Abstract
Ralstonia solanacearum, a widely distributed and economically important plant pathogen, invades the roots of diverse plant hosts from the soil and aggressively colonizes the xylem vessels, causing a lethal wilting known as bacterial wilt disease. By examining bacteria from the xylem vessels of infected plants, we found that R. solanacearum is essentially nonmotile in planta, although it can be highly motile in culture. To determine the role of pathogen motility in this disease, we cloned, characterized, and mutated two genes in the R. solanacearum flagellar biosynthetic pathway. The genes for flagellin, the subunit of the flagellar filament (fliC), and for the flagellar motor switch protein (fliM) were isolated based on their resemblance to these proteins in other bacteria. As is typical for flagellins, the predicted FliC protein had well-conserved N- and C-terminal regions, separated by a divergent central domain. The predicted R. solanacearum FliM closely resembled motor switch proteins from other proteobacteria. Chromosomal mutants lacking fliC or fliM were created by replacing the genes with marked interrupted constructs. Since fliM is embedded in the fliLMNOPQR operon, the aphA cassette was used to make a nonpolar fliM mutation. Both mutants were completely nonmotile on soft agar plates, in minimal broth, and in tomato plants. The fliC mutant lacked flagella altogether; moreover, sheared-cell protein preparations from the fliC mutant lacked a 30-kDa band corresponding to flagellin. The fliM mutant was usually aflagellate, but about 10% of cells had abnormal truncated flagella. In a biologically representative soil-soak inoculation virulence assay, both nonmotile mutants were significantly reduced in the ability to cause disease on tomato plants. However, the fliC mutant had wild-type virulence when it was inoculated directly onto cut tomato petioles, an inoculation method that did not require bacteria to enter the intact host from the soil. These results suggest that swimming motility makes its most important contribution to bacterial wilt virulence in the early stages of host plant invasion and colonization.
Motility solves many of the problems that confront microbes: it allows them to obtain more or better nutrients, avoid toxic substances or unfavorable environments, find a host, and disperse effectively. Many species of bacteria, including most soil-borne species studied to date, can move by swimming, gliding, twitching, or swarming (37, 58). Swimming motility is mediated by flagella, structures consisting of a long, helical filament anchored in the cell envelope by a flexible hook and basal body complex. The flagellar filament is a hollow tube composed of about 20,000 copies of a single protein called flagellin (FliC) polymerized into a complex helix. Rotation of the flagellum is controlled in the basal body by the flagellar motor switch, which is composed of FliG, FliM, and FliN (37). Many additional proteins make up the flagellar basal body and secretion apparatus, which is evolutionarily related to type III secretion systems (22).
The ability to move toward or within a potential host generally confers a significant selective advantage on bacteria pathogenic to animals (46). However, the role of motility in plant-associated microbes is not as well understood (58). Undefined chemically induced nonmotile mutants of the plant pathogens Erwinia amylovora, Pseudomonas phaseolicola, and P. syringae pv. glycinea were significantly reduced in virulence in assays that required bacteria to actively enter the plant, though all three mutants were fully virulent when injected directly into host tissue (7, 29, 47). In contrast, nonmotile mutants of the crown gall pathogen Agrobacterium tumefaciens were fully virulent (10), although another group found that an A. tumefaciens flagellin mutant induced smaller and fewer tumors even when it was inoculated directly onto wounded plant tissue (15). Moreover, a chemically induced nonmotile A. tumefaciens mutant could not cause disease when inoculated in soil, though it was still fully virulent when applied directly to wounded roots (30). Among nonpathogenic plant-associated microbes, swimming motility increased epiphytic fitness of leaf-colonizing strains of P. syringae (27), while nonmotile strains of the free-living nitrogen-fixing bacterium Azospirillum brasilense and of a plant growth-promoting P. fluorescens strain were much less able to colonize host roots (19, 57). Conversely, nonmotile P. fluorescens biocontrol strains colonized wheat and pea roots as well as wild-type motile strains (9, 33). Some, but not all, data suggest that motility increases competitiveness of Rhizobium strains under field conditions (5, 13, 38). These mixed results permit no generalization about the role of motility in plant-microbe interactions, although it is clear that the stringency of the virulence assay and the method of inoculation can dramatically affect the apparent importance of motility.
Ralstonia solanacearum is a soil-borne gram-negative bacterium that causes bacterial wilt disease in over 200 families of plants, including such mainstay crops as potatoes, tomatoes, peanuts, tobacco, bananas, and plantains, as well as many native plant species (31). Losses caused by the disease are known to be enormous but cannot be accurately estimated because of its large but undocumentable impact on subsistence agriculture and because of the abandonment of wilt-susceptible crops in many parts of the world (31). The pathogen invades plant roots through wounds, notably those created by lateral root emergence. Once inside a susceptible host, the bacteria multiply in the cortical tissue before invading the xylem elements. In a matter of hours, the bacteria spread into the crown and stem through the plant's own vascular system, presumably carried along by the transpirational flow (59). Symptoms of bacterial wilt disease include yellowing and wilting, followed by generalized necrosis and death.
R. solanacearum possesses swimming motility mediated by one to four polar flagella (12). In this species, motility is a quantitative trait; at minimal motility, up to 1% of cells are still moving, while only around 60% of cells are moving at any given time in a culture at maximum motility (17). Motility is coregulated with several known virulence factors in a regulatory cascade ultimately controlled by a cell density-responsive LysR-type global regulator called PhcA. In culture, expression of PhcA increases when the population reaches about 107 CFU/ml (23). PhcA then induces expression of the virulence factors extracellular polysaccharide (EPS), endoglucanase, and possibly others via a complex cascade involving additional regulatory elements (25). At the same time that it induces some virulence factor genes, PhcA represses expression of others by reducing the transcription of a two-component regulator called PehSR. PehSR is a positive regulator of plant cell wall-degrading polygalacturonases, which are also virulence factors. In addition, PehSR positively regulates bacterial motility (3). It is hypothesized that during saprophytic life and early in wilt disease development, when bacterial cell density is low and phcA is not expressed, R. solanacearum cells are motile and highly pectolytic. As bacterial populations increase in the host xylem elements, phcA is expressed, inducing production of EPS and other late-stage virulence factors and reducing motility by repressing pehSR expression (49). However, bacterial motility has not been directly measured in the plant during pathogenesis.
The fact that motility is coregulated with known virulence factors suggests that motility may also contribute to virulence. Although phcA mutants are hypermotile and pehR mutants are completely nonmotile, both strains are strikingly affected in virulence; indeed, phcA mutants are avirulent (11). However, since both are pleiotropic regulatory mutants, it is impossible to deduce the importance of motility in bacterial wilt disease from their behavior. Results from other systems suggest that bacterial motility can play an important ecological role in plant-microbe interactions. However, inconsistent and artificial virulence assays make it difficult to compare results from different groups. Further, much of the research in this area is inconclusive because it was conducted with uncharacterized chemical or spontaneous nonmotile mutants that may have been affected in other traits. To more conclusively determine the role of motility in bacterial wilt disease, we constructed two defined, nonpleiotropic nonmotile mutants of R. solanacearum and measured their virulence on tomato plants using two different assays. We also compared the motility of bacteria in host plant xylem vessels with that of bacteria growing in culture.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. R. solanacearum strains were grown at 28°C in CPG broth (32) or on TZC plates containing CPG plus 1.8% agar and 0.05% 2,3,5-triphenyltetrazolium chloride (35). Escherichia coli strains were cultured at 37°C in Luria-Bertani medium (42). Antibiotics were added as required in the following concentrations (milligram per liter): ampicillin, 50; kanamycin, 25; tetracycline, 15; and gentamicin, 25.
TABLE 1.
Bacterial strains and plasmids used in this studya
| Designation | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli | ||
| BL21(DE3) | hsdS gal (λcI ts857 ind1 Sam7 UV5-T7 gene-1) | 53 |
| DH5α | F−endA1 relA φ80 lacZΔM15 hsdR17 supE44 thi-1 recA1 gyrA96 | 28 |
| R. solanacearum | ||
| K60 | Wild type, race 1, biovar 1, tomato isolate | 35 |
| K71 | K60 pehR::Tn3-uidA, Kmr | 3 |
| K701 | K60 fliC::Gmr | This study |
| K724 | K60 fliM::aphA, Kmr | This study |
| Plasmids | ||
| pBS | Apr | Stratagene |
| pLAFR3 | Tcr | 51 |
| pGEM-T Easy | A-T cloning vector, Apr | Promega |
| pUC18K | pUC18 containing aphA cassette, Kmr | 41 |
| pUCGM | Contains Gmr cassette, Gmr | Novagen |
| pGp5Gm | Plasmid containing Tn5, Gmr | E. Stabb, personal communication |
| pHUHFli | 7.0-kb BamHI fragment containing fliC and fliD in pBS SK+, Apr | This study |
| pFliC1 | 0.85-kb fragment containing fliC in pGEM-T Easy, Apr | This study |
| pFliC2 | pFLIC1 with the 0.85-kb Gmr cassette in BalI site of fliC, Gmr Apr | This study |
| pFliC9 | 1.1-kb AvaI fragment containing fliC in pET29b | This study |
| pFliM1 | 7.5-kb HindIII fragment containing fliM in pBS KS+, Apr | This study |
| pFliM2 | 1.1-kb fragment containing fliM in pGEM-T Easy, Apr | This study |
| pFliM3 | pFLIM2 with the 0.85-kb aphA cassette in BsrBRI site of fliM, Apr Kmr | This study |
pBS, pBluescript; Ap, ampicillin; Tc, tetracycline; Gm, gentamicin; Km, kanamycin.
Growth levels of wild-type and mutant strains were compared in Boucher's minimal medium (BMM) broth supplemented with 0.2% glucose (8). Cultures were inoculated in triplicate to an initial optical density at 600 nm of 0.02, and bacterial growth was measured over time as turbidity using a spectrophotometer. To measure growth of wild-type and mutant strains in planta, we infused leaves of 3-month-old tobacco (cv. Bottom Special) with a 108-CFU/ml suspension of bacteria. At various time points, 1-cm2 leaf punches were ground and dilution plated on TZC plates to quantify bacteria; infusions and platings were triplicated. The ability of bacterial strains to elicit a hypersensitive response (HR) in nonhost plants was tested in 2-week-old cucumber cotyledons as previously described (60).
DNA manipulations.
Isolation of plasmid and chromosomal DNA, restriction mapping, cloning, subcloning, Southern blot hybridization, and PCR were performed by standard procedures (6). R. solanacearum and E. coli were transformed by electroporation as described previously (4). DNA sequencing was performed at the University of Wisconsin—Madison Biotechnology Center. Genomic DNA sequence from the closely related R. solanacearum GMI1000 strain was generously provided by Christian Boucher (INRA-Toulouse, Toulouse, France). DNA sequence was analyzed with software from DNASTAR (Madison, Wis.).
Motility assays.
Motility of wild-type and mutant strains was determined by transferring individual colonies to semisolid motility medium plates (36). After 4 to 6 days at 28°C, motile colonies were surrounded by an even white halo or, occasionally by radiating streaks. We also directly observed motility of bacteria by microscopy after growth in BMM broth or in xylem fluid collected from symptomatic plants. Xylem fluid from infected tomato plants was obtained by cutting plant stems with a razor blade above the cotyledons and collecting the extruding xylem fluid with a large-bore pipetter. Motility was determined microscopically within 10 mins of extraction from xylem vessels by counting motile and nonmotile cells in a known volume of fluid on a hemocytometer slide; bacterial densities were confirmed by dilution plating in triplicate.
Cloning and mutagenesis of the fliM and fliC genes.
A nonmotile R. solanacearum transposon Tn5 mutant was created by standard methods using suicide plasmid pGp5Gm; sequencing of the DNA flanking the Tn5 insertion suggested that the fliM gene had been interrupted. Using this sequence and a putative fliM sequence from the GMI1000 genome, we designed primers to amplify an internal 200-bp fragment of fliM from chromosomal DNA of wild-type R. solanacearum strain K60 (5′GGAAAGGTTCCGTTCAGG3′ and 5′TTCGCTGTACTTCTGGAC3′). The resulting amplified fragment was used to probe a K60 genomic library (54). A 7.5-kb HindIII fliM-containing fragment was subcloned from a hybridizing cosmid into pBluescript, creating pFliM1, which was used to sequence fliM (GenBank accession number [hereafter simply GenBank] AF283286). A 1.1-kb subclone was amplified from pFliM1 using primers 5′ATGCTGCTGTCGTCCAAG3′ and 5′TTCAGCATGCGGTTGACG3′ and cloned into pGEM-T Easy to create pFliM2. To make a nonpolar mutation in fliM, the aphA cassette was inserted into the BsrBRI site of pFliM2, creating pFliM3. The resulting construct was sequenced to confirm in-frame insertion of the aphA cassette.
We designed oligonucleotide primers based on a putative fliC sequence from R. solanacearum GMI1000 to amplify an internal fragment of the fliC gene from R. solanacearum strain K60 genomic DNA (5′GTCCCTCAGCCTCAATACCAA3′ and 5′GCCCTTCAGCAGGTTCAGAAT3′). The resulting 0.8-kb amplified fragment was A-T cloned into the pGEM-T Easy vector to create pFliC1; its identity was confirmed by sequencing. A 7.0-kb BamHI fragment containing fliC was subcloned from the K60 genomic library into pBluescript to create pHUHFli, and the region containing fliC was sequenced (GenBank AF283285). To interrupt the fliC gene, we inserted a 0.85-kb SmaI fragment of the pUCGM gentamicin resistance (Gmr) cassette into the BalI site of fliC to create pFliC2. Both the fliM::aphA and fliC::Gmr interrupted gene constructs were introduced into wild-type R. solanacearum strain K60 by homologous recombination as previously described (4). Transformants with appropriate antibiotic resistance phenotypes were screened for loss of motility on semisolid motility medium plates.
Electron microscopy.
Cultures were grown to a density of 107 CFU/ml in BMM broth supplemented with 0.2% glucose. Cells were placed on copper grids, stained with 1% uranyl acetate, and examined with a Philips model EM-120 electron microscope. For each strain, a minimum of 1,000 cells were examined for presence or absence of flagella and, where applicable, for flagellar morphology.
Overexpression and isolation of flagellin.
A 1.1-kb AvaI fragment containing the fliC open reading frame (ORF) and flanking regions was subcloned from pHUHFli into the T7 expression vector pET29b (Novagen) to create pFliC9. This construct was transformed into the E. coli overexpression strain BL21(DE3), and protein expression was induced with 2 mM isopropylthiogalactopyranoside (IPTG) according to Novagen's instructions. Crude flagellin was isolated as sheared bacterial cell extracts as previously described (56). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 12% gels and stained with Coomassie brilliant blue (6).
Virulence assays.
We measured the virulence of R. solanacearum strains on tomato (highly susceptible cv. Bonnie Best). Virulence of strains on unwounded plants was quantified by soil inoculation as previously described (54). In summary, we poured a 50-ml bacterial suspension onto the soil of 15-day-old plants to achieve a final concentration of about 6.25 × 106 CFU/g of soil. Direct petiole inoculation of tomato plants was as previously described (48). Briefly, we cut the petiole of the first true leaf 0.5 cm from the stem and placed 104, 103, or 102 bacteria in a 2-μl volume onto the freshly cut surface. For both assays, inoculum concentrations were determined turbidimetrically and confirmed by dilution plating. Plants were coded and inspected for wilt symptoms daily for 14 days following inoculation by a rater blind to treatment identity. Plants were rated on a disease index scale of 0 to 4 (0, no wilting; 1, 1 to 25% wilting; 2, 26 to 50% wilting; 3, 51 to 75% wilting; and 4, 76 to 100% wilted or dead). Each assay was repeated in three successive trials. Within each trial we treated 16 plants with each strain, yielding a total of 48 plants for each strain.
Analysis.
We analyzed the data together by combining results from all 48 plants treated with a given strain across the three trials. We analyzed the daily disease indices in a combined analysis of variance (ANOVA) using repeated-measure analysis (50). This analysis accounts for the autocorrelation of disease index scored on successive days. Trial and pathogen strain were treated as fixed effects in the factorial design. All analyses were done using Systat version 5.2 (Systat Inc., Evanston, Ill.).
RESULTS
R. solanacearum is virtually nonmotile in planta.
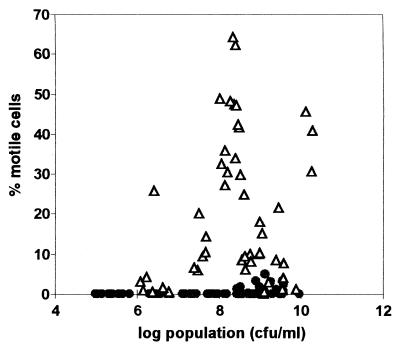
To determine if motility is expressed differentially in plants and in culture, we extracted and quantified bacteria in xylem fluid from infected tomato plants and in BMM broth and viewed them under the microscope. In minimal medium, wild-type strain K60 behaved as previously observed in related race 1 strain AW-1 (17): motility increased with cell density and was greatest (about 60% motile) around 108 CFU/ml before declining with higher cell densities (Fig. 1). Surprisingly, we found that bacteria behaved very differently in host plant xylem vessels. In the plant, R. solanacearum was almost never motile below 5 × 108 CFU/ml and reached maximum motility of only 5% at about 109 CFU/ml (Fig. 1).
FIG. 1.
R. solanacearum motility in culture and in planta. Wild-type strain K60 cells were grown in BMM broth (open triangles) or extracted from midstem xylem vessels of infected tomato plants (closed circles). The percentage of motile cells was determined microscopically by counting motile and nonmotile cells on a hemocytometer slide. Cell density was confirmed by dilution plating in triplicate.
Cloning and analysis of the fliC gene.
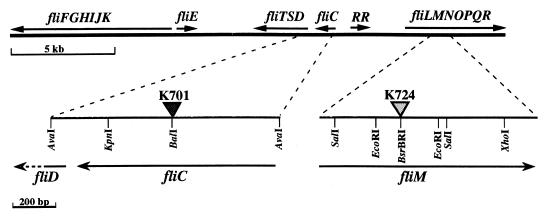
An 850-bp amplified fliC gene fragment hybridized to K60 genomic library cosmid 18-73, which contained several flagellar genes (Fig. 2). The 822-bp fliC ORF encodes a putative protein with a calculated molecular mass of 28,283 Da. The strain K60 FliC is 94% identical at the predicted amino acid level to FliC from the closely related R. solanacearum strain GMI1000 and has at least 35% overall identity with several known flagellins, notably those of Aeromonas caviae (40%; GenBank AF198617), P. fluorescens (40%; GenBank AF034765), and Legionella pneumophila (38%; GenBank X83232). At the N terminus, which is conserved among flagellins from gram-negative bacteria, the first 120 amino acids are over 50% identical to flagellins from Burkholderia pseudomallei (GenBank AF078151) and B. cepacia (GenBank AF080259). The N-terminal region also contains a small, highly conserved 15-amino-acid domain that is over 60% identical to other flagellins (21).
FIG. 2.
Large-scale physical map of R. solanacearum flagellar gene cluster, assembled from related strain GMI1000 genome sequence and wild-type strain K60 genomic library cosmids 19-84 and 18-73. Filled triangle, Gmr gene cassette; stippled triangle, nonpolar aphA mutagenesis cassette; RR, putative response regulator gene of unknown function.
Mutagenesis of the fliC gene.
A construct containing fliC interrupted by a Gmr cassette was recombined into the R. solanacearum strain K60 genome to create K701 (Fig. 2). Southern blot analysis confirmed that K701 contains a single interrupted copy of fliC (data not shown). It is unlikely that other genes are cotranscribed with fliC since there is a stem-loop structure that may function as a factor-independent terminator located 20 bp 3′ of the fliC stop codon and the closest 3′ gene, fliD (encoding HAP2, the flagellar cap/hook-associated protein), is 179 bp away.
Cloning, analysis, and mutagenesis of the fliM gene.
The R. solanacearum K60 fliM gene was identified by sequence flanking the transposon insertion in a nonmotile mutant and by the putative fliM sequence from the GMI1000 genome. A 200-bp PCR fragment internal to fliM hybridized to genomic library cosmid 19-84, which contains fliM and apparently several other genes in the flagellar regulon. The 1,008-bp fliM ORF encodes a putative protein of 335 amino acids with a predicted molecular mass of 38,000 Da. 5′ to fliM lies fliL, with only one base separating the two ORFs. The fliN locus is 20 bp downstream (3′) of fliM; this arrangement suggests that these genes are transcribed in one unit, as in other bacteria, with the promoter located 3′ to the fliL ORF (37). The fliM protein is similar to flagellar motor switch proteins from Pseudomonas putida (43.2% identity; GenBank AF031418) and P. aeruginosa (42.7% identity; GenBank AE004574).
Because fliM is embedded in the fliLMNOPQR operon, we used the aphA cassette to create a nonpolar fliM mutant. This cassette contains a kanamycin resistance marker which is preceded by translation stop codons in all three reading frames and is followed by a consensus ribosome binding site and start codon to ensure continuing translation 3′ of the insertion (41). We cloned the aphA cassette into the BsrBRI site of fliM and recombined this construct into the K60 genome to create K724. Southern blot analysis confirmed that a single copy of fliM was interrupted by the aphA cassette in this strain, and sequencing confirmed that the cassette was inserted properly for continued translation of downstream genes (data not shown).
Phenotypes of nonmotile mutants.
Mutants K701 (fliC) and K724 (fliM) had normal mucoid colony morphology on solid media and grew as well as wild-type strain K60 in minimal media supplemented with 0.2% glucose and in tobacco plant leaves (data not shown). K701 (fliC) caused a normal, wild-type HR when infused into cucumber cotyledons. To directly visualize bacterial motility, cultures of the nonmotile mutants grown in BMM-glucose were compared to wild-type cells under a light microscope. At a cell density of about 108 CFU/ml, when R. solanacearum would normally be expressing greatest motility, up to 60% of the wild-type cells were motile. However, no motile bacteria were ever seen in cultures of strains K701 (fliC) and K724 (fliM) under these conditions. We also examined the motility of bacteria in xylem fluid from infected tomato plants; about 5% of wild-type K60 cells were motile in the xylem fluid (which contained about 109 CFU/ml), but motile K701 or K724 cells were never observed in infected plants. The instability of plasmid vectors in R. solanacearum made it impossible to examine the in planta virulence or motility of K701 or K724 cells complemented with wild-type fliC or fliM genes.
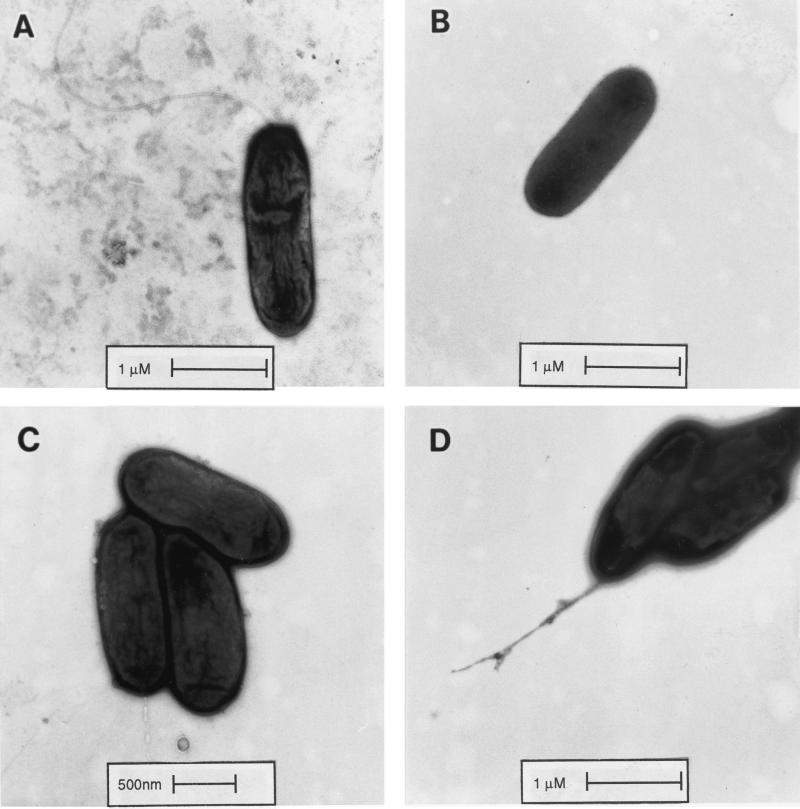
We examined the degree of flagellation and the flagellar morphology of various strains directly by electron microscopy. Approximately 50% of wild-type strain K60 cells had one to five helical polar flagella (Fig. 3A), while the rest were aflagellate, with many broken flagella present. None of the K701 (fliC) mutant cells had flagella, and no broken flagella were present (Fig. 3B). Most of the K724 (fliM) cells were aflagellate as well (Fig. 3C), although abnormal truncated or branched flagella were present on fewer than 10% of the K724 cells, and a few broken flagella were seen in the preparations (Fig. 3D). No flagella on K724 had the long, curved appearance of the wild-type K60 flagella. Nonmotile strain K71 (pehR::gus), which lacks the pehR response regulator that positively controls motility, had no flagellate cells (data not shown).
FIG. 3.
Morphology of wild-type and nonmotile R. solanacearum cells. Cells were grown to 107 CFU/ml, stained with 1% uranyl acetate, and examined by electron microscopy. (A) Wild-type strain K60; (B) K701 (fliC) aflagellate cells; (C) K724 (fliM) aflagellate cells; (D) K724 cells showing abnormal truncated or branched flagella.
Expression and isolation of flagellin.
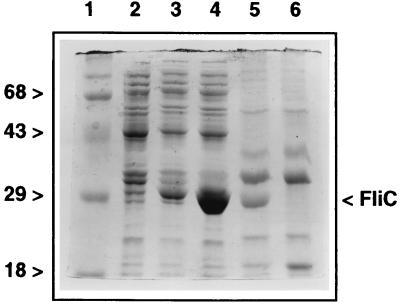
Flagellin is usually present in sheared-cell extracts, the fraction of outer cell-associated protein that can be removed only by high-speed vortexing or blending. Wild-type strain K60 sheared-cell extracts contained an abundant band that closely corresponds to the predicted molecular mass of R. solanacearum flagellin (30 kDa); however, this band was completely absent from sheared-cell extracts of the fliC mutant K701 (Fig. 4, lanes 5 and 6). When the cloned fliC gene was overexpressed in E. coli, a 30-kDa protein band was induced which comigrated with the apparent flagellin from the R. solanacearum sheared-cell extracts (Fig. 4, lane 4).
FIG. 4.
Visualization of flagellin from R. solanacearum and its overexpression in E. coli. Crude flagellin was isolated as sheared-cell extracts. Protein expression was induced in mid-log-phase E. coli BL21(DE3) with 2 mM IPTG. Proteins were separated by SDS-PAGE on 12% gels and stained with Coomassie brilliant blue. Lane 1, molecular weight markers (positions are indicated in kilodaltons); lane 2, E. coli BL21(DE3)(pET29b) induced for 1 h; lane 3, E. coli BL21(DE3)(pFliC9) uninduced; lane 4, E. coli BL21(DE3)(pFliC9) induced for 1 h; lane 5, wild-type strain K60 sheared cell extract; lane 6, K701 (fliC) sheared-cell extract.
Virulence of nonmotile mutants.
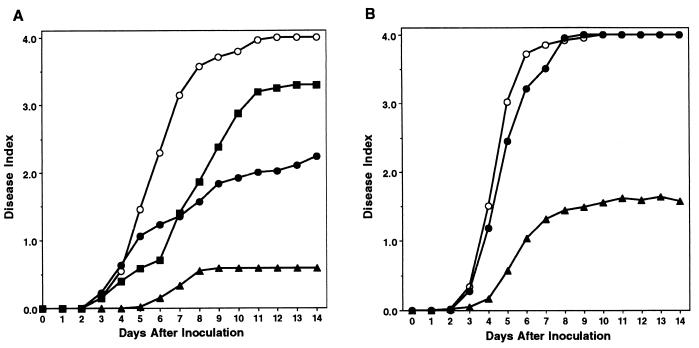
To determine whether the ability to move contributes to invasive bacterial wilt virulence on intact host plants, we inoculated tomatoes with the nonmotile mutants using a natural soil-soak inoculation. Under these conditions, wild-type strain K60 completely wilted nearly all tomato plants by about 8 days after inoculation, with an average disease index of 3.8 (Fig. 5A). Both K701 (fliC) and K724 (fliM) were significantly reduced in virulence, with an average disease index of around 2.4 by the end of the assay. Consistent with previous observations, K71 (pehR), a nonmotile pleiotropic regulatory mutant, had the lowest virulence, with an average disease index of only 1.2 by day 14 postinoculation. Repeated-measures ANOVA demonstrated that the virulence of nonmotile strains K701 (fliC) and K724 (fliM) was significantly different from that of wild-type strain K60 and from that of K71 (pehR) (P < 0.001), but K701 and K724 did not differ significantly from each other by this test.
FIG. 5.
Virulence of R. solanacearum nonmotile mutants on tomato plants by different inoculation methods. Fifteen-day-old tomato plants were inoculated either by soaking the soil to a final bacterial population of about 6.25 × 106 CFU/g (A) or by applying 104 bacteria directly to the cut surface of the first true leaf petiole (B). Plants were rated daily on a disease index scale of 0 to 4; each point represents the mean disease index of 48 plants combined from three separate experiments. (A) Closed triangles, K71 (pehR); closed circles, K701 (fliC); open circles, K60 (wild type); closed squares, K724 (fliM); (B) closed triangles, K71; closed circles, K701; open circles, K60. Repeated-measure ANOVA found that for panel A, the virulence of nonmotile mutants K701 and K724 was significantly different from that of wild-type strain K60 and from that of regulatory mutant K71 (P < 0.001), but the virulence of K701 and K724 was not significantly different. In panel B, the virulence of wild-type and nonmotile strains was not significantly different, though both differed from that of K71 (P < 0.001).
To determine whether a nonmotile mutant could cause disease normally if it did not have to invade and colonize from the soil, we inoculated bacteria directly into the host vascular system via the cut leaf petioles of tomato plants. When 10,000 bacteria were applied to each plant, wild-type strain K60 and K701 (fliC) both wilted all tomatoes by day 10 postinoculation, and K701 was statistically indistinguishable from the wild type. However, K71 (pehR) differed significantly from either K60 or K701 (P < 0.001); K71 wilted only a few of the plants, reaching a final disease index of below 2.0 (Fig. 5B). At lower inoculum levels (1,000 and 100 cells), K60 (wild type) and K701 (fliC) remained indistinguishable from each other in virulence. Maximum disease indices for both strains were 3.8 at 103 CFU/plant and 2.5 at 102 CFU/plant, where about 35% of the inoculated plants remained asymptomatic. pehR regulatory mutant K71 did not cause any detectable disease symptoms at an inoculation level of either 103 or 102 CFU/plant (data not shown).
DISCUSSION
The expression pattern of R. solanacearum virulence genes in culture is well understood, but less is known about how these genes are regulated in the host plant. As bacterial population density increases during growth in broth, the global virulence regulator PhcA induces expression of EPS and endoglucanase while concomitantly reducing polygalacturonase expression and motility by repressing pehSR. In planta studies found that EPS is regulated similarly in tomato plants and in culture (34, 40). However, when we quantified motility of bacteria from the xylem vessels of infected tomato plants, we found that this bacterium was completely nonmotile at low to moderate cell densities in the plant host, becoming only rarely motile (fewer than 5% of cells) around 109 CFU/ml. We note that free bacteria floating in the xylem fluid represent only part of the population in planta; bacteria aggregated in biofilms on vessel walls would not be measured in our assay. However, free bacteria are more likely to be motile than those in biofilms, suggesting that our numbers may actually overrepresent the overall motility of R. solanacearum living in xylem vessels (18).
This result highlights the importance of in planta studies and significantly alters the existing regulatory model. Because pehSR expression is similar in culture and in planta (D. Brown and C. Allen, unpublished results), we suspect the existence of additional regulatory elements intervening between pehSR and the expression of motility. Such elements may respond to conditions inside the plant or to a specific plant signal (which may trigger phosphorylation of the PehR response regulator by the PehS sensor-kinase). Given the complex networks that regulate motility in other systems, additional levels of regulation would not be surprising. Many mammalian pathogens are motile in the external environment or in culture but quickly shed their flagella and become nonmotile once they are inside a host. For example, Bordetella pertussis and B. brochiseptica are motile at temperatures below 37°C, corresponding to conditions outside the host, but at 37°C or above, they become nonmotile and simultaneously produce an array of toxins and other virulence factors (2, 39). Indeed, constitutively motile B. bronchiseptica mutants were avirulent (1), suggesting that motility within the host can be actively disadvantageous to the pathogen.
To separate the effects of the flagellum itself from those of bacterial movement, we attempted to create a paralyzed strain of R. solanacearum (with intact but nonfunctional flagella) by mutating the flagellar motor switch protein gene (fliM). However, most cells of the nonmotile fliM::aphA mutant K724 were aflagellate rather than paralyzed, though a few cells had truncated or branched flagella. The presence of these occasional abnormal flagella in K724 suggests that the aphA cassette did not have a polar effect on downstream genes, since loss of the entire fliL operon generally creates a completely aflagellate phenotype (37).
To determine the importance of pathogen motility in bacterial wilt disease development, we assayed the virulence of nonmotile mutants K701 (fliC) and K724 (fliM) on tomato plants, using a biologically representative soil-soak inoculation that required bacteria to actively find and invade host plant roots from the soil. Under these conditions, the nonmotile mutants caused significantly less disease on tomato plants than the wild-type strain, demonstrating that pathogen motility is necessary for full virulence in R. solanacearum.
Interestingly, the low-virulence phenotype of nonmotile mutant K701 could be rescued by inoculating the strain directly into the tomato stem vascular system. When bacteria were applied to cut leaf petioles, the nonmotile strain was indistinguishable in virulence from the wild-type strain. Indeed, in this assay the nonmotile strain had wild-type virulence even at very low inoculum densities (100 cells per plant). The finding that motility does not appear to be required once the pathogen reaches the stem vasculature is consistent with our observation that wild-type bacteria do not express motility in xylem fluid from the stem. On a biological level, this result demonstrates that flagella make their contribution to virulence before bacteria reach the stem, in the early stages of disease development. Several animal pathogens also depend on motility primarily to invade and colonize hosts. Nonmotile P. aeruginosa cells are much less able to invade mice from burn wounds, and a nonmotile mutant of the fish pathogen Vibrio anguillarum is virtually avirulent in an immersion assay; however, both strains are fully virulent when they are injected directly into the host peritoneal cavity (20, 43).
How might motility contribute to R. solanacearum virulence early in pathogenesis? Swimming motility is obviously necessary for chemotaxis through soil to optimal infection sites on host roots, and from there through the cortex to developing protoxylem tissue. As disease progresses, motility can help the pathogen spread out of infected xylem vessels into adjacent uninfected vessels and xylem parenchyma cells. Flagella may also help bacteria attach to host cells (44, 46); if this is the case, there should be a difference in the ability of wild-type, paralyzed, and aflagellate cells to attach to tomato root surfaces. Finally, swimming motility is usually required for biofilm formation (45, 55). Microscopic observations suggest that R. solanacearum forms biofilms on host xylem vessel walls; these specialized aggregates likely protect bacteria from host defenses and may also contribute to bacterial survival during latent infections and saprophytic life.
The flagellar complex may also play a more direct role in bacterial wilt virulence. Several proteins that assemble flagella are evolutionarily related to type III secretion systems, which inject bacterial virulence factors into plant or animal cells; in Yersinia enterocolitica, the flagellar assembly apparatus itself secretes a key virulence factor, phospholipase (24, 61). At this point we cannot rule out the possibility that the R. solanacearum flagellar apparatus also transports virulence factors.
The flagellum itself can be highly antigenic in mammals, since it is abundant, exposed on the bacterial cell surface, and capable of eliciting a strong host immune response (16, 52). Although plants lack immune systems, there is some evidence that they can recognize and respond to flagellins. Flagellin from an incompatible strain (but not from a compatible strain) of Pseudomonas avenae induces a resistance response in cultured rice cells (14). Synthetic 15-mer oligopeptides derived from the conserved N terminus of P. aeruginosa flagellin elicit defense responses in tomato cell lines and Arabidopsis (21). Interestingly, the corresponding 15 amino acids are less conserved (50% identity) in the plant-associated species A. tumefaciens and Rhizobium meliloti, possibly so that these bacteria do not trigger host defenses (21). We note that R. solanacearum flagellin has only 60% identity to this conserved region, suggesting that this protein may have evolved to avoid host plant recognition. Alternatively, if host plants can recognize R. solanacearum flagellin, avoiding host recognition of flagellin may be one reason why this species loses motility once it is inside a plant host. To distinguish between these two possibilities, we need to determine if purified wild-type flagellin or the isolated 15-mer flagellin oligopeptide can elicit defense responses in tomato and other plant species. Arguing against this possibility is our observation that aflagellate strain K701 bacteria still induced a wild-type HR on the nonhost plant cucumber. However, this result is not conclusive because many factors contribute to HR induction by R. solanacearum, including several proteins injected by the hrp secretion system (26). Thus, even if flagellin is recognized by the host plant, it is not likely to act alone.
We have shown that R. solanacearum needs motility to effectively invade and colonize host plants. Many questions still remain about the process and function of bacterial movement in bacterial wilt disease development. A paralyzed mutant should allow us to separate the effects of bacterial motility from those of the flagellum itself, such as attachment and stimulation of host defenses. We found that R. solanacearum is effectively nonmotile in host xylem vessels, suggesting the existence of a regulatory structure that responds to a plant signal. Reporter gene analyses should help define the regulation of motility in this species.
ACKNOWLEDGMENTS
We thank Christian Boucher and Stephane Genin (INRA-Toulouse, France) for generous prepublication access to genomic sequence data from R. solanacearum strain GMI1000, and we thank Donald Waller for statistical advice. We are also grateful to Jo Handelsman, Tom German, and Heidi Goodrich-Blair for useful discussions.
This research was supported by the National Science Foundation (IBN-9973372) and by the University of Wisconsin College of Agricultural and Life Sciences.
REFERENCES
- 1.Akerley B J, Cotter P A, Miller J F. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell. 1995;80:611–620. doi: 10.1016/0092-8674(95)90515-4. [DOI] [PubMed] [Google Scholar]
- 2.Akerley B J, Miller J F. Flagellin gene transcription in Bordetella bronchiseptica is regulated by the BvgAS virulence control system. J Bacteriol. 1993;175:3468–3479. doi: 10.1128/jb.175.11.3468-3479.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen C, Gay J, Simon-Buela L. A regulatory locus, pehSR, controls polygalacturonase production and other virulence functions in Ralstonia solanacearum. Mol Plant-Microbe Interact. 1997;10:1054–1064. doi: 10.1094/MPMI.1997.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 4.Allen C, Huang Y, Sequeira L. Cloning of genes affecting polygalacturonase production in Pseudomonas solanacearum. Mol Plant-Microbe Interact. 1991;4:147–154. [Google Scholar]
- 5.Ames P, Bergman K. Competitive advantage provided by bacterial motility in the formation of nodules by Rhizobium meliloti. J Bacteriol. 1981;148:728–729. doi: 10.1128/jb.148.2.728-729.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. Short protocols in molecular biology. 3rd ed. New York, N.Y: John Wiley & Sons; 1995. [Google Scholar]
- 7.Bayot R G, Ries S M. Role of motility in apple blossom infection by Erwinia amylovora and studies of fire blight control with attractant and repellent compounds. Phytopathology. 1986;76:441–445. [Google Scholar]
- 8.Boucher C, Barberis P, Trigalet A, Demery D. Transposon mutagenesis of Pseudomonas solanacearum: isolation of Tn5-induced avirulent mutants. J Gen Microbiol. 1985;131:2449–2457. [Google Scholar]
- 9.Bowers J H, Parke J L. Colonization of pea (Pisum sativum L.) taproots by Pseudomonas fluorescens: effect of soil temperature and bacterial motility. Soil Biol Biochem. 1993;25:1693–1701. [Google Scholar]
- 10.Bradley D E, Douglas C J, Peshon J. Flagella-specific bacteriophages of Agrobacterium tumefaciens: demonstration of virulence of nonmotile mutants. Can J Microbiol. 1984;30:676–681. doi: 10.1139/m84-101. [DOI] [PubMed] [Google Scholar]
- 11.Brumbley S M, Denny T P. Cloning of wild-type Pseudomonas solanacearum phcA, a gene that when mutated alters expression of multiple traits that contribute to virulence. J Bacteriol. 1990;172:5677–5685. doi: 10.1128/jb.172.10.5677-5685.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buchanan R E, Gibbons N E, editors. Bergey's manual of determinative bacteriology. 8th ed. Baltimore, Md: The Williams & Wilkins Co.; 1974. [Google Scholar]
- 13.Catlow H Y, Glenn A R, Dilworth M J. The use of transposon-induced nonmotile mutants in assessing the significance of motility of Rhizobium leguminosarum bv trifolii for movement in soils. Soil Biol Biochem. 1990;22:331–336. [Google Scholar]
- 14.Che F-S, Nakajima Y, Tanaka N, Iwano M, Yoshida T, Takayama S, Kadota I, Isogai A. Flagellin from an incompatible strain of Pseudomonas avenae induces a resistance response in cultured rice cells. J Biol Chem. 2000;275:32347–32356. doi: 10.1074/jbc.M004796200. [DOI] [PubMed] [Google Scholar]
- 15.Chesnokova O, Coutinho J, Khan I, Mikhail M, Kado C. Characterization of flagella genes of Agrobacterium tumefaciens, and the effect of a bald strain on virulence. Mol Microbiol. 1997;23:579–590. doi: 10.1046/j.1365-2958.1997.d01-1875.x. [DOI] [PubMed] [Google Scholar]
- 16.Ciacci-Woodwine F, Blomfield I C, Richardson S H, Mizel S B. Salmonella flagellin induces tumor necrosis factor alpha in a human promonocytic cell line. Infect Immun. 1998;66:1127–1134. doi: 10.1128/iai.66.3.1127-1134.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clough S J, Flavier A B, Schell M A, Denny T P. Differential expression of virulence genes and motility in Ralstonia (Pseudomonas) solanacearum during exponential growth. Appl Environ Microbiol. 1997;63:844–850. doi: 10.1128/aem.63.3.844-850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costerton J W, Stewart P, Greenberg E P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 19.deWeger L A, vanderVlugt C I, Wijfjies A, Bakker P A, Shippers B, Lugtenberg B. Flagella of a plant growth-stimulating Pseudomonas fluorescens strain are required for colonization of potato roots. J Bacteriol. 1987;169:2769–2773. doi: 10.1128/jb.169.6.2769-2773.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drake D, Montie T C. Flagella, motility, and invasive virulence of Pseudomonas aeruginosa. J Gen Microbiol. 1988;134:43–52. doi: 10.1099/00221287-134-1-43. [DOI] [PubMed] [Google Scholar]
- 21.Felix G, Duran J D, Volko S, Boller T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999;18:265–276. doi: 10.1046/j.1365-313x.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- 22.Fernandez L A, Berenguer J. Secretion and assembly of regular surface structures in Gram-negative bacteria. FEMS Microbiol Rev. 2000;24:21–44. doi: 10.1111/j.1574-6976.2000.tb00531.x. [DOI] [PubMed] [Google Scholar]
- 23.Flavier A, Clough S, Schell M, Denny T. Identification of 3-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Mol Microbiol. 1997;26:251–259. doi: 10.1046/j.1365-2958.1997.5661945.x. [DOI] [PubMed] [Google Scholar]
- 24.Galan J E, Collmer A. Type III secretion machines: bacterial devices for protein delivery into host cells. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 25.Garg R P, Huang J, Yindeeyoungyeon W, Denny T P, Schell M A. Multicomponent transcriptional regulation at the complex promoter of the exopolysaccharide I biosynthetic operon of Ralstonia solanacearum. J Bacteriol. 2000;182:6659–6666. doi: 10.1128/jb.182.23.6659-6666.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gueneron M, Timmers A, Boucher C, Arlat M. Two novel proteins, PopB, which has functional nuclear localization signals, and PopC, which has a large leucine-rich repeat domain, are secreted through the Hrp-secretion apparatus of Ralstonia solanacearum. Mol Microbiol. 2000;36:261–277. doi: 10.1046/j.1365-2958.2000.01870.x. [DOI] [PubMed] [Google Scholar]
- 27.Haefele D M, Lindow S E. Flagellar motility confers epiphytic fitness advantages upon Pseudomonas syringae. Appl Environ Microbiol. 1987;53:2528–2533. doi: 10.1128/aem.53.10.2528-2533.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 29.Hatterman D R, Ries S M. Motility of Pseudomonas syringae pv. glycinea and its role in infection. Phytopathology. 1989;79:284–289. [Google Scholar]
- 30.Hawes M C, Smith L Y. Requirement for chemotaxis in pathogenicity of Agrobacterium tumefaciens on roots of soil-grown pea plants. J Bacteriol. 1989;171:5668–5671. doi: 10.1128/jb.171.10.5668-5671.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayward A C. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu Rev Phytopathol. 1991;29:65–87. doi: 10.1146/annurev.py.29.090191.000433. [DOI] [PubMed] [Google Scholar]
- 32.Hendrick C, Sequeira L. -Lipopolysaccharide-defective mutants of the wilt pathogen Pseudomonas solanacearum. Appl Environ Microbiol. 1984;48:94–101. doi: 10.1128/aem.48.1.94-101.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howie W J, Cook R J, Weller D M. Effects of soil matric potential and cell motility on wheat root colonization by fluorescent pseudomonads suppressive to take-all. Phytopathology. 1987;77:286–292. [Google Scholar]
- 34.Kang Y, Saile E, Schell M A, Denny T P. Quantitative immunofluorescence of regulated eps gene expression in single cells of Ralstonia solanacearum. Appl Environ Microbiol. 1999;65:2356–2362. doi: 10.1128/aem.65.6.2356-2362.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelman A. The relationship of pathogenicity of Pseudomonas solanacearum to colony appearance in a tetrazolium medium. Phytopathology. 1954;44:693–695. [Google Scholar]
- 36.Kelman A, Hruschka J. The role of motility and aerotaxis in the selective increase of avirulent bacteria in still broth cultures of Pseudomonas solanacearum. J Gen Microbiol. 1973;76:177–188. doi: 10.1099/00221287-76-1-177. [DOI] [PubMed] [Google Scholar]
- 37.MacNab R M. Flagella and motility. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. (ed.), Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 123–145. [Google Scholar]
- 38.Malek W. The role of motility in the efficiency of nodulation by Rhizobium meliloti. Arch Microbiol. 1992;158:26–28. [Google Scholar]
- 39.Manson M D, Armitage J P, Hoch J A, MacNab R M. Bacterial locomotion and signal transduction. J Bacteriol. 1998;180:1009–1022. doi: 10.1128/jb.180.5.1009-1022.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGarvey J A, Denny T P, Schell M A. Spatial-temporal and quantitative analysis of growth and EPSI production by Ralstonia solanacearum in resistant and susceptible tomato cultivars. Phytopathology. 1999;89:1233–1239. doi: 10.1094/PHYTO.1999.89.12.1233. [DOI] [PubMed] [Google Scholar]
- 41.Menard R, Sansonetti P J, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1992. [Google Scholar]
- 43.Milton D, O'Toole R, Horstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. J Bacteriol. 1996;178:1310–1319. doi: 10.1128/jb.178.5.1310-1319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ormonde P, Horstedt P, O'Toole R, Milton D. Role of motility in adherence to and invasion of a fish cell line by Vibrio anguillarum. J Bacteriol. 2000;182:2326–2328. doi: 10.1128/jb.182.8.2326-2328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Toole G, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 46.Otteman K M, Miller J F. Roles for motility in bacterial-host interactions. Mol Microbiol. 1997;24:1109–1117. doi: 10.1046/j.1365-2958.1997.4281787.x. [DOI] [PubMed] [Google Scholar]
- 47.Panapoulos N J, Schroth M N. Role of flagellar motility in the invasion of bean leaves by Pseudomonas phaseolicola. Phytopathology. 1974;64:1389–1397. [Google Scholar]
- 48.Saile E, McGarvey J, Schell M, Denny T. Role of extracellular polysaccharide and endoglucanase in root invasion and colonization of tomato plants by Ralstonia solanacearum. Phytopathology. 1997;87:1264–1271. doi: 10.1094/PHYTO.1997.87.12.1264. [DOI] [PubMed] [Google Scholar]
- 49.Schell M A. To be or not to be: how Pseudomonas solanacearum decides whether or not to express virulence genes. Eur J Plant Pathol. 1996;102:459–469. [Google Scholar]
- 50.Sokal R R, Rohlf F J. Biometry. W. H. San Francisco, Calif: Freeman and Co.; 1981. [Google Scholar]
- 51.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1986;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steiner T S, Nataro J P, Poteet-Smith C E, Smith J A, Guerrant R L. Enteroaggregative Escherichia coli expresses a novel flagellin that causes IL-8 release from intestinal epithelial cells. J Clin Investig. 2000;105:1769–1777. doi: 10.1172/JCI8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Studier F W, Moffat B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 54.Tans-Kersten J, Guan Y, Allen C. Ralstonia solanacearum pectinmethylesterase is required for growth on methylated pectin but not for bacterial wilt virulence. Appl Environ Microbiol. 1998;64:4918–4923. doi: 10.1128/aem.64.12.4918-4923.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tolker-Nielsen T, Brinch U C, Ragas P C, Andersen J B, Jacobsen C S, Molin S. Development and dynamics of Pseudomonas sp. biofilms. J Bacteriol. 2000;182:6482–6489. doi: 10.1128/jb.182.22.6482-6489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Totten P A, Lory S. Characterization of the type a flagellin gene from Pseudomonas aeruginosa PAK. J Bacteriol. 1990;172:7188–7199. doi: 10.1128/jb.172.12.7188-7199.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.VandeBroek A, Lambrecht M, Vanderleyden J. Bacterial chemotactic motility is important for the initiation of wheat root colonization by Azospirillum brasiliense. Microbiology. 1998;144:2599–2606. doi: 10.1099/00221287-144-9-2599. [DOI] [PubMed] [Google Scholar]
- 58.VandeBroek A, Vanderleyden J. The role of bacterial motility, chemotaxis, and attachment in bacteria-plant interactions. Mol Plant-Microbe Interact. 1995;8:800–810. [Google Scholar]
- 59.Vasse J, Frey P, Trigalet A. Microscopic studies of intercellular infection and protoxylem invasion of tomato roots by Pseudomonas solanacearum. Mol Plant-Microbe Interact. 1995;8:241–251. [Google Scholar]
- 60.Xu P, Leong S, Sequeira L. Molecular cloning of genes that specify virulence in Pseudomonas solanacearum. J Bacteriol. 1988;170:617–622. doi: 10.1128/jb.170.2.617-622.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Young G, Schmiel D, Miller V L. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein secretion system. Proc Natl Acad Sci USA. 1999;96:6456–6461. doi: 10.1073/pnas.96.11.6456. [DOI] [PMC free article] [PubMed] [Google Scholar]