Abstract
Background
The COVID-19 pandemic was met with strict containment measures. We hypothesized that societal infection control measures would impact the number of hospital admissions for respiratory tract infections, as well as, the spectrum of pathogens detected in patients with suspected community acquired pneumonia (CAP).
Methods
This study is based on aggregated surveillance data from electronic health records of patients admitted to the hospitals in Bergen Hospital Trust from January 2017 through June 2021, as well as, two prospective studies of patients with suspected CAP conducted prior to and during the COVID-19 pandemic (pre-COVID cohort versus COVID cohort, respectively). In the prospective cohorts, microbiological detections were ascertained by comprehensive PCR-testing in lower respiratory tract specimens. Mann–Whitney’s U test was used to analyse continuous variables. Fisher’s exact test was used for analysing categorical data. The number of admissions before and during the outbreak of SARS-CoV-2 was compared using two-sample t-tests on logarithmic transformed values.
Results
Admissions for respiratory tract infections declined after the outbreak of SARS-CoV-2 (p < 0.001). The pre-COVID and the COVID cohorts comprised 96 and 80 patients, respectively. The proportion of viruses detected in the COVID cohort was significantly lower compared with the pre-COVID cohort [21% vs 36%, difference of 14%, 95% CI 4% to 26%; p = 0.012], and the proportion of bacterial- and viral co-detections was less than half in the COVID cohort compared with the pre-COVID cohort (19% vs 45%, difference of 26%, 95% CI 13% to 41%; p < 0.001). The proportion of bacteria detected was similar (p = 0.162), however, a difference in the bacterial spectrum was observed in the two cohorts. Haemophilus influenzae was the most frequent bacterial detection in both cohorts, followed by Streptococcus pneumoniae in the pre-COVID and Staphylococcus aureus in the COVID cohort.
Conclusion
During the first year of the COVID-19 pandemic, the number of admissions with pneumonia and the microbiological detections in patients with suspected CAP, differed from the preceding year. This suggests that infection control measures related to COVID-19 restrictions have an overall and specific impact on respiratory tract infections, beyond reducing the spread of SARS-CoV-2.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12879-022-07732-5.
Keywords: COVID-19, SARS-CoV-2, Respiratory tract infections, Community acquired pneumonia, FilmArray pneumonia panel, Epidemiology, Molecular testing
Introduction
Community acquired pneumonia (CAP) is a leading cause of hospital admissions and mortality in all age groups and most parts of the world [1–3]. Our understanding of CAP has evolved in the last years. The introduction of PCR-based methods for detecting viruses and bacteria in respiratory specimens has shown a large proportion of bacterial-/viral coinfections and pure viral infections [4–6]. Several studies have demonstrated a close interaction between different viral and bacterial pathogens, especially for coinfections with Streptococcus pneumoniae and influenza virus [7–10].
The global pandemic of SARS-Coronavirus-2 (SARS-CoV-2) with corresponding coronavirus disease 2019 (COVID-19) was met with different containment strategies within countries, to reduce the spread of the virus. Most European countries, including Norway, introduced during March 2020 strict public infection control measures. These included imposing social distancing, prohibiting social gatherings, use of face masks, encouraging work from home solutions, increased border control and the closure of kindergartens, schools, and universities. The extent of lockdown implemented during the outbreak of COVID-19 is unprecedented. Recent studies have shown that such measures not only decreased the transmission of SARS-CoV-2, but also contributed to a massive reduction of circulating seasonal viruses [11–13]. We hypothesized that societal infection control measures would impact the number of hospital admissions for respiratory tract infections (RTIs), as well as, the spectrum of pathogens detected in patients with suspected CAP. Thus, we analysed both aggregated patient admission data (2017–2021) to compute numbers admitted with respiratory symptoms to the emergency department (ED), and furthermore studied the detection rates for common respiratory pathogens in two cohorts with acute community acquired RTIs, recruited before and during the COVID-19 pandemic at our tertiary care hospital. To our knowledge, this is the first study to both analyse admission data and systematically compare microbiological detections by syndromic PCR-based testing of lower respiratory tract samples in adult patients hospitalized with community acquired RTIs admitted before and during the outbreak of SARS-CoV-2.
Methods
Patients and study design
This study consists of a retrospective study of aggregated patient data for patients admitted to the hospitals in Bergen Hospital Trust from January 2017 through June 2021, as well as, patients with suspected CAP included from two prospective cohort studies.
Retrospective evaluation of hospital admissions
Using information from electronic health records, we captured information on the total number of admissions to the ED per month from January 2017 to June 2021, for the hospitals in Bergen Hospital Trust, including the number of patients with acute respiratory tract infections belonging to the five defined subgroups of International Classification of Diseases 10th revision (ICD-10) diagnostic codes: upper respiratory tract infections (J00–06 and J36); infections with influenza (J09–J11) other lower respiratory tract infections (J12 and J16–22); bacterial pneumonia (J13–J15); and obstructive lung diseases (J44–J46).
Prospective cohort studies
Data were selected from two studies conducted in two consecutive winter seasons (2019/2020 and 2020/2021) at Haukeland University Hospital, a tertiary care referral centre in Bergen, Norway. The first from a cohort study with prospectively recruited patients with suspected CAP, enrolled between December 2nd 2019 and February 17th 2020 (pre-COVID cohort) [14]. The second from a cohort of prospectively enrolled patients with suspected CAP from an ongoing randomized controlled trial (RCT) (NCT04660084), recruited between September 25th 2020 and May 31st 2021 (COVID cohort). Patients with suspected CAP at admission (retrospectively diagnosed as CAP or other lower RTIs) and a specimen from the lower respiratory tract at admission were consecutively selected from the two cohorts.
The first study was conducted as a feasibility study to inform the design of the RCT from where the second patient group was selected. The RCT aims to evaluate the clinical impact of rapid diagnostic methods on antibiotic use and outcome. The inclusion- and exclusion criteria for both cohort studies were the same. Patients were eligible for inclusion if they were ≥ 18 years, presenting to the ED with a suspicion of CAP and fulfilling at least two of the following criteria: new or worsening cough; new or worsening expectoration of sputum; new or worsening dyspnoea; haemoptysis; pleuritic chest pain; radiological evidence of pneumonia; abnormalities on chest auscultation and/or percussion; fever (≥ 38.0 °C). Exclusion criteria were cystic fibrosis, severe bronchiectasis, hospitalization within the last 14 days prior to admission, a palliative approach (defined as life expectancy below 2 weeks), or if the patient was not willing or able to provide a lower respiratory tract sample.
Data collection and sampling
Patients were enrolled on weekdays between 08:00 a.m. and 09:00 p.m. Relevant baseline information was collected by study nurses or investigating physicians through a structured interview. Symptoms and findings upon clinical examinations were recorded. Data pertaining to treatment and results from laboratory tests and medical imaging were obtained from electronic medical records and charts. Data were registered in an electronic case report form (eCRF) from VieDoc™ (Viedoc Technologies, Uppsala, Sweden).
Microbiological sampling and methods
At inclusion, a lower respiratory tract sample for the BioFire® FilmArray® Pneumonia panel plus (FAP plus) (bioMérieux S.A., Marcy-l’Etoile, France) and standard culture was obtained from all patients. Depending on clinical symptoms, vital signs, and medical history, either spontaneous sputum, or sputum induced by either nebulized isotonic (0.9%) or hypertonic (5.8%) saline was collected. Patients with known obstructive lung disease and patients with hypoxemia or signs of airway obstruction upon physical examination, were additionally treated with a bronchodilator (salbutamol and/or ipratropium bromide) prior to sampling. If sputum induction was unsuccessful, endotracheal aspiration was performed.
The FAP plus is a commercial automated multiplex PCR panel for the detection of 27 bacteria and viruses as well as seven genetic markers of antibiotic resistance, validated for lower respiratory tract samples [15] (see Additional file 1). The standard diagnostic methods (SDs) included culture of respiratory tract samples and blood according to current guidelines (adapted from [16]). Nasopharyngeal and/or oropharyngeal swabs were examined by an in-house real-time PCR test to detect respiratory viruses and atypical bacteria. SDs also included rapid tests; the pneumococcal urine antigen test (Quidel Corporation, San Diego, US) and a point of care test (POC) for influenza virus A and B (ID NOW™, Illinois, US). The latter was only available for the pre-COVID cohort. Blood culture results deemed as contamination by the microbiologists, were not counted. Any additional tests requested by the treating physician were noted and counted as part of SDs.
Statistical analysis
Descriptive statistics for continuous variables are reported as median with interquartile range (IQR). Mann–Whitney’s U test was used to analyse continuous variables. Fisher’s exact test was used for analysing categorical data, by use of contingency tables. The number of admissions for acute RTIs or other acute respiratory complaints before and during the outbreak of SARS-CoV-2 was compared using two-sample t-tests on logarithmic transformed values. Percentage changes with 95% confidence intervals (95% CIs) were obtained after back-transformation. A two tailed p-value ≤ 0.05 was considered statistically significant for all analyses. The statistics were performed using IBM SPSS Statistics (version 26.0; Armonk, NY, US), the statistical environment R (Vienna, Austria), GraphPad Prism (GraphPad Software, La Jolla, CA, USA) and the GraphPad QuickCalcs Web site: https://www.graphpad.com/quickcalcs/contingency1/ (last accessed 2nd May 2022).
Ethics
The two prospective patient cohorts were selected from a study approved by the Regional Committee for Medical and Health Research Ethics in South East Norway (REK ID: 31935) and performed in accordance with the Declaration of Helsinki. Written informed consent was obtained from all participants or from their legal guardian/close relative (in case of altered consciousness or confusion) at the time of recruitment. Regarding data extracted from the electronic heath records, only aggregated anonymous patient data was used and consent was deemed unnecessary by the Regional Committee for Medical and Health Research Ethics in Western Norway (REK ID: 221336).
Results
Hospital admissions before and during the COVID-19 pandemic
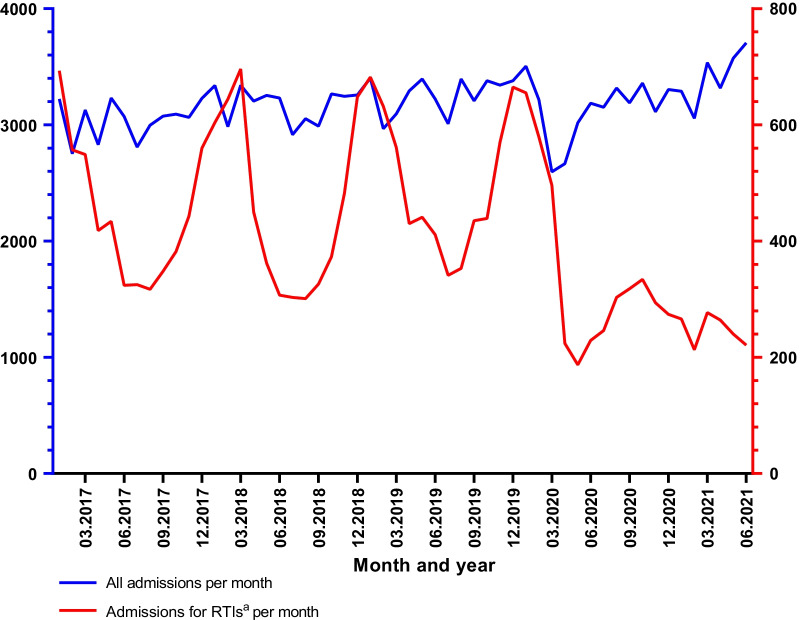
A total of 22,870 patients were discharged with a primary diagnosis of acute RTI or other acute respiratory symptoms from January 2017 to June 2021. There were 5391 admissions in 2017, 5585 in 2018, 6108 in 2019, 4271 in 2020 and 1515 in the first 6 months of 2021. An overview of the number of visits per month to the ED in Bergen Hospital Trust is shown in Fig. 1. There was a total of 6684 admissions with upper RTIs, 1413 with influenza, 4560 with other lower RTIs (LRTIs), 5020 with bacterial pneumonia and 5193 with obstructive lung disease. The median number of admissions with RTIs and acute respiratory complaints per month was 447 (370–594) before the pandemic and 268 (236–309) after March 2020 (p < 0.001), while the total number of admissions remained stable (p = 0.672) (Table 1). The reduction in admission rate was observed for all subcategories (upper and lower RTIs, obstructive lung disease, and bacterial pneumonia) (p < 0.001). For influenza virus, 59 of the 1413 influenza cases were admitted during the COVID period, and 55 of these were admitted during March 2020.
Fig. 1.
Number of visits to the emergency department. The total number of emergency department visits to the Bergen Hospital Trust per month displayed in blue. The number of patients admitted for acute RTIsa is displayed in red. RTIs respiratory tract infections, ICD International Classification of Diseases. aAcute RTIs or other acute respiratory symptoms defined as ICD-10 primary diagnosis of J00–06, J12–J122, J36 and J44–J46
Table 1.
Number of admissions per month before and during the outbreak of SARS-CoV-2
| Diagnosis | Jan. 2017–Febr. 2020 | March 2020–June 2021 | Difference (95% CI) | p-value |
|---|---|---|---|---|
| Upper RTI | 137 (100–170) | 83 (72–100) | − 37.0% (− 48.0% to − 23.6%) | < 0.001 |
| Other lower RTI | 80 (62–134) | 49 (34–60) | − -46.7% (− 58.8% to − 31.2%) | < 0.001 |
| Bacterial pneumonia | 105 (94–121) | 62 (53–67) | − 43.7% (− 50.1% to − 63.4%) | < 0.001 |
| Obstructive lung disease | 101 (92–116) | 77 (72–87) | − 24.8% (− 32.3% to − 16.6%) | < 0.001 |
| Total number of RTIs/acute respiratory complaints | 447 (370–594) | 268 (236–309) | − 41.0% (− 49.2% to − 31.5%) | < 0.001 |
| All admissions | 3219 (3053–3292) | 3239 (3085–3338) | 1.1% (− 4.1% to 6.6%) | 0.672 |
Number of admissions before the COVID-19 pandemic compared with number of admissions during the COVID-19 pandemic. Numbers are given as median admissions per month with interquartile range. P-values were calculated with two sample t-tests on logarithm-transformed values. Percentage changes with 95% CIs were obtained after back-transformation
Jan January, Febr February, 95% CI 95% confidence interval, RTIs respiratory tract infections
The microbial spectrum of hospitalized RTIs before and during the COVID-19 pandemic
Patient characteristics of the prospective pre-COVID and COVID cohorts
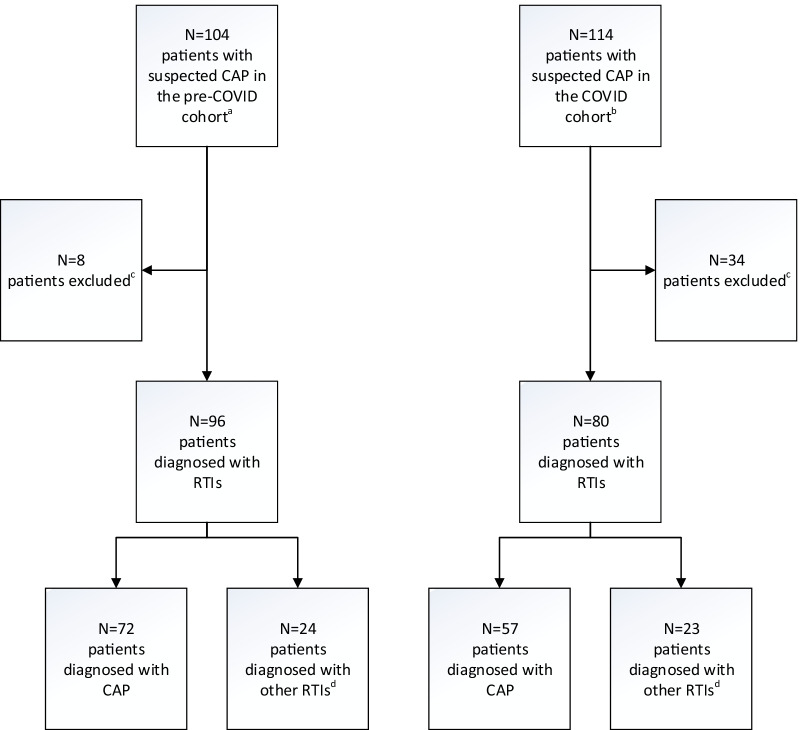
A total of 176 patients with suspected CAP (i.e., confirmed CAP or other lower RTIs) were included from the two cohort studies. Ninety-six patients were from the pre-COVID cohort and included before the outbreak of SARS-CoV-2 in Norway, while 80 patients were included during the COVID-19 pandemic in Norway (COVID cohort) (Fig. 2). A lower respiratory tract specimen was obtained from all patients: 82% (144/176) by sputum induction; 9% (16/176) by spontaneous expectoration; and 9% (16/176) by endotracheal aspiration. All lower respiratory tract specimens were cultured and 95% (168/176) were also analysed by the FAP plus. Blood cultures were performed in 99% (175/176) of the patients, in-house PCR testing for 95% (167/176) and a pneumococcal urine antigen test for 71% (125/176) of the patients. The patient characteristics are shown in Table 2.
Fig. 2.
Cohort study flowchart. CAP community acquired pneumonia, RTI respiratory tract infection, COPD chronic obstructive pulmonary disease. aInclusion before the COVID-19 pandemic (between December 2nd 2019 and February 17th 2020). bInclusion during the COVID-19 pandemic (between September 25th 2020 and May 31st 2021). cPatients were excluded due to other diagnoses, most frequently non-infectious exacerbation of COPD; heart failure; other infection; and pulmonary embolism. di.e. exacerbation of COPD/asthma other lower respiratory tract infections
Table 2.
The patient characteristics of the prospective pre-COVID and COVID cohorts (n = 176)
| Pre-COVID cohort (n = 96) | COVID cohort (n = 80) | P-value | |
|---|---|---|---|
| A: Baseline characteristics | |||
| Demography | |||
| Age | 73 (59–80) | 73 (58–79) | 0.966 |
| Female | 52 (54) | 32 (40) | 0.070 |
| Male | 44 (46) | 48 (60) | 0.070 |
| Comorbidity | |||
| Cardiovascular disease | 48 (50) | 42 (53) | 0.764 |
| Diabetes mellitus | 11 (11) | 11 (14) | 0.655 |
| Asthma/COPD | 38 (40) | 42 (53) | 0.096 |
| Kidney disease | 15 (16) | 7 (9) | 0.252 |
| Previous smoker | 43 (45) | 47 (59) | 0.071 |
| Current smoker | 20 (21) | 16 (20) | > 0.999 |
| Vaccine status | |||
| Influenza virusa | 58 (60) | 45 (56) | 0.646 |
| Pneumococcal | 28 (29) | 34 (43) | 0.081 |
| B: Severity and outcome | |||
| Severity scoreb | |||
| CURB-65 | 1.0 (1.0–2.0) | 1.0 (1.0–2.0) | 0.296 |
| PSI | 93 (71–111)c | 91 (65–114) | 0.864 |
| Outcome | |||
| Length of stay (days) | 3.1 (2.0–5.0) | 3.1 (2.0–6.1) | 0.747 |
| HDU or ICU admission | 10 (10) | 10 (13) | 0.812 |
| Case fatality rate | |||
| In-hospital | 1 (1) | 0 (0) | > 0.999 |
| 30 days | 1 (1) | 1 (1) | > 0.999 |
| 60 days | 4 (4) | 1 (1) | 0.378 |
Data shown as count (%) or median (IQR). P-values are calculated with Mann-Whitney’s U test and Fisher’s exact test, comparing the pre-COVID cohort with the COVID cohort
COPD chronic obstructive pulmonary disease, CURB-65 confusion, urea, respiratory rate, blood pressure, age ≥ 65 years, PSI pneumonia severity index, HDU high dependency unit, ICU intensive care unit, IQR interquartile range, CAP community acquired pneumonia
aVaccinated for influenza virus with the latest vaccine
bOnly performed in for CAP patients
cMissing for five CAP patients
Microbiological findings
There was a significant decrease in detection rates for viruses in patients with suspected CAP in the COVID cohort compared with the pre-COVID cohort [21% (24/112) vs 36% (62/174), difference of 14%, 95% CI 4% to 26%; p = 0.012]. The proportion of detected bacteria remained stable between the two cohorts; 71% (79/112) in the COVID cohort vs 62% (108/174) in the pre-COVID cohort (difference of 8%, 95% CI − 3% to 20%; p = 0.162).
Figure 3 shows the distribution of patients with viral, bacterial, and bacterial- and viral co-detections in the two cohorts. The proportion of patients with multiple detections, i.e., multiple bacteria or bacterial- and viral co-infections, was lower in the COVID cohort compared with the pre-COVID cohort [33% (26/80) vs 55% (53/96), difference of 23%, 95% CI 9% to 38%; p = 0.0037]. Indeed, the proportion of patients with bacterial- and viral co-detections in the COVID cohort was less than half compared with the pre-COVID cohort [19% (15/80) vs 45% (43/96), difference of 26%, 95% CI 13% to 41%; p = 0.0004]. Further, the number of patients with a viral detection (solely or in combination with another microbe) in the COVID cohort was also considerably lower compared with the pre-COVID cohort [26% (21/80) vs 58% (56/96), difference of 32%, 95% CI 19% to 47%; p < 0.0001].
Fig. 3.
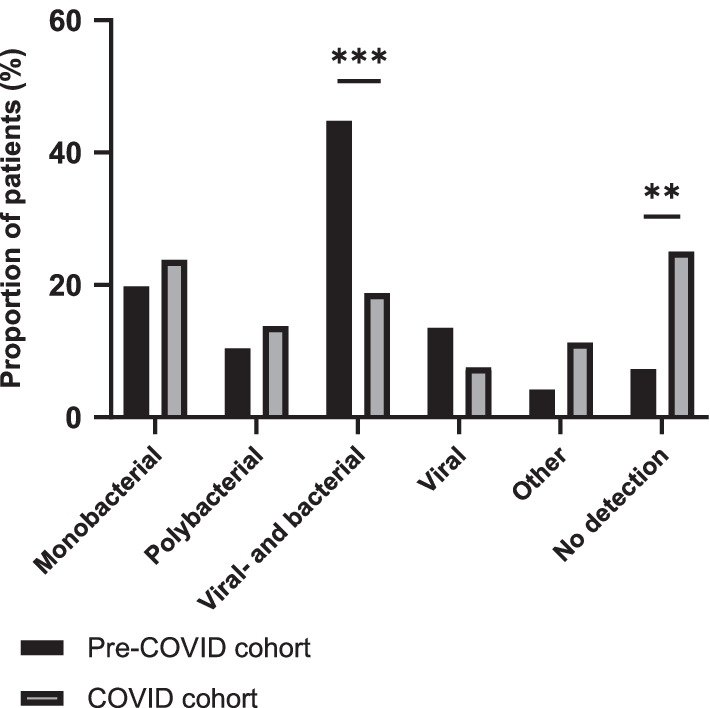
Proportion of patients stratified by microbiological detection categories. Proportion of 96 patients included before the COVID-19 pandemic (pre-COVID cohort) and 80 patients included during the COVID-19 pandemic (COVID cohort), stratified by microbiological detection categories. P-values are calculated with Fisher’s exact test. **P ≤ 0.01; ***P ≤ 0.001
The overview of the microbiological findings is listed in Table 3, stratified by cohort. The most frequent viral detections in the pre-COVID cohort were influenza A virus (29%) followed by human metapneumovirus (17%), while the corresponding detections in the COVID cohort were rhino-enterovirus (15%) followed by SARS-CoV-2 (13%). Haemophilus influenzae was the most frequent bacterial detection in both cohorts (36% and 26%, respectively). Streptococcus pneumoniae (25%) followed H. influenzae in the pre-COVID cohort, while Staphylococcus aureus increased significantly and was the second most common detection in the COVID cohort (23%).
Table 3.
Overview and comparison of microbiological detections in two cohorts of patients with respiratory tract infections
| Microbes | Acute respiratory tract infections | |||
|---|---|---|---|---|
| Pre-COVID cohort (n = 96) | COVID cohort (n = 80) | Difference in proportion (95% CI) | p-value | |
| Number of detections | Number of detections | |||
| Viruses | 62 | 24 | – | – |
| Influenza A virus | 29 (30) | 0 | − 30% (− 41% to − 20%) | < 0.0001 |
| Human metapneumovirus | 16 (17) | 0 | − 17% (− 26% to − 8%) | < 0.0001 |
| Rhino-/enterovirus | 3 (3) | 12 (15) | 12% (1% to 21%) | 0.0061 |
| Coronavirus (SARS-CoV-2) | 0 | 10 (13) | 13% (3% to 20%) | 0.0003 |
| Respiratory syncytial virus | 7 (7) | 0 | − 7% (− 15% to 0%) | 0.0164 |
| Coronavirus (229E, OC43, HKU1, NL63) | 5 (5) | 0 | − 5% (− 12% to 1%) | 0.0641 |
| Parainfluenza virus | 2 (2) | 1 (1) | − 1% (− 7% to 6%) | > 0.9999 |
| Adenovirus | 0 | 1 (1) | 1% (− 5% to 6%) | 0.4545 |
| Bacteria | 108 | 79 | – | – |
| H. influenzae | 35 (36) | 21 (26) | − 10% (− 24% to 4%) | 0.1934 |
| S. pneumoniae | 24 (25) | 12 (15) | − 10% (− 22% to 3%) | 0.1331 |
| S. aureus | 7 (7) | 18 (23) | 15% (4% to 26%) | 0.0048 |
| M. catarrhalis | 11 (11) | 8 (10) | − 1% (− 11% to 9%) | 0.8114 |
| E. coli | 8 (8) | 3 (4) | − 5% (− 13% to 4%) | 0.3490 |
| S. agalactiae | 6 (6) | 5 (6) | 0% (− 9% to 8%) | > 0.9999 |
| P. aeruginosa | 3 (3) | 2 (3) | 1% (− 7% to 7%) | > 0.9999 |
| K. pneumoniae | 3 (3) | 1 (1) | − 2% (− 8% to 5%) | 0.6270 |
| S. marcescens | 3 (3) | 1 (1) | − 2% (− 8% to 5%) | 0.6270 |
| E. cloacae complex | 1 (1) | 2 (3) | 2% (− 6% to 7%) | 0.5916 |
| Proteus spp. | 2 (2) | 1 (1) | − 1% (− 7% to 6%) | > 0.9999 |
| K. oxytoca | 1 (1) | 1 (1) | 0% (− 6% to 6%) | > 0.9999 |
| M. pneumoniae | 2 (2) | 0 | − 2% (− 8% to 4%) | 0.5013 |
| K. variicola | 0 | 2 (3) | 3% (− 5% to 8%) | 0.2052 |
| A. calcoaceticus–A. baumanii complex | 1 (1) | 0 | − 1% (− 6% to 5%) | > 0.9999 |
| L. pneumophila | 1 (1) | 0 | − 1% (− 6% to 5%) | > 0.9999 |
| S. maltophilia | 0 | 1 (1) | 1% (− 5% to 6%) | 0.4545 |
| S. pyogenes | 0 | 1 (1) | 1% (− 5% to 6%) | 0.4545 |
| Other detections: | 4 | 9 | – | – |
| C. albicans | 3 (3) | 8 (10) | 7% (− 3% to 15%) | 0.1144 |
| P. jirovecii | 1 (1) | 1 (1) | 0% (− 6% to 6%) | > 0.9999 |
Microbiological detections in patients admitted with acute respiratory tract infection and who were able to provide a sample from the lower respiratory tract. Ninety-six patients from the pre-COVID cohort (included before the COVID-19 pandemic) are compared with 80 patients from the COVID cohort (included during the COVID-19 pandemic). Data are shown as counts with percent in brackets. The percentage was calculated as proportion of patients in the respective cohorts. P-values are calculated by using Fisher’s exact test
Streptococcus pneumoniae was detected more frequently in combination with a viral pathogen in the pre-COVID cohort compared with the COVID cohort [79% (19/24) vs 42% (5/12), difference of 38%, 95% CI 1% to 66%; p = 0.0576], moreover influenza A virus was detected in 54% (13/24) of patients with S. pneumoniae (pre-COVID cohort). In the COVID cohort, S. aureus was detected more frequently in patients with SARS-CoV-2 compared to patients without SARS-CoV-2 [60% (6/10) vs 17% (12/70), difference of 43% 95% CI 8% to 70%; p = 0.0071].
Discussion
The Norwegian COVID-19 restrictions were comprehensive and intrusive, but comparable to other European countries [17]. During the inclusion period of our COVID cohort, strict national restrictions were still in place. This likely resulted in important lifestyle changes and an increased awareness in the general population on measures to avoid respiratory tract infections, especially in combination with continued widespread use of hand disinfection fluids and face masks. Based on retrospective surveillance of ICD-10 codes, we demonstrated a large reduction in patients admitted to our hospital with acute respiratory infections, corresponding to the period with imposed COVID-19 restrictions. The reduction was specific for acute respiratory diseases as there was no reduction in the total number of admissions. Code-based surveillance of microbiological data has several pit-falls, including physician adherence to coding-practices and an unawareness of the diagnostic repertoire performed during hospitalization. Consequently, high-quality, microbiological data cannot be collected through code-based surveillance data. This study is one of the first to compare the microbiological aetiology of lower respiratory tract infections in patients with suspected CAP included from two prospective hospital cohorts, one recruited just prior to the outbreak of the COVID-19 pandemic and the other during the pandemic. The microbiological aetiology was rigorously ascertained by a comprehensive molecular pneumonia panel combined with conventional methods. Our results confirm recent reports of substantially decreased viral detections in patients with acute respiratory tract infections after the outbreak of the COVID-19 pandemic compared to earlier years. Although, the proportion of detected bacteria remained stable in both cohorts, a difference was observed in the bacterial spectrum in the two cohorts. Further, a significant reduction in hospital admissions with acute respiratory tract infections, including bacterial pneumonia, was observed during the first year of the pandemic compared to the previous years.
The role of viruses in CAP have been increasingly recognized in the last decades. The introduction of PCR-based methods capable of rapidly and accurately detecting viral pathogens, has led to a great increase of viral detections in CAP patients, either alone or in combination with bacteria [4–6, 9, 18–20]. The high proportion of viral detections in our pre-COVID cohort, is consistent with these findings. However, the results from our COVID cohort, show a marked reduction in viral detections. Recent studies have shown a similar reduction of non-SARS-CoV-2 respiratory viruses during the pandemic compared to previous years [11–13, 21]. Specifically, the prevalence of influenza and respiratory syncytial virus (RS-virus) are shown to be almost negligible. The drop in viral detections is shown to coincide with COVID-19 control measures introduced at the start of the pandemic, and probably is a direct result of reduced person to person spread of pathogens [11–13, 21]. Interestingly, rhinoviruses showed a completely different trend with an increased rate of detection in the COVID cohort. This has also been observed in other studies, and correlates with the easing of social distancing and the reopening of schools after the summer in 2020 [22, 23].
We observed a shift in the microbial patterns of detected bacteria in the COVID cohort, including, fewer bacterial- and viral co-detections. Notably, there was a reduced proportion of patients with detected S. pneumoniae, H. influenzae or a combination of these. A large study analysing surveillance data from 26 countries and territories across six continents on invasive disease due to S. pneumoniae, H. influenzae and Neisseria meningitidis before and during the outbreak of SARS-CoV-2, showed a substantial and sustained reduction of hospital reported invasive disease for these pathogens [24]. Although these findings were based on all invasive diseases, we find it likely that they also reflect a decrease in respiratory infections.
The development of LRTIs, including bacterial CAP, is complex and still not completely understood. Our data indicate a reduction of detected bacterial pathogens with a known potential to transmit by respiratory droplets, like S. pneumoniae and H. influenzae after the outbreak of SARS-CoV-2. This can be interpreted in support of the hypothesis that person-to-person transmission of microbes is an important cause of LRTIs for both viruses and bacteria. However, the proportion of detected S. aureus, which also has the potential for person to person spread, increased in our COVID cohort. It is known that S. aureus tend to colonize the upper respiratory tract of adults more frequently than S. pneumoniae and H. influenzae and this might explain why S. aureus did not decrease in the same manner [25–29]. Nasal carriage of S. aureus is strongly associated with infection and clinical studies consistently describe a significantly greater risk of bacteremia among carriers [30]. Societal restrictions, as imposed during the pandemic, might therefore play a lesser role in reducing S. aureus invasive infections, than they do for other pathogens.
Many respiratory tract pathogens colonize the upper respiratory tract, especially among the children and elderly, without causing an infection [26–29, 31]. Any change within the host or the environment, such as a change in circulating viruses, antimicrobial treatment or reduced person-to-person spread of microbes, could potentially alter the conditions for colonization and thereby the subsequent risk of infection. We found that S. aureus was found more frequently in patients with detected SARS-CoV-2 compared with patients without (60% vs 17%, p = 0.0071). An interaction between viral and bacterial respiratory pathogens in CAP has been discussed for many years [7, 10, 32–37]. It is believed that a bacterial CAP was the most frequent cause of death during the influenza pandemic of 1918–19 [38]. Several reports have shown that influenza virus, by several complex interactions, can increase the potential of S. pneumoniae both as a colonizer and as a pathogen [10, 32, 34, 35, 37]. In our pre-COVID cohort, 79% (19/24) of all detected S. pneumoniae were found in combination with a viral pathogen, most frequently with influenza virus.
In relation, data from the Norwegian Cause of Death Registry, show a decrease in age-adjusted death rate of LRTIs, including CAP, in 2020 compared with 2010–2019 [39]. This indicates that the COVID-19 restrictions and dramatic decrease in circulating viral pathogens had an effect not only on the spectrum of bacterial patterns detected, but also in reducing the overall incidence of bacterial CAP.
The major strength of our work is that microbiological data were collected from two prospective studies with an attention to detail and accuracy of lower respiratory tract sampling. Lower respiratory tract samples were collected from all patients and tested with a comprehensive multiplex molecular panel in addition to standard methods in most patients. The study has limitations; the inclusion of CAP patients at a single hospital in Norway, a limited sample size and enrolment of patients restricted to fixed hours during weekdays. We excluded patients that were unable to provide a lower respiratory tract sample, e.g. due to confusion, severe hypoxemia, need of assisted ventilation, or a non-productive cough; symptoms which are often found in patients with COVID-19. In addition, in Norway, patients with mild COVID-19 were often treated outside the hospitals at designated COVID-19 wards. Finally, concerning surveillance data from electronic health records, ICD-10 diagnoses are registered by the treating physician and the primary diagnosis may have been influenced by the pandemic. Nevertheless, national surveillance data show that influenza, invasive pneumococcal disease, and systemic disease caused by H. influenzae have decreased [40].
In conclusion, admissions with pneumonia and microbiological detections in patients with suspected CAP during the first year of the COVID-19 pandemic, differed from the previous year, suggesting that infection control measures related to COVID-19 restrictions are effective beyond reducing the spread of SARS-CoV-2. The number of detected viruses declined, and accordingly, the proportion of patients with bacterial- and viral co-detections decreased. Furthermore, we observed a change in both the proportion and pattern of certain bacterial detections, implying that presence of viruses may facilitate colonization and infection by certain types of bacteria and play an important role in the etiopathogenesis of CAP.
Supplementary Information
Additional file 1. BioFire® FilmArray® Pneumonia panel plus (FAP plus) targets.
Acknowledgements
We gratefully acknowledge the CAPNOR study group, the study nurses, the staff at the Emergency Care Clinic, and staff at the Department of Microbiology. Last, we thank all the enrolled patients.
CAPNOR study group: Rune O. Bjørneklett (Emergency Care Clinic, Haukeland University Hospital, Bergen, Norway; Department of Clinical Medicine, University of Bergen, Bergen, Norway), Tristan W. Clark (School of Clinical and Experimental Sciences, Faculty of Medicine, University of Southampton, Southampton, UK), Marit H. Ebbesen (Department of Microbiology, Haukeland University Hospital, Bergen, Norway), Daniel Faurholt-Jepsen (Department of Clinical Science, Bergen Integrated Diagnostic Stewardship cluster, University of Bergen, Bergen, Norway; Department of Infectious Diseases, Rigshospitalet, Denmark), Harleen M. S. Grewal (Department of Clinical Science, Bergen Integrated Diagnostic Stewardship cluster, University of Bergen, Bergen, Norway; Department of Microbiology, Haukeland University Hospital, Bergen, Norway), Lars Heggelund (Department of Clinical Science, Bergen Integrated Diagnostic Stewardship cluster, University of Bergen, Bergen, Norway; Department of Internal Medicine, Vestre Viken Hospital Trust, Drammen, Norway), Siri T. Knoop (Department of Clinical Science, Bergen Integrated Diagnostic Stewardship cluster, University of Bergen, Bergen, Norway; Department of Microbiology, Haukeland University Hospital, Bergen, Norway), Øyvind Kommedal (Department of Clinical Science, Bergen Integrated Diagnostic Stewardship cluster, University of Bergen, Bergen, Norway; Department of Microbiology, Haukeland University Hospital, Bergen, Norway), Dagfinn L. Markussen (Emergency Care Clinic, Haukeland University Hospital, Bergen, Norway), Pernille Ravn (Department of Internal Medicine, Section for Infectious Diseases, Herlev and Gentofte Hospital, Hellerup, Denmark), Christian Ritz (Department of Clinical Science, Bergen Integrated Diagnostic Stewardship cluster, University of Bergen, Bergen, Norway; National Institute of Public Health, University of Southern Denmark, Copenhagen, Denmark), Sondre Serigstad (Emergency Care Clinic, Haukeland University Hospital, Bergen, Norway; Department of Clinical Medicine, University of Bergen, Bergen, Norway; Department of Clinical Science, Bergen Integrated Diagnostic Stewardship cluster, University of Bergen, Bergen, Norway), Elling Ulvestad (Department of Clinical Science, Bergen Integrated Diagnostic Stewardship cluster, University of Bergen, Bergen, Norway; Department of Microbiology, Haukeland University Hospital, Bergen, Norway), Cornelis H. van Werkhoven (Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, Utrecht, The Netherlands).
Abbreviations
- CAP
Community acquired pneumonia
- PCR
Polymerase chain reaction
- SARS-CoV-2
SARS-Coronavirus-2
- COVID-19
Coronavirus disease 2019
- RTI
Respiratory tract infection
- ED
Emergency department
- ICD-10
International Classification of Diseases 10th revision
- RCT
Randomized controlled trial
- eCRF
Electronic case report form
- FAP plus
BioFire® FilmArray® Pneumonia panel plus
- SDs
Standard diagnostic methods
- IQR
Interquartile range
- 95% CI
95% confidence interval
- LRTIs
Lower respiratory tract infections
Author contributions
SS, DLM, STK, and ROB coordinated patient recruitment and follow-up. MHE and HMSG supervised the microbiology studies. SS and DLM wrote the first draft of the manuscript with contribution from STK. All authors (SS, DLM, CR, MHE, STK, ØK, LH, EU, ROB and HMSG) have provided feedback to the first draft of the manuscript. CR reviewed the statistical analysis. HMSG had primary responsibility for the final content of the manuscript. All authors read and approved the final manuscript.
Funding
Open access funding provided by University of Bergen. This work was supported financially by the Research Council of Norway (NORCAP; 288718), the Trond Mohn Foundation (COVID-19 CAPNOR; TMS2020TMT07 and RESPNOR; BFS2019TMT06), the University of Bergen and Haukeland University Hospital. The funders had no role in designing the study and no role in collection, analysis and interpretation of data, nor in writing the manuscript.
Availability of data and materials
The datasets used and analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The two prospective patient cohorts were selected from a study approved by the Regional Committee for Medical and Health Research Ethics in South East Norway (REK ID: 31935) and performed in accordance with the Declaration of Helsinki. Written informed consent was obtained from all participants or from their legal guardian/close relative (in case of altered consciousness or confusion) at the time of recruitment. Regarding data extracted from the electronic heath records, only aggregated anonymous patient data was used and consent was deemed unnecessary by the Regional Committee for Medical and Health Research Ethics in Western Norway (REK ID: 221336).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sondre Serigstad, Email: sondre.serigstad@helse-bergen.no.
Harleen M. S. Grewal, Email: harleen.grewal@uib.no
the CAPNOR study group:
Tristan W. Clark, Daniel Faurholt-Jepsen, Pernille Ravn, and Cornelis H. van Werkhoven
References
- 1.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nair H, Simoes EA, Rudan I, Gessner BD, Azziz-Baumgartner E, Zhang JSF, et al. Global and regional burden of hospital admissions for severe acute lower respiratory infections in young children in 2010: a systematic analysis. Lancet. 2013;381(9875):1380–1390. doi: 10.1016/S0140-6736(12)61901-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torres A, Cilloniz C, Blasi F, Chalmers JD, Gaillat J, Dartois N, et al. Burden of pneumococcal community-acquired pneumonia in adults across Europe: a literature review. Respir Med. 2018;137:6–13. doi: 10.1016/j.rmed.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Clark TW, Medina MJ, Batham S, Curran MD, Parmar S, Nicholson KG. Adults hospitalised with acute respiratory illness rarely have detectable bacteria in the absence of COPD or pneumonia; viral infection predominates in a large prospective UK sample. J Infect. 2014;69(5):507–515. doi: 10.1016/j.jinf.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jain S, Self WH, Wunderink RG, Fakhran S, Balk R, Bramley AM, et al. Community-acquired pneumonia requiring hospitalization among US adults. N Engl J Med. 2015;373(5):415–427. doi: 10.1056/NEJMoa1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holter JC, Muller F, Bjorang O, Samdal HH, Marthinsen JB, Jenum PA, et al. Etiology of community-acquired pneumonia and diagnostic yields of microbiological methods: a 3-year prospective study in Norway. BMC Infect Dis. 2015;15:64. doi: 10.1186/s12879-015-0803-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosch AA, Biesbroek G, Trzcinski K, Sanders EA, Bogaert D. Viral and bacterial interactions in the upper respiratory tract. PLoS Pathog. 2013;9(1):e1003057. doi: 10.1371/journal.ppat.1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chertow DS, Memoli MJ. Bacterial coinfection in influenza: a grand rounds review. JAMA. 2013;309(3):275–282. doi: 10.1001/jama.2012.194139. [DOI] [PubMed] [Google Scholar]
- 9.Ruuskanen O, Lahti E, Jennings LC, Murdoch DR. Viral pneumonia. Lancet. 2011;377(9773):1264–1275. doi: 10.1016/S0140-6736(10)61459-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rynda-Apple A, Robinson KM, Alcorn JF. Influenza and bacterial superinfection: illuminating the immunologic mechanisms of disease. Infect Immun. 2015;83(10):3764–3770. doi: 10.1128/IAI.00298-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poole S, Brendish NJ, Clark TW. SARS-CoV-2 has displaced other seasonal respiratory viruses: results from a prospective cohort study. J Infect. 2020;81(6):966–972. doi: 10.1016/j.jinf.2020.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Redlberger-Fritz M, Kundi M, Aberle SW, Puchhammer-Stockl E. Significant impact of nationwide SARS-CoV-2 lockdown measures on the circulation of other respiratory virus infections in Austria. J Clin Virol. 2021;137:104795. doi: 10.1016/j.jcv.2021.104795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park S, Michelow IC, Choe YJ. Shifting patterns of respiratory virus activity following social distancing measures for coronavirus disease 2019 in South Korea. J Infect Dis. 2021;224(11):1900–1906. doi: 10.1093/infdis/jiab231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Serigstad S, Markussen D, Grewal HMS, Ebbesen M, Kommedal O, Heggelund L, et al. Rapid syndromic PCR testing in patients with respiratory tract infections reduces time to results and improves microbial yield. Sci Rep. 2022;12(1):326. doi: 10.1038/s41598-021-03741-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biomérieux. BioFire® FilmArray Pneumonia plus. https://www.biomerieux-diagnostics.com/biofire-filmarray-pneumonia-panel. Accessed 2 May 2022.
- 16.Leber AM. Clinical microbiology procedures handbook. 4. Washington, DC: ASM Press; 2016. [Google Scholar]
- 17.Timeline: news from Norwegian ministries about the coronavirus disease Covid-19—regjeringen.no. https://www.regjeringen.no/en/topics/koronavirus-covid-19/timeline-for-news-from-norwegian-ministries-about-the-coronavirus-disease-covid-19/id2692402/. Accessed 2 May 2022.
- 18.Jennings LC, Anderson TP, Beynon KA, Chua A, Laing RT, Werno AM, et al. Incidence and characteristics of viral community-acquired pneumonia in adults. Thorax. 2008;63(1):42–48. doi: 10.1136/thx.2006.075077. [DOI] [PubMed] [Google Scholar]
- 19.Karhu J, Ala-Kokko TI, Vuorinen T, Ohtonen P, Syrjala H. Lower respiratory tract virus findings in mechanically ventilated patients with severe community-acquired pneumonia. Clin Infect Dis. 2014;59(1):62–70. doi: 10.1093/cid/ciu237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Self WH, Williams DJ, Zhu Y, Ampofo K, Pavia AT, Chappell JD, et al. Respiratory viral detection in children and adults: comparing asymptomatic controls and patients with community-acquired pneumonia. J Infect Dis. 2016;213(4):584–591. doi: 10.1093/infdis/jiv323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varela FH, Scotta MC, Polese-Bonatto M, Sartor ITS, Ferreira CF, Fernandes IR, et al. Absence of detection of RSV and influenza during the COVID-19 pandemic in a Brazilian cohort: likely role of lower transmission in the community. J Glob Health. 2021;11:05007. doi: 10.7189/jogh.11.05007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poole S, Brendish NJ, Tanner AR, Clark TW. Physical distancing in schools for SARS-CoV-2 and the resurgence of rhinovirus. Lancet Respir Med. 2020;8(12):e92–e93. doi: 10.1016/S2213-2600(20)30502-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu D, Lu J, Sun Z, Cao L, Zeng Q, Liu Q, et al. Rhinovirus remains prevalent in school teenagers during fight against COVID-19 pandemic. Immun Inflamm Dis. 2021;9(1):76–79. doi: 10.1002/iid3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brueggemann AB, Jansen van Rensburg MJ, Shaw D, McCarthy ND, Jolley KA, Maiden MCJ, et al. Changes in the incidence of invasive disease due to Streptococcus pneumoniae, Haemophilus influenzae, and Neisseria meningitidis during the COVID-19 pandemic in 26 countries and territories in the invasive respiratory infection surveillance initiative: a prospective analysis of surveillance data. Lancet Digit Health. 2021;3(6):e360–e370. doi: 10.1016/S2589-7500(21)00077-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stralin K, Tornqvist E, Kaltoft MS, Olcen P, Holmberg H. Etiologic diagnosis of adult bacterial pneumonia by culture and PCR applied to respiratory tract samples. J Clin Microbiol. 2006;44(2):643–645. doi: 10.1128/JCM.44.2.643-645.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.den Heijer CD, van Bijnen EM, Paget WJ, Pringle M, Goossens H, Bruggeman CA, et al. Prevalence and resistance of commensal Staphylococcus aureus, including meticillin-resistant S. aureus, in nine European countries: a cross-sectional study. Lancet Infect Dis. 2013;13(5):409–415. doi: 10.1016/S1473-3099(13)70036-7. [DOI] [PubMed] [Google Scholar]
- 27.Drayss M, Claus H, Hubert K, Thiel K, Berger A, Sing A, et al. Asymptomatic carriage of Neisseria meningitidis, Haemophilus influenzae, Streptococcus pneumoniae, group A Streptococcus and Staphylococcus aureus among adults aged 65 years and older. PLoS ONE. 2019;14(2):e0212052. doi: 10.1371/journal.pone.0212052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gunnarsson RK, Holm SE, Soderstrom M. The prevalence of potential pathogenic bacteria in nasopharyngeal samples from healthy children and adults. Scand J Prim Health Care. 1998;16(1):13–17. doi: 10.1080/028134398750003340. [DOI] [PubMed] [Google Scholar]
- 29.Zanella RC, Brandileone MCC, Almeida SCG, de Lemos APS, Sacchi CT, Goncalves CR, et al. Nasopharyngeal carriage of Streptococcus pneumoniae, Haemophilus influenzae, and Staphylococcus aureus in a Brazilian elderly cohort. PLoS ONE. 2019;14(8):e0221525. doi: 10.1371/journal.pone.0221525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakr A, Brégeon F, Mège J-L, Rolain J-M, Blin O. Staphylococcus aureus nasal colonization: an update on mechanisms, epidemiology, risk factors, and subsequent infections. Front Microbiol. 2018;9:2419. doi: 10.3389/fmicb.2018.02419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vestrheim DF, Hoiby EA, Aaberge IS, Caugant DA. Impact of a pneumococcal conjugate vaccination program on carriage among children in Norway. Clin Vaccine Immunol. 2010;17(3):325–334. doi: 10.1128/CVI.00435-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Domenech de Cellès M, Arduin H, Lévy-Bruhl D, Georges S, Souty C, Guillemot D, et al. Unraveling the seasonal epidemiology of pneumococcus. Proc Natl Acad Sci. 2019;116(5):1802. doi: 10.1073/pnas.1812388116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeannoel M, Casalegno J-S, Ottmann M, Badiou C, Dumitrescu O, Lina B, et al. Synergistic effects of influenza and Staphylococcus aureus toxins on inflammation activation and cytotoxicity in human monocytic cell lines. Toxins (Basel) 2018;10(7):286. doi: 10.3390/toxins10070286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCullers JA. Do specific virus–bacteria pairings drive clinical outcomes of pneumonia? Clin Microbiol Infect. 2013;19(2):113–118. doi: 10.1111/1469-0691.12093. [DOI] [PubMed] [Google Scholar]
- 35.McCullers JA. Insights into the interaction between influenza virus and pneumococcus. Clin Microbiol Rev. 2006;19(3):571–582. doi: 10.1128/CMR.00058-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mulcahy ME, McLoughlin RM. Staphylococcus aureus and influenza A virus: partners in coinfection. MBio. 2016;7(6):e02068-16. doi: 10.1128/mBio.02068-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murdoch DR, Jennings LC. Association of respiratory virus activity and environmental factors with the incidence of invasive pneumococcal disease. J Infect. 2009;58(1):37–46. doi: 10.1016/j.jinf.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 38.Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis. 2008;198(7):962–970. doi: 10.1086/591708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Numbers from the Norwegian cause of death registry. https://www.fhi.no/hn/helseregistre-og-registre/dodsarsaksregisteret/tall-fra-dodsarsaksregisteret-for-2020/. Accessed 2 May 2022.
- 40.The Norwegian surveillance system for communicable diseases (MSIS). http://www.msis.no/. Accessed 2 May 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. BioFire® FilmArray® Pneumonia panel plus (FAP plus) targets.
Data Availability Statement
The datasets used and analysed during the current study are available from the corresponding author on reasonable request.