. Proc Natl Acad Sci USA. 2012;109(24):E1513-E1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nguyen TH, Liu J, Lombroso PJ. Striatal enriched phosphatase 61 dephosphorylates Fyn at phosphotyrosine 420. J Biol Chem. 2002;277(27):24274-24279. [DOI] [PubMed] [Google Scholar]
- 73.Kiss T, Fayet-Lebaron E, Jády BE. Box H/ACA small ribonucleoproteins. Mol Cell. 2010;37(5):597-606. [DOI] [PubMed] [Google Scholar]
- 74.Chlon TM, Stepanchick E, Hershberger CE, et al. Germline DDX41 mutations cause ineffective hematopoiesis and myelodysplasia. Cell Stem Cell. 2021;28(11):1966-1981.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6): 553-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chowdhury R, Yeoh KK, Tian YM, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12(5):463-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Laukka T, Myllykoski M, Looper RE, Koivunen P. Cancer-associated 2-oxoglutarate analogues modify histone methylation by inhibiting histone lysine demethylases. J Mol Biol. 2018;430 (18 18 Pt B):3081-3092. [DOI] [PubMed] [Google Scholar]
- 78.Lu C, Ward PS, Kapoor GS, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Falini B, Bolli N, Shan J, et al. Both carboxy-terminus NES motif and mutated tryptophan(s) are crucial for aberrant nuclear export of nucleophosmin leukemic mutants in NPMc+ AML. Blood. 2006;107(11): 4514-4523. [DOI] [PubMed] [Google Scholar]
- 81.Brunetti L, Gundry MC, Sorcini D, et al. Mutant NPM1 maintains the leukemic state through HOX expression. Cancer Cell. 2018;34(3):499-512.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stone RM, Mandrekar SJ, Sanford BL, et al. Midostaurin plus chemotherapy for acute myeloid leukemia with a FLT3 mutation. N Engl J Med. 2017;377(5):454-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Propper DJ, McDonald AC, Man A, et al. Phase I and pharmacokinetic study of PKC412, an inhibitor of protein kinase C. J Clin Oncol. 2001;19(5):1485-1492. [DOI] [PubMed] [Google Scholar]
- 84.Manley PW, Caravatti G, Furet P, et al. Comparison of the kinase profile of midostaurin (Rydapt) with that of its predominant metabolites and the potential relevance of some newly identified targets to leukemia therapy. Biochemistry. 2018; 57(38):5576-5590. [DOI] [PubMed] [Google Scholar]
- 85.Robinson LJ, Xue J, Corey SJ. Src family tyrosine kinases are activated by Flt3 and are involved in the proliferative effects of leukemia-associated Flt3 mutations. Exp Hematol. 2005;33(4):469-479. [DOI] [PubMed] [Google Scholar]
- 86.Leischner H, Albers C, Grundler R, et al. SRC is a signaling mediator in FLT3-ITD- but not in FLT3-TKD-positive AML. Blood. 2012; 119(17):4026-4033. [DOI] [PubMed] [Google Scholar]
- 87.Pugliese GM, Latini S, Massacci G, Perfetto L, Sacco F. Combining mass spectrometry-based phosphoproteomics with a network-based approach to reveal FLT3-dependent mechanisms of chemoresistance. Proteomes. 2021;9(2):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cucchi DGJ, Van Alphen C, Zweegman S, et al. Phosphoproteomic characterization of primary AML samples and relevance for response toward FLT3-inhibitors. HemaSphere. 2021;5(7):e606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schaab C, Oppermann FS, Klammer M, et al. Global phosphoproteome analysis of human bone marrow reveals predictive phosphorylation markers for the treatment of acute myeloid leukemia with quizartinib. Leukemia. 2014;28(3):716-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.