Abstract
The opportunistic pathogenic yeast Candida albicans exhibits growth phase-dependent changes in cell surface hydrophobicity, which has been correlated with adhesion to host tissues. Cell wall proteins that might contribute to the cell surface hydrophobicity phenotype were released by limited glucanase digestion. These proteins were initially characterized by their rates of retention during hydrophobic interaction chromatography–high-performance liquid chromatography and used as immunogens for monoclonal antibody production. The present work describes the cloning and functional analysis of a C. albicans gene encoding a 38-kDa protein recognized by the monoclonal antibody 6C5-H4CA. The 6C5-H4CA antigen was resolved by two-dimensional electrophoresis, and a partial protein sequence was determined by mass spectrometry analysis of tryptic fragments. The obtained peptides were used to identify the gene sequence from the unannotated C. albicans DNA database. The antibody epitope was provisionally mapped by peptide display panning, and a peptide sequence matching the epitope was identified in the gene sequence. The gene sequence encodes a novel open reading frame (ORF) of unknown function that is highly similar to several other C. albicans ORFs and to a single Saccharomyces cerevisiae ORF. Knockout of the gene resulted in a decrease in measurable cell surface hydrophobicity and in adhesion of C. albicans to fibronectin. The results suggest that the 38-kDa protein is a hydrophobic surface protein that meditates binding to host target proteins.
Cell surface hydrophobicity (CSH) has a central role in the pathogenesis of the opportunistic fungal pathogen Candida albicans. Hydrophobic cells, compared to hydrophilic cells, exhibit greater adherence to epithelial and endothelial cells and extracellular matrix proteins, appear to be more resistant to killing by phagocytes, and are more virulent in mice (2, 12, 16, 26, 28). C. albicans is unique among Candida species in that CSH status varies in response to different environmental conditions and growth phases (17). Within the laboratory setting, populations of C. albicans cells can be switched between the hydrophobic and hydrophilic phenotypes by simply changing the growth temperature. The degree of outer chain mannosylation of cell wall proteins may play a key role in regulating the switch between the two phenotypes (25), but the factors that actually confer the hydrophobic phenotype are unclear.
Previous work has identified several specific surface antigens that appear to contribute to CSH and affect cell attachment to host targets (10, 11, 24, 25, 26). Identification of proteins that might contribute to the CSH phenotype has been accomplished by partial cell wall digestion to release minimally covalently linked proteins and proteins that are noncovalently trapped within the wall matrix. Extracts containing candidate proteins were then separated by high-performance liquid chromatography–hydrophobic interaction chromatography to obtain fractions enriched in proteins with a greater hydrophobic character (10, 11). These fractions have been used as immunogens for monoclonal and polyclonal serum generation (10, 26). Using these antibodies, we have identified several proteins that contribute to CSH and affect cell adherence to host targets in static and flow binding assays (12, 26). However, all of the evidence to date indicating the role of CSH in adhesion and pathogenesis has been equivocal, because it is possible that factors determining CSH might not have a direct effect in these assays. It is important, therefore, to further characterize the molecular nature of the genes and proteins that are responsible for CSH.
One candidate surface antigen is a 38-kDa protein recognized by the monoclonal antibody (MAb) 6C5-H4CA. Indirect immunofluorescence on unpermeabilized cells using MAb 6C5-H4CA shows weak surface localization, with a stronger signal on the more hydrophobic pseudohyphae and germ tubes (unpublished observation). Both the MAb (10, 12, 26) and peptides derived from the putative antibody epitope (unpublished observation) are effective in partially blocking cell binding in static assays and homotypic and heterotypic attachment in shear adhesion assays. These results suggest that the 38-kDa protein may have a role in hydrophobic cell attachment. Western blot analysis of other pathogenic yeast species suggested that although the MAb 6C5-H4CA antigen could be weakly detected in some of the other species, the strongest signal by far was seen in C. albicans cells (reference 26 and unpublished results). Previous characterization of the MAb 6C5-H4CA antigen shed little light on the molecular identity of the protein. The 38-kDa antigen recognized by polyclonal sera raised against hydrophobic proteins is poorly glycosylated, if at all, and its hydrophobic nature is supported by the observation that the antigen spontaneously autoaggregates upon concentration and dialysis (9, 11). The lack of detectable glycosylation suggests that the antigen is not covalently attached to the cell wall but may instead be immobilized in the meshwork of the wall. The present work was begun in order to precisely characterize the epitope of the antibody, to identify the gene encoding the antigen, and to address more rigorously the role of surface hydrophobic molecules in pathogenesis and in conferring surface hydrophobicity on C. albicans.
MATERIALS AND METHODS
Media and strains.
C. albicans strain LGH1095 cells (15) were grown in YNB medium (0.65% yeast nitrogen base [Difco] plus 2% glucose) at either 23 or 37°C. C. albicans uridine-auxotrophic strain CAI4 cells (the kind gift of William Fonzi, Georgetown University) were used for transformations and were grown in either YPD medium (1% Difco yeast extract, 2% Difco Bacto Peptone, 2% glucose) or YNB medium. Saccharomyces cerevisiae strain YPH250 (MATa ura3-52 lys2-801 ade2-101 trp1Δ1 his3-Δ200 leu2-Δ1; American Type Culture Collection) was grown on YPD medium or on synthetic complete (SC) medium (14).
Yeast transformation was carried out by the lithium acetate method according to the modified protocol described by Wilson (29). Escherichia coli strains were grown on Luria-Bertani medium plus appropriate antibiotics at 37°C.
Random peptide panning.
The MAb 6C5-H4CA was used to pan a random peptide display library (Flitrx Random Peptide Display Library; Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. The library consists of random 12-amino-acid segments presented as a loop on plasmid-borne thioredoxin-flagellin fusion proteins, which are incorporated into flagella on the surface of recombinant E. coli (23). Library pools were adsorbed to antibody-coated enzyme-linked immunosorbent assay plates to bind immunoreactive clones, which were then gently eluted and amplified by brief growth in medium. Amplified cultures were then used for subsequent rounds of panning as before. After five rounds of panning, cultures were plated on Luria-Bertani medium plus ampicillin to single-colony density and individual colonies were tested for MAb 6C5-H4CA immunoreactivity by Western blotting of whole-cell lysates. Plasmid DNA from immunoreactive clones was purified and sequenced. Cassettes containing the random peptides were PCR amplified using primers flanking the thioredoxin subunit for subsequent in-frame cloning into a glutathione S-transferase (GST) vector for bacterial expression.
Protein purification and mass spectrometry.
Protein was extracted from C. albicans strain LGH1095 cells grown in YNB medium at 23°C by rapidly freezing cell pellets in liquid nitrogen and then mechanically breaking the frozen cell pellets with an electric coffee grinder (electric coffee mill, model 203B; Krups, Peoria, Ill.), the basin of which had been cooled by grinding dry ice (27). The method was chosen since it releases a significantly larger amount of the antigen than the limited lyticase digest procedure used to characterize the antigen previously (11). Insoluble material from broken cells was removed by centrifugation for 15 min at 5,000 × g. Soluble material released from broken cells was initially precipitated by addition of ammonium sulfate to 50% saturation, and the pellet was dissolved in 0.05 M NaPO4, pH 7.2. This material was loaded onto a hydrophobic interaction chromatography column (Butyl Sepharose; Pharmacia) equilibrated with 1.7 M ammonium sulfate–50 mM NaPO4. Fractions were eluted with a step gradient (0.68 M ammonium sulfate–50 mM NaPO4; 0.34 M ammonium sulfate–50 mM NaPO4; 50 mM NaPO4 alone) of decreasing ammonium sulfate concentrations (9, 10). Fractions with significant MAb 6C5-H4CA immunoreactivity were subsequently pooled and concentrated using an Ultrafree Biomax 5 centrifuge concentrator (Millipore, Bedford, Mass.). Concentrated material was analyzed by two-dimensional electrophoretic separation using first dimension isoelectric focusing (ampholyte mixture of 1.5% pH 3 to 10 Biolytes, 0.5% pH 4 to 6 Biolytes, 0.5% pH 5 to 8 Biolytes; Bio-Rad, Hercules, Calif.) and a second dimension 12.5% denaturing acrylamide gel. Coomassie-stained gels were aligned with Western blottings of duplicate gels to identify protein spots reactive with the antibody. Spots were excised from the gel and treated with trypsin in the gel slice. Tryptic fragments extracted from the polyacrylamide were analyzed by liquid chromatography-ion trap mass spectrometry to determine molecular masses of individual peptides and to identify peptide ion spectra (University of Virginia Biomedical Research Facility). Peptide masses obtained from this analysis were used to search the unannotated C. albicans DNA database (http://www-sequence.stanford.edu/group/candida) for potential open reading frames encoding proteins with these peptides, using the Seaquest algorithm (30). Peptides yielding sequence interpretable spectra were used to search the same database with the Genetics Computer Group (GCG; Madison, Wis.) module TFASTA.
Cloning of CSH1.
The putative CSH1 gene sequence was used to design PCR primers flanking the open reading frame for subsequent amplification using strain LGH1095 genomic DNA as a template. The obtained PCR product was then sequenced to verify identity and was cloned in frame into the GST fusion vector pGEX-KG (13). E. coli transformants carrying the GST-CSH1p fusion plasmid were analyzed by Western blot analysis for MAb 6C5-H4CA reactivity.
GST fusion protein purification.
Large amounts of GST fusion proteins (FliTrx random peptide sequences cloned into the vector pGEX-KG) were purified from E. coli lysates following induction of fusion protein expression in exponential-phase cultures with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Lysates were made by suspending bacterial pellets in B-PER bacterial protein extraction reagent (Pierce) and were clarified by centrifugation. GST fusion proteins were purified by batch adsorption to glutathione-agarose (Sigma) and elution with 5 mM reduced glutathione. GST fusions to whole CSH1p formed insoluble inclusion bodies under standard E. coli growth conditions and were therefore partially purified as inclusion bodies using B-PER reagent according to the manufacturer's instructions.
Knockout generation.
C. albicans knockout of the CSH1 gene was generated using the PCR-based adaptation (29) of the sequential URA-Blaster technique (7). PCR primers were designed to amplify the mini-URA cassette in plasmid pDDB57, tailed with an additional 65 nucleotides of sequence flanking the open reading frame of the gene to be knocked out (forward primer, CAACTACAAATAAC TACAAC T TAAC T T TAAC TAAAAAAAAAAACAGG TCAATCGATAAATCAAGAT TT TCCCAGTCACGACGT T; reverse primer, TTTTGTAAT TCATATC TAGACC TAATAAAGATAAACGAT TAGCAAAGC TAAAC TACATAACAAACTGTGGAATTGTGAGCGGATA). The presence of PCR product was verified by gel electrophoresis of an aliquot of the reaction mixture, and 10 μg of PCR product was used to transform strain CAI4 cells by following the protocol described by Wilson (29). URA+ transformants were grown on uridine-deficient medium (YNB), and proper genomic insertion of the transforming cassette was determined by a PCR-based analysis of transformant colonies by using a combination of primer pairs internal to the wild-type locus and to the URA cassette. Heterozygous transformants were reverted to the ura− phenotype by counterselection on medium containing 5-fluororotic acid and were retransformed as described above with fresh PCR product to knock out the second allele of the gene. Presumptive homozygous knockouts were verified by PCR analysis and by Western blotting.
Knockout phenotypic characterization.
A knockout clone was assessed for the following properties: (i) relative cell surface hydrophobicity when grown at 23 or 37°C, (ii) population doubling time, (iii) efficiency of germination (yeast-to-hypha transition), and (iv) adhesion to fibronectin.
(i) Cell surface hydrophobicity.
CSH of C. albicans strains was measured by the hydrophobic microsphere-binding assay as described previously (28). In brief, stationary-phase cells were washed in double-distilled water and counted and were then mixed with a suspension of latex beads. After a brief incubation at room temperature, cells were counted microscopically and assessed as either hydrophobic (three or more beads bound per cell) or hydrophilic (fewer than three beads per cell).
(ii) Doubling time determination.
Exponential-phase doubling times of C. albicans strains grown in YPD medium at 37°C were measured by dilution of overnight cultures of strains into fresh medium and counting of cell sphere units as opposed to cell clumps at time intervals. The population doubling time was calculated between time points where the culture demonstrated exponential growth kinetics.
(iii) Induction of germination.
Overnight cultures of CAI4 or csh1/csh1 strains grown in YPD medium at 37°C were used as an inoculum into prewarmed (37°C) RPMI 1640 medium (Sigma) at a cell density of 2 × 106 cells/ml. Samples were removed at 30-min intervals and examined microscopically. Duplicate samples of 100 cells were counted and assessed for cell morphology and scored as normal, pseudohyphal (constricted at hyphal neck with nonparallel walls), or germ tube (no constriction at neck with parallel walls). Percent germination was calculated as the number of pseudohyphae or germ tubes per 100 cells. Culture inocula demonstrated greater than 97% normal morphology initially. Fifty percent culture germination was determined by plotting the percentage of germinated cells for each time point over 3 h and interpolating the time needed for 50% of the culture to show a change in morphology.
(iv) Adhesion to fibronectin.
The cell adhesion assay was carried out as described previously (26, 28). A total of 200 washed cells from stationary-phase cultures of hydrophobic and hydrophilic cells were added and allowed to bind to fibronectin-coated 48-well tissue culture plates. Nonadherent cells were gently washed away, and the wells of the plate were overlaid with 45°C corn meal agar medium and allowed to solidify. Plates were incubated overnight to allow the adherent cells to form colonies during overnight incubation. Relative adhesion was expressed as the percent decrease in adhesion with the knockout in comparison to adhesion with the parent strain. This was calculated by normalizing colonies in quadruplicate wells for each cell growth condition to the number of colonies on a plate that received no washing (total colonies) and then calculating the percent change in adhesion between the parent strain and the knockout strain: 100 × (mean number of knockout colonies − mean number of parent colonies)/number of parent colonies. The significance of changes in adhesion for different cell types was determined by using a paired two sample for means t test.
Expression in S. cerevisiae.
The PCR product containing the CSH1 open reading frame was cloned into the vector pUNI10 (21) and was recombined in vitro using Cre recombinase with the S. cerevisiae shuttle vector p1223 (21). The recombination event places the cloned open reading frame under the control of the GAL promoter contained in p1223. The recombinant plasmid was transformed into strain YPH and selected on SC-ura medium. Expression of the C. albicans CSH1 gene was induced in cultures grown in the presence of 2% galactose and was repressed in cultures grown in the presence of 2% glucose. Cell lysates of yeasts for Western blotting were generated by vortexing cell pellets from 5-ml liquid cultures with glass beads for 30 s and boiling the broken cells with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer.
RESULTS
Epitope sequence determination.
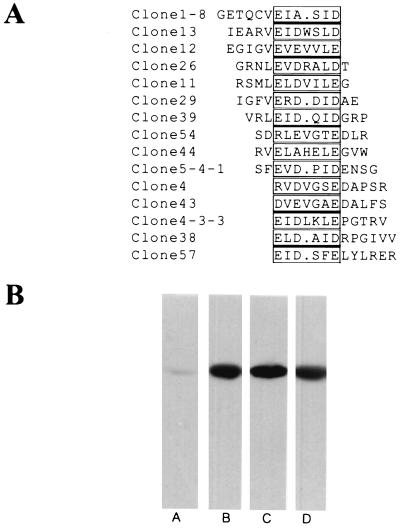
The putative epitope of MAb 6C5-H4CA was determined by panning a random peptide display library. Conceptual translations of DNA sequences corresponding to the random peptides were aligned using the GCG module PILEUP, and a consensus peptide sequence was identified (Fig. 1A). To further verify that MAb 6C5-H4CA recognizes positive clones, thioredoxin peptide loops were subcloned by PCR into the GST fusion vector pGEX-KG (13) for fusion protein purification. Three candidate purified GST fusion proteins chosen from our pilot screening were strongly recognized by the antibody (Fig. 1B), whereas a randomly chosen clone was nonreactive towards the antibody.
FIG. 1.
Putative epitope of MAb 6C5-H4CA antigen. (A) The conceptual translation of the DNA sequence encoding random peptides was aligned using the GCG module PILEUP, and the putative epitope of the antigen was determined. (B) Immunoreactivity of soluble GST fusion proteins was tested on glutathione-agarose-purified material by Western blotting. Lane A, an arbitrarily chosen clone from the fifth round of panning that did not react with MAb 6C5-H4CA; lanes B, C, and D, immunopositive clones 1–8, 4–3-3, and 5–4-1 from panel A, respectively.
CSH1 gene identification.
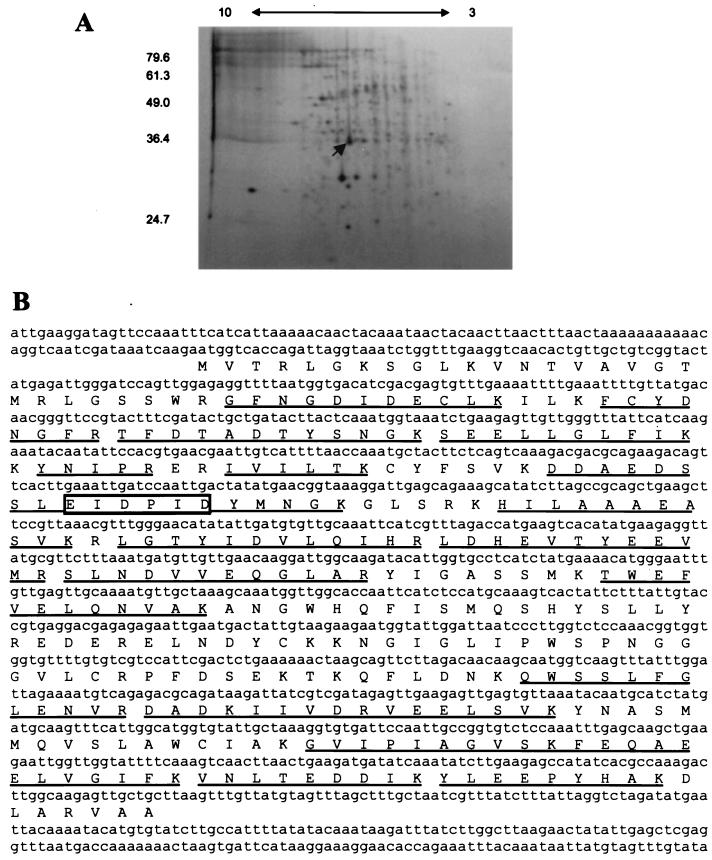
To identify and clone the gene encoding the protein recognized by MAb 6C5-H4CA, the antigen was isolated to apparent homogeneity by two-dimensional gel electrophoresis (Fig. 2B) followed by mass spectrometry. Comparison of mass spectrometry-derived masses of tryptic fragments with the predicted masses of peptides identified from the unannotated C. albicans DNA database (http://www-sequence.stanford.edu/group/candida) indicated a single open reading frame capable of generating the tryptic fragments observed (Fig. 2A), which we herein designated the CSH1 gene. The putative open reading frame encodes a protein with a molecular mass of 38 kDa and with an isoelectric point of approximately 6.0 in accordance with the observed characteristics of the protein isolated from yeast cell walls. A GST fusion protein containing the identified open reading frame was constructed, and this construct strongly reacted with 6C5-H4CA. Furthermore, a six-amino-acid sequence closely resembling the consensus epitope identified in Fig. 1 was seen in the putative genomic sequence. Hydrophilicity was calculated according to the algorithm described by Kyte and Doolittle (20) using the GCG module PEPTIDESTRUCTURE with a window of either four or seven residues. In either analysis, no extensive hydrophobic segments were seen.
FIG. 2.
(A) Purification scheme for MAb 6C5-H4CA antigen isolation. Soluble material released from mechanically broken C. albicans cells was separated by two-dimensional PAGE separation. Coomassie-stained gels were aligned with Western blottings of duplicate gels to identify protein spots reactive with the antibody. The blot was stained with colloidal gold to label protein and was subsequently used for antibody detection. The arrow indicates the protein spot that was recovered for mass spectrometric sequence determination. Molecular masses (in kilodaltons) are shown on the left. The pH gradient of the first dimension is shown on the top. (B) Mass spectrometric analysis of MAb 6C5-H4CA antigen. The DNA sequence of a C. albicans gene product identified by mass spectrometric analysis of the MAb 6C5-H4CA antigen is presented here, with single-letter amino acid symbols of the translated open reading frame underneath. Tryptic fragments identified by mass spectrometry are underlined, and a sequence matching that of the antibody epitope (Fig. 1) is boxed.
Comparing CSH1 and other microbial gene products.
A comparison of the CSH1 sequence with the nonredundant protein database in GenBank indicated that the protein closely resembles a number of microbial gene products. A dendrogram indicating pairwise comparisons between the similar gene products was constructed (Fig. 3). The unannotated C. albicans DNA database contains at least six other paralogs that closely resemble the CSH1 gene product. It is unknown how many of these represent truly distinct gene products which are functionally expressed, unexpressed, or redundant gene duplications or different alleles of homologous loci on sister chromosomes of the obligate diploid C. albicans. Percent similarity between gene products varies from greater than 65% within an organism to between 30 and 40% between organisms. All of the gene products had approximately the same molecular mass. None of the other C. albicans gene products had an amino acid sequence that appeared to closely match the MAb 6C5-H4CA consensus epitope of EIDPID to allow recognition, though the panned epitope consensus sequence is not rigid. However, definitive analysis of the immunoreactivity of C. albicans paralogs towards MAb 6C5-H4CA awaits the cloning and in vitro expression of their gene products. The families of similar C. albicans gene sequences resemble a family of putative aryl alcohol dehydrogenase (AAD) gene sequences from S. cerevisiae (6), with a single divergent sequence (YPL088w) sharing the greatest degree of similarity to the C. albicans sequences. An additional sequence of unknown function from E. coli (GenBank accession no. P77735) also shares significant sequence similarity with the C. albicans sequences.
FIG. 3.
The CSH1 gene product defines a family of related gene products in C. albicans and S. cerevisiae. The C. albicans sequences are referred to by open reading frame designations assigned to them in assembly 6 of the C. albicans database (http://www-sequence.stanford.edu/group/candida). The gene that we refer to as the CSH1 gene is designated orf6.4130 in the database. The CSH1 gene closely resembles six other C. albicans open reading frames, as well as a single S. cerevisiae gene (YPL088w). The S. cerevisiae gene and the C. albicans genes share similarity with a family of putative AADs from S. cerevisiae (AAD genes; YFL057c), as well as a single gene of unknown function from E. coli (p77735). Data are as follows: residues, number of amino acids; M, calculated molecular mass; IFP, calculated isoelectric point of each open reading frame.
Generation and phenotypic analysis of a CSH1 knockout.
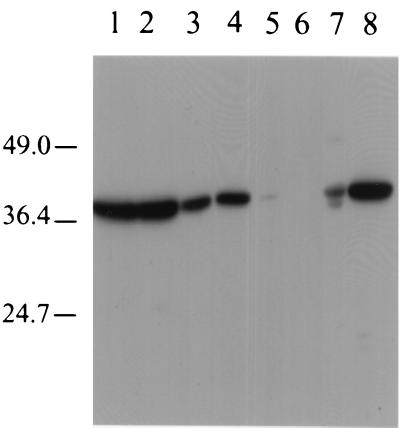
A homozygous knockout strain was generated in cells of strain CAI4 by using an adaptation of the commonly used URA-Blaster technique (7), and a basic phenotypic analysis of the knockout with respect to strain CAI4 was undertaken. One knockout strain which lacks a 38-kDa band on Western blottings of cell lysates was generated (Fig. 4, lane 5), further supporting the contention that the cloned gene sequence encodes the MAb 6C5-H4CA antigen. The absence of immunoreactivity in the knockout strain also suggests that any expressed paralog gene products are unable to be recognized by the antibody. Heterologous expression of the CSH1 gene in S. cerevisiae results in the acquisition of an antigen-reactive band of the correct molecular weight on Western blottings of cell lysates (Fig. 4, lane 7). The knockout strain showed no significant detriment in growth rate or in germination but did show a reproducible decrease in CSH relative to that of the parent strain at 23 and 37°C (Table 1). The knockout strain, when grown at 23°C, also demonstrated statistically significantly decreased levels of adhesion to fibronectin in a static adhesion assay (Table 2). The knockout strain, when grown at 37°C, also showed a decreased measure of adhesion to fibronectin; however, this decrease was judged as being not significant during statistical analysis.
FIG. 4.
Analysis of CSH1 expression in C. albicans and S. cerevisiae strains. Whole-cell lysates were prepared by glass bead breakage, followed by boiling in SDS-PAGE sample buffer plus 2-mercaptoethanol. Samples were loaded onto and run on 12% PAGE gels and were subsequently transferred to nitrocellulose for testing for MAb 6C5-H4CA reactivity by Western blotting. Lane 1, C. albicans strain CAI4; lanes 2, 3, and 4, CAI4-derivative CSH1/csh1 single knockouts; lane 5, CAI4-derivative csh1/csh1 double knockout; lane 6, S. cerevisiae cells expressing CSH1 grown in glucose (expression repressed); lane 7, S. cerevisiae cells expressing CSH1 grown in galactose (expression induced); lane 8, C. albicans strain LGH1095. Molecular masses (in kilodaltons) are shown on the left.
TABLE 1.
Summary of phenotypic characterization of a CSH1 knockout strain
| Strain | Doubling time (min) (mean ± SD)a | 50% culture germination (min) (mean ± SD)b | CSH (mean ± SD)c
|
|
|---|---|---|---|---|
| 23°C | 37°C | |||
| CAI4 | 68 ± 0.2 | 90 ± 2.1 | 96.6 ± 2.9 | 52.8 ± 20.3 |
| csh1/csh1 | 79 ± 0.0 | 86 ± 5.7 | 26.3 ± 13.2 | 7.3 ± 4.6 |
Doubling time represents estimates (in minutes) of the generation time of duplicate exponentially growing liquid cultures.
Percent culture germination was observed at timed intervals, and the time where 50% of the cells in a given sample microscopically demonstrated pseudohyphal or germ tube morphology was extrapolated from the time course.
Relative CSH of individual cells from cultures at a given temperature is presented as the percentage of cells in a sample showing the hydrophobic phenotype. CSH measurements represent samples counted in triplicate in each experiment, and each experiment was repeated at least six times.
TABLE 2.
Percent decrease in adhesion of a csh1/csh1 strain relative to CAI4
| Well coating | Temp of incubation | % Decreasea (mean ± SD) | P valueb |
|---|---|---|---|
| Fibronectin | 23°C | −72.4 ± 12.5 | <0.01 |
| Fibronectin | 37°C | −59.1 ± 26.5 | >0.1 |
Percent decrease was calculated as described in Materials and Methods, plus a standard deviation measurement representing the calculated standard deviation for the mean adherent cells of the knockout strain at a given temperature divided by the mean percent adherent cells for the parent strain.
The P value denoting the statistical significance of the difference between mean adhesion of parent strain in comparison to the knockout was calculated with a paired t test.
DISCUSSION
The cell wall is the primary mediator of interactions between the fungal cell and host substrates. To date, many workers have described individual gene products that affect fungal cell adherence and virulence in both in vitro and in vivo analyses (for review, see reference 5). The majority of these gene products have been identified by virtue of their binding to specific host proteins, such as extracellular matrix components, in in vitro assays. Further characterization has, in many cases, led to the cloning and knockout of individual C. albicans gene products, which typically supports the role of a given gene in adhesion. The number of gene products reported to have significant roles in adhesion and virulence of C. albicans, however, would seem to argue that the effects are too strain specific or assay dependent to have such a significant role. It is likely that in vivo adhesion of fungi is a pleiotropic phenomenon, due to the combined effect of a number of gene products showing cell surface localization.
Cell surface hydrophobicity is proposed to be a general and possibly specific mechanism whereby fungal cells attach to host tissues and the fidelity of attachment is maintained. One possibility is that exposed hydrophobic proteins directly bind to extracellular matrix proteins. Alternatively, CSH may mediate attachment by facilitating and maintaining specific receptor-ligand interactions. Significantly, published reports describing putative virulence and adhesion factors in C. albicans used cells grown under conditions that promote hydrophobic cell growth (3, 4, 8, 19). A particular virulence determinant is either absent or reduced when cells are hydrophilic. This consistency suggests that hydrophobicity may be a key ancillary factor in potentiating virulence. To date, a variety of yeast cell wall antigens have been described as possessing some degree of hydrophobic character, typically by virtue of their retention by hydrophobic interaction chromatography (1, 18, 22, 24, 26). Further analysis of these gene products in adhesion and virulence and their relative contribution to the CSH phenotype for the most part awaits the functional description of genetic knockout and overexpression studies in comparison with other known mediators of adhesion.
The present work describes the cloning and initial functional analysis of a novel C. albicans gene product that has been implicated in cell adhesion. The gene encoding the MAb 6C5-H4CA antigen of C. albicans is one member of a family of related gene products of unknown function. All members of the family are proteins with masses of approximately 35 to 40 kDa and show extensive sequence similarity. It is possible that several of these genes may have allelic differences with the same gene in the parent strain. The primary protein sequence of the CSH1 gene offers few clues as to its cellular function; however, the gene family does show significant homology to a family of putative AADs in S. cerevisiae. The panel of AAD genes has individually and combinatorially been knocked out as part of the sequential functional analysis of the S. cerevisiae genome (6). Surprisingly, loss of function of all of the AAD genes resulted in no obvious phenotypic differences from wild-type strains, complicating easy interpretation of possible functions for the gene family. The CSH1 gene product also shares similar levels of homology with the cytoplasmically localized β subunit of vertebrate voltage-gated potassium channels and more broadly to the oxido-reductase protein superfamily. No corresponding α subunit for the potassium channel sharing similar levels of homology can be identified in the C. albicans or S. cerevisiae genome.
When analyzed by computer modeling methods, the CSH1 gene product does not demonstrate any particular exposed hydrophobic characteristic when using analysis windows set at standard values. These methods do not predict the surface accessibility of short stretches of hydrophobic residues (e.g., two or three residues) and their contribution to the overall hydrophobic behavior of the whole protein. The CSH1p protein does behave as a hydrophobic protein when analyzed by hydrophobic interaction chromatography. The in vivo contribution of the CSH1p protein to the hydrophobic cellular phenotype could be either direct if its hydrophobic character is significant or indirect through interactions with other hydrophobicity-conferring gene products. Further biochemical analysis of purified CSH1p and genetic supplementation in heterologous strains and in C. albicans knockouts would shed some light on the authentic role of CSH1p in affecting cell surface hydrophobicity.
The primary gene sequence suggests that the mature protein is localized to the cytoplasm as opposed to within the cell wall, as the open reading frame lacks a signal sequence directing insertion into the secretory pathway. The observation that a significantly greater quantity of CSH1p is released by mechanical breakage of cells in comparison to limited glucanase digestion (data not shown) indirectly supports this localization. However, there is a growing body of literature detailing the partial surface expression of a wide variety of proteins that would be predicted to show cytoplasmic localization on the basis of sequence analysis (reviewed in reference 5). In the case of CSH1p, several pieces of evidence support the presence of the protein in the wall. First, readily detectable levels of the protein are released from cells by limited lyticase digestion that maintains cell integrity (10, 26). Second, indirect immunofluorescence on intact cells demonstrates at least partial surface localization (unpublished observations). Third, the MAb 6C5-H4CA is effective in blocking adhesion of yeast cells both to extracellular matrix-coated plastic and to epithelial cells (references 12 and 26 and unpublished observations). Finally, at least some portion of CSH1p is accessible to membrane-impermeant protein biotinylation reagents, and labeled CSH1p can be precipitated from cell lysates by immobilized avidin and recognized on Western blottings (unpublished observations.)
In light of the ambiguous physiologic role of CSH1p that is indicated by loss of function of putative S. cerevisiae paralogs and the predicted cytoplasmic expression of the C. albicans gene product, elimination of the C. albicans gene was anticipated to have minimal effects. Knockout of the CSH1 gene was undertaken ultimately to address the potential contribution of the antigen in mediating fungal cell adhesion to tissue. It was anticipated that the redundancy of the CSH1 gene family identified in the C. albicans DNA database might lead to an apparent lack of an obviously different phenotype in csh1/csh1 knockouts relative to those of parent strains. On the other hand, the reproducible effect of the MAb on blocking adhesion argued that loss of CSH1 gene expression might have a significant phenotypic effect. Loss of function of the gene product does have little effect on the overall cell growth rate and viability and does not block the yeast-to-hypha transition that is important in C. albicans species determination. However, knockout of the gene does have a significant and reproducible effect in lowering the number of and relative degree of hydrophobic cells under conditions that promote cell surface hydrophobicity.
Given the large number of fungal cell adhesion molecules reported to date, what distinguishes the CSH1 gene is its ability to affect a broad population phenomenon, namely cell surface hydrophobicity. Involvement of the CSH status of a given fungal cell in attachment and virulence is presently circumstantial and potentially a secondary effect. However, it is apparent that the correlation between hydrophobicity and virulence is significant, and as found here, a specific CSH gene product can be implicated in cell adherence. The identification of the molecular mechanism of cell surface hydrophobicity and the ability to knock out specific gene products allow us to further address the role of these gene products in fungal cell attachment.
ACKNOWLEDGMENTS
We thank Jean Wu and Elizabeth Baker for technical assistance. C. albicans strain CAI4 was kindly provided by William Fonzi. Plasmid pDDB57 for constructing URA-blaster cassettes was generously provided by Aaron Mitchell. Sequence data for C. albicans was obtained from the Stanford DNA Sequencing and Technology Center website (http://www-sequence.stanford.edu/group/candida).
Sequencing of C. albicans was accomplished with the support of the NIDR and the Burroughs Wellcome Fund. This work was supported by PHS grants RO1AI31048 and RO1AI43997 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Alexandre H, Blanchet S, Charpentier C. Identification of a 49-kDa hydrophobic cell wall mannoprotein present in velum yeast which may be implicated in velum formation. FEMS Microbiol Lett. 2000;185:147–150. doi: 10.1111/j.1574-6968.2000.tb09053.x. [DOI] [PubMed] [Google Scholar]
- 2.Antley P P, Hazen K C. Role of yeast cell growth temperature on Candida albicans virulence in mice. Infect Immun. 1988;56:2884–2890. doi: 10.1128/iai.56.11.2884-2890.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouchara J-P, Tronchin G, Annaix V, Robert R, Senet J-M. Laminin receptors on Candida albicans germ tubes. Infect Immun. 1990;58:48–54. doi: 10.1128/iai.58.1.48-54.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calderone R A, Linehan L, Wadsworth E, Sandberg A L. Identification of C3d receptors on Candida albicans. Infect Immun. 1988;56:252–258. doi: 10.1128/iai.56.1.252-258.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaffin W L, Lopez-Ribot J L, Casanova M, Gozalbo D, Martinez J P. Cell wall and secreted proteins of Candida albicans: identification, function, and expression. Microbiol Mol Biol Rev. 1998;62:130–180. doi: 10.1128/mmbr.62.1.130-180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delneri D, Gardner D C, Bruschi C V, Oliver S G. Disruption of seven hypothetical aryl alcohol dehydrogenase genes from Saccharomyces cerevisiae and construction of a multiple knockout strain. Yeast. 1999;15:1681–1689. doi: 10.1002/(SICI)1097-0061(199911)15:15<1681::AID-YEA486>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 7.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gale C A, Bendel C M, McClellan M, Hauser M, Becker J M, Berman J, Hostetter M K. Linkage of adhesion, filamentous growth, and virulence in Candida albicans to a single gene, INT1. Science. 1998;279:1355–1358. doi: 10.1126/science.279.5355.1355. [DOI] [PubMed] [Google Scholar]
- 9.Glee P M, Hazen K C. Aggregation of hydrophobic cell wall proteins of Candida albicans. Colloids Surf. 1995;5:181–188. [Google Scholar]
- 10.Glee P M, Sundstrom P, Hazen K C. Expression of surface hydrophobic proteins by Candida albicans in vivo. Infect Immun. 1995;63:1373–1379. doi: 10.1128/iai.63.4.1373-1379.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glee P M, Masuoka J, Ozier W T, Hazen K C. Presence of multiple laminin- and fibronectin-binding proteins in cell wall extracts of Candida albicans: influence of dialysis. J Med Vet Mycol. 1996;34:57–62. doi: 10.1080/02681219680000091. [DOI] [PubMed] [Google Scholar]
- 12.Glee P M, Cutler J E, Benson E E, Bargatze R F, Hazen K C. Inhibition of hydrophobic protein-mediated Candida albicans attachment to endothelial cells during physiologic shear flow. Infect Immun. 2001;69:779–786. doi: 10.1128/IAI.69.5.2815-2820.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guan K, Dixon J E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 14.Guthrie C, Fink G R, editors. Methods in enzymology. 194. Guide to yeast genetics and molecular biology. New York, N.Y: Academic Press; 1990. [PubMed] [Google Scholar]
- 15.Hazen K C, Hazen B W. A polystyrene microsphere assay for detecting surface hydrophobicity variations within Candida albicans populations. J Microbol Methods. 1987;6:289–299. [Google Scholar]
- 16.Hazen K C, Glee P M. Cell surface hydrophobicity and medically important fungi. Curr Top Med Mycol. 1995;6:1–31. [PubMed] [Google Scholar]
- 17.Hazen K C, Wu J G, Masuoka J. Comparison of hydrophobic properties between Candida albicans and Candida dubliniensis. Infect Immun. 2001;69:2815–2820. doi: 10.1128/IAI.69.2.779-786.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoyer L L, Hecht J E. The ALS5 gene of Candida albicans and analysis of the Als5p N-terminal domain. Yeast. 2001;18:49–60. doi: 10.1002/1097-0061(200101)18:1<49::AID-YEA646>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 19.Klotz S A, Rutten M J, Smith R L, Babcock S R, Cunningham M D. Adherence of Candida albicans to immobilized extracellular matrix proteins is mediated by calcium-dependent surface glycoproteins. Microb Pathog. 1993;14:133–147. doi: 10.1006/mpat.1993.1014. [DOI] [PubMed] [Google Scholar]
- 20.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 21.Liu Q, Li M Z, Liu D, Elledge S J. Rapid construction of recombinant DNA by the univector plasmid-fusion system. Methods Enzymol. 2000;328:530–549. doi: 10.1016/s0076-6879(00)28417-6. [DOI] [PubMed] [Google Scholar]
- 22.Lopez-Ribot J L, Casanova M, Martinez J P, Sentandreu R. Characterization of cell wall proteins of yeast and hydrophobic mycelial cells of Candida albicans. Infect Immun. 1991;59:2324–2332. doi: 10.1128/iai.59.7.2324-2332.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu Z, Murray K S, VanCleave V, LaVallie E R, Stahl M L, McCoy J M. Expression of thioredoxin random peptide libraries on the Escherichia coli cell surface as functional fusions to flagellin: a system designed for exploring protein-protein interactions. Bio/Technology. 1995;13:366–372. doi: 10.1038/nbt0495-366. [DOI] [PubMed] [Google Scholar]
- 24.Marot-Leblond A, Robert R, Loiseau O, Apaire-Marchais V, Senet J-M. Hydrophobic and hydrophilic cell surface (glyco)proteinic components of Candida albicans. J Med Mycol. 2000;10:115–122. [Google Scholar]
- 25.Masuoka J, Hazen K C. Cell wall protein mannosylation determines Candida albicans cell surface hydrophobicity. Microbiology. 1997;143:3015–3021. doi: 10.1099/00221287-143-9-3015. [DOI] [PubMed] [Google Scholar]
- 26.Masuoka J, Wu G, Glee P M, Hazen K C. Inhibition of Candida albicans attachment to extracellular matrix by antibodies that recognize hydrophobic cell wall proteins. FEMS Immunol Med Microbiol. 1999;24:421–429. doi: 10.1111/j.1574-695X.1999.tb01314.x. [DOI] [PubMed] [Google Scholar]
- 27.Shultz M C, Hockman D J, Harkness T A, Garinther W I, Altheim B A. Chromatin assembly in a yeast whole cell extract. Proc Natl Acad Sci USA. 1997;94:9034–9039. doi: 10.1073/pnas.94.17.9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silva T M J, Glee P M, Hazen K C. Influence of cell surface hydrophobicity on attachment of Candida albicans to extracellular matrix proteins. J Med Vet Mycol. 1995;33:117–122. [PubMed] [Google Scholar]
- 29.Wilson R B, Davis D, Enloe B M, Mitchell A P. A recyclable Candida albicans URA3 cassette for PCR product-directed disruptions. Yeast. 2000;16:65–70. doi: 10.1002/(SICI)1097-0061(20000115)16:1<65::AID-YEA508>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 30.Yates J R, Eng J K, McCormack A L, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]