Abstract
The sdeK gene is essential to the Myxococcus xanthus developmental process. We reported previously, based on sequence analysis (A. G. Garza, J. S. Pollack, B. Z. Harris, A. Lee, I. M. Keseler, E. F. Licking, and M. Singer, J. Bacteriol. 180:4628–4637, 1998), that SdeK appears to be a histidine kinase. In the present study, we have conducted both biochemical and genetic analyses to test the hypothesis that SdeK is a histidine kinase. An SdeK fusion protein containing an N-terminal polyhistidine tag (His-SdeK) displays the biochemical characteristics of a histidine kinase. Furthermore, histidine 286 of SdeK, the putative site of phosphorylation, is required for both in vitro and in vivo protein activity. The results of these assays have led us to conclude that SdeK is indeed a histidine kinase. The developmental phenotype of a ΔsdeK1 strain could not be rescued by codevelopment with wild-type cells, indicating that the defect is not due to the mutant's inability to produce an extracellular signal. Furthermore, the ΔsdeK1 mutant was found to produce both A- and C-signal, based on A-factor and codevelopment assays with a csgA mutant, respectively. The expression patterns of several Tn5lacZ transcriptional fusions were examined in the ΔsdeK1-null background, and we found that all C-signal-dependent fusions assayed also required SdeK for full expression. Our results indicate that SdeK is a histidine kinase that is part of a signal transduction pathway which, in concert with the C-signal transduction pathway, controls the activation of developmental-gene expression required to progress past the aggregation stage.
The gram-negative soil bacterium Myxococcus xanthus undergoes multicellular development upon nutrient deprivation. The developmental process requires the coordinated effort of approximately 105 cells and culminates in the formation of the multicellular fruiting body (3). Within the fruiting body, rod-shaped vegetative cells differentiate into environmentally resistant, metabolically quiescent myxospores.
The transition from a colony of vegetatively growing rods to a spore-filled fruiting body requires cell-cell communication to coordinate changes in cell behavior and movement. These changes are facilitated by the production of several cell-derived extracellular signals (2, 6, 21). Production and reception of these signals result in a signal transduction cascade that directs the coordinated expression of specific developmentally regulated genes. Thus far, three signaling systems, the A-, C-, and E-signaling pathways, have been described in detail for M. xanthus (2, 6). To gain a better understanding of the relationship between the various signaling systems and gene expression, a battery of developmentally regulated Tn5lac fusions has been used to examine the dependence of gene expression on each signaling system (14–16, 20). Such analyses have allowed the identification of genetic regulatory circuits used by M. xanthus to control developmental-gene expression. Additionally, this type of genetic approach has allowed the placement of additional genes, such as those responsible for the reception and transduction of the various signals, onto the genetic regulatory circuits.
A-signaling mutants are blocked early in development (about 1 to 2 h poststarvation), and as such, expression of most developmentally regulated Tn5lacZ fusions is impaired. A-signal is a mixture of amino acids and peptides, believed to be produced by extracellular proteolysis, that acts as a quorum sensor (21). Defective C-signaling results in a developmental block at approximately 6 to 8 h post-starvation initiation, and activation of Tn5lac fusions expressed after this time point is diminished (15). C-signal activity requires the presence of a cell surface-associated protein, CsgA, which shares a high degree of sequence similarity and identity with members of the short-chain alcohol dehydrogenase family (22). CsgA is encoded by the csgA gene (7), and csgA mutants fail to aggregate or to sporulate properly (30). Furthermore, csgA mutants do not form the synchronous cell waves, or ripples, that are characteristic of developing cells. Little is known about how C-signal is transmitted. Transduction of C-signal does require FruA, a two-component response regulator (4). The phenotype of cells carrying a fruA mutation is similar to that of csgA mutant cells (32). Thus far, FruA is the only proposed component of the C-signal transduction pathway.
Previous work on early developmental-gene expression has identified a bifurcation early in the M. xanthus developmental pathway. One branch, designated the population starvation branch (31), requires the A-signal quorum sensor for expression of its genes. The genes on the second branch, designated the cellular starvation branch (31), require only the starvation initiation signal (p)ppGpp for expression. By studying mutants, it has been determined that both branches are required for fruiting-body development and sporulation, indicating that the branches eventually converge. Mutations in specific genes in either branch lead to developmental arrest. One such gene, which is on the cellular starvation branch, is sdeK (5, 16, 17), a proposed histidine kinase-encoding gene. Here we provide direct evidence that sdeK encodes a histidine kinase, and we further define the role that sdeK plays in developmental signal transduction. We also propose that SdeK may be the link between the cellular and population pathways and that the SdeK signal transduction pathway works in concert with the C-signal transduction pathway to regulate gene expression through the aggregation stage in M. xanthus development.
MATERIALS AND METHODS
Bacterial strains, transductions, and plasmids.
A complete list of strains and plasmids used in this study is shown in Table 1. Plasmids were propagated in Escherichia coli DH5α (8). M. xanthus DK1622, which is wild type for both fruiting-body formation and sporulation, was chosen as the wild-type strain for this study (12). Strain MS1512 (DK1622 ΔsdeK1) was constructed as described previously for the DK101 derivative MS1503 (5). Myxophages Mx4 ts18 ts27 hrm and Mx8 clp2, which have been described previously (1, 24), were used to transduce the Tn5lacZ transcriptional fusions employed in this study from their parental strains (16) into MS1512. Myxophage Mx8 was used to transduce csgA::Tn5-132 (18) from DK5216 into MS1523. Plasmids pJEF39 and pJEF40 were introduced into MS1512 via electroporation as described previously (28). The resulting kanamycin-resistant transformants were screened by Southern blot analysis (29) for confirmation of a single insertion of the appropriate plasmid at the correct chromosomal location, within the sdeK locus.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5-α | 8 | |
| M. xanthusa | ||
| DK1622 | Wild type | 12 |
| DK2630 | csgA741Ω1519 (Tn5) | 29 |
| DK4292 | Ω4400 (Tn5lac) | 16 |
| DK4294 | Ω4406 (Tn5lac) | 16 |
| DK4296 | Ω4445 (Tn5lac) | 16 |
| DK4300 | sdeK4408::Tn5lac (Ω4408) | 16 |
| DK4323 | sglA1 asgA476 Ω4521 (Tn5lac) | 16 |
| DK4368 | Ω4403 (Tn5lac) | 16 |
| DK4469 | Ω4469 (Tn5lac) | 16 |
| DK4521 | Ω4521 (Tn5lac) | 16 |
| DK5204 | Ω4435 (Tn5lac) | 16 |
| DK5206 | Ω4455 (Tn5lac) | 16 |
| DK5247 | csgA::Tn5-132 (Tetr) Ω4400 (Tn5lac) | 15 |
| DK5279 | Ω4414 (Tn5lac) | 16 |
| DK5287 | csgA::Tn5-132 (Tetr) Ω4414 (Tn5lac) | 15 |
| MS1512 | ΔsdeK1 | This study |
| MS1519 | ΔsdeK1 Ω4400 (Tn5lac) | This study |
| MS1520 | ΔsdeK1 Ω4406 (Tn5lac) | This study |
| MS1522 | ΔsdeK1 Ω4435 (Tn5lac) | This study |
| MS1523 | ΔsdeK1 Ω4414 (Tn5lac) | This study |
| MS1524 | ΔsdeK1 Ω4455 (Tn5lac) | This study |
| MS1526 | ΔsdeK1::pJEF39 tandem duplication; sdeK+ ΔsdeK1 | This study |
| MS1527 | ΔsdeK1::pJEF40 tandem duplication; sdeKH286A ΔsdeK1 | This study |
| Plasmids | ||
| pBluescript SKII | Ampr | Stratagene |
| pBGS18 | Kanr | 33 |
| pTrcHisB | Ampr | Invitrogen |
| psdeK1 | Ampr; pBluescript SKII (cut with XmaI-HindIII) containing 1.6-kb AgeI-HindII fragment with sdeK ORF | This study |
| psdeK2 | Ampr; pTrcHisB (cut with BamHI-HindIII) containing ΩsdeK ORF on a 1.6-kb BamHI-HindIII fragment | This study |
| psdeK2.5 | Ampr; psdeK2 with codon substitution (GCG→CAC) in sdeK ORF so as to produce His-SdeKH286A | This study |
| pJEF39 | Kanr; pBGS18 (cut with SmaI-HindIII) containing sdeK ORF with 1 kb of upstream DNA and 500 bp of downstream DNA on a 3.1-kb StuI-HindIII fragment | This study |
| pJEF40 | Kanr; pJEF39 with codon substitution (GCG→CAC) in sdeK to encode SdeKH286A | This study |
All M. xanthus strains are derivatives of DK1622.
Media for growth, transductions, and development.
The M. xanthus strains were grown with vigorous shaking at 32°C in CTT broth (1% Casitone [Difco Laboratories], 10 mM Tris-HCl [pH 7.6], 1 mM KH2PO4, 8 mM MgSO4) or on CTT plates containing 1.5% agar (Difco). CTT plates were supplemented with 40 μg of kanamycin monosulfate (Sigma) per ml or 12.5 μg of oxytetracycline (Sigma) per ml as required. Myxospores and transduced cells were suspended in CTT soft agar (CTT broth containing 0.7% agar) prior to being plated. E. coli DH5α was grown with shaking at 37°C in Luria broth or on Luria-Bertani agar medium as described previously (29). Luria broth and Luria-Bertani agar medium were supplemented with 40 μg of kanamycin monosulfate or 50 μg of ampicillin (Sigma) per ml as needed. Stocks of the generalized transducing phages Mx4 and Mx8 were prepared on donor cells grown with vigorous shaking at 32°C in CYE broth (1% Casitone, 0.5% yeast extract [Difco Laboratories], 8 mM MgSO4). M. xanthus development was conducted at 32°C on TPM agar medium, which is composed of TPM buffer (10 mM Tris-HCl [pH 7.6], 1 mM KH2PO4, 8 mM MgSO4) and 1.5% agar.
Development, sporulation efficiencies, rippling assays, and β-galactosidase expression.
Vegetatively growing M. xanthus cells were harvested during mid-exponential phase by centrifugation (7,700 × g for 5 min) and plated for development as described previously (16). Progress of fruiting-body development was observed visually by using a dissecting microscope (Wild-Heerburg). Determination of sporulation efficiencies was conducted as described previously (34).
For rippling experiments, cells were grown to exponential phase and plated on CF agar [10 mM Tris-HCl (pH 7.6), 8 mM MgSO4, 1 mM potassium phosphate, 0.2 mg of (NH4)2SO4/ml, 150 mg of Casitone/ml, 1.5% Difco agar] as described previously (23). Cells were observed visually for rippling behavior between 16 and 20 h after being plated.
M. xanthus cells plated for development as described above were harvested as described previously, and β-galactosidase activity (1 U being defined as the amount of enzyme required to produce 1 nmol of o-nitrophenol min−1 mg of protein−1) assays were performed on quick-frozen cell extracts as described previously (16).
Developmental rescue experiments.
Wild-type DK1622 cells and the isogenic ΔsdeK1 derivatives were prepared for developmental assay as described above. Cells suspended in TPM medium were mixed in equal proportions, applied to TPM agar in 20-μl aliquots, and incubated at 32°C for 5 days.
A- and C-factor assays.
A-factor assays were conducted, as described previously, by the in situ method (16). One unit of A-factor is defined as the amount required to produce 1 U of β-galactosidase activity above background (21). C-factor assays were performed by a developmental mixing protocol. Cells defective for C-signal production (DK2630) were mixed with an equal proportion of DK1512 (ΔsdeK1) cells, and the cell mixture was plated for development as described above. Germinated spores (100-colony sample size) were then scored for kanamycin resistance, to determine lineage, by patching colonies onto CTT plates containing kanamycin.
Overproduction and purification of His-SdeK.
His-SdeK was overproduced using the plasmid pTrcHisB (Invitrogen). The putative sdeK open reading frame (ORF) was cloned into pTrcHisB as a 1.6-kb BamHI-HindIII fragment from psdeK1 to form psdeK2 (Table 1). An additional 33 bases at the 5′ end of the putative sdeK ORF were included in the clone because of restriction site availability. As a result, SdeK contained an additional 11 amino acids at its N terminus. The overexpressed SdeK contained an N-terminal polyhistidine tag. E. coli cells containing the plasmid psdeK2 were grown as described above and induced to express His-SdeK by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) when culture turbidity reached an optical density at 600 nm of 0.3. Protein was overproduced for 3 h, at which point the cells were pelleted (1,000 × g for 10 min) and stored at −20°C.
Frozen cell pellets were thawed at room temperature, and the cells were resuspended in STET buffer (50 mM Tris [pH 7.7], 8% sucrose, 5% Triton X-100, 50 mM EDTA, 1 mM dithiothreitol [DTT]) supplemented with 500 μM phenylmethylsulfonyl fluoride just prior to cell lysis. Cells were lysed via three passes at 13,000 lb/in2 through a French pressure cell (American Instruments Company). Cell debris was pelleted in a Sorvall MC12C microcentrifuge at 12,000 rpm for 5 min, and the overexpressed protein was found in the pellet in the form of inclusion bodies. The pellet was partially solubilized by 30 min of incubation, with shaking, in a solution containing 2 M guanidinium hydrochloride (Sigma) and 100 mM Tris (final pH, 8.2). The insoluble material was pelleted and solubilized in a solution consisting of 4 M guanidinium hydrochloride, 100 mM Tris, and 100 mM DTT (final pH, 8.2) by 30 min of shaking at room temperature. The protein solution was brought to pH 3.0 by the addition of 1 M hydrochloric acid and dialyzed overnight in 100 mM acetic acid with 1 mM DTT (25). The protein was stored frozen in 100-μl aliquots at −80°C until needed.
Phosphorylation assays.
Frozen aliquots of His-SdeK or His-SdeKH286A protein were thawed to room temperature and diluted 10-fold in 20 mM Tris (pH 8.7). The protein was incubated 1 h on ice to allow refolding. The phosphorylation assays were performed with a 30-μl reaction volume containing 50 mM Tris (pH 7.6), 50 mM KCl, 10 mM MgCl2, 1 mM DTT, and 0.1 mM EDTA. Refolded protein was added to a final concentration of 0.5 μM. The reaction was initiated by the addition of 30 μCi of [γ-32P]ATP (6,000 Ci/mmol; NEN Life Science Products, Inc.). The reaction mixture was incubated at room temperature for 10 min, and the reaction was stopped by addition of 0.2 volume of 5× protein loading buffer (250 mM Tris [pH 6.8], 10% glycerol, 0.02% bromophenol blue, 1% sodium dodecyl sulfate [SDS], 150 mM β-mercaptoethanol). The samples were heated for 3 min at 55°C and subsequently loaded onto an SDS–12% polyacrylamide gel. Electrophoresis was performed at a constant 200 V for approximately 1 h. The gel was subsequently soaked in approximately 15 ml of SDS running buffer (100 mM Tris base, 800 mM glycine, 35 mM SDS) twice, for 20 min each time, to remove any unincorporated [γ-32P]ATP. Labeled protein was detected with a PhosphorImager (Storm 840; Molecular Dynamics).
MBP-EnvZ (10) was used as the positive control for the phosphorylation reactions. The MBP-EnvZ phosphorylation reaction was performed as described above, with the following changes. Protein stored in 50% glycerol at −20°C was added to a final concentration of 0.4 μM. Unlabeled ATP (Sigma) was added to the reaction mixture to a final concentration of 830 μM. The labeling reaction was initiated by the addition of 10 μCi of [γ-32P]ATP.
Site-directed mutagenesis.
The plasmid psdeK2 (Table 1) was used as the template for mutagenesis. The single-stranded primers used to generate His-SdeKH286A and SdeKH286A within psdeK2.5 and pJEF40, respectively, were primer 1 (5′-GGCGTGCTGGGCGCGGACCTGGGCAATCC-3′) and primer 2 (5′-GGATTGCCCAGGTCCGCGCCCAGCACGCC-3′) (Fisher). Mutagenesis was carried out by the method described by the manufacturer of the ExSite kit (Stratagene).
RESULTS
SdeK is a histidine kinase.
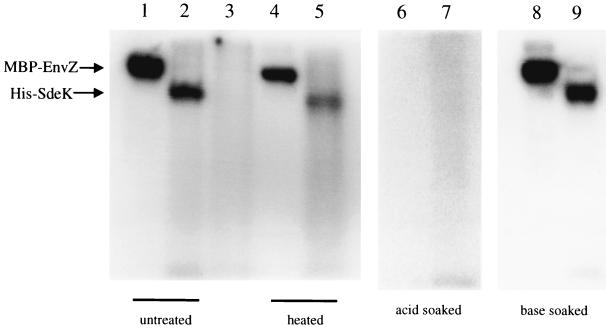
As reported previously, the ORF contains the Ω4408 Tn5lacZ insertion, which disrupts a gene that encodes a histidine kinase, as determined by sequence analysis (5). The defining characteristic of histidine kinases is an autophosphorylation activity in which ATP is used to phosphorylate a conserved histidine residue (9, 27). To determine whether SdeK possesses this activity, an N-terminal polyhistidine-tagged SdeK fusion protein (His-SdeK) was constructed and purified from E. coli DH5α. When overexpressed in E. coli, the His-SdeK fusion protein formed inclusion bodies, which were then purified, solubilized, and refolded as described in Materials and Methods. The His-SdeK purification scheme yielded a protein preparation of approximately 70 to 80% purity, as determined by Coomassie staining of the polyacrylamide gel after electrophoresis (data not shown). The soluble, refolded protein preparation was then assayed for autophosphorylation in the presence of [γ-32P]ATP. A parallel assay was performed on MBP-EnvZ (a gift from M. Igo) as a positive control. Figure 1 shows that both the His-SdeK fusion protein (lane 2) and the MBP-EnvZ control (lane 1) were labeled in the presence of [γ-32P]ATP.
FIG. 1.
His-SdeK is a histidine kinase. Phosphorylation reactions with MBP-EnvZ (lane 1), His-SdeK (lane 2), and His-SdeKH286A (lane 3) are described in Materials and Methods. H268 is required for phosphorylation of His-SdeK (lane 3). Two sets of MBP-EnvZ and His-SdeK phosphorylation reactions were performed; one product set was incubated for 10 min at 100°C prior to electrophoresis (lanes 4 and 5). MBP-EnvZ (lane 4) and His-SdeK (lane 5) were stable to 10 min of heat treatment at 100°C (compare lanes 1 and 2 with lanes 4 and 5, respectively). Phosphoproteins were visualized by phosphorimaging. Stability of the phosphorylated forms of His-SdeK and MBP-EnvZ to acid and to base treatment was assayed (lanes 6 through 9). After phosphorylation and electrophoresis, two identical gels were treated with 0.2 M HCl (lanes 6 and 7) or 1.0 M NaOH (lanes 8 and 9).
Phosphorylated His-SdeK displays lability characteristics indicative of a histidine phosphate label.
The stability of the phosphorylated His-SdeK fusion protein was assayed at a high temperature and under acidic and basic conditions to determine whether the phosphorylated His-SdeK protein was phosphorylated on a histidine residue. Phosphohistidyl residues are heat and acid labile but base stable, while phosphoester groups, such as phosphoserines and phosphotyrosines, are heat and acid stable but base labile (1a, 27). Heating of phosphorylated His-SdeK at 100°C for 10 min resulted in a significant decrease in phosphorylation activity, as determined by phosphorimaging (Fig. 1, lane 5). The loss of phosphorylated protein was not due to heat-induced protein degradation (data not shown). Furthermore, after incubation of an SDS-polyacrylamide gel containing phosphorylated His-SdeK for 30 min at 50°C in pH 4 buffer, phosphorylated protein was no longer detectable by phosphorimaging (Fig. 1, lane 7). Coomassie staining of the gel indicated that the loss of labeled protein did not result from protein degradation. Incubation in basic buffer (see Materials and Methods) yielded no change in intensity of the labeled His-SdeK (Fig. 1, lane 9). A phosphorylated MBP-EnvZ control displayed similar characteristics in each experiment (Fig. 1, lanes 4, 6, and 8).
H286 in SdeK is required for activity.
It was determined, based on protein sequence alignment, that the putative site of autophosphorylation of SdeK is H286 (5). To test whether this histidine is required for autophosphorylation of His-SdeK, the residue was changed to an alanine by site-directed mutagenesis (see Materials and Methods). The gene encoding the SdeK mutant protein in which an alanine is substituted for the histidine at position 286 (SdeKH286A) is carried on the plasmid psdeK2.5 (Table 1). Sequencing of psdeK2.5 verified that no additional mutations were introduced during the site-directed mutagenesis. The corresponding fusion protein, His-SdeKH286A, was then expressed, purified, and assayed for activity in parallel with the wild-type His-SdeK fusion. Equal amounts of wild-type and mutant protein were assayed in each reaction. The His-SdeKH286A fusion protein exhibited no detectable autophosphorylation activity (Fig. 1, compare lanes 2 and 3). Thus, H286 is essential for His-SdeK activity.
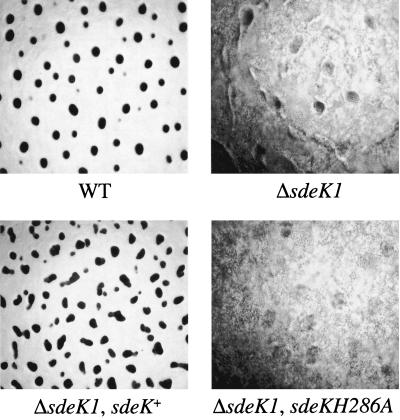
As a further test of the importance of H286 in SdeK, the sdeKH286A mutant was assayed for its ability to complement the developmental and sporulation defects of MS1512, a strain that carries a deletion in sdeK (ΔsdeK1). The plasmid pJEF40, which carries a 3-kb fragment containing the sdeKH286A ORF and the chromosomal region required for its full expression (5), was used to construct a tandem duplication at the sdeK locus, yielding strain MS1527. A single insertion of the plasmid into the correct chromosomal locus was confirmed by Southern analysis (data not shown). As a control, plasmid pJEF39, carrying a fragment that was identical with the exception of containing the wild-type sdeK locus, was also analyzed for complementation of the ΔsdeK1 phenotype. The ΔsdeK1 phenotype persisted in MS1527 (<104 spores/ml) (Fig. 2), while the wild-type copy was able to complement the developmental block (Fig. 2) and sporulation defect (108 spores/ml). Together, these studies provide in vivo evidence that H286 is essential for SdeK activity.
FIG. 2.
The putative phosphorylation site, H286, is required for in vivo activity of SdeK. M. xanthus strains DK1622 (wild type [WT]), MS1512 (ΔsdeK1), MS1527(ΔsdeK1 sdeK+), and MS1526 (ΔsdeK1 sdeKH286A) were developed for 3 days on TPM agar. Photographs were taken 72 h after development initiation. Magnification, ×96.
Is sdeK required for extracellular signaling?
If the defects in fruiting-body formation and sporulation in the sdeK mutant strain are caused by a deficiency in extracellular signal production, then codevelopment with wild-type cells should rescue the developmental phenotype. Isogenic wild-type DK1622 cells and DK4300 (sdeK::Tn5lac) cells were mixed at a 1:1 ratio and allowed to codevelop on TPM agar. After 3 days, spores were harvested and assayed for viability. To determine whether any of the spores were derived from DK4300 cells, 100 colonies were scored on CTT plates containing kanamycin. None of the 100 colonies scored after codevelopment was kanamycin resistant, indicating that there was no rescue of the DK4300 sporulation defect by codevelopment with wild-type cells.
The ΔsdeK mutant produces both A- and C-signal.
While the above-described experiments demonstrated that the phenotype of an sdeK-null mutant cannot be rescued extracellularly, the experiments did not specifically address whether ΔsdeK1 cells are defective in the production of specific extracellular signals. Therefore, we assayed the ability of the ΔsdeK1 mutant to produce A- and C-signal. A-signal production by wild-type, asgA476, and ΔsdeK1 cells was assayed directly by measuring the amount of A-factor released into the cell suspension (20, 28). Conditioned medium was assayed for A-factor activity by using the A-factor-dependent Ω4521 Tn5lacZ gene fusion as a reporter as previously described (20, 28). The results, shown in Table 2, indicated that the levels of A-factor produced by ΔsdeK1 cells were equivalent to those produced by wild-type cells, thereby demonstrating that ΔsdeK1 cells are capable of producing A-factor.
TABLE 2.
SdeK is not required for A-signal productiona
| Strain assayed | A-signal produced (U) |
|---|---|
| Wild type | 508 ± 47 |
| asgA476 | 45 ± 3 |
| ΔsdeK1 | 505 ± 83 |
Wild-type, ΔsdeK1 mutant, and asgA476 mutant strains were tested for the ability to produce A-factor as described in Materials and Methods. Values are averages of data from three independent experiments ± standard deviations.
To determine whether ΔsdeK1 cells are able to produce C-factor, the ΔsdeK1 mutant was tested for its ability to ripple. The rippling phenomenon associated with early development is known to require C-signaling (23). When ΔsdeK1 cells were plated for development on CF agar, they were observed to ripple to the same extent and in the same time period as wild-type cells (data not shown). As a further test of C-signal production by the ΔsdeK1 cells, the mutant cells were assayed for the ability to rescue the development and sporulation phenotype of a csgA mutant. Isogenic DK1622 wild-type and ΔsdeK1 cells were codeveloped with the kanamycin-resistant DK1622 csgA strain DK2630 (csgA741). The ΔsdeK1 strain was able to rescue development of the csgA741 strain during coincubation. The sporulation efficiency of each mixture after 3 days of development is presented in Table 3. When ΔsdeK1 cells were codeveloped with csgA741 cells, the csgA741 cells sporulated at or near wild-type levels. To determine whether any of these viable spores were derived from the ΔsdeK1 strain as a result of developmental rescue by the csgA741 mutant, colonies were scored on CTT plates containing kanamycin. None of the 100 colonies scored after codevelopment of the ΔsdeK1 and csgA741 strains was kanamycin resistant, indicating that the ΔsdeK1 cells rescued the csgA741 cells for development and sporulation, not vice versa. These data demonstrate that ΔsdeK1 cells are able to produce C-signal.
TABLE 3.
The sdeK mutant rescues a csgA mutant in codevelopment assaysa
| Strain(s) plated for development | Total no. of viable spores/mlb | No. of viable csgA741 spores/ml |
|---|---|---|
| Wild type | 1.5 × 108 | NAc |
| csgA741 | 3.2 × 104 | 3.2 × 104 |
| ΔsdeK1 | 4.0 × 103 | NA |
| Wild type + csgA741 (1:1) | 9.4 × 107 | 5.8 × 107 |
| ΔsdeK1 + csgA741 (1:1) | 1.1 × 108 | 1.1 × 108 |
Spores were collected after 3 days of codevelopment as described in Materials and Methods. Spores were plated on CTT, and germinated cells were patched onto CTT plates containing kanamycin to determine strain derivation, as described in Materials and Methods.
Standard deviations were between 15 and 23%.
NA, not applicable.
SdeK is required for activation of developmental-gene expression.
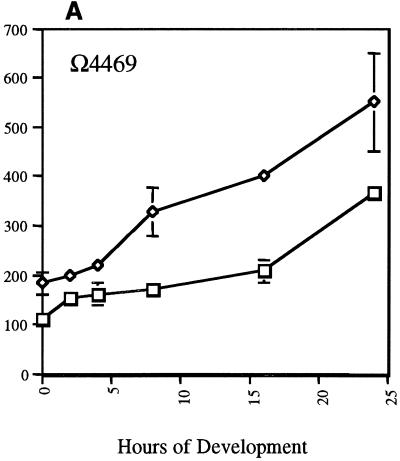
To further define the role played by SdeK in M. xanthus development and to determine the timing of its action, expression of several Tn5lac reporter fusions in the ΔsdeK1 mutant background was studied. Nine developmentally regulated Tn5lacZ fusions were transduced into the ΔsdeK1 strain. All transductants were verified for single-copy insertion at the appropriate location by Southern analysis prior to use. Isogenic wild-type and ΔsdeK1 strains carrying each of the nine Tn5lac reporter fusions individually were assayed for developmental expression of each fusion. Two of the fusions, Ω4455 and Ω4469, like sdeK, lie on the cellular starvation pathway branch and require only the starvation initiation signal for expression. They are expressed independently of A- and C-signal. The Ω4455 fusion, which is activated between 1 and 2 h poststarvation, is expressed at wild-type levels in the ΔsdeK1 background (data not shown), while Ω4469 expression, which is activated at approximately 4 h poststarvation, is increased about twofold over wild-type levels (Fig. 3A).
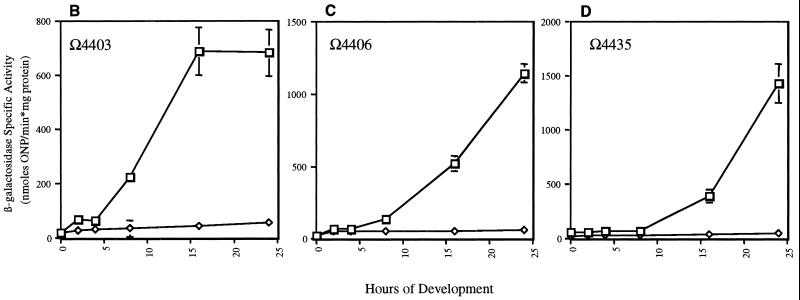
FIG. 3.
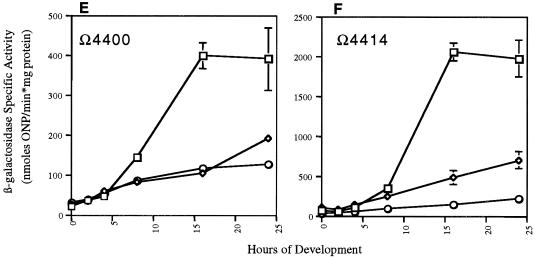
Effects of ΔsdeK1-null genotype on expression of Tn5lac fusions Ω4469 (A-signal-independent fusion) (A), Ω4403 (C-signal-dependent fusion) (B), Ω4406 (C-signal-dependent fusion) (C), Ω4435 (C-signal-dependent fusion) (D), Ω4400 (partially C-signal dependent fusion) (E), and Ω4414 (partially C-signal dependent fusion) (F). Tn5lacZ fusions were transduced into ΔsdeK1 strain MS1512 as described in Materials and Methods. Cells were plated for development, and mean β-galactosidase specific activities were determined from experiments done in triplicate at various time points as described in Materials and Methods. Error bars represent standard deviations of the means. Each triplicate experiment was conducted two or three times, and the data presented are from one representative experiment. Wild type, □; ΔsdeK1, ⋄; csgA, ○. ONP, o-nitrophenol.
The next fusions examined, Ω4445 and Ω4521, lie on the population starvation branch and require both (p)ppGpp and A-signal for expression. Both fusions are expressed at wild-type levels throughout development. This is consistent with the fact that ΔsdeK1 cells are able to produce A-signal. These data indicate that these cells are able to receive and transduce A-signal, at least up until the time at which these fusions are expressed.
The final set of fusions tested, Ω4400, Ω4403, Ω4406, Ω4414, and Ω4435, requires starvation initiation signal and both A- and C-signal for full expression. The expression patterns of these fusions in the ΔsdeK1 background fall into two classes. Class I fusions, which include Ω4403, Ω4406, and Ω4435, are essentially not expressed in the ΔsdeK1 mutant (Fig. 3B to D, respectively), indicating that sdeK is absolutely required for activation of these genes. Expression of the class II fusions, which include Ω4400 and Ω4414, is only partially affected by the ΔsdeK1 mutation. Expression of Ω4400 in the ΔsdeK1 background is decreased approximately fourfold in comparison to wild-type expression (Fig. 3E). Similarly, a fourfold decrease in expression of the Ω4414 fusion in the ΔsdeK1 background in comparison to wild-type expression was observed (Fig. 3F), which is similar to previous results (19).
The partial dependence observed for Ω4400 and Ω4414 expression in the sdeK-null background was also seen when expression of these fusions was examined in a csgA-null background (15). So that a direct comparison could be made, we assayed Ω4400 and Ω4414 expression in the csgA-null background ourselves. We found that the Ω4400 expression pattern in the csgA mutant background was similar to that seen in the ΔsdeK1 background (Fig. 3E). Expression of the Ω4414 fusion in the csgA background was decreased approximately eight- and twofold in comparison to the wild-type and ΔsdeK1 backgrounds, respectively (Fig. 3F). These data suggest that the ΔsdeK1 mutant may be defective in C-signal reception and/or transduction.
DISCUSSION
The sdeK gene was originally defined by the Ω4408 Tn5lacZ transcriptional fusion. We previously reported that sdeK expression is dependent on (p)ppGpp and becomes activated when cells begin to enter stationary phase (5), which resulted in its classification as a starvation-dependent gene. Although sdeK is expressed immediately upon starvation, disruption of sdeK results in both a block in M. xanthus development approximately 6 to 8 h poststarvation at the early aggregation stage and a strong sporulation defect (5, 15). Analysis of the sdeK sequence led to the conclusion that sdeK encodes a histidine kinase. Here we have presented biochemical and genetic data demonstrating that SdeK is a histidine kinase and have further characterized the role that SdeK plays in M. xanthus development.
SdeK shares a high degree of sequence similarity and identity with the PhoR family of histidine kinases (5). The feature that best defines a histidine kinase is its ability to autophosphorylate on a conserved histidine residue (9, 27). We constructed an SdeK fusion protein with a polyhistidine tag at its N terminus (His-SdeK) and found that it was phosphorylated when incubated with [γ-32P]ATP. Based on sequence analysis of SdeK, H286 was identified as the putative phosphorylation site. When this residue was changed to an alanine by site-directed mutagenesis, the protein was no longer phosphorylated in vitro, nor was it able to complement the ΔsdeK1 phenotype. Thus, H286 is required for in vitro phosphorylation of His-SdeK and in vivo function of SdeK. Phosphorylated His-Sdek also shows the heat and acid lability and base stability indicative of histidine kinases. Analysis of these data has led us to conclude that SdeK is a histidine kinase.
SdeK and C-signal both exert control over a similar set of genes.
The fact that M. xanthus development does not progress beyond the aggregation stage (6 to 8 h poststarvation) when sdeK is disrupted indicates that SdeK plays a critical role in development. We demonstrated that sdeK cells are able to produce both A- and C-signal to wild-type levels and cannot be rescued extracellularly when codeveloped with wild-type cells. These data indicate that SdeK is not involved in the production of extracellular signals.
Although it was evident that SdeK is not required for signal production, the role of sdeK in signal reception and transmission was not clear. To address this issue, we assayed the effect of ΔsdeK1 on the expression of three classes of Tn5lac fusions: two starvation dependent and A-signal independent, two A-signal dependent, and five C-signal dependent. The studies presented in this paper demonstrated two effects of sdeK on developmental-gene expression. First, sdeK may play a negative role in the regulation of some starvation-dependent and A-signal-independent fusions, as determined based on the twofold increase in expression of Ω4469 in the ΔsdeK1 background. Although this is only a twofold effect, it is possible that the SdeK signal transduction pathway modulates a negative regulator of Ω4469 gene expression. Alternatively, the effect could be indirect, due to a general disruption of the early developmental program by ΔsdeK1. The significance of the increase in Ω4469 expression is difficult to ascertain at this time and will be a focus of the continued study of this gene.
Second, sdeK was found to be required for the expression of all C-signal-dependent genes examined. Expression of three of these fusions, Ω4403, Ω4406, and Ω4435, was ostensibly eliminated in the ΔsdeK1 strain. All three fusions have previously been shown to be absolutely dependent on C-signal for expression (15). The Ω4400 fusion in the ΔsdeK1 background was expressed at approximately 25% of wild-type levels, which is the same decrease observed for this fusion in an isogenic csgA mutant (Fig. 3E). This is consistent with reports from other laboratories stating that expression of Ω4400 is only partially dependent on C-signal (15). Finally, expression of the Ω4414 fusion, which defines the devTRS locus, was decreased about fourfold in the ΔsdeK1 background and eightfold in the csgA-null background (Fig. 3F). Although the observed effect on devTRS expression by a csgA mutation was stronger than that observed previously, expression was still induced approximately fivefold over vegetative levels. This is consistent with the conclusion that devTRS expression is partially dependent on csgA (15).
What role does the SdeK signal transduction pathway play in development?
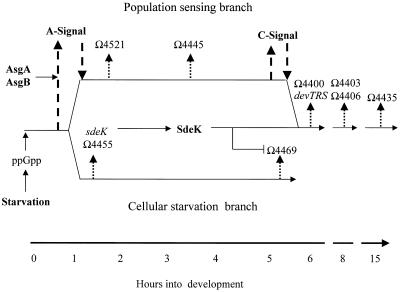
We have previously proposed that M. xanthus cells monitor their nutritional status by monitoring their translation efficiency via relA and the stringent response (31). Our current model for the regulation of early developmental-gene expression is depicted in Fig. 4. Upon activation of the developmental program, a bifurcation in the pathway occurs. One branch, designated the population-sensing branch, leads to A-signal production and reception; in addition, this branch controls production and reception of C-signal, a second extracellular signaling system required for aggregation (13, 23). The second branch acts independently of any extracellular signals and only requires starvation for activation of gene expression (31). This branch has been designated the cellular starvation branch. Genes under the control of this branch include Ω4469 and sdeK (31). Based on the fusion data presented in this paper, we confirm that these two branches converge at approximately 6 to 8 h post-development initiation (15). The question now becomes the following: what is the mechanism by which SdeK and its signal transduction pathway interact with the C-signaling pathway to alter gene expression at this point in development?
FIG. 4.
Model for early developmental-gene expression. Development is initiated by amino acid limitation, which activates the stringent response mediated by the ribosome-associated protein RelA. RelA activation induces an increase in the level of (p)ppGpp, which activates the cellular starvation pathway and, in conjunction with AsgA and AsgB, leads to the production of A-signal (upward-pointing, broadly dashed arrow on left) and activation of the population-sensing pathway. C-signal, a second extracellular signal required for regulation of coordinated cell movement and gene expression, lies on the population-sensing branch and is depicted by the upward-pointing, broadly dashed arrow on the right. Genes expressed along the two different branches are designated by thinner, more narrowly dashed arrows at the time of activation. The two branches merge at approximately 6 h, at which point both C-signal (downward-pointing, broadly dashed arrow on right) and SdeK become necessary for gene expression. The model also shows the inhibitory effect (bent tipless arrow) of SdeK on the expression of Ω4469.
One possible model holds that the SdeK and C-signaling signal transduction pathways are integrated through a common regulator. Previous studies have shown that C-signal reception and transmission occur through the response regulator FruA (4, 26, 31). It is possible that SdeK modulates the phosphorylation state of FruA and, therefore, the activity of FruA. However, this does not appear to be the case, based on two experiments. First, we examined whether phosphorylated His-SdeK could phosphorylate FruA in vitro. No phosphotransfer was observed using either purified FruA or extracts from an E. coli strain that overproduces FruA (data not shown). Second, Ellehauge and Søgaard-Anderson examined whether a constitutively active form of FruA (FruAD59E) could bypass, and therefore suppress, the ΔSdeK1 phenotype. Phenotypically, the resulting double mutant (ΔSdeK1 FruAD59E) behaves like the ΔsdeK1 parent (E. Ellehauge and L. Søgaard-Anderson, personal communication). The constitutively active form of FruA does, however, complement the developmental and sporulation defects of a fruA mutant strain (4). These results indicate that FruA is not the SdeK cognate response regulator and that SdeK acts either downstream or independently of FruA to control developmental-gene expression.
An alternative model holds that the SdeK and the C-signaling pathways coordinately control gene expression by acting independently on genes expressed relatively early in development (e.g., Ω4400 and devTRS). The separate C-signaling and SdeK signal transduction pathways might subsequently merge to form a single pathway, thus affecting expression of subsequently expressed genes (e.g., Ω4403, Ω4406, and Ω4435) via a common regulatory component. There is circumstantial evidence implicating the devTRS operon as a possible candidate for the gene(s) that codes for this common factor. Julien et al. have reported that the abolishment of expression of several Tn5lac fusions after devTRS is expressed in a devTRS-null background (11).
It is clear that the identification of the SdeK cognate response regulator would significantly aid our understanding of how the SdeK signal transduction pathway and the C-signal pathway coordinately regulate developmental-gene expression at the 6- to 8-h juncture. We have used a variety of biochemical approaches in an attempt to identify the SdeK cognate response regulator; however, these approaches have not been successful. Genetic methods are presently being employed to identify this gene.
ACKNOWLEDGMENTS
We thank M. Igo for the gift of MBP-EnvZ and for advice on the phosphorylation assays and E. Baldwin for advice on His-SdeK purification. We also thank T. Powers for critical reading of the manuscript.
This work was supported in part by a National Institutes of Health grant (GM54592) to M.S.
REFERENCES
- 1.Campos J M, Geisselsoder J, Zusman D R. Isolation of bacteriophage Mx4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978;11:167–178. doi: 10.1016/0022-2836(78)90431-x. [DOI] [PubMed] [Google Scholar]
- 1a.Cullen P J, Bowman W C, Kranz R G. In vitro reconstitution and characterization of the Rhodobacter capsulatus NtrB and NtrC two-component system. J Biol Chem. 1996;271:6530–6536. doi: 10.1074/jbc.271.11.6530. [DOI] [PubMed] [Google Scholar]
- 2.Downard J, Ramaswamy S V, Kil K-S. Identification of esg, a genetic locus involved in cell-cell signaling during Myxococcus xanthus development. J Bacteriol. 1993;175:7762–7770. doi: 10.1128/jb.175.24.7762-7770.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dworkin M, Kaiser D. Cell interactions in myxobacterial growth and development. Science. 1985;230:18–24. doi: 10.1126/science.3929384. [DOI] [PubMed] [Google Scholar]
- 4.Ellehauge E, Norregaard-Madsen M, Søgaard-Andersen L. The FruA signal transduction protein provides a checkpoint for the temporal co-ordination of inter-cellular signals in Myxococcus xanthus development. Mol Microbiol. 1998;30:807–817. doi: 10.1046/j.1365-2958.1998.01113.x. [DOI] [PubMed] [Google Scholar]
- 5.Garza A G, Pollack J S, Harris B Z, Lee A, Keseler I M, Licking E F, Singer M. SdeK is required for early fruiting body development in Myxococcus xanthus. J Bacteriol. 1998;180:4628–4637. doi: 10.1128/jb.180.17.4628-4637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hagen D C, Bretscher A P, Kaiser D. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev Biol. 1978;64:284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- 7.Hagen T J, Shimkets L J. Nucleotide sequence and transcription products of the csg locus of Myxococcus xanthus. J Bacteriol. 1990;172:15–23. doi: 10.1128/jb.172.1.15-23.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–560. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 9.Hoch J A. Two-component and phosphorelay signal transduction. Curr Opin Microbiol. 2000;3:165–170. doi: 10.1016/s1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 10.Huang K J, Igo M M. Identification of the bases in the ompF regulatory region, which interact with the transcription factor OmpR. J Mol Biol. 1996;262:615–628. doi: 10.1006/jmbi.1996.0540. [DOI] [PubMed] [Google Scholar]
- 11.Julien B, Kaiser D, Garza A. Spatial control of cell differentiation in Myxococcus xanthus. Proc Natl Acad Sci USA. 2000;97:9098–9103. doi: 10.1073/pnas.97.16.9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci USA. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S K, Kaiser D. C-factor: a cell-cell signaling protein required for fruiting body morphogenesis of Myxococcus xanthus. Cell. 1990;61:19–26. doi: 10.1016/0092-8674(90)90211-v. [DOI] [PubMed] [Google Scholar]
- 14.Kroos L, Kaiser D. Construction of Tn5lac, a transposon that fuses lacZ expression to exogenous promoters, and its introduction into Myxococcus xanthus. Proc Natl Acad Sci USA. 1984;81:5816–5820. doi: 10.1073/pnas.81.18.5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kroos L, Kaiser D. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1987;1:840–854. doi: 10.1101/gad.1.8.840. [DOI] [PubMed] [Google Scholar]
- 16.Kroos L, Kuspa A, Kaiser D. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev Biol. 1986;117:252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- 17.Kroos L, Kuspa A, Kaiser D. Defects in fruiting body development caused by Tn5 lac insertions in Myxococcus xanthus. J Bacteriol. 1990;172:484–487. doi: 10.1128/jb.172.1.484-487.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuner J, Kaiser D. Introduction of transposon Tn5 into Myxococcus for analysis of developmental and other nonselectable mutants. Proc Natl Acad Sci USA. 1981;78:425–429. doi: 10.1073/pnas.78.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuspa A. Ph.D. thesis. Stanford, Calif: Department of Biochemistry, Stanford University Medical School; 1986. [Google Scholar]
- 20.Kuspa A, Kroos L, Kaiser D. Intercellular signaling is required for developmental gene expression in Myxococcus xanthus. Dev Biol. 1986;117:267–276. doi: 10.1016/0012-1606(86)90369-6. [DOI] [PubMed] [Google Scholar]
- 21.Kuspa A, Plamann L, Kaiser D. Identification of heat-stable A-factor from Myxococcus xanthus. J Bacteriol. 1992;174:3319–3326. doi: 10.1128/jb.174.10.3319-3326.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee K, Shimkets L J. Cloning and characterization of the socA locus which restores development to Myxococcus xanthus C-signaling mutants. J Bacteriol. 1994;176:2200–2209. doi: 10.1128/jb.176.8.2200-2209.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S-F, Lee B, Shimkets L J. CsgA expression entrains Myxococcus xanthus development. Genes Dev. 1992;6:401–410. doi: 10.1101/gad.6.3.401. [DOI] [PubMed] [Google Scholar]
- 24.Martin S, Sodergren T, Mauda T, Kaiser D. Systematic isolation of transducing phages for Myxococcus xanthus. Virology. 1978;88:44–53. doi: 10.1016/0042-6822(78)90108-3. [DOI] [PubMed] [Google Scholar]
- 25.Martson F A O, Hartley D L. Solubilization of protein aggregates. Methods Enzymol. 1990;182:264–276. doi: 10.1016/0076-6879(90)82022-t. [DOI] [PubMed] [Google Scholar]
- 26.Ogawa M, Fujitani S, Mao X H, Inouye S, Komano T. FruA, a putative transcription factor essential for the development of Myxococcus xanthus. Mol Microbiol. 1996;22:757–767. doi: 10.1046/j.1365-2958.1996.d01-1725.x. [DOI] [PubMed] [Google Scholar]
- 27.Parkinson J K, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 28.Plamann L, Kuspa A, Kaiser D. Proteins that rescue A-signal-defective mutants of Myxococcus xanthus. J Bacteriol. 1992;174:3311–3318. doi: 10.1128/jb.174.10.3311-3318.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Shimkets L J, Kaiser D. Induction of coordinated movement of Myxococcus xanthus cells. J Bacteriol. 1982;152:451–461. doi: 10.1128/jb.152.1.451-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singer M, Kaiser D. Ectopic production of guanosine penta- and tetraphosphate can initiate early developmental gene expression in Myxococcus xanthus. Genes Dev. 1995;9:1633–1644. doi: 10.1101/gad.9.13.1633. [DOI] [PubMed] [Google Scholar]
- 32.Søgaard-Andersen L, Kaiser D. C factor, a cell surface-associated intercellular signaling protein, stimulates the Frz signal transduction system in Myxococcus xanthus. Proc Natl Acad Sci USA. 1996;93:2675–2679. doi: 10.1073/pnas.93.7.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spratt B G, Hedge P J, Heesen S T, Edelman A, Broome-Smith J K. Kanamycin-resistant vectors that are analogs of plasmids pUC8, pUC9, pEMBL8, and pEMBL9. Gene. 1986;41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- 34.Thöny-Meyer L, Kaiser D. devRS, an autoregulated and essential genetic locus for fruiting body development in Myxococcus xanthus. J Bacteriol. 1993;175:7450–7462. doi: 10.1128/jb.175.22.7450-7462.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]