Abstract
Respiratory reduction of nitrate to nitrite is the first key step in the denitrification process that leads to nitrate loss from soils. In Paracoccus pantotrophus, the enzyme system that catalyzes this reaction is encoded by the narKGHJI gene cluster. Expression of this cluster is maximal under anaerobic conditions in the presence of nitrate. Upstream from narK is narR, a gene encoding a member of the FNR family of transcriptional activators. narR is transcribed divergently from the other nar genes. Mutational analysis reveals that NarR is required for maximal expression of the membrane-bound nitrate reductase genes and narK but has no other regulatory function related to denitrification. NarR is shown to require nitrate and/or nitrite is order to activate gene expression. The N-terminal region of the protein lacks the cysteine residues that are required for formation of an oxygen-sensitive iron-sulfur cluster in some other members of the FNR family. Also, NarR lacks a crucial residue involved in interactions of this family of regulators with the ς70 subunit of RNA polymerase, indicating that a different mechanism is used to promote transcription. narR is also found in Paracoccus denitrificans, indicating that this species contains at least three FNR homologues.
In environments depleted of oxygen, facultatively anaerobic bacteria are capable of expressing alternative respiratory enzymes so that electron acceptors other than oxygen may be utilized. Many bacteria use nitrate as an electron acceptor in the absence of oxygen (8). Organisms such as Escherichia coli reduce nitrate to nitrite and subsequently ammonia, whereas the denitrifying bacteria reduce nitrate via nitrite to the gaseous products nitric oxide, nitrous oxide, and finally dinitrogen gas. The four reactions of denitrification are linked to the membrane-associated electron transport chain such that the transfer of electrons to the reductases for nitrate, nitrite, nitric oxide, and nitrous oxide is associated with the generation of proton motive force and hence the conservation of energy in the form of ATP.
Although respiration can continue under anaerobic conditions at the expense of nitrate, the P/2e− ratio during denitrification is lower than during oxygen respiration. Bacteria therefore tend to respire oxygen in preference to nitrate, and this process is regulated at the level of transcription such that anaerobic metabolic apparatus is down-regulated in the presence of oxygen. The regulation of metabolism in response to oxygen is best understood in E. coli, in which a global regulator, FNR, activates transcription of the genes necessary for anaerobic respiration only when oxygen is depleted (32). The activation of FNR is mediated via the formation of an oxygen-sensitive Fe4S4 cluster which depends on four cysteine residues, three of which are found toward the N terminus of the FNR protein. Breakdown of the Fe4S4 cluster in the presence of oxygen prevents transcriptional activation of the FNR regulon under aerobic conditions (16).
FNR-like regulators have been identified in control of anaerobic metabolism in a number of diverse organisms with different nutritional requirements. In E. coli, global regulation of anaerobic metabolism is governed by a single FNR-like regulator. The regulation of anaerobic metabolism in denitrification has been studied in Paracoccus denitrificans, Rhodobacter sphaeroides, Pseudomonas stutzeri, and Pseudomonas aeruginosa (3, 34, 35, 36, 37, 39). In each case, control of anaerobic metabolism is regulated by multiple FNR homologues, some of which contain the Cys residues that are necessary for oxygen-sensitive Fe4S4 cluster formation and some of which lack these residues such that their sensory substrate(s) is less clear.
In this paper we report the identification and characterization of an fnr-like gene (narR) which specifically regulates nitrate reductase expression in the denitrifier Paracoccus pantotrophus. We have also identified narR in P. denitrificans, making it the third member of the fnr family to be identified in that species. The genetic organization and mode of control of nitrate reductase (nar) in Paracoccus species differs from that found in other nitrate reducers so far analyzed and reflects the diverse strategies that are employed by organisms to regulate gene expression. The sensing mechanism and regulatory specificities of NarR are discussed.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this work are listed in Table 1. Paracoccus strains were grown in Luria-Bertani (LB) medium or in a defined mineral salts medium (28) supplemented with 20 mM succinate as the carbon and energy source. Aerobic growth was achieved in 5 ml of growth medium in 25-ml Universal flasks or in 20 ml of medium in 250-ml conical flasks, which were incubated in a rotary shaker at 250 rpm and 37°C. Anaerobic growth was carried out in 25 ml of growth medium in 25-ml Universal flasks incubated stationary at 37°C or in 5-ml test tubes capped with Suba-Seals and sparged with nitrogen prior to inoculation in order to ensure anaerobic conditions. For anaerobic growth, cultures were supplemented with the electron acceptor sodium nitrate (20 mM), sodium nitrite (3 mM), or nitrous oxide (growth medium was sparged with N2O to give a 25 mM saturated solution). E. coli strains were grown in LB medium in 5-ml aerobic cultures, incubated as described for Paracoccus strains. Media for antibiotic-resistant strains were supplemented with the antibiotics rifampin (50 μg/ml), kanamycin (50 μg/ml), spectinomycin (50 μg/ml), streptomycin (20 μg/ml), and ampicillin (100 μg/ml), are appropriate. E. coli strains resistant to tetracycline were grown with 12 μg of the antibiotic per ml, whereas tetracycline-resistant P. pantotrophus strains were grown with 1 μg of tetracycline per ml. Growth on solid media used liquid growth medium supplemented with 1.5% bacteriological agar.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Genotype and description | Source or reference |
|---|---|---|
| Escherichia coli DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi1 relA1 (general cloning vehicle) | Gibco BRL |
| E. coli S17-1 | thi pro hsdR recA RP4-2 integrated Tc::Mu Km::Tn7 (for conjugative transfer of mobilizable plasmids into Paracoccus strains) | 30 |
| E. coli TOP10F′ | F′ (Tetr) (mrr-hsdRMS-mcrBC) lacZΔM15 rpsL (Smr) endA1 | Invitrogen |
| Paracoccus pantotrophus Rifr | Spontaneously Rifr strain of P. pantotrophus, Smr (formerly Thiosphaera pantotropha) | 28 |
| P. denitrificans Pd1222 | Rifr Spr strain of P. denitrificans | 12 |
| P. pantotrophus nar R::Ω | Contains disrupted chromosomal copy of narR | This work 38 |
| Plasmids | ||
| pUC18 | Ampr | 38 |
| pBluescript | Ampr | Stratagene |
| pCR2.1 | Vector for cloning Taq-amplified products with A tails | Invitrogen |
| pHP45Ω | Contains Ω cassette (Smr Spr) | 27 |
| pARO181 | Contains oriT necessary for conjugative transfer from E. coli into Paracoccus strains; incapable of replication in Paracoccus sp.; Kanr | 26 |
| SuperCos123 | Cosmid containing P. pantotrophus nar region | This work |
| pJWB7 | Subcloned 7-kb EcoRI fragment from SuperCos123 | This work |
| pCRTPnarR | Amplified region containing narR from P. pantotrophus cloned into pCR2.1 | This work |
| pCRPDnarR | Amplified region containing narR from P. denitrificans cloned into pCR2.1 | This work |
| pBSTPnarR | narR region from pCRTPnarR cloned into pBluescript | This work |
| pBSTPnarR::Ω | pBSTPnarR with Ω cassette inserted into narR | This work |
| pAROTPnarR::Ω | Disrupted narR gene from pBSTPnarR::Ω cloned into pARO181 | This work |
| pRK415 | Broad-host-range vector, Tetr | 15 |
| pRKnarR | narR cloned into pRK415 | This work |
| pMP220 | lacZ promoter probe vector, Tetr | 31 |
| pMPnarKpro | Transcriptional fusion of narK promoter to lacZ | This work |
| pMPnarRpro | Transcriptional fusion of narR promoter to lacZ | This work |
Nucleic acid manipulation and sequencing.
A library of P. pantotrophus chromosomal DNA in SuperCos (Stratagene) was screened with the narH gene (one of the structural genes of the membrane-bound nitrate reductase narGHJI) from P. pantotrophus. SuperCos123, which hybridized strongly with narH, was digested with EcoRI, and the resultant fragments were cloned into pUC18. The 7-kb EcoRI fragment cloned in pJWB7 contained part of narG and the region upstream from this gene (Fig. 1). The complete sequence of this fragment was achieved using sequence-derived primers. DNA sequencing was carried out using an ABI373A automatic DNA sequencer (Applied Biosystems).
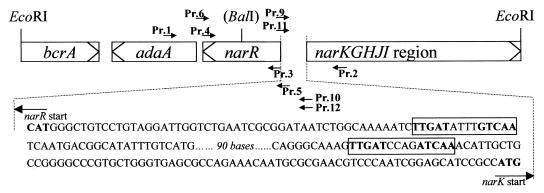
FIG. 1.
narR and flanking regions. The 7-kb region containing narR, part of the narKGHJI region, adaA, and bcrA is shown. Unique EcoRI sites and the BalI site (not unique) used for insertional mutagenesis of narR are shown. The DNA regions to which oligonucleotide primers were designed for use in this work are shown as Pr.1 to Pr.6 and Pr.9 to Pr.12, and the arrows indicate the 5′-to-3′ direction of those primers. The region between narR and narK is expanded, and the consensus FNR binding sites are boxed and in bold.
Oligonucleotide primers were designed for amplification of narR and parts of the flanking genes narK and adaA (Fig. 1). Primer 1 (5′-AACGTCCCGGGGTCGTCGCC-3′) is complementary to part of adaA; primer 2 (5′-ACCGCCCAGATCTGCGCCAC-3′) is complementary to part of narK. A 2.3-kb amplification product was obtained using a colony of P. pantotrophus as the DNA template (35 cycles of amplification; 30 s of denaturation at 94°C, 1 min of annealing at 60°C, and 3-min extension at 72°C) and using Taq polymerase from Promega. This product was cloned into pCR2.1; an EcoRI fragment containing narR was excised and inserted into the EcoRI site of pBluescript. Subsequently, the Ω cassette encoding resistance to spectinomycin was inserted into a unique BalI site within the narR gene, and the EcoRI fragment containing the disrupted copy of narR was cloned into pARO181 to yield a vector conferring kanamycin and spectinomycin resistance, pAROTPnarR::Ω. This vector was transformed into E. coli S17-1 and transferred into P. pantotrophus via conjugative mating in order to allow allelic exchange of the disrupted copy of narR with the wild-type copy of the gene. Transconjugants in which the Ω cassette had become incorporated into the recipient chromosome were spectinomycin resistant. Clones in which a second recombination event had occurred, giving rise to a single Ω-disrupted copy of narR, were spectinomycin resistant but kanamycin sensitive. Double crossovers were further confirmed by colony PCR (35 cycles of amplification; 30 s of denaturation at 94°C, 1 min of annealing at 60°C, and 4-min extension at 72°C) using oligonucleotide primers 3 (5′-CCCATGCGCGAAGAGGAC-3′) and 4 (5′-CCGCTTGCGCTCAAATCC-3′), which anneal at the 5′ and 3′ ends, respectively, of narR. Wild-type strains gave an amplification product of 700 bp, whereas in double crossovers the size of the product was increased to 2.7 kb due to the insertion of the Ω cassette.
The gene narR was amplified with primers 5 (5′-GCGCTGCAGACCAATCCTAC-3′) and 6 (5′-CTCGGATCCGGCCGTCAGGGG-3′). Primer 5 anneals upstream from the putative ribosome binding site before the start of the coding region of narR and contains an engineered PstI site. Primer 6 is complementary to the region just beyond the stop codon at the end of narR and contains an engineered BamHI site. The 700-bp product of this amplification was digested with PstI and BamHI and cloned into broad-host-range vector pRK415, yielding pRKnarR. pRKnarR was transferred into P. pantotrophus strains by conjugative mating.
Degenerate oligonucleotide primers 7 (5′-TNGAYGAYCAYCCNATG-3′) and 8 (5′-ARRTANCCRTCNGCNCC-3′) (N = A, T, G, or C; Y = C or T; R = G or A) were designed to be complementary to the known sequences of the transcriptional regulator narL. Thirty-five cycles of amplification with 30 s of denaturation at 94°C, 1 min of annealing at 45°C, and 1-min extension at 72°C should yield a 300-bp product if narL is present.
The promoter region for narK was amplified with primers 9 (5′-GTCGAATTCGCGCATGGGCTGTCC-3′) and 10 (5′-GGGTCTAGAATGGCGGATGCTCCGATT-3′). The promoter region for narR was amplified with primers 11 (5′-TCTTCTAGAATGGGCTGTCCTGTAGG-3′) and 12 (5′-AAGGAATTCGGGTCCGGCATGGCGGA-3′). The products were digested with XbaI and EcoRI and cloned into promoter probe vector pMP220 (31), which had also been digested with XbaI and EcoRI. This yielded plasmids pMPnarKpro and pMPnarRpro, constructs in which the lacZ gene from pMP220 has been placed under control of the narK and narR promoters, respectively. In each case the translational start site of narR or narK has been destroyed such that the constructs are transcriptional fusions. The plasmid constructs were introduced into P. pantotrophus strains by conjugative transfer from E. coli S17-1. β-Galactosidase activity was measured by the method of Miller (20).
Analysis of denitrification.
Whole-cell assays for nitrate and nitrite reductases were carried out in a Perspex chamber fitted with a Clark-type oxygen electrode (Rank Brothers, Bottisham, United Kingdom) at 30°C after the cell suspension had become anaerobic, as described previously (22). Nitrite concentration was estimated by the method of Nicholas and Nason (24). When used, the final concentration of Triton X-100 was 0.02% (vol/vol). NO reductase activity in intact cells was measured in anaerobic suspensions of cells using an iso-NO electrode (World Precision Instruments, Stevenage, United Kingdom).
Periplasmic extracts were prepared as described previously (21); the procedure for membrane isolation was the same except that the spheroplasts were lysed by resuspension in 10 mM Tris-Cl (pH 8.0) followed by the addition of a few grains of DNase I and incubation at 37°C for 30 min to solubilize the pellet. Membranes were collected by centrifugation at 12,000 × g and 4°C for 5 min, followed by resuspension in 1% (wt/vol) sodium dodecyl sulfate (SDS) prepared in 10 mM Tris-Cl (pH 8.0). Extracts were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and stained for covalently bound heme (13). Proteins were blotted onto polyvinylidene difluoride membranes, which were hybridized with antibodies raised to nitrous oxide reductase from P. pantotrophus (a kind gift from Ben C. Berks, University of East Anglia, Norwich, United Kingdom) and pseudoazurin (from P. pantotrophus [23]).
Nitrate and chlorate reductase activities were determined using reduced methyl viologen as an electron donor and measuring its oxidation spectrophotometrically at a wavelength of 600 nm (11). For these assays, intact cells were subjected to sonication at 4°C until cell breakage had occurred.
Nucleotide sequence accession number.
The P. pantotrophus DNA sequence of the region discussed in this paper has been deposited in the GenBank database under accession no. AF295359.
RESULTS
Sequence analysis of the nar cluster.
Transposon mutagenesis was previously used to identify the narH gene of P. pantotrophus (6), leading to the sequence of the narHJI genes (9). We have extended this sequence to characterize a 7-kb region of DNA (Fig. 1) that contains part of nitrate reductase structural gene narG (10), a gene which consists of two narK-like transporters (25) which are fused into a single open reading frame, open reading frame narR, and genes identified as adaA and bcrA by similarity to these genes in other organisms. adaA encodes the enzyme Ada, which catalyzes transfer of methyl groups from methylated DNA as part of the adaptive response in E. coli (29). bcrA encodes a transmembrane protein which confers resistance to the antibiotic bicyclomycin (7). These latter genes are highly unlikely to have any direct function in denitrification, and therefore they mark the end of the nar cluster.
The predicted translation product of narR is a 234-amino-acid polypeptide. BLASTP was used to analyze the similarity of NarR to known protein sequences. The top 20 hits using this method were members of the FNR family of transcriptional regulators. Sequence identity was distributed throughout the length of the sequences, but there is a very high degree of similarity towards the C terminus; a 27-amino-acid stretch (from 198 to 224, numbering in alignment) displays 70% identity between NarR and NNR from P. denitrificans (Fig. 2). This C-terminal region is predicted to contain the helix-turn-helix region of the proteins, which is the domain binding to the DNA during transcriptional activation. The high degree of identity in this region between NarR and other FNR homologues strongly suggests that NarR binds to the same consensus DNA sequence as do the other FNR homologues (i.e., TTGA [T/C]) (32). BLAST was repeated with a truncated version of NarR in which the helix-turn-helix region had been removed. The most similar protein was found to be DnrE, an FNR homologue from Pseudomonas stutzeri (37). The overall degree of identity was only 22%.
FIG. 2.
Alignment of members of the FNR family. An alignment created by the Genetics Computer Group program Pileup is shown. The sequences used were DnrE and DnrD from Pseudomonas stutzeri, NNR from P. denitrificans, NarR from P. pantotrophus, and FNR from E. coli. Residues conserved among at least four of the five aligned sequences are shown in bold. The predicted helix-turn-helix DNA binding domain is marked. The cysteine residues that are responsible for binding the oxygen-responsive iron-sulfur cluster in FNR are marked with asterisks. Glycine residue 85 (FNR numbering), which is involved in making contact with RNA polymerase (in all of the FNR homologues except NarR), is marked with a cross.
It is striking that a glycine conserved between the other members of the family (G85 of the FNR sequence) is absent from the NarR sequence (Fig. 2). This glycine has been shown to be crucial for contact of FNR with the ς70 subunit of RNA polymerase (4, 19), and its absence indicates that the mechanism for transcriptional activation by NarR is independent of this particular protein-protein contact.
narR and narK are divergently transcribed. Between the two protein coding regions is a stretch of 271 bases. Within this region there are two pairs of FNR consensus half sites (Fig. 1), appropriately spaced to allow dimers of FNR, NNR, and/or NarR to bind and potentially regulate the expression of narR and narK. In E. coli, the regulation of narGHJI expression is controlled by FNR and also by nitrate/nitrite concentration via two-component regulatory systems designated NarXL and NarQP (33). NarL and NarP bind to the promoter region at consensus NarL recognition heptamers TAC(C/T)N(A/C)T. A search for such consensus sequences in the promoter region between coding regions for narR and narK and in the first 500 bases of the coding regions of these two genes revealed no such NarL heptamers. This finding and the absence of narXL immediately upstream from the narK sequence (as found in other nar clusters that have been sequenced) suggest that there is no nitrate/nitrite regulation of nar expression via NarXL in P. pantotrophus.
Primers were designed to conserved regions of narL from E. coli, Pseudomonas stutzeri, and Pseudomonas aeruginosa. Thirty-five cycles of amplification using these primers with colonies of E. coli, Pseudomonas stutzeri, Pseudomonas aeruginosa, and P. pantotrophus as templates gave products of the right size for the first three organisms but yielded no product with P. pantotrophus as the template, further indicating the absence of narXL from this organism. Furthermore, narL from Pseudomonas aeruginosa did not hybridize with genomic DNA from P. pantotrophus, as judged by Southern blotting (not shown).
P. denitrificans also possesses a narR gene.
A thermal cycling experiment identical to that described in Materials and Methods was carried out using oligonucleotide primers 1 and 2, except that a colony of P. denitrificans Pd1222 was used as the template. This procedure yielded a 2.3-kb fragment of DNA. The amplified product was cloned into pCR2.1 to yield pCRPDnarR, and sequence for the region was obtained using M13 forward and reverse primers and custom-made primers. The 2.3-kb fragment from P. denitrificans is highly homologous to the adaA-narR-narK region from P. pantotrophus, demonstrating that P. denitrificans also possesses a narR gene and hence is capable of synthesizing three FNR-like proteins: NarR plus two other FNR homologues previously identified in this species (FnrP and NNR [34]). The coding regions of adaA, narR, and narK are highly conserved between P. pantotrophus and P. denitrificans. The region between narR and narK in P. denitrificans contains the FNR consensus binding sites as found in P. pantotrophus and also lacks any discernible heptamers for binding NarL.
Analysis of a P. pantotrophus narR null mutant.
A narR-deficient strain was constructed by insertional disruption of the gene with an Ω cassette (see Materials and Methods). Growth rates of the wild-type and narR mutant strains grown aerobically in LB medium or anaerobically in minimal medium with nitrite or nitrous oxide as the electron acceptor were comparable, but the narR mutant failed to grow appreciably in minimal medium with nitrate as the sole electron acceptor.
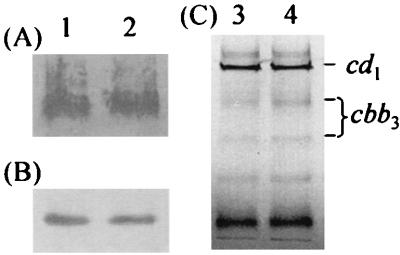
Activities of nitrate, nitrite, and nitric oxide reductases were measured in intact cells grown anaerobically on nitrite. There were no significant differences in activities of the nitrite and nitric oxide reductases between the wild-type and narR mutant strains, but the rate of nitrate reduction in the narR mutant strain was 10 to 15% of the rate in the wild type (Table 2). Expression of other proteins associated with denitrification, namely, the nitrous oxide reductase and small copper-containing electron transport protein pseudoazurin (which is known to be anaerobically inducible [23]), were estimated from Western blots. There did not appear to be any significant difference in levels of expression of these proteins between wild-type and narR mutant strains (Fig. 3). Extracts of wild-type and narR mutant strains were separated by SDS-PAGE and stained for heme, revealing no significant differences in intensity of c-heme-containing proteins between the strains, including the cytochrome cd1 nitrite reductase and c-heme-containing subunits of the cytochrome cbb3 oxidase (Fig. 3).
TABLE 2.
Denitrification activities in P. pantotrophus wild type and narR mutant
| Prepn | Activity (U nmol min−1 mg of protein−1) in:
|
|
|---|---|---|
| P. pantotrophus wild type | P. pantotrophus narR::Ω | |
| Intact cells | ||
| Nitrate reduction | 85 | 10 |
| Nitrate reduction + 0.02% Triton X-100 | 95 | 10 |
| Nitrite reduction | 370 | 330 |
| Nitric oxide reduction | 2,400 | 1,900 |
| Total-cell extract | ||
| Nitrate reductase | 390 | 100 |
| Chlorate reductase | 1,500 | 180 |
FIG. 3.
Expression of respiratory proteins in P. pantotrophus wild type and narR::Ω. Strains were grown anaerobically with nitrite as the electron acceptor; 20-ml cultures were harvested by centrifugation, resuspended in 1 ml of 10 mM Tris-HCl (pH 8), and sonicated. Extracts were separated by SDS-PAGE (20 μg of protein in each lane) and Western blotted with antibodies raised to nitrous oxide reductase (A) and pseudoazurin (B) or stained for proteins containing covalently attached heme (C). Heme proteins cytochrome cd1 nitrite reductase and the c-heme-containing subunits of cytochrome cbb3 oxidase are marked. Lanes 1 and 3 contain extracts from P. pantotrophus wild type; lanes 2 and 4 contain extracts from P. pantotrophus narR::Ω.
Expressed divergently from narR is a set of nar genes, narK, narG, narH, narJ, and narI. narK is required for transport of nitrogen oxyanions across the cytoplasmic membrane, whereas the other four genes are required for production of the nitrate reductase enzyme itself. Given that there is an FNR consensus sequence upstream from the first of these genes (narK), it is not clear whether the lesion in nitrate reduction in the narR mutant is due to the absence of a transporter for nitrogen oxyanions and/or the absence of a nitrate reductase enzyme itself. Measurement of nitrate reduction in intact cells was repeated in the presence of Triton X-100, which allows free passage of nitrate and nitrite across the membrane and therefore removes the need for a specific transport protein (2). This treatment did not lead to an increase in nitrate reduction in either the wild type or the narR mutant strain (Table 2), indicating that the narR mutant is defective in reduction of nitrate and not just nitrate transport. Reduced levels of nitrate reductase expression were confirmed by measuring nitrate and chlorate reductase activity in cell extracts of wild-type and narR mutant strains of P. pantotrophus (Table 2) grown anaerobically in the presence of nitrite. The substrate chlorate was used routinely, since chlorate reductase activity differentiates between membrane-bound nitrate reductase (NAR) and periplasmic nitrate reductase (NAP), because only NAR can use chlorate as a substrate (5).
Complementation of narR disruption.
A copy of narR including its putative ribosome binding site, but lacking its promoter, was cloned into broad-host-range vector pRK415 (15), with narR oriented to allow its expression from the lac promoter of the plasmid. The resultant plasmid (pRKnarR) was transferred into the narR mutant strain of P. pantotrophus in an attempt to complement the mutant phenotype. Indeed, the introduction of the plasmid did complement the lesion in growth on nitrate. Growth rates anaerobically with nitrate were as follows: P. pantotrophus wild type, μ = 0.355 h−1; P. pantotrophus narR::Ω, no growth; P. pantotrophus (pRKnarR), μ = 0.306 h−1; and P. pantotrophus narR::Ω(pRKnarR), μ = 0.300 h−1. This demonstrated that (i) the lesion in the narR strain was not due to a polar effect on the genes downstream of narR and (ii) expression of narR from the lac promoter of pRK415 can be achieved in P. pantotrophus.
Given that the lac promoter is not controlled by variation in the availability of oxygen, we decided to test whether the complemented strain might express nitrate reductase under oxic conditions as a consequence of the constitutive expression of narR. We found that the expression of nitrate reductase was heavily repressed during aerobic growth (Table 3) of the narR strain complemented with the plasmid-borne copy of narR, indicating that induction of nitrate reductase expression by NarR is not simply a consequence of NarR expression itself (i.e., NarR-dependent expression is controlled biochemically by some sensory mechanism, rather than by a hierarchical genetic mechanism such as an FNR-dependent expression of narR).
TABLE 3.
Chlorate and nitrate reductase activities in P. pantotrophus extracts
| Electron acceptor(s) | Activity (nmol min−1 mg of protein−1)
|
|||
|---|---|---|---|---|
| Chlorate reductase
|
Nitrate reductase
|
|||
| P. pantotrophus wild type | P. pantotrophus narR::Ω | P. pantotrophus wild type | P. pantotrophus narR::Ω | |
| NO3− | 3,600 | NDa | ||
| NO2− | 1,000 | 110 | ||
| N2O | 190 | 60 | ||
| N2O + NO3− | 2,100 | 65 | ||
| N2O + NO2− | 1,700 | 85 | ||
| O2 | <2 | <2 | ||
| O2 + NO3− | 10 | <2 | 65 | 70 |
| O2 + NO2− | <2 | <2 | ||
ND, not done. Activity could not be measured in P. pantotrophus narR::Ω which was unable to grow with nitrate as the sole electron acceptor.
To test possible sensory substrates of NarR, we examined the effects of the presence of various electron acceptors on nitrate reductase expression in a strain expressing narR constitutively [P. pantotrophus narR::Ω(pRKnarR)] and a strain unable to express narR (P. pantotrophus narR::Ω) as a control. Nitrate reductase expression was heavily repressed under aerobic growth conditions in both strains (Table 3). The only aerobic growth condition under which nitrate reductase expression could be detected was in P. pantotrophus narR::Ω(pRKnarR) when the medium was supplemented with nitrate. This nitrate-dependent induction of membrane-bound nitrate reductase expression was narR dependent, since there was no expression of the nitrate reductase in the uncomplemented narR mutant strain. In strains with or without narR, there were higher levels of nitrate reductase expression under anaerobic growth conditions than under aerobic growth conditions. The key difference was that in the strain containing the plasmid-borne copy of narR, both nitrate and nitrite induced expression of membrane-bound nitrate reductase (Table 3). This was found to be the case if the nitrogen oxyanions were used as the sole electron acceptor during growth or whether they were added as supplements to bacteria growing with nitrous oxide as the electron acceptor. The basal anaerobic rate of nitrate reductase activity, as exemplified by the rate after growth on nitrous oxide, is of the same order of magnitude in P. pantotrophus narR::Ω(pRKnarR) as in P. pantotrophus narR::Ω, indicating that the anaerobic induction of narKGHJI gene expression is independent of narR, but achieving maximal expression of the nitrate reductase, which is required to support anaerobic growth on nitrate, is dependent on NarR and requires the presence of nitrate and/or nitrite. Aerobic expression of NAP is unaffected by the presence or absence of a functional copy of narR, as shown by the similar nitrate (but not chlorate) reductase rates in aerobically grown strains (Table 3).
Some of those transcriptional regulators most similar to NarR have been found to regulate gene expression in response to NO (19, 34, 36). For this reason, we investigated whether the dependence of NarR activity on nitrate and/or nitrite was indirect and whether it was due to the accumulation of NO during the metabolism of nitrate and nitrite. P. pantotrophus narR::Ω(pRKnarR) was grown anaerobically on nitrous oxide as the sole electron acceptor. Cultures grown overnight were treated with NO-releasing compounds S-nitrosoglutathione and sodium nitroprusside at concentrations ranging from 100 nM to 10 μM; the cultures were harvested after 3 h, and total-cell extracts were examined for chlorate reductase activity. Neither of these chemicals was able to elicit an increase in NAR activity. An alternative approach was to treat cultures of P. pantotrophus narR::Ω(pRKnarR) grown anaerobically on nitrate with 2-phenyl-4,4,5,5,-tetramethylimidazoline-1-oxyl-3-oxide (PTIO), which reacts with NO (1) and will hence remove the NO formed by denitrification. Treatment with 100 μM PTIO did not decrease the nitrate-dependent induction of chlorate reductase (NAR) expression. As a control, the effect of PTIO on nitrite reductase expression, which is dependent on NO activation of NNR, was examined. As expected, we found that PTIO decreased the expression of nitrite reductase (not shown).
narR and narK promoter activity.
To analyze the impact of environmental conditions and the genotype of P. pantotrophus strains on the promoter activity of the region between narR and narK, we monitored the activities of promoters for narK and narR by using transcriptional fusions of the promoters to lacZ.
The narK promoter is induced under anaerobic denitrifying conditions, compared to aerobic conditions, in wild-type P. pantotrophus (Fig. 4A). This induction of narK expression is almost completely obliterated in the narR mutant strain, demonstrating that NarR is necessary for maximal transcription of narK. When aerobically grown and denitrifying cultures were compared, transcription from the narR promoter was found to be repressed under denitrifying conditions (Fig. 4B). Furthermore, higher β-galactosidase activity was measured in the narR mutant strain than in the wild type. This indicates that the narR promoter is autoregulated by NarR and repressed by anaerobic conditions, possibly through the action of FnrP.
FIG. 4.
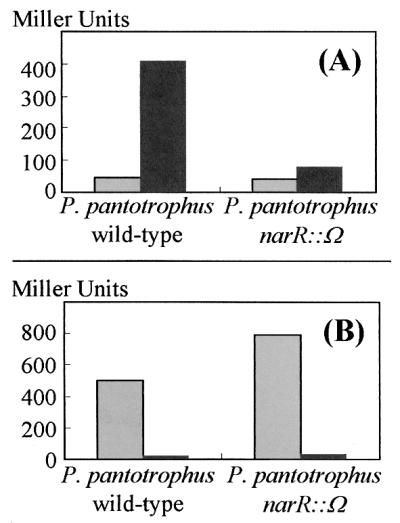
Regulation of narK and narR promoter activities. Expression of the promoters for narK and narR was assessed in P. pantotrophus wild type and P. pantotrophus narR::Ω bearing plasmids pMPnarKpro (A) and pMPnarRpro (B). Activity is expressed in Miller units for the cultures grown under aerobic conditions (grey bars) and cultures grown under denitrifying conditions with nitrite as the electron acceptor (black bars).
Northern blots yielded patterns of expression of the narR and narK genes similar to those found by analysis of lacZ fusions (not shown).
DISCUSSION
In this paper we have shown the new gene narR to be present in P. pantotrophus and in P. denitrificans. This is now the third member of the FNR family of transcriptional regulators found in the latter organism. Why are so many of these regulators required? And more specifically, how do the functions of these regulators differ? We address this question in terms of five specific questions.
What does NarR regulate?
Mutants of P. pantotrophus incapable of synthesizing narR have been constructed, and it has been clearly shown that the only effect of such mutations on denitrification is on the expression of the apparatus necessary for nitrate reduction via NAR. The expression of narGHJI, encoding the membrane-bound nitrate reductase, as judged by nitrate reductase activity at the enzyme and intact cell levels, was 10 to 50 times lower in the narR mutant strain than in the wild type after anaerobic growth in the presence of nitrate (or nitrite). Using lacZ fusions, we were also able to show that narK transcription is drastically reduced in the narR mutant under denitrifying conditions. Also, NarR negatively autoregulates its own expression (see “How is narR regulated” below). We were unable to find any effect of NarR on other parts of the denitrification apparatus or any other respiratory components. The activity of NAP under aerobic conditions in both wild-type and narR mutant strains demonstrates that NarR is not required for assembly of the molybdenum cofactor found in both NAP and NAR.
Are there other regulators of NAR?
narGHJI expression in E. coli is controlled by FNR (32) and by a pair of nitrate/nitrite-responsive two-component regulators, NarXL and NarPQ (33). In E. coli, narXL is located upstream from narK (25). There is no homologue of narXL to be found in an equivalent location in P. pantotrophus; furthermore, we were unable to amplify a narL homologue by using degenerate primers or to identify narL by Southern blotting. Also, in the absence of narR there is no induction of nitrate reductase expression in response to nitrate or nitrite. Furthermore, no NarL recognition heptamers were found in the promoter region for narK and narR. Taken together, the evidence suggests that P. pantotrophus (unlike some other denitrifiers such as Pseudomonas aeruginosa [accession no. AF112870] and Pseudomonas stutzeri [14]) lacks a narXL system for controlling nitrate reductase expression.
In P. denitrificans, a mutant incapable of expressing fnrP has been found to have ∼30% of the NAR activity of the wild type (35), indicating that this anaerobic transcription activator is involved in induction of the nar structural genes. This is consistent with our finding that in the absence of narR there is still anaerobic induction of NAR expression; in fact, after anaerobic growth with nitrous oxide as the sole electron acceptor, there is little difference in NAR activity between the wild type and narR mutant. We expect that this anaerobic induction is due to FnrP in P. pantotrophus. There is an FNR box upstream from the start of narK which may be the site of binding of both FnrP and NarR during induction of nar gene expression. The very high rate of NAR activity in the wild type compared to the narR mutant (after growth with nitrate present) clearly indicates that NarR is the most important activator for maximal nar expression in P. pantotrophus.
The anaerobic induction of NAR after growth on nitrous oxide as the sole electron donor leads us to reassess some work that we carried out a number of years ago to investigate expression of the denitrification apparatus after growth on nitrous oxide (23). In that work, we found that there was a low level of expression of nitrite reductase, nitric oxide reductase, and pseudoazurin after growth on nitrous oxide, suggesting that these conditions are somehow similar to aerobic growth conditions. We explained this in terms of the high reduction potential of N2O/N2 couple causing an oxidation of the respiratory chain that simulates aerobic conditions. However, it seems clear now that the lack of expression under these conditions was more likely due to the absence of nitric oxide which is required for activation of NNR, which in turn induces expression of nitrite and nitric oxide reductases.
How is promoter specificity conferred?
The mechanisms governing specificity of given FNR homologues for particular promoters is something of a conundrum. The helix-turn-helix regions of the FNR homologues of Paracoccus are all very similar, suggesting that they bind to similar consensus sequences. However, the absence of G85 from NarR, which is vital for the formation of a contact with the ς70 subunit of RNA polymerase (4, 19), indicates that differences in contacts with RNA polymerase may control the promoter specificities of NarR compared to the other FNR homologues in Paracoccus species. Clearly this needs to be assessed experimentally to be confirmed or refuted. The N-terminal region of NarR bears a low level of similarity to other members of the FNR family, indicating that NarR may have a different fold and hence distinct surfaces to interact with the polymerase, and thus activate gene expression.
How is narR regulated?
We considered it possible that induction of gene expression by NarR may be simply a consequence of the amount of NarR available to bind to the narK promoter; i.e., NarR is part of a regulatory cascade, in which it is induced by a global regulator (such as FnrP). However, promoter analysis indicated that narR negatively autoregulates its own expression and is repressed under anaerobic conditions by another regulator (possibly fnrP) (Fig. 4B). This clearly demonstrates that the induction of expression of the nar structural genes by NarR is not a consequence of the level of expression of NarR, but that there is some biochemical signal to which NarR responds. This supposition was also lent support by the finding that constitutive expression of narR (in strains bearing pRKnarR) did not lead to aerobic expression of NAR.
What does NarR sense?
Like DnrE and DnrD from Pseudomonas stutzeri (37) and NNR from P. denitrificans and R. sphaeroides (34, 36) (and several other FNR homologues), NarR lacks the N-terminal cysteines that have been shown to be required for Fe4S4 cluster formation and hence oxygen sensing in FNR from E. coli (16). It has been found that the NNR proteins sense NO (probably indirectly) in order to regulate gene expression (34, 36). We examined whether chemicals that release NO (S-nitrosoglutathione and sodium nitroprusside) and that sequester NO (PTIO) had any effect on NarR-dependent expression but could find no evidence to support this. However, it was clear that under anaerobic conditions, nitrate and nitrite can both cause NarR to induce gene expression. Given that this activation of NarR does not occur when the bacteria are grown under aerobic conditions (aerobically there is a very minor NarR-dependent induction of NAR expression in the presence of nitrate but not nitrite), it is probable that the effect of nitrate and nitrite on NarR activity is indirect and may depend on an unknown anaerobically induced factor.
To conclude, NarR is necessary to achieve maximal expression of nitrate reductase under anaerobic conditions in the presence of nitrate. The cost of maintenance of a gene for this regulator is small compared to the cost that would be incurred by the production of larger amounts of nitrate reductase under all anaerobic conditions, rather than just those conditions under which the enzyme is needed, i.e., in the absence of oxygen, when the substrate nitrate is available.
ACKNOWLEDGMENTS
This work was supported by Biotechnology and Biological Sciences Research Council (BBSRC) grant P10251, awarded to J.W.B.M., D.J.R. and S.J.F.
REFERENCES
- 1.Akaike T, Yoshida M, Miyamoto Y, Sato K, Kohno M, Sasamoto K, Miyazaki K, Ueda S, Maeda H. Antagonistic action of imidazolineoxyl N-oxides against endothelium derived relaxation factor/NO through a radical reaction. Biochemistry. 1993;32:827–832. doi: 10.1021/bi00054a013. [DOI] [PubMed] [Google Scholar]
- 2.Alefounder P R, Ferguson S J. The location of dissimilatory nitrite reductase and the control of dissimilatory nitrate reductase by oxygen in Paracoccus denitrificans. Biochem J. 1980;192:231–240. doi: 10.1042/bj1920231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arai H, Kodama T, Igarashi Y. Cascade regulation of the two CRP/FNR-related transcriptional regulators (ANR and DNR) and the denitrification enzymes in Pseudomonas aeruginosa. Mol Microbiol. 1997;25:1141–1148. doi: 10.1046/j.1365-2958.1997.5431906.x. [DOI] [PubMed] [Google Scholar]
- 4.Bell A, Busby S. Location and orientation of an activating region in the Escherichia coli transcription factor, FNR. Mol Microbiol. 1994;11:383–390. doi: 10.1111/j.1365-2958.1994.tb00318.x. [DOI] [PubMed] [Google Scholar]
- 5.Bell L C, Richardson D J, Ferguson S J. Periplasmic and membrane-bound nitrate reductases in Thiosphaera pantotropha: the periplasmic enzyme catalyses the first step in aerobic denitrification. FEBS Lett. 1990;265:85–87. doi: 10.1016/0014-5793(90)80889-q. [DOI] [PubMed] [Google Scholar]
- 6.Bell L C, Page M D, Berks B C, Richardson D J, Ferguson S J. Insertion of transposon Tn5 into a structural gene of the membrane-bound nitrate reductase of Thiosphaera pantotropha results in anaerobic overexpression of periplasmic nitrate reductase activity. J Gen Microbiol. 1993;139:3205–3214. doi: 10.1099/00221287-139-12-3205. [DOI] [PubMed] [Google Scholar]
- 7.Bentley J, Hyatt L S, Ainley K, Parish J H, Herbert R B, White G R. Cloning and sequence analysis of an Escherichia coli gene conferring bicyclomycin resistance. Gene. 1993;127:117–120. doi: 10.1016/0378-1119(93)90625-d. [DOI] [PubMed] [Google Scholar]
- 8.Berks B C, Ferguson S J, Moir J W B, Richardson D J. Enzymes and associated electron transport systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. Biochim Biophys Acta. 1995;1232:97–173. doi: 10.1016/0005-2728(95)00092-5. [DOI] [PubMed] [Google Scholar]
- 9.Berks B C, Page M D, Richardson D J, Reilly A, Cavill A, Outen F, Ferguson S J. Sequence analysis of subunits of the membrane-bound nitrate reductase from a denitrifying bacterium: the integral membrane subunit provides a prototype for the dihaem electron carrying arm of a redoc loop. Mol Microbiol. 1995;15:319–331. doi: 10.1111/j.1365-2958.1995.tb02246.x. [DOI] [PubMed] [Google Scholar]
- 10.Blasco F, Iobbi C, Giordano G, Chippaux M, Bonnefoy V. Nitrate reductase of Escherichia coli: completion of the nucleotide sequence of the nar operon and reassessment of the role of the α and β subunits in iron binding and electron transfer. Mol Gen Genet. 1989;218:249–256. doi: 10.1007/BF00331275. [DOI] [PubMed] [Google Scholar]
- 11.Craske A, Ferguson S J. The respiratory nitrate reductase from Paracoccus denitrificans. Molecular characterization and kinetic properties. Eur J Biochem. 1986;158:429–436. doi: 10.1111/j.1432-1033.1986.tb09771.x. [DOI] [PubMed] [Google Scholar]
- 12.De Vries G E, Harms N, Hoogendijk J, Stouthamer A H. Isolation and characterization of Paracoccus denitrificans mutants with increased conjugation frequencies and pleiotropic loss of a n(GATC)n DNA-modifying property. Arch Microbiol. 1989;152:52–57. [Google Scholar]
- 13.Goodhew C F, Brown K R, Pettigrew G W. Haem staining in gels, a useful tool in the study of bacterial c-type cytochromes. Biochim Biophys Acta. 1986;852:288–294. [Google Scholar]
- 14.Hartig E, Schiek U, Vollack K-U, Zumft W G. Nitrate and nitrite control of respiratory nitrate reduction in denitrifying Pseudomonas stutzeri by a two-component regulatory system homologous to NarXL of Escherichia coli. J Bacteriol. 1999;181:3658–3665. doi: 10.1128/jb.181.12.3658-3665.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 16.Kiley P J, Beinert H. Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol Rev. 1999;22:341–352. doi: 10.1111/j.1574-6976.1998.tb00375.x. [DOI] [PubMed] [Google Scholar]
- 17.Kolb A, Busby S, Buc H, Garges S, Adhya S. Transcriptional regulation by cAMP and its receptor protein. Annu Rev Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- 18.Kwiatkowski A V, Shapleigh J P. Requirement of nitric oxide for induction of genes whose products are involved in nitric oxide metabolism in Rhodobacter sphaeroides 2.4.3. J Bacteriol. 1996;178:4958–4964. doi: 10.1128/jb.178.16.4958-4964.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lonetto M A, Rhodius V, Lamberg K, Kiley P, Busby S, Gross C. Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase sigma70 subunit. J Mol Biol. 1998;284:1353–1365. doi: 10.1006/jmbi.1998.2268. [DOI] [PubMed] [Google Scholar]
- 20.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 21.Moir J W B, Baratta D, Richardson D J, Ferguson S J. The purification of a cd1-type nitrite reductase and the absence of a copper-type nitrite reductase from the aerobic denitrifier Thiosphaera pantotropha; the role of pseudoazurin as an electron donor. Eur J Biochem. 1993;212:377–385. doi: 10.1111/j.1432-1033.1993.tb17672.x. [DOI] [PubMed] [Google Scholar]
- 22.Moir J W B, Ferguson S J. Properties of a Paracoccus denitrificans mutant deleted in cytochrome c550 indicate that a copper protein can substitute for this cytochrome in electron transport to nitrite, nitric oxide and nitrous oxide. Microbiology. 1994;140:389–397. [Google Scholar]
- 23.Moir J W B, Richardson D J, Ferguson S J. The expression of redox proteins of denitrification in Thiosphaera pantotropha grown with oxygen, nitrate and nitrous oxide as electron acceptors. Arch Microbiol. 1995;164:43–49. [Google Scholar]
- 24.Nicholas D J D, Nason A. Determination of nitrite and nitrate. Methods Enzymol. 1957;3:981–984. [Google Scholar]
- 25.Noji S, Nohno T, Saito T, Taniguchi S. The narK gene product participates in nitrate transport induced in Escherichia coli nitrate-respiring cells. FEBS Lett. 1989;252:139–143. doi: 10.1016/0014-5793(89)80906-8. [DOI] [PubMed] [Google Scholar]
- 26.Parke D. Construction of mobilizable vectors derived from plasmids RP4, pUC18 and pUC19. Gene. 1990;93:135–137. doi: 10.1016/0378-1119(90)90147-j. [DOI] [PubMed] [Google Scholar]
- 27.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 28.Robertson L A, Kuenen J G. Thiosphaera pantotropha gen. nov. sp. nov., a facultatively anaerobic, facultatively autotrophic sulfur bacterium. J Gen Microbiol. 1983;129:2847–2855. [Google Scholar]
- 29.Sakumi K, Sekiguchi M. Regulation of expression of the ada gene controlling the adaptive response. J Mol Biol. 1989;205:373–385. doi: 10.1016/0022-2836(89)90348-3. [DOI] [PubMed] [Google Scholar]
- 30.Simon R, Priefer U, Puhler A. A broad host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 31.Spaink H P, Okker R J H, Wijffelman C A, Pees E, Lugtenberg B J J. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol Biol. 1987;9:27–39. doi: 10.1007/BF00017984. [DOI] [PubMed] [Google Scholar]
- 32.Spiro S. The FNR family of transcriptional regulators. Antonie Leeuwenhoek. 1994;66:23–36. doi: 10.1007/BF00871630. [DOI] [PubMed] [Google Scholar]
- 33.Stewart V. Regulation of nitrate and nitrite reductase synthesis in enterobacteria. Antonie Leeuwenhoek. 1994;66:37–45. doi: 10.1007/BF00871631. [DOI] [PubMed] [Google Scholar]
- 34.Tosques I E, Shi J, Shapleigh J P. Cloning and characterization of nnrR, whose product is required for expression of proteins involved in nitric oxide metabolism in Rhodobacter sphaeroides 2.4.3. J Bacteriol. 1996;178:4958–4964. doi: 10.1128/jb.178.16.4958-4964.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Spanning R J M, DeBoer A P N, Reijnders W N M, Westerhoff H V, Stouthamer A H, Van der Oost J. FnrP and NNR of Paracoccus denitrificans are both members of the FNR family of transcriptional activators but have distinct roles in respiratory adaptation in response to oxygen limitation. Mol Microbiol. 1997;23:893–907. doi: 10.1046/j.1365-2958.1997.2801638.x. [DOI] [PubMed] [Google Scholar]
- 36.Van Spanning R J M, Houben E, Reijnders W N M, Spiro S, Westerhoff H V, Saunders N. Nitric oxide is a signal for NNR-mediated transcription activation in Paracoccus denitrificans. J Bacteriol. 1999;181:4129–4132. doi: 10.1128/jb.181.13.4129-4132.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vollack K U, Hartig E, Korner H, Zumft W G. Multiple transcription factors of the FNR family in denitrifying Pseudomonas stutzeri: characterization of four fnr-like genes, regulatory responses and cognate metabolic processes. Mol Microbiol. 1999;31:1681–1694. doi: 10.1046/j.1365-2958.1999.01302.x. [DOI] [PubMed] [Google Scholar]
- 38.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 39.Zeilstra-Ryalls J H, Kaplan S. Role of fnrL gene in photosystem gene expression and photosynthetic growth of Rhodobacter sphaeroides 2.4.1. J Bacteriol. 1998;180:1496–1503. doi: 10.1128/jb.180.6.1496-1503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]