Abstract
The task-evoked positive BOLD response (PBR) to a unilateral visual hemi-field stimulation is often accompanied by robust and sustained contralateral as well as ipsilateral negative BOLD responses (NBRs) in the visual cortex. The signal characteristics and the neural and/or vascular mechanisms that underlie these two types of NBRs are not completely understood. In this paper, we investigated the properties of these two types of NBRs. We first demonstrated the linearity of both NBRs with respect to stimulus duration. Next, we showed that the hemodynamic response functions (HRFs) of the two NBRs were similar to each other, but significantly different from that of the PBR. Moreover, the subject-wise expressions of the two NBRs were tightly coupled to the degree that the correlation between the two NBRs was significantly higher than the correlation between each NBR and the PBR. However, the activation patterns of the two NBRs did not show a high level of interhemispheric spatial similarity, and the functional connectivity between them was not different than the interhemispheric functional connectivity between the NBRs and PBR. Finally, while attention did modulate both NBRs, the attention-related changes in their HRFs were similar. Our findings suggest that the two NBRs might be generated through common neural and/or vascular mechanisms involving distal/deep brain regions that project to the two hemispheres.
Keywords: Functional neuroimaging, Functional magnetic resonance imaging (fMRI), Negative BOLD response, Visual cortex, hemodynamic response function
1. Introduction
Functional magnetic resonance imaging (fMRI) is the most commonly used modality for in-vivo, and non-invasive functional mapping of the human brain based on endogenous blood oxygenation level depended (BOLD) contrast (Ogawa et al., 1992). A focal task/stimulus-evoked neuronal activation usually generates a localized change in the MR signal of the activated area relative to its pre-stimulus period. The shape and properties of this change in MR signal, henceforth referred to as positive BOLD response (PBR), have been thoroughly investigated and reported as the hemodynamic response function (HRF) of the BOLD signal (Menon et al., 1995; Mandeville et al., 1999). In addition to the PBR, fMRI responses in the opposite direction, which are often referred to as negative BOLD response (NBR), have been observed in regions adjacent and remote to the PBR regions (Shmuel et al., 2002; Smith et al., 2004).
While much progress has been made towards characterizing the shape of PBRs hemodynamics and their underlying mechanisms, the properties of NBRs and their corresponding underlying mechanisms are yet to be fully characterized (Logothetis, 2008; Liu et al., 2011). Numerous studies have detected NBR during stimulation of sensory and motor cortices from regions in the vicinity of the PBR (Shmuel et al., 2002, 2006) and in ipsilateral regions relative to a unilateral stimulus (Shmuel et al., 2003; Stefanovic et al., 2004; Smith eta l., 2004; Kastrup et al., 2008; Mullinger et al., 2014). However, there is no consensus on the origins and properties of these two types of NBRs. Several hypotheses have been made with regard to the mechanisms contributing to NBRs: (1) Decreases in neuronal activity due to local neuronal inhibition or decreases in afferent neuronal input (Smith et al., 2000, 2004; Shmuel et al., 2002, 2006; Stefanovic et al., 2004); (2) Passive reduction of cerebral blood flow (CBF) in regions adjacent to PBR, due to shared upstream arterial supply (blood steal), independent of changes in neuronal activity and/or neural control signals (Shmuel et al., 2002; Harel et al., 2002; Hu et al., 2015); (3) Active reduction of CBF (blood flow control) due to neural control signals that cause contraction of smooth muscles surrounding arteries and arterioles supplying the NBR regions, to ensure adequate supply to the areas of demand (Smith et al., 2004), and (4) Venous backpressure due to limited drainage capacity of venous compartments in response to massive increase in CBF to the PBR region (Shmuel et al., 2006; Boas et al., 2008; Goense et al., 2012). However, none of these mechanisms has been proposed and/or evaluated considering concurrent observation of NBRs in proximity to PBR and in the opposite hemisphere during unilateral sensory stimulation. While some studies have used the observed NBR in the ipsilateral hemisphere as evidence to invalidate the blood steal hypothesis (mechanism 2), since the two hemispheres have distinct, and quasi-independent arterial supply (Shmuel et al., 2003; Smith et al., 2004; Mullinger et al., 2014), we postulate that inducing concurrent NBR in two opposite hemispheres has not been used to its full capacity to provide evidence in support or against of the existing hypotheses.
Since both contralateral NBR (cNBR) from the vicinity of PBR, and ipsilateral NBR (iNBR) from the opposite hemisphere, can be simultaneously induced and detected during unilateral visual hemi-field stimulation, it is natural to ask whether a common or distinct neural/vascular mechanism/s underlies the cNBR and iNBR? Furthermore, if both NBRs are generated through a common mechanism, then it is important to know whether PBR is the underlying force for their simultaneous generation. The answer to these questions is of paramount importance because they could provide strong evidence for remote but active blood flow control (mechanism 3) and against two of the aforementioned hypotheses (blood steal, and venous backpressure). It essentially determines whether the two NBRs are regulated locally and independently within each hemisphere’s visual cortex, through major inter-hemispheric callosal pathway connecting homologues regions, or through the reported inter-hemispheric functional connectivity in visual cortex, or remotely through an intervening distal region with access to both hemispheres. By investigating the temporal and spatial relationships between these two types of NBRs and comparing those with the relationship of each NBR with PBR, we aim to provide evidence for distal regulation and against local regulatory mechanisms, such as inter-hemispheric synchrony through major callosal pathway or functional connectivity.
We start by investigating the linearity of the two NBRs in respect to stimulus duration since it is the pre-requisite for most of the statistical analysis conducted in fMRI. Even the most basic HRF extraction techniques using deconvolution (i.e. finite impulse response (FIR)) is based on the linearity assumption. Linearity of NBR with respect to stimulus duration, to our knowledge, has not been shown previously and without its validation our extracted HRFs for NBR and brain statistical maps for NBRs will not be valid. Once linearity was established, we extracted the HRFs of both NBRs to investigate any difference between their magnitudes and dynamics and compare them with the HRF of the PBR. Furthermore, we investigated the association between subject-wise expression of the two NBRs and compared it with the relationship of each NBR with PBR. Our results indicate that the HRFs of two NBRs obtained from the two hemispheres have similar amplitudes and dynamics that are significantly different than the magnitude and dynamics of the HRF of the PBR. Furthermore, the subject-wise expressions of the two NBRs are significantly more correlated with each other than each one with that of the PBR. The high inter-hemispheric similarity of the magnitudes, dynamics and inter-subject variabilities of the two NBRs motivates the examination of the role of interhemispheric callosal pathway, or functional connectivity for facilitating such similarity. Thus, we first computed interhemispheric spatial similarities of the regions showing NBRs and compared it to the interhemispheric spatial similarity of the iNBR with PBR. Then, to rule out the effect of interhemispheric functional connectivity in inducing coherent NBRs in two hemispheres, we computed the inter-hemispheric functional connectivity of the regions showing NBRs and compared it with the inter-hemispheric functional connectivity of the iNBR with PBR. Neither interhemispheric callosal pathway nor interhemispheric functional connectivity explained the observed synchrony between two NBRs, providing evidence for remote blood flow control.
Finally, we studied the performance correlates of the two NBRs and whether the two NBRs were modulated by attention. These experiments are motivated by our recent findings that the NBRs detected in the default mode network (DMN) regions are attention-specific and are correlated with performance (Parker and Razlighi 2019). Any difference between the cNBR and iNBR in the attention and/or performance-association highlights the possibility of having a separate underlying neural and/or vascular mechanism. Our findings suggest that the cNBR and iNBR share a common neural and/or vascular mechanism which is different not only from the ones underlying PBR, but also from the mechanism underlying NBR detected in the DMN, and possibly regulated by a distant brain structure that projects to the two hemispheres and actively controls blood flow distribution of the cerebral cortex.
2. Methods
2.1. Participants
Twenty-seven healthy young subjects (mean age ± SD = 25.11 ± 3.24, female/male = 20/7, all right-handed) were recruited using random market mailing approach within 50-mile radius of Columbia University Irving Medical Center and were compensated for their time spent partaking in this study. All subjects gave their informed written consent prior to the scanning sessions. The experimental design of our study and the recruitment process were approved by Columbia University institutional review board.
2.2. fMRI experimental design
We employed an event-related fMRI experimental task design to effectively examine the timing and the shape of the extracted HRFs. The task consisted of two ongoing stimuli: (1) An almost square checker-board with alternating luminance between 25% and 44% of maximum luminance, spatial frequency of 1.17 cycle per degree, and contrast reversal rate (temporal frequency) of 6 Hz was presented to the right or left of the screen (with 50% of maximum luminance) approximately between 2.29° and 9.14° angle, as shown in Fig. 1 b; and (2) An alternating tone paradigm played on either the right or the left ear via MR compatible earphones, with alternating frequency (pure tones at 698.46 Hz and 440.00 Hz with a 0.1 s duration of each tone). The two sensory stimuli were presented with random onsets and durations, sampled from a uniform continuous distribution in the range of 1.0–5.0 s; visual and auditory stimuli timings were sampled independently. Overlaps between visual and audio stimuli were allowed, however, bilateral overlapping presentation of the same sensory stimulus (left-visual with right-visual or left-auditory with right-auditory) was prohibited. The data were collected in 2 runs; in the first run, subjects were instructed to attend only one sensory modality (i.e., either visual or tonal) while ignoring the other. In the second run, they were instructed to attend the other sensory modality. Each scan consisted of 120 events: 60 events for visual and 60 for auditory stimulus; for each modality, 30 events on the right and 30 events on the left side spaced at inter-stimulus-intervals drawn from an exponential distribution (center=5.6, λ=9.8). Monitoring attention was achieved by asking the subjects to press a button twice with their right/left index finger (depending on the lateralization of the attended stimulus) as soon as the attended stimulus terminated. These responses were recorded during the scans. Moreover, subjects were required to constantly fix their gaze on a green minuscule fixation spot in the center of the screen for the entire period of the scan. Subjects were given feedback on their incorrect or out-of-time (>3 s) responses (if any) via change in the color of the fixation spot to red. Eye fixation was monitored at all time during scans using an eye-tracking system (EyeLink 1000 Plus). If subjects excessively deviated their eye’s fixation from the central dot, we stopped the scan, instructed the participants again, and repeated the scan. However, this happened only during two fMRI runs. Subjects were first trained outside of the scanner to learn and perform the task comfortably and accurately (almost 100% correct) in short training runs. All subjects learned the task correctly. An example of the timing and the design of the task is illustrated in Fig. 1.
Fig. 1.
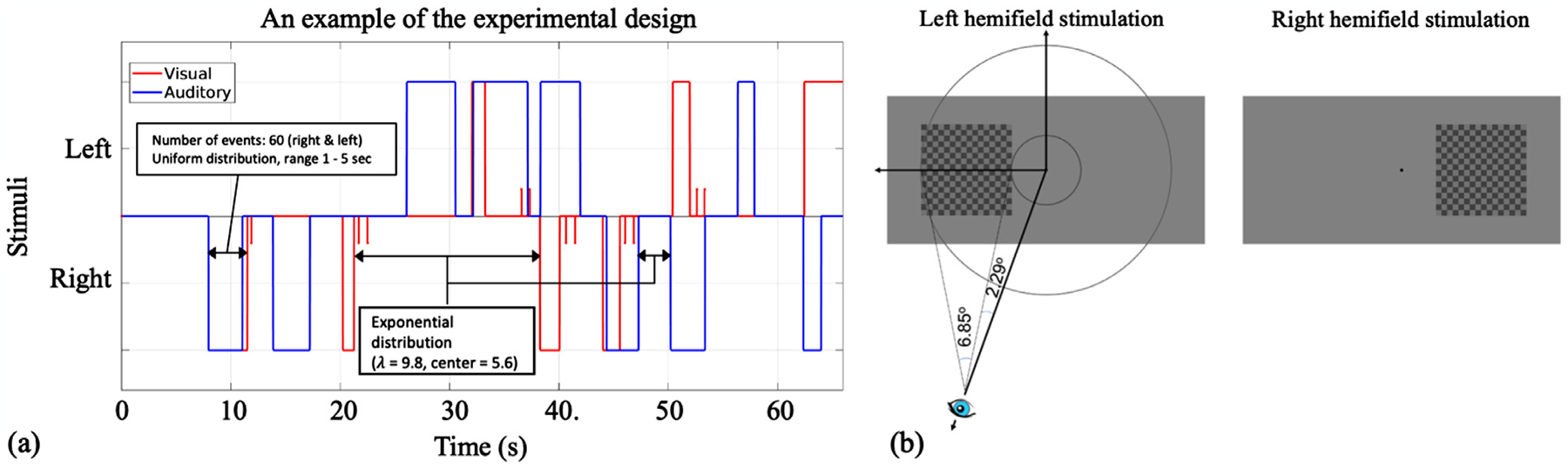
(a) Illustration of a segment of the time-course of the visual and audio event related task. The blue line shows the timing of the auditory stimuli, the red line shows the timing of the visual stimuli, and the red stick line shows the subject responses (i.e., each time they press the button). In this sample demonstration, subjects were requested to attend to the visual stimulus and ignore the auditory stimulus. This is evident by the response pattern as the button is pressed twice as soon as the attended stimulus (i.e., visual stimulus) is terminated. (b) Flashing checkerboard visual stimulus was presented on the left or right hemifield, between 2.29 and 9.14 °extending over 6.85 degrees.
2.3. MRI acquisition parameters
Imaging was carried out using a 3 Tesla Siemens Magnetom Prisma scanner equipped with an 80 mT/m gradient system with a T2*-weighted echo-planar imaging (EPI) sequence using interleaved slice acquisition (TR/TE = 1000/30 ms; flip angle = 62°; field of view (FOV) = 200 × 200 mm; bandwidth = 1852 Hz/px; multiband factor = 4, matrix size = 100 × 100; voxel size = 2 ×2 ×2 mm; 64 axial slices with alternating phase encoding directions of anterior-posterior or posterior-anterior). Subjects were positioned supine inside the magnet bore wearing noise isolating MR safe earbuds to listen to the auditory stimuli. The flashing checkerboard visual stimuli presented within a square aligned to the horizontal meridian were projected onto a back-projection screen in the scanner. Each task-based fMRI (tb-fMRI) scan lasted 8 min (480 vol.). A resting-state fMRI (rs-fMRI) scan with the same duration and parameters was also acquired for every participant. To prevent any bias in visual-attending/tonal-attending task modalities, we alternated the order of fMRI acquisitions between; (1) visual-attending, resting-state, and tonal-attending; (2) tonal-attending, resting-state, and visual-attending; for each participant. An accompanying anatomical T1-weighted structural image was collected for localization and registration purposes during the same MRI session (TR/TE = 2300/2.32 ms; flip angle = 8°; field of view (FOV) = 256 ×256 mm; matrix size = 256 × 256; voxel size = 1 ×1 ×1 mm; 196 sagittal slices).
2.4. Preprocessing of fMRI data
The fMRI data were processed using FSL (V5.0.7) and in-house-developed packages. The preprocessing pipeline for the task-based fMRI data is illustrated in Fig. 2. Briefly, slice timing correction was applied to the raw fMRI timeseries to account for the difference in the acquisition delay between slices (Parker et al., 2017; Parker and Razlighi, 2019). At the same time, motion parameters were estimated on raw fMRI scans using rigid-body registrations performed on all the volumes in reference to the first volume. Additionally, the first volume was extracted from another fMRI scans with opposite phase encoding directions to estimate the geometric distortion correction field using the susceptibility-induced distortions correction technique topup (Andersson et al., 2003) provided in the FSL software package (Smith et al., 2004). Then, the estimated motion parameters and geometric distortion field were combined and applied to the slice timing corrected fMRI time-series to get the distortion and motion-corrected fMRI time-series. In order to enhance the precision in the localization of the statistically significant responsive voxels, no spatial smoothing was applied. To remove the slow scanner drift, a temporal high-pass filter (> 0.01 Hz) was applied to the fMRI timeseries. The pre-processed fMRI data were modeled with 6 predictors (except for the linearity analysis, which will be discussed later) for the presented stimuli and subject responses (i.e., visual left, visual right, audio left, audio right, motor-response left, motor-response right). The predictors were obtained by convolving the visual, auditory, and motor boxcar timings with the canonical double-gamma HRF. First-level analysis was performed via multiple regression using an in-house-developed software package in Python. Similar pre-processing was performed on the rs-fMRI data by applying slice-timing correction, spatial realignment, geometric distortion correction, and high-pass filtering (>0.01 Hz). Structural scans were processed using FreeSurfer (Fischl et al., 2002; Fischl et al., 2004).
Fig. 2.
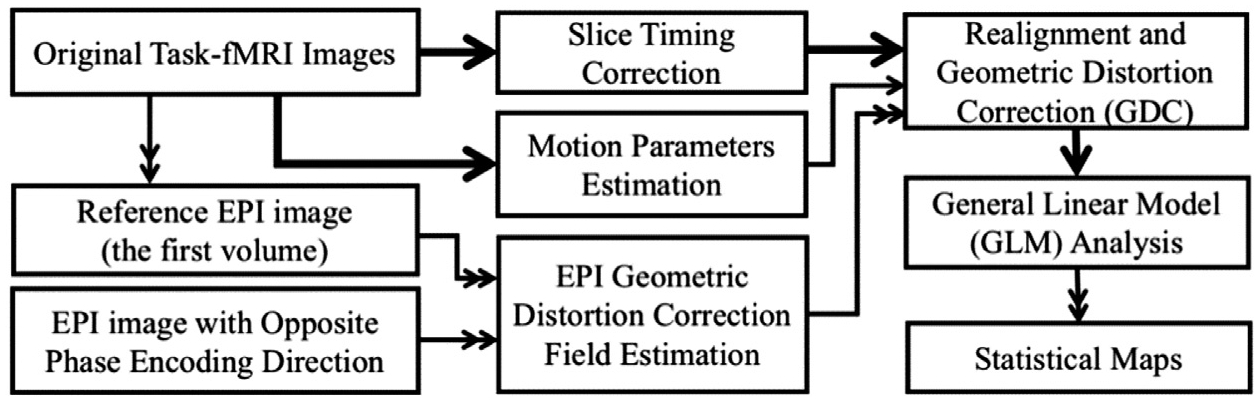
The preprocessing pipeline for task-based fMRI data. The thick arrows show the transfer of 4D fMRI data, the double thin arrow shows the transfer of the 3D data, and the thin arrow shows the transfer of the parameters.
2.5. Linearity of the cNBR and iNBR
In this experiment we aimed to assess the linearity of the both NBR with regards to the change in the duration of the stimulation. The stimuli were categorized into four distinct categories with respect to their duration (i.e., category 1 included stimuli with durations between 1 and 2 s and in the same way category 2, 3, and 4 included stimuli with durations between 2 and 3, 3 to 4, and 4 to 5 s, respectively). The time series of the preprocessed fMRI data were standardized by subtracting the mean and dividing by the standard deviation. To prevent any bias in voxel selection towards voxels that randomly exhibit the linear property, we employed bootstrapping technique where the trials were randomly split in half for voxel selection (training) and BOLD response extraction (testing). For a given iteration, the training trials’ boxcar (convolved with a canonical HRF) were used in a general linear modeling (GLM) analysis to obtain the significantly responsive voxels (|t| > 3). Once the voxels were selected, the training trials were disregarded and only the remaining trials (testing) were used for assessing the linearity in the selected voxels. The amplitude of each trial in the testing set was also normalized with regard to its duration to make sure it has a unit area under the curve in each trial’s boxcar. This was done to maintain the same amplitude in the GLM predictors of all stimulus durations, which makes it possible to assess the increase in the magnitude of the BOLD response using the β-coefficient of the GLM analysis. Each trial in the test set was then assigned to one of the four different regressors according to their categorized stimuli durations. Next, four separate GLM analyses were performed using one of the four regressors; Each one estimating the BOLD response magnitude for each duration category. The bootstrapping process was repeated with 500 iterations, and the β-coefficient of each category was averaged separately for the cNBR and iNBR across all iterations and right and left visual stimuli to compute the corresponding BOLD response amplitude. The subject-wise mean β-coefficient (amplitudes) for the four duration categories were then plotted against the corresponding duration categories. We performed a least squares line regression analysis along with t-test of the slope to check for significance of the linear relationship (if existed). As a sanity check, we also used this method to check the linearity of the PBR.
2.6. Generating regions of interest (ROIs)
Throughout this work, instead of performing the formal group level analysis in standard space, we performed the analysis in the subjects’ native space (unless explicitly noted otherwise). Analysis in native space increases the precision of localizing ROIs in each subject compared to the regular group level analysis in the standard space. The latter often introduces spatial misalignments across different subjects due to lack of precision in non-linear registrations (Klein et al., 2009; Razlighi et al., 2014; Liu et al., 2017). Hence, for each individual subject, two ROIs per hemisphere were generated. The first ROI was generated for each hemisphere, by selecting the voxels within the visual cortex that showed statistically significant PBR (activation) during the unilateral visual stimulation using the z-statistical map derived from first-level analysis (z > 4). The subject-wise mask of the visual cortex was obtained using FreeSurfer. The first ROI was then dilated by a spherical kernel with 10 mm radius - resulting in a large ribbon surrounding it in each hemisphere - to delineate the neighboring voxels required for identifying the NBR in the vicinity of the activated regions (cNBR). The union of the two masks on the opposite hemisphere (due to visual stimulation of the opposite hemi-field) was used as the ROI to identify voxels with iNBR. Any intersection with the inferior parietal and precuneus regions was eliminated from the two generated ROIs, since they might be part of the NBR of the DMN which may have different characteristics and/or underlying mechanism. Finally, in order to eliminate any possible misalignment or interhemispheric leakage, the two bilateral ROIs were constrained to be within each brain hemisphere (hemisphere mask delineated by using FreeSurfer software). This was also done to avoid any possible overlap between the ROIs from the two hemispheres.
2.7. Hemodynamic response functions of cNBR and iNBR
The goal of this experiment was to extract and compare the shapes of the HRFs of the two task-evoked cNBR and iNBR obtained from the visual cortex. Since we are investigating the shape of the HRF, voxel selection cannot be performed using a canonical double Gamma HRF. This is to prevent any bias toward a pre-selected HRF shape. Hence, we used a set of optimized basis functions known as FMRIB’s linear optimal basis set (FLOBS) to perform our voxel selection for this experiment (Woolrich et al., 2004). The FLOBS technique does not impose a fix timing and/or dynamic for the BOLD response; Thus, voxels exhibiting response with unique shape and dynamics can be selected as responsive voxels for a given stimuli timing. We selected the significantly responsive voxels according to the F statistics |F|>6 within the predefined ROI masks. An initial response function for each voxel was computed by a weighted average of the FLOBS basis functions according to the estimated β-coefficient in the first-level FLOBS GLM analysis. Then, we integrated the initial response function for the first 10 s interval to obtain the sign of the area under the curve, and we classified the significantly responsive voxels into either positive or negative responsive voxels. Once the negatively and significantly responsive voxels are identified within the generated ROIs, the time courses of the selected voxels were given to the FIR deconvolution technique, which to our knowledge is the least constrained approach for extracting the shape of the impulse response function in fMRI (Goutte et al., 2000). We extracted the voxel-wise HRFs of each subject for cNBR and iNBR for both attended and unattended stimuli. Subsequently, the extracted HRFs were averaged across all responses to the right and left visual stimuli to obtain the mean subject-wise HRFs for cNBR and iNBR for both attended and unattended stimuli (i.e., , , , and ). For comparison purposes, the positive BOLD HRF was also extracted using the same procedure and the time-series were converted to percent change (subtracted by the mean and then divided by the mean).
We subtracted the mean signal of the pre-stimulus period (time points within 5 s before the onset of the stimulus) from the subject-wise HRF. Then, we up-sampled the subject-wise HRF with sinc-interpolation. In order to compare the magnitudes and dynamics of the extracted HRFs for cNBR, and iNBR, as well as for PBR, the subject-wise extracted HRFs were used to obtain onset time, time to peak, amplitude, falling edge time, and time to undershoot of the BOLD responses. As shown in Fig. 6 A for illustrating the PBR case, time to peak was defined as the first instance where the HRF reached its maximum magnitude starting from time zero (stimulus onset). The onset time was defined as the first time point where the HRF exceeded 10% of the maximum amplitude. The falling edge time was defined as the time duration between the peak time and the time at which the HRF reached back to its estimated average pre-stimulus magnitude. The time to undershoot was defined as the time that it took the HRF to reach its maximum opposite magnitude after falling back to the baseline, starting from time zero. For each characteristic, we removed measurements outside three standard deviations from the mean to eliminate the influence of outliers. Finally, Student’s t-test was used to test for the significance of the difference (if any) between magnitude, time to peak, onset time, falling edge time, and time to undershoot of the HRFs obtained for cNBR, and iNBR, as well as for PBR
Fig. 6.
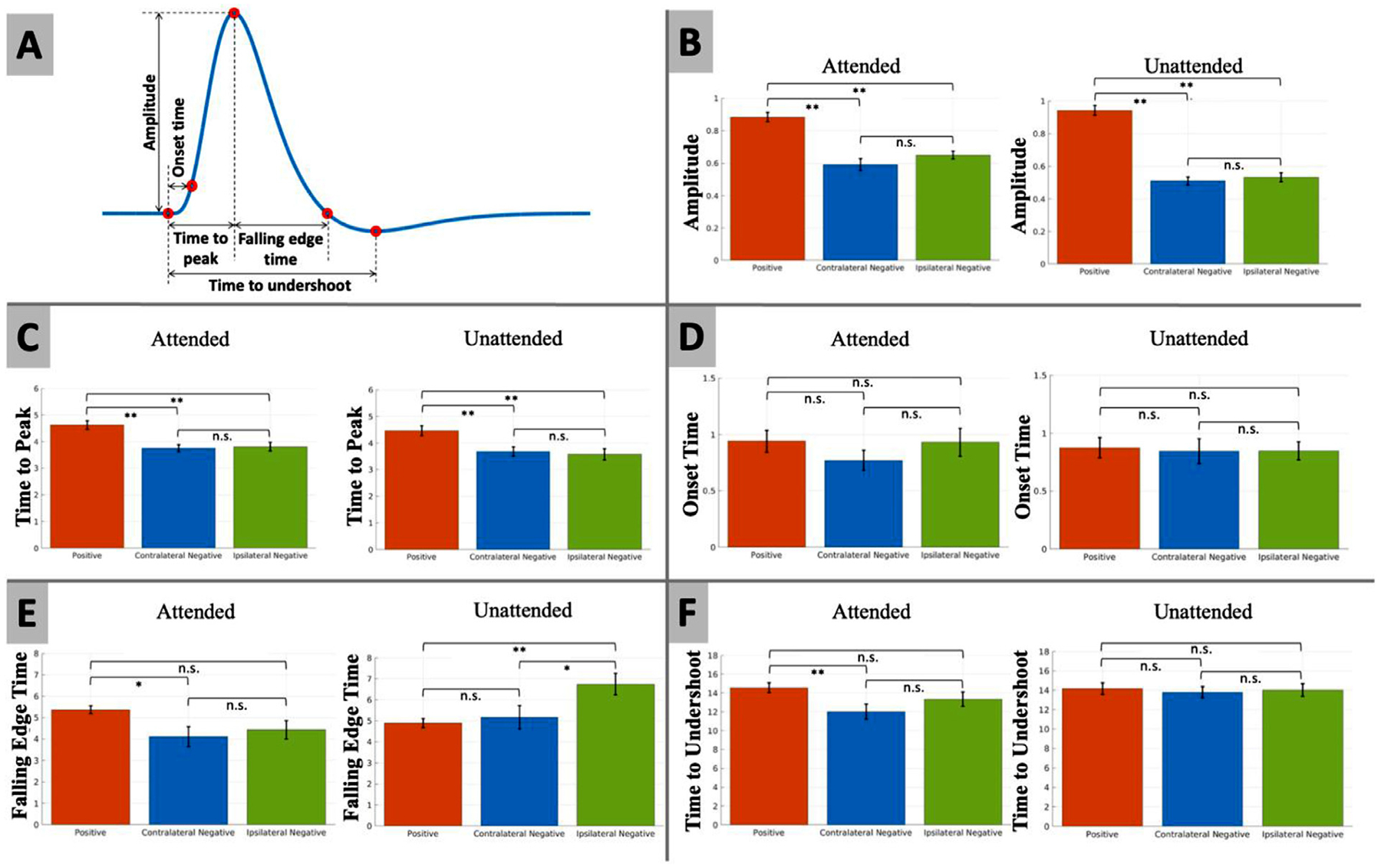
(A) The definition of amplitude, time to peak, onset time, falling edge time, and time to undershoot of the BOLD responses as illustrated for the PBR case. (B–F) Student’s t-test of statistically significant difference between the (B) amplitude in percent change, (C) time to peak, (D) onset time, (E) falling edge time, and (F) time to undershoot of the two NBRs and PBR using sinc-interpolation up-sampled subject-wise HRF. Error bars represent the standard error of the mean. Statistically significant differences are marked with asterisk symbols (* p < 0.05; ** p < 0.01; uncorrected).
2.8. Subject-wise expressions of cNBR and iNBR
The aim of this analysis was to assess the relationship between the subject-wise expression of different BOLD responses, which could potentially hint at the underlying driver of NBRs. Using the previously generated ROI masks, the statistically significant voxels were selected inside each ROI using a z-statistics threshold of above +4 and below −3 for the positive and negative BOLD responses, respectively (not corrected for multiple comparisons). The NBR is usually a weaker signal than PBR, hence, we chose a slightly lower threshold for NBR to add more negatively responsive voxels. The subject-wise expression of cNBR and iNBR were calculated by averaging the β-coefficient from the significantly responding voxels to the right and left visual stimuli. For comparison, subject-wise expressions of PBR were also obtained similarly. For each pair of the BOLD responses (i.e., PBR vs cNBR, PBR vs iNBR, and cNBR vs iNBR), Pearson correlation coefficient (PCC) was used to assess the association (if any) between the BOLD amplitudes separately for the attended and unattended cases. Outliers were removed based on median absolute deviation and PCC are computed again to make sure the results were not influenced by outliers. A statistical test was conducted to test for significant difference in the correlation (use the absolute value of PCC) between pairs of the BOLD responses.
2.9. Functional connectivity between cNBR and iNBR
In this analysis, we aimed to investigate the inter-hemispheric functional connectivity in the visual cortex between regions showing BOLD responses during unilateral visual stimulation (i.e., iNBR vs PBR, and iNBR vs cNBR). The similarly pre-processed rs-fMRI data were aligned to the tb-fMRI space using 6 degrees of freedom intra-subject registration (Jenkinson et al., 2002). A binary mask of the regions with significant PBR, iNBR, and cNBR was intersected with each participant’s left/right hemisphere’s visual cortex mask (delineated by using the FreeSurfer software) to obtain the mean time-series for the regions with PBR and NBRs from rs-fMRI data. The PCC was used to compute functional connectivity between the regions. Any significant difference between functional connectivity was assessed with the Student’s t-test.
2.10. Interhemispheric spatial similarities of the cNBR and iNBR
The goal of this analysis was to investigate the topographical similarities between the spatial patterns of the BOLD responses elicited during unilateral visual stimulation and observed in the two hemispheres (i.e., iNBR vs PBR, and iNBR vs cNBR). Similar to the HRF extraction analysis, our spatial similarity analysis was within previously generated ROIs that exceeded the selected significance threshold (i.e., |F| > 6) in the first-level FLOBS GLM analysis. Voxels were classified into positively and negatively responsive voxels based on the area under the curve during the first 10 s interval of the HRF. Spatial pattern similarity was measured by Dice similarity coefficient (DSC) between regions showing significant cNBR and iNBR, and for comparison, between iNBR and PBR. In order to compute the spatial pattern similarity between regions located in two different hemispheres, these ROI masks were transferred into MNI standard space. To this end, the first EPI volumes from the fMRI data (as the reference volume in preprocessing) were registered to their T1-weighted structural image using a rigid-body transformation and normalized mutual information. Then, each subject’s T1-weighted structural image was registered to the standard MNI space using Landmark Guided Region Based Spatial Normalization (LG-RBSN) (He and Razlighi, 2022). The combination of the rigid-body transformation and the non-linear warping field was used to transfer the obtained ROI masks into the MNI space. Next, the transferred ROI masks were flipped around the mid-sagittal plane (i.e., mirrored) to make it possible to use DSC overlap measurement to quantify the spatial pattern similarities between iNBR and PBR, as well as iNBR and cNBR. To assess for significance in the difference (if any) between the DSC of iNBR vs PBR, and iNBR vs cNBR, t-test was used across the obtained DSC measurements.
2.11. Behavioral correlates of the cNBR and iNBR
To assess the relationship between the NBRs and task performance, we used subject-wise median response-time as a measurement of performance, and subject-wise expression of NBRs as the strength of BOLD response. For comparison, we also investigated the relationship between PBR and performance using the same method. The response-time was defined as the time interval between the end of the attended stimulus and the instance when the first corresponding button was pressed by subjects. Using PCC, we tested for any association between the median response-time of each subject and the corresponding expression for PBR, cNBR, and iNBR.
2.12. Attention dependency of the cNBR and iNBR
In this analysis, the goal was to check the effect of attention on the negative BOLD signals. We used the difference in the amplitudes and dynamics of the previously extracted HRFs between the attended and unattended/ignored stimuli as a measure of attention dependency. Student t-test was used to assess the significance in the difference (if any) between the attended and unattended cases at every second after the start of the stimulus. Multiple comparisons correction was performed using Bonferroni correction method.
3. Results
The visual stimulation induced statistically significant PBR in lower visual areas within the occipital lobe of all subjects. In addition, sustained, statistically significant NBRs ipsilateral and contralateral to the visual stimuli were observed in all subjects, for both attended and unattended cases (details in supplementary material Table S2). Participants responded correctly in 98.0/96.0% of the visual/audio stimulus presentations, where a correct response is defined as a button press within 3 s of the attended stimulus termination. Fig. 3 illustrates the spatial pattern of a typical subject’s PBR, cNBR, and iNBR to unilateral right hemifield presentation of attended visual stimuli using z-statistics overlaid on three orthogonal slices of the participant T1-weighted structural MRI image.
Fig. 3.
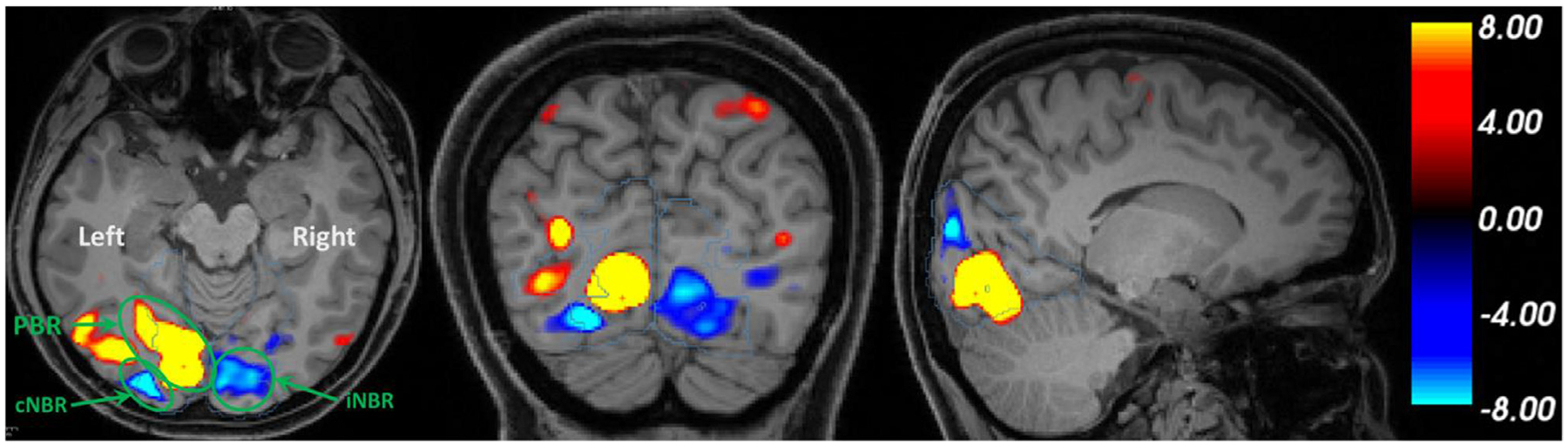
Demonstration of activation/deactivation in response to right visual hemifield stimulation from a single subject in the attended condition. Positive BOLD responses are color-coded with red/yellow and negative BOLD responses are color-coded with blue/cyan. The unilateral visual stimulation induced robust PBR in the contralateral (relative to the stimuli) visual cortex accompanied by a robust cNBR in its vicinity, and a robust iNBR in the opposite hemisphere. Note that spatial smoothing is carried out here only for better illustration.
We did not impose any overlap between the timings of the visual and auditory stimuli. The event-related designs of the two sensory stimuli were independent, with the timing of the stimuli randomized separately for visual and auditory stimuli. Thus, this design resulted in some overlapping visual and auditory stimulations, as seen in Fig. 1. To make sure the overlapping stimuli had negligible influence on our results, we categorized the stimuli according to their temporal overlap between each visual stimulus and the left or right auditory stimuli. Based on this, the visual stimuli trials were classified into two classes: (1) high visual and auditory stimuli overlap (overlap larger than 50%) and (2) low visual and auditory stimuli overlap (overlap smaller than 50%). We performed Student’s t-test between the mean response time of these two classes’ trials across subjects and found that there was no significant difference (t = −0.76; p > 0.45) between them. In addition, there was no significant difference between the bimodal stimuli overlapping rate between left and right presentations (t = 1.54; p > 0.13). The mean bimodal stimuli overlapping rates of the left and right visual stimuli presentations were 0.63 and 0.61, respectively.
On average participants’ gaze deviations from the center of the screen were 1.39 ± 1.58 (mean ± std) degree. In more than 71% of the time their gaze deviations were less than 1°. In 11% of the time, the deviations were between 1 and 2°, and in 17% of the time, the deviations were higher than 2°. There was no significant difference between the participants’ gaze deviation during the left versus right visual stimulation as well as during the 5 s pre-stimulus period. We also did not see any correlation between the detected BOLD response parameters and the gaze deviations. Details of these analyses are given in the supplementary materials.
3.1. Stimulus duration linearly scales the magnitude of cNBR and iNBR
In this experiment, we aimed to assess the relationship between the magnitude of the task-evoked negative BOLD signals (i.e., cNBR and iNBR) and the stimulus duration. To this end, we averaged the β-coefficient of the first-level GLM analysis fitted separately for each categorized stimuli duration. Results of the right and left visual stimulations were also averaged for this analysis. We then assessed the relationships between the mean β-coefficient and the categorized stimuli durations using linear regression. Fig. 4 a–c show the averaged β-coefficient as the BOLD response amplitude in terms of stimulus duration for cNBR, iNBR, and PBR, respectively. As illustrated in Fig. 4a and b, the amplitudes of both cNBR and iNBR showed significant linear relationships with the stimulus duration (cNBR: β= −0.312, p < 0.009; iNBR: β= −0.169, p < 0.017) ranging from 1 to 4 s. These results provide evidence for linear relationships between the NBR amplitude and stimulation duration, suggesting that the longer a stimulus duration is, the higher is the magnitude of both induced NBRs in the visual cortex (i.e., the amplitudes get more negative). Since the linearity of PBR has been already shown (Boynton et al., 1996), as a sanity check, we applied the same test of linearity to the PBR (β = 0.845, p < 6.116e-4; Fig. 4c), confirming the linear relationship in the PBR for the same range of the stimulus durations.
Fig. 4.
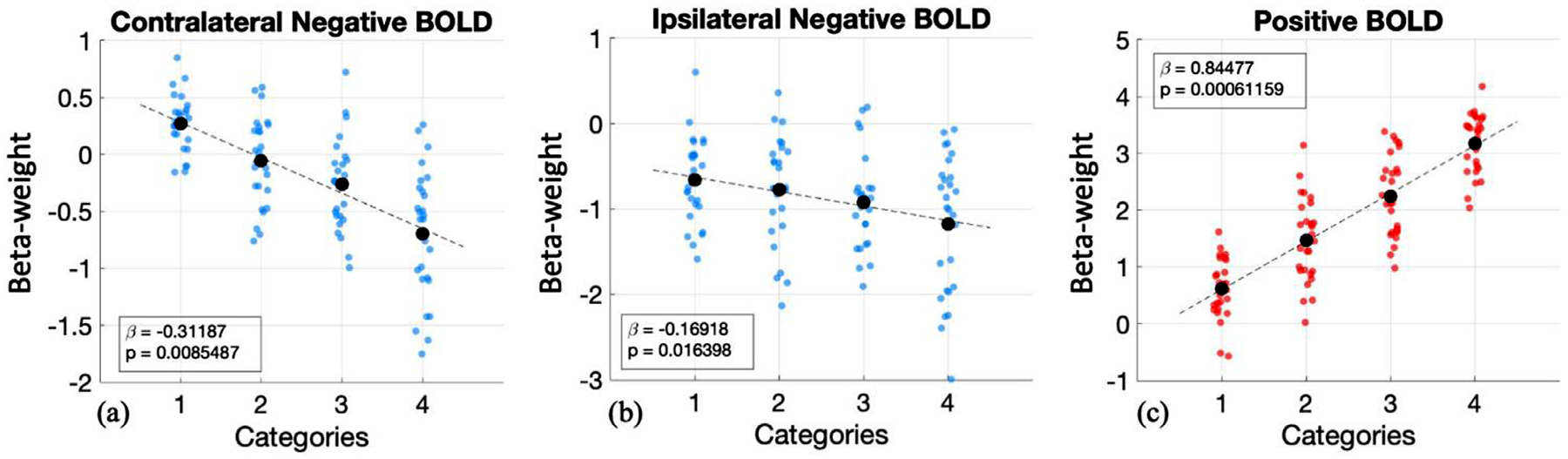
Linearity of the (a) cNBR, (b) iNBR, and (c) PBR with respect to stimulation duration. The blue and red dots show, respectively, the mean amplitudes for negative and positive BOLD responses of each subject. The black dots indicate the mean amplitude of the BOLD responses (i.e., averaged over all subjects) for each duration category. The black dashed lines represent the regression lines. The absolute values of the mean cNBR and iNBR amplitudes scale linearly with the stimulus duration. Please note that the timeseries have zero mean and unit standard deviation for this analysis, thus the relative change between categories carries the information and not the β-coefficient value at each category. As a sanity check for the method, we also applied the same method to the PBR. As expected, the PBR scales linearly with the stimulus duration.
3.2. The cNBR and iNBR have similar HRFs
The goal of this experiment was to compare the magnitudes and dynamics of the HRFs obtained for the two NBRs. As mentioned in the methods section, we used the FIR deconvolution technique to extract the HRFs associated with the cNBR, iNBR, and PBR. The extracted HRFs for the attended and unattended cases are shown in Fig. 5a and b, respectively. As is evident in these figures, the overall shapes of cNBR and iNBR are closely similar in both attended and unattended conditions. To quantify this similarity, we tested for any statistically significant difference between the onset time, time to peak, amplitudes, falling edge time and time to undershoot of the two NBRs with subject-wise up-sampled HRF. As shown in Fig. 6 and Table 1, there was no significant difference between the cNBR and iNRB regarding their onset time in both the attended (Δt = −0.1601 s, p > 0.3770) and the unattended conditions (Δt = −0.0040 s, p > 0.9827). Similarly, there was no significant difference in the time to peak of cNBR and iNBR in the attended (Δt = −0.0580 s, p > 0.7813) and unattended (Δt = 0.1015 s, p > 0.7104) conditions, suggesting that the cNBR and iNBR HRFs start to rise and reach to their peak magnitude at approximately the same time. We also observed no significant differences between the amplitudes of the two NBRs in both the attended (Δ = −0.0578, p > 0.1969) and the unattended (Δ = −0.0219, p > 0.5512) condition. We did not find any significant difference in the falling edge time of the two NBRs in the attended (Δt = −0.3181 s, p > 0.6289) condition, whereas there was a significant difference in the unattended (Δt = −1.5770 s, p < 0.0472) condition; However, this difference did not survive multiple comparison correction. Finally, no significant difference was observed between the cNBR and iNBR in terms of their time to undershoot for either of the attentional conditions (attended: Δt = −1.3289 s, p > 0.2427; unattended: Δt = −0.2268 s, p > 0.8018). These findings suggest that there is no significant difference in the magnitude and dynamics of the HRFs extracted for the iNBR and cNBR after multiple comparisons correction. This evidence suggests that common mechanism/s possibly underlie the contralateral and ipsilateral NBRs while they are induced in two separate hemispheres of the human visual cortex.
Fig. 5.
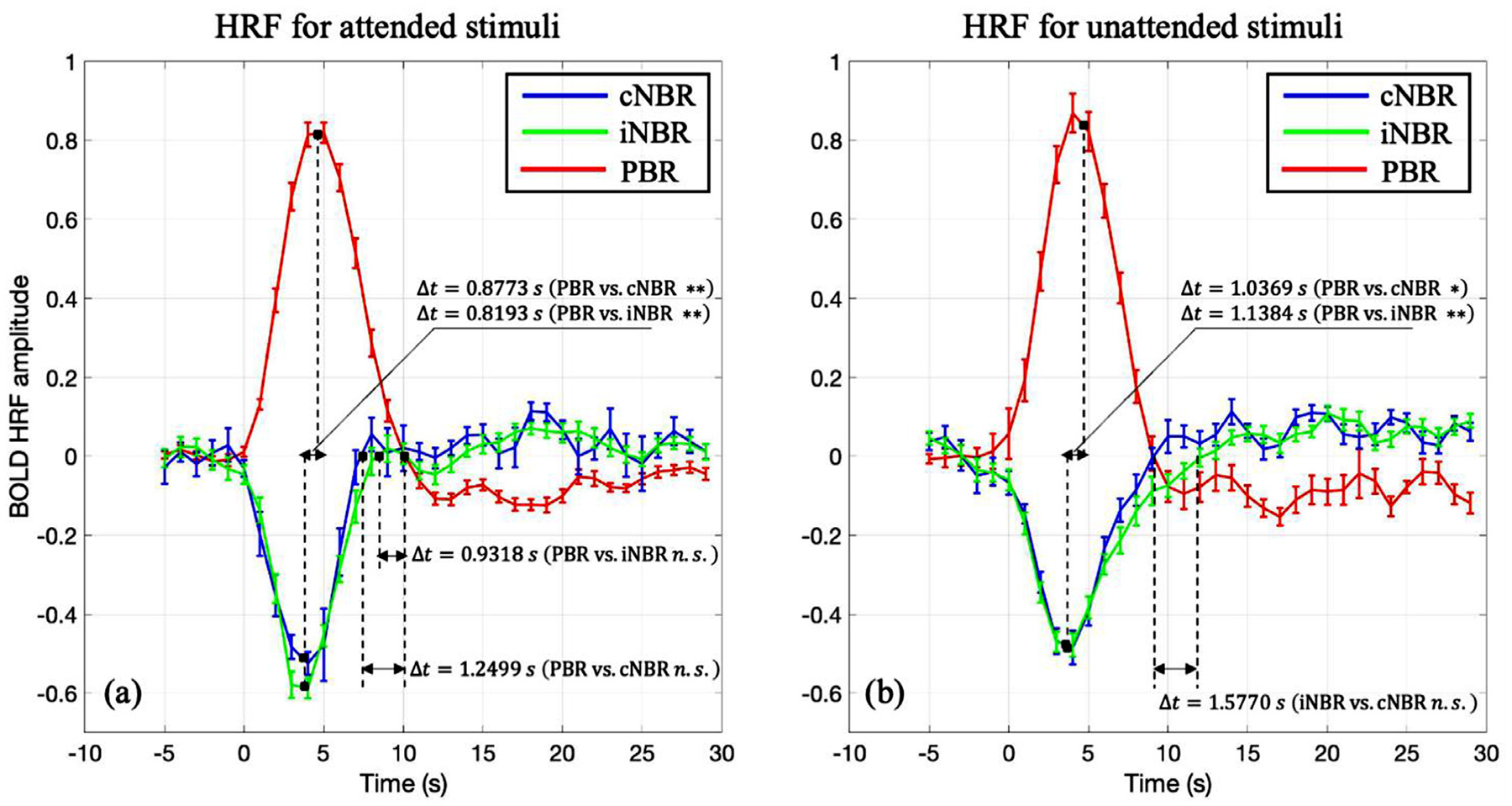
The HRFs of the positive BOLD, and contralateral/ipsilateral negative BOLD responses to (a) attended and (b) unattended visual stimuli. The curves are adjusted based on average of the HRF for 5 s prior to the stimulus onset. The unit of the magnitudes are percent changes (subtracted by mean and then divided by mean). Error bars represent the standard error of the mean. As is evident here, the two negative HRFs are closely similar in terms of their overall dynamics and amplitudes (* p < 0.05; ** p < 0.01; Bonferroni correction).
Table 1.
Comparison of the HRFs obtained for the cNBR and iNBR (cNBR vs. iNBR; * p < 0.05; uncorrected).
| Measurements | Attended | Unattended |
|---|---|---|
| Onset time | Δt = −0.1601 s, p > 0.3770 | Δt = −0.0040 s, p > 0.9827 |
| Time to peak | Δt = −0.0580 s, p > 0.7813 | Δt = 0.1015 s, p > 0.7104 |
| Amplitude | Δ = −0.0578, p > 0.1969 | Δ = −0.0219, p > 0.5512 |
| Falling edge time | Δt = −0.3181 s, p > 0.6289 | Δt = −1.5770 s, p < 0.0472 (*) |
| Time to undershoot | Δt = −1.3289 s, p > 0.2427 | Δt = −0.2268 s, p > 0.8018 |
To assess for any differences between the magnitudes and dynamics of the positive and negative BOLD HRFs, we compared the two negative BOLD HRFs to the positive one (Fig. 6 and Table 2). In the attended condition, the positive BOLD HRF reached its peak magnitude almost one second later than both negative BOLD HRFs (PBR vs. cNBR: Δt = 0.8773 s, p < 0.0001; PBR vs. iNBR: Δt = 0.8193 s, p < 0.0008). The amplitude of the positive BOLD HRF was significantly larger than the amplitudes of the two negative BOLD HRFs (PBR vs. cNBR: Δ = 0.2914, p < 8.2659e-08; PBR vs. iNBR: Δ = 0.2336, p < 8.7143e-08). Furthermore, the PBR’s HRF returned to baseline slower than the cNBR’s HRF (Δt = 1.2499 s, p < 0.0182). The time to undershoot was significantly larger for the PBR’s HRF compared to the cNBR’s HRF (Δt = 2.5450 s, p < 0.0098). Similar to attended case, in the unattended condition, we observed the same trend in time to peak (PBR vs. cNBR: Δt = 1.0369 s, p < 0.0037; PBR vs. iNBR: Δt = 1.1384 s, p < 0.0031). The amplitude of the positive BOLD HRF was significantly larger than the magnitudes of the two negative BOLD HRFs’ (PBR vs. cNBR: Δ = 0.4315, p < 3.6432e-15; PBR vs. iNBR: Δ = 0.4095, p < 1.5076e-13). Unlike the results in the attended condition, there was significant difference in the falling edge time only between PBR’s HRF and iNBR’s HRF (Δt = −1.8553 s, p < 0.0017). Finally, the PBR’s HRF time to undershoot was not significantly different than the two negative BOLD HRFs (PBR vs cNBR: Δt = 0.3628 s, p > 0.6663; PBR vs iNBR: Δt = 0.1360 s, p > 0.8792).
Table 2.
Comparison of the HRFs obtained for the PBR and NBRs (* p < 0.05; ** p < 0.01; uncorrected).
| Measurements | Attended | Unattended | |
|---|---|---|---|
| Onset time | PBR vs. cNBR | Δt = 0.1703 s, p > 0.2384 | Δt = 0.0301 s, p > 0.8619 |
| PBR vs. iNBR | Δt = 0.0102 s, p > 0.9539 | Δt = 0.0261 s, p > 0.8575 | |
| Time to peak | PBR vs. cNBR | Δt = 0.8773 s, p < 0.0001 (**) | Δt = 1.0369 s, p < 0.0037 (**) |
| PBR vs. iNBR | Δt = 0.8193 s, p < 0.0008 (**) | Δt = 1.1384 s, p < 0.0031 (**) | |
| Amplitude | PBR vs. cNBR | Δ = 0.2914, p < 8.2659e-08 (**) | Δ = 0.4315, p < 3.6432e-15 (**) |
| PBR vs. iNBR | Δ = 0.2336, p < 8.7143e-08 (**) | Δ = 0.4095, p < 1.5076e-13 (**) | |
| Falling edge time | PBR vs. cNBR | Δt = 1.2499 s, p < 0.0182 (*) | Δt = −0.2783 s, p > 0.6386 |
| PBR vs. iNBR | Δt = 0.9318 s, p > 0.0547 | Δt = −1.8553 s, p < 0.0017 (**) | |
| Time to undershoot | PBR vs. cNBR | Δt = 2.5450 s, p < 0.0098 (**) | Δt = 0.3628 s, p > 0.6663 |
| PBR vs. iNBR | Δt = 1.2161 s, p > 0.1999 | Δt = t = 0.1360 s, p > 0.8792 |
3.3. Subject-wise expressions of cNBR and iNBR are tightly coupled
The primary goals of this experiment were to investigate whether the PBR observed in the visual cortex is the underlying force behind either or both cNBR and iNBR. To address this question, we examined the strength of the relationship between the PBR and both the cNBR and iNBR, and compared it with the strength of their own relationship (between two NBRs). We compared the subject-wise expressions of the BOLD responses that were computed by averaging the β-coefficient of the significant voxels within the generated ROIs. If PBR was the underlying force to induce cNBR and/or iNBR, then one would expect that the subject-wise expression of the PBR has a higher correlation with the expressions of cNBR and/or iNBR, than the corresponding correlation between the two NBRs’ subject-wise expressions. On the other hand, if the iNBR was correlated more strongly with cNBR than PBR, then it is less likely that PBR would be the solo driving force of the iNBR or cNBR. Fig. 7 shows the scatter plots of the subject-wise mean expressions of (1) iNBR versus cNBR, (2) PBR versus cNBR, and 3) PBR versus iNBR, for both attended (a–c) and unattended (d–f) unilateral visual hemifield stimulation. As depicted in Fig. 7, the subject-wise expressions of the cNBR and iNBR were highly correlated with each other in both the attended (r = 0.8580, p < 1.0605e-08) and unattended (r = 0.8693, p < 8.2796e-09) conditions. The PBR was also shown to be significantly associated with cNBR (attended: r = −0.6408, p < 4.2085e-4, and unattended: r = −0.6466, p < 3.5784e-4) and iNBR (attended: r = −0.6131, p < 8.6863e-4, and unattended: r = −0.7032, p < 6.1550e-5). Although the cNBR and PBR were obtained from adjacent regions located in the same hemisphere, the cNBR showed significantly higher correlation with the iNBR (in the opposite hemisphere) than with the PBR (attended: z = 1.8037, p < 0.0357, and unattended: z = 1.9016, p < 0.0287; one-tailed), suggesting that the PBR is less likely to directly associate with either of the two NBRs. These findings suggest that the two NBRs might not be driven exclusively by the PBR. Together with the results in the previous section, they indicate that the two NBRs have a common underlying neural and/or vascular mechanisms which might be different than the ones that give rise to PBR.
Fig. 7.
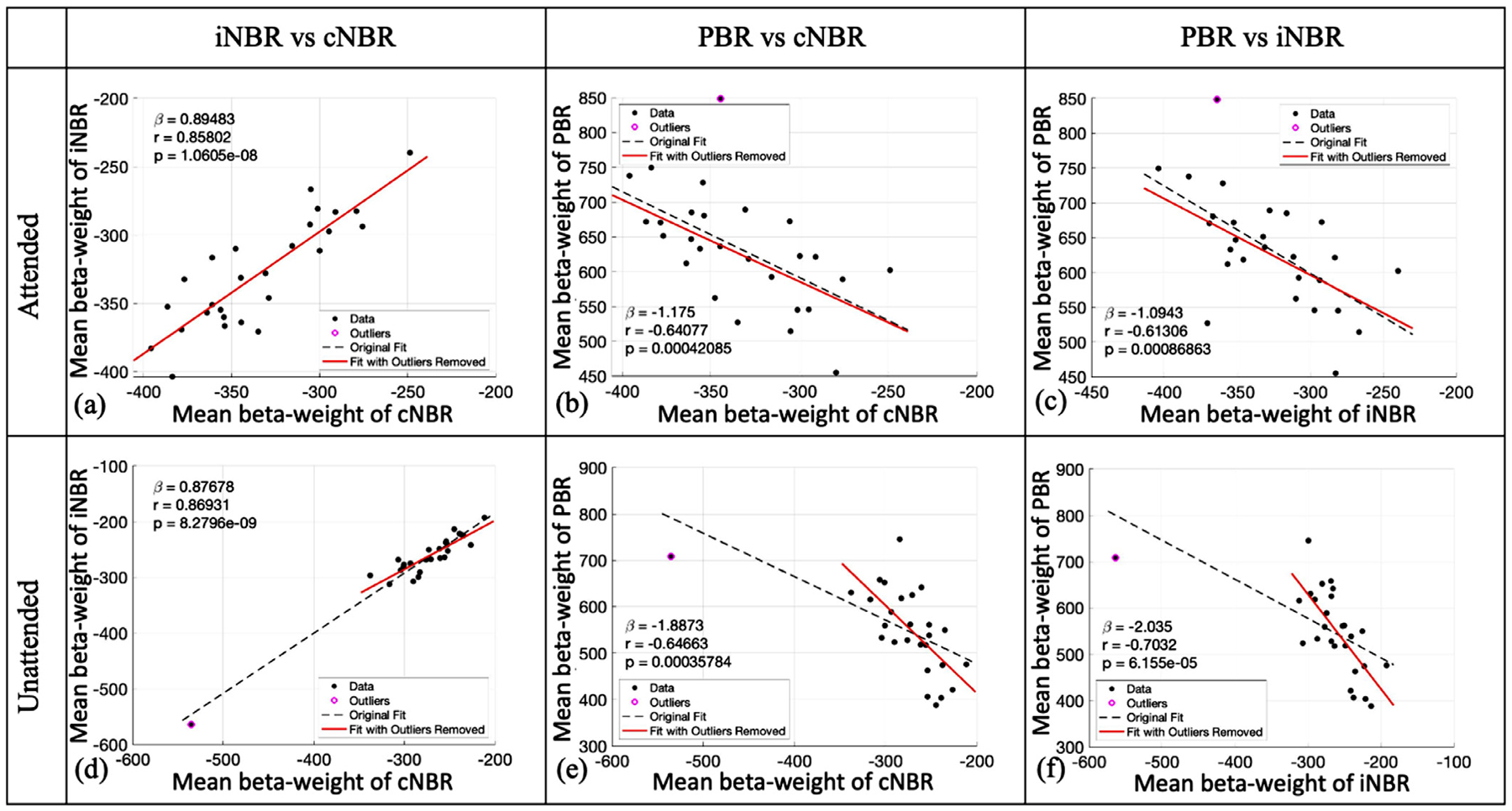
Correlation of the mean amplitudes of the BOLD responses to the visual stimuli in the attended (a–c) and unattended (d–f) conditions. The value of the slop, Pearson correlation coefficient, and the p-value of PCC are presented for each case. The dashed black lines represent the regression lines with all the data, and the solid red lines represent the regression lines with data after outliers were removed. As is depicted, regardless of the attention condition, the subject-wise expression of the iNBR and cNBR are significantly more correlated with each other than each one with the PBR.
3.4. Functional connectivity cannot account for the high coupling between cNBR and iNBR
Results in previous sections provided evidence that cNBR and iNBR are more significantly coupled to each other (similar HRFs and highly correlated subject-wise expressions) than each one with the PBR, highlighting the possibility of having similar underlying mechanism. In this experiment, we investigated whether the inter-hemispheric functional connectivity in the visual cortex could account for observing such high coupling between the two NBRs. While the regions with PBR and NBR responded to the external stimuli with opposite direction, they showed a strikingly high positive resting-state functional connectivity in all participants. Furthermore, there were no significant differences in the interhemispheric functional connectivity of the regions with NBRs (iNBR and cNBR) versus the interhemispheric functional connectivity of the iNBR with regions of PBR (|t| < 0.38, p > 0.70; two hemifields data aggregated; see Fig. 8 for more details). In other words, the strong coupling between the two NBRs cannot be explained by the existing interhemispheric functional connectivity between them since the same level of functional connectivity holds between iNBR and PBR. The same relationship holds when the unilateral stimuli presented in the left hemifield (|t| < 0.24, p > 0.81), and also when the stimuli presented in the right hemifield (|t| < 0.50, p > 0.61), as shown in Fig. 8. Furthermore, it should be noted that the relationship between the subject-wise expression of PBR and iNBR is in the opposite direction of the relationship between their FC. In other words, in the resting-state, increasing MR signal in the regions with PBR is associated with increase of the MR signal in the regions with NBR in the other hemisphere, whereas in the task-evoked BOLD signal, the increase in MR signal (PBR) is associated with a decrease in MR signal (iNBR) in the other hemisphere.
Fig. 8.
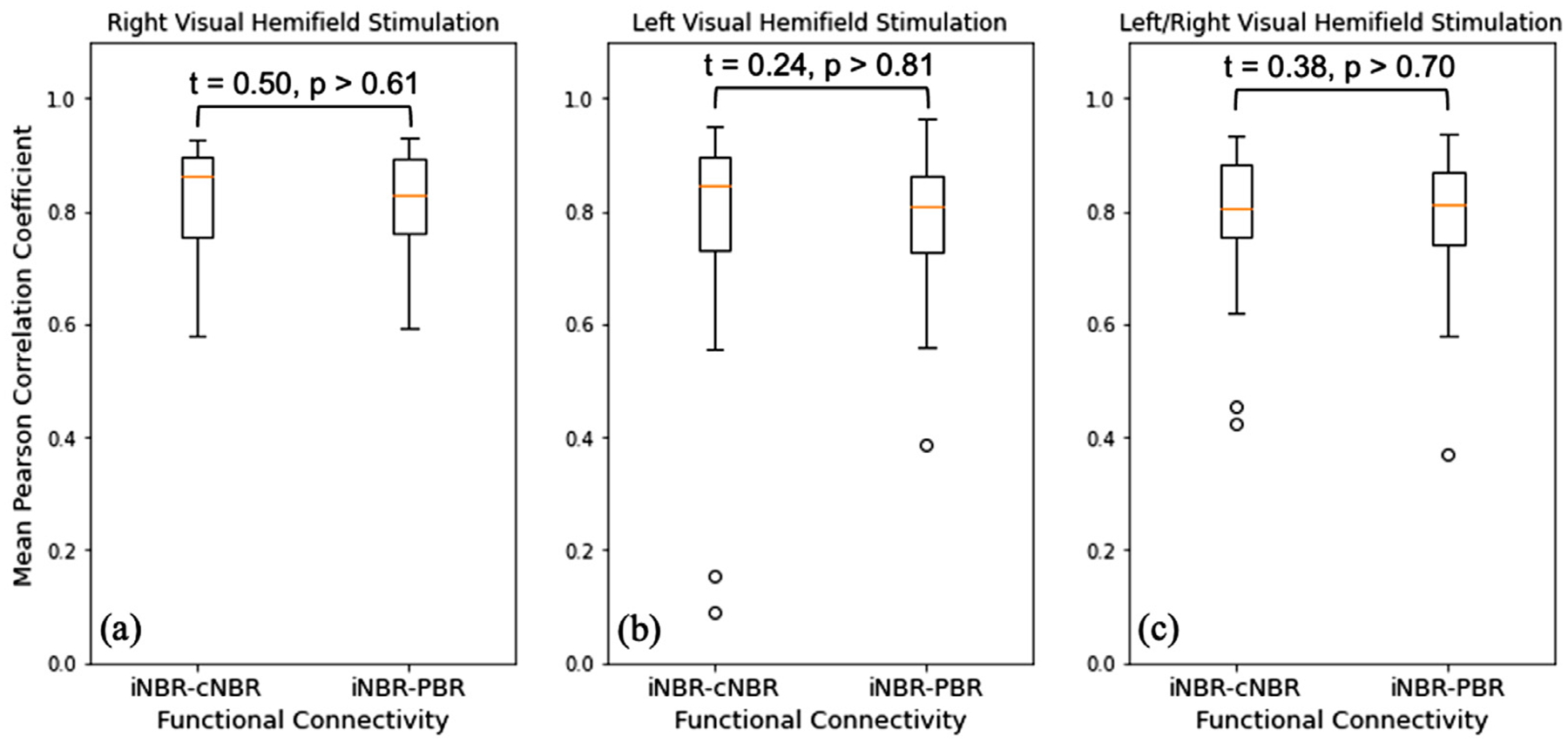
Subject-wise interhemispheric functional connectivity results between regions with iNBR and cNBR are depicted using Pearson correlation coefficient and a boxplot, alongside the subject-wise functional connectivity between regions with iNBR and PBR for (a) right visual hemifield stimulation, (b) left visual hemifield stimulation, and (c) the mean of the two functional connectivity measures. The results of the group differences are also presented in each plot using Student t-test.
3.5. The spatial pattern of the iNBR is more similar to the PBR’s pattern than to the cNBR’s pattern
Considering the substantially high correlation between the subject-wise expressions of iNBR and cNBR, and their similar HRFs, it is natural to examine whether the major callosal pathways which often connect interhemispheric homotopic regions can facilitate the creation of the two synchronized NBRs in the opposing hemisphere. Alternatively, the PBR could also be the force underlying the iNBR through these interhemispheric pathways, as it has been postulated in the past (Bocci T et al., 2014; Fabri et al., 2011; Schäfer et al., 2012). To this end, the interhemispheric spatial pattern similarities between the BOLD signals (both positive and negative) in the two hemispheres were quantified using DSC. If the interhemispheric callosal pathway facilitates such high coupling between the iNBR and cNBR, then the spatial patterns of the observed cNBR and iNBR should be approximately symmetric around the mid-sagittal plane; this can be estimated by the level of interhemispheric overlap. On the other hand, if the PBR was the underlying force for the iNBR through the callosal pathway, then the PBR and iNBR should demonstrate significantly higher interhemispheric spatial similarity. To visualize the interhemispheric spatial similarity between PBR, cNBR, and iNBR, Fig. 9 shows the spatial patterns of the BOLD responses in the two hemispheres of a single subject, projected on the inflated surface of the cerebral cortex, and subsequently on the spherical surface, during the attended unilateral visual stimulation. The approximate boundaries of different visual areas are delineated using a FreeSurfer extension (Benson et al 2014) for this specific subject, to qualitatively examine any relationship between the extent of the iNBR and cNBR and retinotopic regions of the visual cortex. Since we did not perform actual retinotopic mapping in our fMRI task, we did not anticipate to see any quantitatively verified relationship.
Fig. 9.
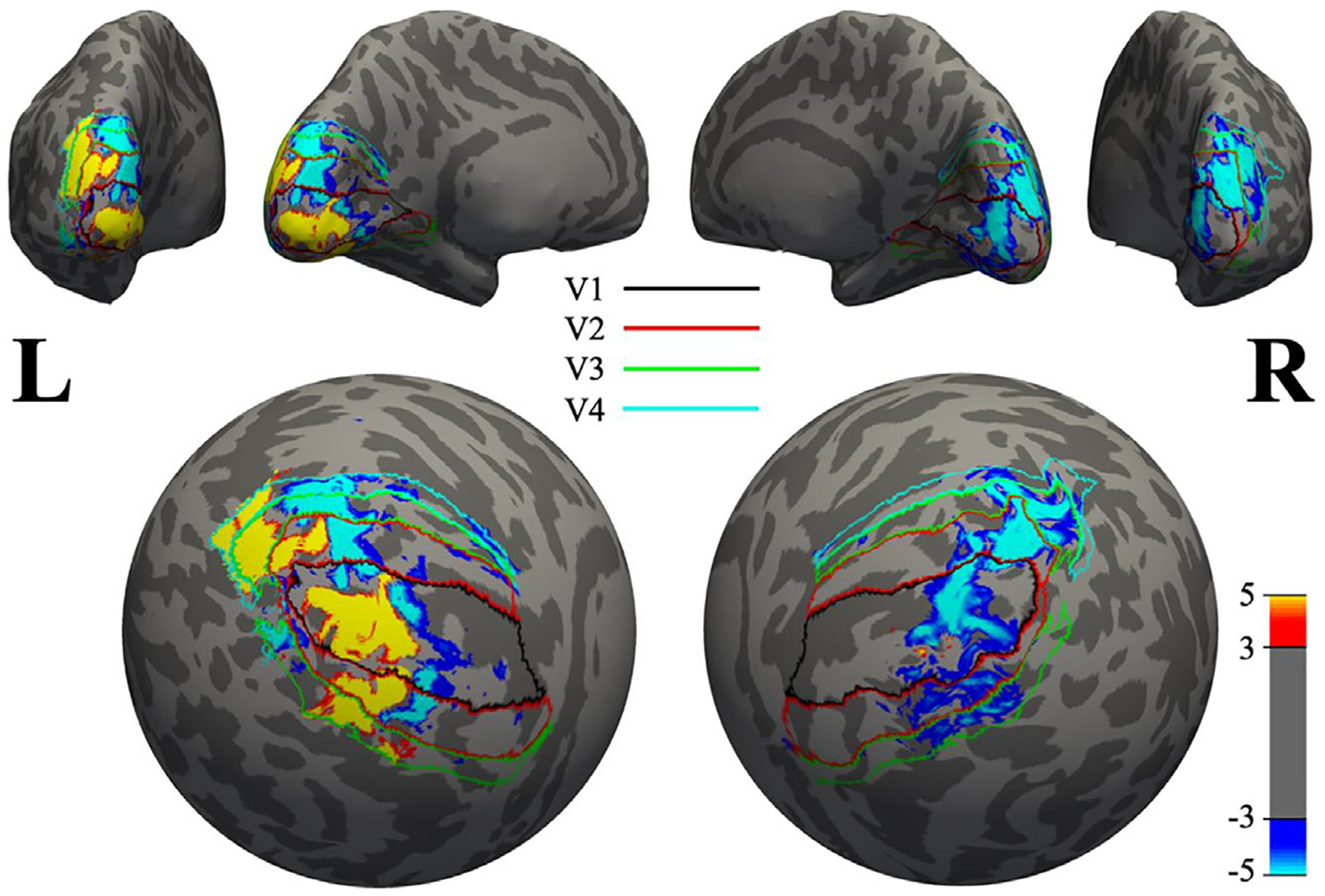
Spatial distribution of the PBR, cNBR, and iNBR overlaid on a sphere (inflated brain). Boundaries of different visual areas V1 to V4 are depicted using different colors. As is demonstrated here, the iNBR has a higher spatial correspondence to the PBR than to cNBR. Note that spatial smoothing is carried out only for improved visualization.
The interhemispheric spatial similarity between regions of iNBR and cNBR was very low (DSC = 0.0532 ± 0.0324; mean ± SD) and even significantly lower (p < 0.0084) than the interhemispheric spatial similarity between regions of iNBR and PBR (DSC = 0.0720 ± 0.0402; mean ± SD) for the attended condition. This result suggests that it is unlikely that the coupling of the two NBRs is facilitated through interhemispheric structural connection such as callosal pathways. Therefore, different regulatory mechanism, possibly involving distal regions with access to both hemispheres are more likely to be responsible for such high coupling between two NBRs.
3.6. Performance correlates and attention dependency of the cNBR and iNBR
The aim of this analysis was to examine whether either the cNBR or the iNBR has any significant association with the task performance. We previously reported that the NBRs detected from the DMN are attention-specific and are correlated with task performance (Parker and Razlighi 2019). Here we aim to: (1) examine whether the same characteristics hold for NBRs detected from ipsilateral and/or contralateral visual cortex, (2) assess whether attention modulates both the iNBR and cNBR in a similar manner, and (3) compare the possible dependence of the cNBR and iNBR on attention to the corresponding dependence of the PBR. Using PCC between subject-wise expression of BOLD signal and median response-time, we found that the expression of the PBR, cNBR and iNBR did not show any statistically significant relationship with performance (details in supplementary results). We next assessed whether attending or ignoring a visual stimulus modulates the magnitudes and dynamics of the cNBR and/or iNBR in the visual cortex. We found that attention increased the magnitude of the NBRs’ HRFs around 3, 4 s following the onset of stimulus, and shortened the return time to the baseline (details in supplementary results). The results indicate that unlike the NBR detected in the DMN, the iNBR and cNBR are not attention specific however attention modulates both iNBR and cNBR similarly suggesting common underlying mechanisms.
4. Discussion
In this paper, we investigated the characteristics of the NBRs elicited simultaneously in the human visual cortex contralateral and ipsilateral to a unilateral visual stimulation. To the best of our knowledge, investigating the spatial and temporal properties of the cNBR and iNBR due to the same visual stimulation has not been reported before. First, we demonstrated that the magnitudes of both NBRs increased linearly with the duration of the visual stimulus. While the linearity of PBR with respect to the stimulus duration has been previously investigated (Boynton et al., 1996; Dale and Buckner, 1997; Huettel et al., 2000), investigations of such linearity in the NBR are rare (see (Shmuel et al., 2002) for cNBR in visual cortex, and (Klingner et al., 2010) for iNBR in somatosensory cortex). To the best of our knowledge, assessment of linearity with respect to stimulus duration for simultaneous iNBR and cNBR has not been reported previously. Demonstrating the linearity is of paramount importance since most of the existing fMRI data analysis methods to extract the BOLD response, including the GLM and FIR deconvolution methods used in this work, are conditioned on an underlying linearity assumption.
Next, we found no significant difference between the amplitudes and dynamics of the HRFs extracted from the cNBR and iNBR after multiple comparison correction. In contrast, we found significant differences in both amplitudes and dynamics of the PBR HRFs and the two NBRs. Similar to our findings, Shmuel et al. demonstrated the difference between the falling edge time of the PBR and cNBR in the visual cortex (Shmuel et al. 2002). Liu et al. reported differences in the onset and falling edge times of the iNBR and PBR in the somatosensory cortex (Liu et al. 2011). Furthermore, consistent with the results of (Shmuel et al., 2002), we also found significant correlations between the expression of the cNBR and PBR. However, strikingly, the subject-wise expression of cNBR was correlated with iNBR (obtained from the ipsilateral hemisphere) with a significantly higher correlation than with the PBR (obtained from adjacent regions), which has not been reported before. The fact that the HRF of the cNBR is different than that of the proximal PBR but similar to iNBR detected from the opposite hemisphere suggests that there is a common force underlying both NBRs; However, the PBR is less likely to be the underlying force, since the subject-wise expressions of the two NBRs showed significantly higher correlation than the correlation between each NBR with the PBR. Furthermore, our results suggest that while there is a strong interhemispheric functional connectivity between regions of iNBR and cNBR, the same level of functional connectivity also exists between the regions of iNBR and PBR; thus the functional connectivity is unlikely to be the underlying common mechanism for such strong coupling between the two NBRs. Interestingly, we demonstrated and reported a new finding that the iNBR regions have a very small interhemispheric topographical spatial correspondence with the PBR regions (<8% on DSC) as well as the cNBR regions (<6% on DSC) in the opposite hemisphere. While the spatial correspondence of iNBR was significantly higher with PBR than with cNBR, they were both too small for being considered homotopic regions. This finding suggests that the two NBRs detected in the two different hemispheres and showing almost identical magnitudes and dynamics cannot be regulated by the interhemispheric callosal pathways. If they were regulated by the interhemispheric callosal connections, one would expect that the spatial similarity between the two NBRs was higher than the one between iNBR and PBR regions.
We also found no significant relationship between task performance and NBRs while PBR showed a non-significant correlation trend with the response-time. While behavioral correlates of PBR have been shown previously (Pessoa et al., 2002), reports on the behavioral correlates of the NBR are inconsistent. Kastrup et al., reported significant correlation between the task performance and the iNBR in human somatosensory cortex during unilateral median nerve stimulation (Kastrup et al., 2008). However, Schäfer et al. (2012) did not find statistically significant correlation under the same conditions. In our experiment, the two NBRs showed no significant correlation with the task performance. This could possibly be due to the lower magnitude of the NBRs (thus lower signal to noise ratio) relative to the PBR. Therefore, a study with a larger number of subjects is warranted to address this question in the future.
Finally, we showed that attending to visual stimuli, slightly but significantly increased the peak magnitude of the ipsilateral NBR. This finding is consistent with the existing works mostly reporting an enhancement in the overall magnitude of the NBR with attention (Bressler et al., 2013; Heinemann et al., 2009; Müller et al., 2004). However, in our results, the peak amplitude of PBR did not change with attention. This is in contrast to previous reports on attentional modulation of the PBR (Bressler et al., 2013; Buracas and Boynton, 2007; Gandhi et al., 1999). Furthermore, by analyzing each time point of the extracted HRFs, we found significant attentional modulations on the HRFs of cNBR in the return to baseline around 8 s after stimulus onset. Similarly, by comparing the dynamics of the HRFs, we also found significant attentional modulations in the HRFs of iNBR in the falling edge time. Fig. S2b and S2c showed release from inhibition-like time-courses in the negative BOLD signals in the attended condition. Specifically, the falling edge following the cessation of the stimulus in the attended condition takes place earlier than its counterpart in the unattended condition. This attentional modulation in human visual cortex is similar to the release from inhibition demonstrated in macaque monkey visual cortex by (Shmuel et al., 2006), in the negative neurophysiological response associated with the NBR. Together, these findings highlight the different influence of attention on the PBR in comparison to NBRs.
The human brain hemodynamic response to any extrinsic stimulation is often more negative than positive. However, depending on the regions where the NBRs are detected, unlike PBR, the neuronal and/or vascular mechanism underlying NBR might be different. For instance, the NBR often observed in the DMN may have completely different mechanism than the one observed in the sensory or motor cortices. Our main focus in this study was on the NBRs detected in the visual cortex. We have previously reported the attention specificity of the NBR from the DMN regions, and its association with task performance (Parker et al., 2019). While in the current study we observed that attention slightly modulated the magnitude of the NBR in the visual cortex, the disengagement from the task did not completely eliminate neither of the iNBR nor cNBR, as it did for the NBR detected from DMN regions. In addition, in contrast to the correlation of task performance with the NBR in the DMN, the subject-wise expression of the NBR in the visual cortex was not correlated with task performance. All together, these findings suggest that NBRs extracted from different regions of the brain may have different underlying neuronal and/or vascular mechanisms. Therefore, it is important for future studies to identify the type of NBR they are investigating to remove any confusion about their results.
Our moderate size pool of samples may seem insufficient to detect any differences between the magnitudes and dynamics of the NBRs HRF. While we cannot rule out this possibility, we emphasize that with the same sample size we did detect significant differences between the magnitudes and dynamics of the two NBRs when compared with the PBR HRFs. Therefore, even if there is any significant difference between the HRF of the two NBRs, it should be substantially smaller than the difference observed between NBRs and PBR HRFs. Furthermore, we performed power analysis which showed that we have more than 85% power to detect significant differences between the amplitudes of the two NBR HRFs for differences as small as half a size of differences observed between the two NBRs and the PBR. On the same note, one might also argue that the lack of correlation between the NBRs (as well as PBR) and performance might be due to the limited sample size of this study. Again, we emphasize that with similar sample size we were able to show previously that the NBR detected in the DMN was correlated with performance (Parker and Razlighi, 2019). Future replication of our findings with a higher number of samples is warranted to ultimately rule out the effect of moderate sample size on our findings.
Recently, Zhou et al. (2018) reported a non-linearity in the PBR for stimuli duration ranging from 17 ms to 533 ms. Their results do not contradict our findings since our stimuli durations are longer and our study focus is on NBR; However, their results call for future studies to assess the linearity of the NBRs for stimuli duration shorter than 1 s. While we used bootstrapping to prevent any bias in our voxel selection for evaluating the linearity of the NBRs, and we normalized each trial regressor to have unit area under the curve, we feel that our task design is not optimal to test the linearity. Thus, we cannot completely rule out the possibility of having different HRF shape in response to different stimuli durations. Future studies with long inter-stimulus-interval (>20 s) are required to completely address this concern. Finally, since we were only interested in the laterality of the visual and auditory stimulations in this study, we did not use a retinotopically-oriented visual task. While our preliminary results did not reveal any association between the NBRs and different visual area, we believe that future studies with optimal retinotopic mapping are required to examine the existence of any association between the NBRs and different visual areas.
4.1. On the origin of NBR
Several hypotheses have been made with regard to the mechanisms underlying NBRs. Here, we discuss how our findings are in agreement or disagreement with or suggest modifications to the existing hypotheses. As was reported previously by (Shmuel et al., 2003; Smith et al., 2004; Chen et al., 2005; Kastrup et al., 2008; Liu et al., 2011; Schäfer et al., 2012; Mullinger et al., 2014), passive vascular/blood steal hypothesis (Harel et al., 2002; Boas et al., 2008) cannot account for the observed task-evoked iNBR in the ipsilateral hemisphere, because the arterial systems of the two hemispheres are quasi-independent.
The venous back-pressure hypothesis (Shmuel et al., 2006; Boas et al., 2008; Goense et al., 2012) assumes that the increase in CBF to the PBR region increases the pressure in adjacent draining veins, thus slowing down the draining of blood from NBR regions adjacent to the PBR. However, for the venous back-pressure to be responsible for iNBR detected in the opposite hemisphere, the increased pressure needs to be generated at the level of the sagittal sinus. This means that the compliance of the sagittal sinus has to be sufficiently sensitive to react to the partial activation of the visual cortex. In addition, one might expect that any changes in the pressure of the sagittal sinus should evenly affect the ipsilateral hemisphere’s regions adjacent to the sinus, which is not the case for the observed iNBR in this study.
Suppression of neuronal activity (Shmuel et al., 2002, 2006; Smith et al., 2004; Schäfer et al., 2012; Mullisnger et al., 2014) could be responsible for observing the almost identical bilateral NBRs (i.e., iNBR and cNBR) facilitated by the existence of major interhemispheric pathways through the corpus callosum, as reported previously (Bocci T et al., 2014; Fabri et al., 2011; Schäfer et al., 2012). However, our findings indicate that the regions with iNBR and cNBR do not have the required homotopic spatial pattern similarity to be connected with major callosal pathways, making the suppression of neuronal activity hypothesis less likely to be the only force underlying the NBRs. Another possibility is that the region showing PBR induces the two NBRs via afferent signal of similar magnitude to both hemispheres to suppress activity in the unstimulated visual regions. However, neither iNBR nor cNBR covers the entire unstimulated visual areas (see Figs. 3 and 9). In addition, any similar or different retinotopic regions on the two hemispheres can be stimulated simultaneously and the associated PBR can be detected in the corresponding retinotopic regions. Therefore, it is uncertain how the visual cortex in one hemisphere would know whether the other hemisphere’s visual cortex or even which retinotopic region of it has been stimulated at the same time to exclude it. Having said that we cannot rule out the possibility that the neural suppression is the underlying force for the two NBRs. In fact, we believe it is not even possible to rule out the neural suppression hypothesis using fMRI alone. Nevertheless, we have shown evidence that if neuronal suppression is the underlying mechanism, it is possible that this mechanism is similar for both NBRs and that it tend to be regulated through distal regions with long projections to the two hemispheres.
Finally, a neurally controlled active blood regulation mechanism, as proposed in the “blood flow control” hypothesis (Smith et al., 2004), raises the possibility of having highly developed neural system for controlling and redistributing CBF throughout the entire brain. Our results are supportive of this hypothesis and indicate that at least the blood flow to the two hemispheres’ visual cortices is regulated by distal/deep brain structure that projects to the two hemispheres. If such a system exists, then it should have the capacity to tightly control the CBF across the two hemispheres, to generate highly coupled BOLD responses with similar magnitudes and dynamics as we have shown in this study. It has been previously reported that small clusters of neurons in the brain stem, basal forebrain, and thalamus have long projections of unmyelinated axons that are directly or indirectly (through GABA interneurons) innervate intracortical arterioles (Cipolla, 2009). Neuromodulators such as noradrenaline, serotonin, dopamine, and acetylcholine from these neurons are shown to influence CBF. For instance, stimulation of cells with acetylcholine in the basal forebrain dilated intracortical vessels within the cortex but did not alter the upstream pial arteries, causing drastic changes in CBF distribution throughout the whole brain (Iadecola et al., 1997). Altogether, this evidence and our results suggest that the brain CBF distribution is not driven entirely by activated or deactivated neurons but might be rather strategically coordinated and regulated through a separate subsystem with access to both hemispheres. However, the existence of such a system should not be considered as evidence against the neuronal suppression hypothesis. Instead, we believe that it is unlikely that this system would restrain the blood flow to a brain region without any level of neuronal suppression. Future studies are warranted to investigate the mechanisms that harmonize this system-level regulation of the distribution of the blood flow in the brain and the brain’s neural activation/suppression.
5. Conclusion
Our findings provide evidence that the two task-evoked NBRs due to unilateral visual stimulation are linearly related to the stimulus duration. Their extracted HRFs have similar magnitude and dynamics, while showing differences compared to the HRF extracted from PBR. The subject-wise expression of the cNBR is tightly coupled with that of the iNBR in the opposite hemisphere and significantly less correlated with the PBR detected in its vicinity. Inversely, the inter-hemispheric similarity of the spatial patterns of the iNBR and the PBR is higher than that of the iNBR and cNBR. Neither of the PBR nor the two NBRs predict the task performance. Moreover, while attention did not alter the timings and peak magnitude of the PBR, both magnitude and return to baseline of the iNBR and cNBR (although not significant for cNBR) were modulated by attention. Our findings suggest that common neural and/or vascular mechanisms underlie the cNBR and iNBR. These mechanisms are possibly different from the mechanisms that give rise to the PBR, and they might involve subcortical brain structures with projections to both hemispheres.
Supplementary Material
Acknowledgments
This work was supported by the NIH-NIA under Grant K01 AG044467 and Grant R01 AG057962.
Footnotes
Declaration of Competing Interest
None
Credit authorship contribution statement
Hengda He: Conceptualization, Methodology, Software, Formal analysis, Data curation, Investigation, Visualization, Writing – review & editing. Nabil Ettehadi: Conceptualization, Investigation, Writing – review & editing. Amir Shmuel: Conceptualization, Writing – review & editing. Qolamreza R. Razlighi: Conceptualization, Methodology, Data curation, Investigation, Funding acquisition, Resources, Project administration, Visualization, Writing – review & editing, Supervision.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi: 10.1016/j.neuroimage.2022.119440.
Data and code availability statement
The datasets in the current study are available from the corresponding author on reasonable request. FSL (V5.0.7) (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/) and in-house-developed packages were used in the preprocessing of fMRI data. Structural scans were processed using FreeSurfer (V5.3) (Fischl et al., 2002; Fischl et al., 2004). Slice timing correction was applied using Filter-Shift (Parker et al., 2017). Spatial normalization was performed using LG-RBSN (He and Razlighi, 2022). First-level analysis was performed via multiple regression using an in-house-developed software package in Python. The code for Filter-Shift and LG-RBSN have been shared on our laboratory website (https://qnlab.weill.cornell.edu/research/pre-processing-fmri-data) as well as our laboratory github repository page (https://github.com/QuantitativeNeuroimagingLaboratory).
References
- Andersson JLR, Skare S, Ashburner J, 2003. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. Neuroimage 20 (2), 870–888. [DOI] [PubMed] [Google Scholar]
- Benson NC, Butt OH, Brainard DH, Aguirre GK, 2014. Correction of distortion in flattened representations of the cortical surface allows prediction of V1–V3 functional organization from anatomy. PLoS Comput. Biol 10 (3), e1003538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boas DA, Jones SR, Devor A, Huppert TJ, Dale AM, 2008. A vascular anatomical network model of the spatio-temporal response to brain activation. Neuroimage 40, 1116–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocci T, Pietrasanta M, Cerri C, Restani L, Caleo M, Sartucci F, 2014. Visual callosal connections: role in visual processing in health and disease. Rev. Neurosci 25 (1), 113–127. [DOI] [PubMed] [Google Scholar]
- Boynton GM, Engel SA, Glover GH, Heeger DJ, 1996. Linear systems analysis of functional magnetic resonance imaging in human V1. J. Neurosci 16 (13), 4207–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressler DW, Fortenbaugh FC, Robertson LC, Silver MA, 2013. Visual spatial attention enhances the amplitude of positive and negative fMRI responses to visual stimulation in an eccentricity-dependent manner. Vis. Res 85, 104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buracas GT, Boynton GM, 2007. The effect of spatial attention on contrast response functions in human visual cortex. J. Neurosci 27 (1), 93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Tyler CW, Liu CL, Wang YH, 2005. Lateral modulation of BOLD activation in unstimulated regions of the human visual cortex. Neuroimage 24 (3), 802–809. [DOI] [PubMed] [Google Scholar]
- Cipolla MJ, 2009. The cerebral circulation. Integr. Syst. Physiol. Mol. Funct 1 (1), 1–59. [Google Scholar]
- Dale AM, Buckner RL, 1997. Selective averaging of rapidly presented individual trials using fMRI. Hum. Brain Mapp 5 (5), 329–340. [DOI] [PubMed] [Google Scholar]
- Fabri M, Polonara G, Mascioli G, Salvolini U, Manzoni T, 2011. Topographical organization of human corpus callosum: an fMRI mapping study. Brain Res. 1370, 99–111. [DOI] [PubMed] [Google Scholar]
- Fischl B, et al. , 2002. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33, 341–355. [DOI] [PubMed] [Google Scholar]
- Fischl B, et al. , 2004. Automatically parcellating the human cerebral cortex. Cereb. Cortex 14, 11–22. [DOI] [PubMed] [Google Scholar]
- Gandhi SP, Heeger DJ, Boynton GM, 1999. Spatial attention affects brain activity in human primary visual cortex. Proc. Natl. Acad. Sci 96 (6), 3314–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goense J, Merkle H, Logothetis NK, 2012. High-resolution fMRI reveals laminar differences in neurovascular coupling between positive and negative BOLD responses. Neuron 76, 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goutte C, Nielsen FA, Hansen KH, 2000. Modeling the hemodynamic response in fMRI using smooth FIR filters. IEEE Trans. Med. Imaging 19 (12), 1188–1201. [DOI] [PubMed] [Google Scholar]
- Harel N, Lee SP, Nagaoka T, Kim DS, Kim SG, 2002. Origin of negative blood oxygenation level–dependent fMRI signals. J. Cereb. Blood Flow Metab 22, 908–917. [DOI] [PubMed] [Google Scholar]
- He H, Razlighi QR, 2022. Landmark-guided region-based spatial normalization for functional magnetic resonance imaging. Hum. Brain Mapp 43 (11), 3524–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann L, Kleinschmidt A, Müller NG, 2009. Exploring BOLD changes during spatial attention in non-stimulated visual cortex. PLoS One 4 (5), e5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Huang L, 2015. Negative hemodynamic response in the cortex: evidence opposing neuronal deactivation revealed via optical imaging and electrophysiological recording. J. Neurophysiol. 114 (4), 2152–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettel SA, McCarthy G, 2000. Evidence for a refractory period in the hemodynamic response to visual stimuli as measured by MRI. Neuroimage 11 (5), 547–553. [DOI] [PubMed] [Google Scholar]
- Iadecola C, Yang G, Ebner TJ, Chen G, 1997. Local and propagated vascular responses evoked by focal synaptic activity in cerebellar cortex. J. Neurophysiol 78 (2), 651–659. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Bannister PR, Brady JM, Smith SM, 2002. Improved optimisation for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17 (2), 825–841. [DOI] [PubMed] [Google Scholar]
- Schäfer K, Blankenburg F, Kupers R, Grüner JM, Law I, Lauritzen M, Larsson HB, 2012. Negative BOLD signal changes in ipsilateral primary somatosensory cortex are associated with perfusion decreases and behavioral evidence for functional inhibition. Neuroimage 59 (4), 3119–3127. [DOI] [PubMed] [Google Scholar]
- Kastrup A, Baudewig J, Schnaudigel S, Huonker R, Becker L, Sohns JM, Dechent P, Klingner C, Witte OW, 2008. Behavioral correlates of negative BOLD signal changes in the primary somatosensory cortex. Neuroimage 41 (4), 1364–1371. [DOI] [PubMed] [Google Scholar]
- Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, Christensen GE, Collins DL, Gee J, Hellier P, Song JH, 2009. Evaluation of 14 non-linear deformation algorithms applied to human brain MRI registration. Neuroimage 46 (3), 786–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingner CM, Hasler C, Brodoehl S, Witte OW, 2010. Dependence of the negative BOLD response on somatosensory stimulus intensity. Neuroimage 53 (1), 189–195. [DOI] [PubMed] [Google Scholar]
- Liu Y, Shen H, Zhou Z, Hu D, 2011. Sustained negative BOLD response in human fMRI finger tapping task. PLoS One 6 (8), e23839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Gerraty RT, Grinband J, Parker D, Razlighi QR, 2017. Brain atrophy can introduce age-related differences in BOLD response. Hum. Brain Mapp 38 (7), 3402–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logothetis NK, 2008. What we can do and what we cannot do with fMRI. Nature 453 (7197), 869. [DOI] [PubMed] [Google Scholar]
- Mandeville JB, Marota JJ, 1999. Vascular filters of functional MRI: spatial localization using BOLD and CBV contrast. Magn. Reson. Med 42 (3), 591–598 An Official Journal of the International Society for Magnetic Resonance in Medicine. [DOI] [PubMed] [Google Scholar]
- Menon RS, Ogawa S, Hu X, Strupp JP, Anderson P, Uǧurbil K, 1995. BOLD based functional MRI at 4 Tesla includes a capillary bed contribution: echo-planar imaging correlates with previous optical imaging using intrinsic signals. Magn. Reson. Med 33 (3), 453–459. [DOI] [PubMed] [Google Scholar]
- Müller NG, Kleinschmidt A, 2004. The attentional ‘spotlight’s’ penumbra: center-surround modulation in striate cortex. Neuroreport 15 (6), 977–980. [DOI] [PubMed] [Google Scholar]
- Mullinger KJ, Mayhew SD, Bagshaw AP, Bowtell R, Francis ST, 2014. Evidence that the negative BOLD response is neuronal in origin: a simultaneous EEG–BOLD–CBF study in humans. Neuroimage 94, 263–274. [DOI] [PubMed] [Google Scholar]
- Ogawa S, Tank DW, Menon R, Ellermann JM, Kim SG, Merkle H, Ugurbil K, 1992. Intrinsic signal changes accompanying sensory stimulation: functional brain mapping with magnetic resonance imaging. Proc. Natl. Acad. Sci 89 (13), 5951–5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D, Liu X, Razlighi QR, 2017. Optimal slice timing correction and its interaction with fMRI parameters and artifacts. Med. Image Anal 35, 434–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker DB, Razlighi QR, 2019. Task-evoked negative BOLD response and functional connectivity in the default mode network are representative of two overlapping but separate neurophysiological processes. Sci. Rep 9, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessoa L, Gutierrez E, Bandettini PA, Ungerleider LG, 2002. Neural correlates of visual working memory: fMRI amplitude predicts task performance. Neuron 35 (5), 975–987. [DOI] [PubMed] [Google Scholar]
- Razlighi QR, Habeck C, Steffener J, Gazes Y, Zahodne LB, MacKay-Brandt A, Stern Y, 2014. Unilateral disruptions in the default network with aging in native space. Brain Behav. 4 (2), 143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmuel A, Yacoub E, Pfeuffer J, Van de Moortele PF, Adriany G, Hu X, Ugurbil K, 2002. Sustained negative BOLD, blood flow and oxygen consumption response and its coupling to the positive response in the human brain. Neuron 36 (6), 1195–1210. [DOI] [PubMed] [Google Scholar]
- Shmuel A, Augath M, Rounis E, Logothetois NK, Smirnakis S, 2003. Negative BOLD response Ipsi-lateral to the visual stimulus: origin is not blood stealing. In: Proceedings of the 9th International Conference of the Organization for Human Brain Mapping. New York, NY, USA 9:309, NeuuroImage (19)2. [Google Scholar]
- Shmuel A, Augath M, Oeltermann A, Logothetis NK, 2006. Negative functional MRI response correlates with decreases in neuronal activity in monkey visual area V1. Nat. Neurosci 9 (4), 569–577. [DOI] [PubMed] [Google Scholar]
- Smith AT, Singh KD, Greenlee MW, 2000. Attentional suppression of activity in the human visual cortex. Neuroreport 11 (2), 271–278. [DOI] [PubMed] [Google Scholar]
- Smith AT, Williams AL, Singh KD, 2004. Negative BOLD in the visual cortex: evidence against blood stealing. Hum. Brain Mapp 21 (4), 213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy R, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM, 2004. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 23 (S1), 208–219. [DOI] [PubMed] [Google Scholar]
- Stefanovic B, Warnking JM, Pike GB, 2004. Hemodynamic and metabolic responses to neuronal inhibition. Neuroimage 22 (2), 771–778. [DOI] [PubMed] [Google Scholar]
- Woolrich MW, Behrens TE, Smith SM, 2004. Constrained linear basis sets for HRF modelling using Variational Bayes. Neuroimage 21 (4), 1748–1761. [DOI] [PubMed] [Google Scholar]
- Zhou J, Benson NC, Kay KN, Winawer J, 2018. Compressive temporal summation in human visual cortex. J. Neurosci 38 (3), 691–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets in the current study are available from the corresponding author on reasonable request. FSL (V5.0.7) (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/) and in-house-developed packages were used in the preprocessing of fMRI data. Structural scans were processed using FreeSurfer (V5.3) (Fischl et al., 2002; Fischl et al., 2004). Slice timing correction was applied using Filter-Shift (Parker et al., 2017). Spatial normalization was performed using LG-RBSN (He and Razlighi, 2022). First-level analysis was performed via multiple regression using an in-house-developed software package in Python. The code for Filter-Shift and LG-RBSN have been shared on our laboratory website (https://qnlab.weill.cornell.edu/research/pre-processing-fmri-data) as well as our laboratory github repository page (https://github.com/QuantitativeNeuroimagingLaboratory).


