Abstract
The transcription of 16 genes encoding 12 peptidases (pepC, pepN, pepX, pepP, pepA, pepF2, pepDA1, pepDA2, pepQ, pepT, pepM, and pepO1), PI and PIII proteinases (prtP1 and prtP3), and three transport systems (dtpT, dtpP, and opp-pepO1) of Lactococcus lactis MG1363 was analyzed in response to different environmental factors. Promoter fusions with luciferase reporter genes and/or mRNA analysis were used to study the effects of sugar sources, growth at 37°C, and peptide supply on the transcription of these genes. Only transcription of the pepP gene is modulated by the source of sugar. The presence of potential catabolite-responsive element (CRE) boxes in its promoter region suggests that expression of this gene is directly controlled by catabolic repression. Elevated temperature had no significant effect on the level of transcription of these genes. prtP1, prtP3, pepC, pepN, pepX, and the opp-pepO1 operon are the most highly expressed genes in chemically defined medium, and their expression is repressed 5- to 150-fold by addition of peptide sources such as Casitone in the medium. Moreover, the transcription of prtP1, prtP3, pepC, pepN, and the opp-pepO1 operon is repressed two- to eight-fold by the dipeptides leucylproline and prolylleucine. The transcription of pepDA2 might also be repressed by the peptide sources, but this effect is not observed on the regulation of dtpT, pepP, pepA, pepF2, pepDA1, pepQ, pepT, pepM, and the dtpP operon. The significance of these results with respect to the functions of different components of the proteolytic system in L. lactis are discussed.
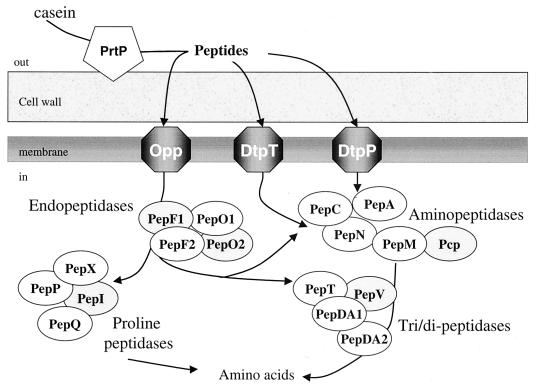
Proteolysis in bacteria plays a central role in turnover, maturation, and regulation of proteins and in assimilation of extracellular proteins and peptides (17). Lactic acid bacteria that are isolated from many dairy products, such as cheeses and yogurts, generally possess and efficient proteolytic system to break down caseins, the main proteins in milk, into the amino acids necessary for their growth (26). This function sometimes limits the growth of lactic acid bacteria in milk since these bacteria have multiple amino acid auxotrophies (5, 44). Casein breakdown products (peptides, amino acids and derivatives of amino acids) also contribute to the formation of flavor and texture of the fermented milk products. The proteolytic system of lactococci is one of the best documented. The biological and genetic properties of the majority of the enzymes involved in this system have been recently reviewed (6, 29). This system is composed of (i) an extracellular proteinase, (ii) peptide transport systems, and (iii) intracellular peptidases (Fig. 1). The degradation of milk proteins is initiated by an extracellular proteinase, PrtP, that is bound to the cell wall. Two types of proteinase, PI and PIII, have been characterized in Lactococcus lactis subsp. cremoris on the basis of the casein degradation pattern (25, 57). In L. lactis, peptides produced by the proteinase are internalized by three transporters. The Opp system takes up oligopeptides of 4 to 18 residues, while DtpT and DtpP transport hydrophilic and hydrophobic di- and tripeptides, respectively (12, 29). Internalized peptides are further hydrolyzed by several intracellular peptidases that are classified depending on their cleavage specificity. Six aminopeptidases (PepN, PepC, PepP, PepX, PepA, and Pcp) generate dipeptides and free amino acids by cleaving the N-terminal end of oligopeptides. Endopeptidases such as PepO1, PepO2, PepF1, and PepF2 cleave internal peptide bonds of oligopeptides, and several other peptidases, such as PepV, PepQ, and PepT, cleave di- or tripeptides (29, 40, 42, 46). PepN and PepC aminopeptidases, PepV dipeptidase, and PepT tripeptidase display a low substrate specificity (4, 19, 40, 55), while PepA glutamyl aminopeptidase liberates N-terminal Glu and Asp residues (31). PepQ prolidase, PepP aminopeptidase P, PepX X-prolyl-dipeptidyl aminopeptidase, and PepI proline iminopeptidase are found in hydrolyzing peptides containing proline residues (1, 3, 38, 45). The pyrrolidone carboxylyl peptidase (Pcp) specifically cleaves N-terminal pyrrolidone carboxylyl residues of peptides (11). Last, we have found in the genome of L. lactis IL 1403 genes for potential peptidases such as PepDA1 and PepDA2 that share sequence similarity with the PepD dipeptidase of Lactobacillus helveticus and PepM, showing sequence similarity with the methionyl peptidase of Escherichia coli (2).
FIG. 1.
Schematic representation of the L. lactis proteolytic system. The cell wall proteinase (pentagon), three transport systems (hexagon), and 18 intracellular peptidases (oval) are represented in their relative locations in the cell. Peptidases are classified on the basis of their cleavage specificity. White and grey ovals represent peptidases that were included and not included, respectively, in this study.
Although functional analysis of peptidase genes has been systematically carried out, regulation of expression of the various components of the proteolytic pathway is still poorly documented. The regulation of the plasmid-encoded cell wall proteinase PrtP is the best known among the components of this system. Early experiments showed that in several strains, the synthesis of the cell wall proteinase is reduced during growth in rich media compared to milk medium (23). Moreover, proteinase activity in L. lactis subsp. cremoris AM1 is repressed after the addition of peptides to the growth medium and is increased in the stationary phase (10). Transcription of the extracellular proteinase gene of L. lactis SK11 has been analyzed by prtP-gusA gene fusion (36). A 10-fold repression of initiation of transcription was observed by adding a complex peptide mixture to the medium. Moreover, peptide-dependent regulation was examined by adding specific peptides to the growth medium. Out of 12 di- and tripeptides tested, only leucylproline (LP) and prolylleucine (PL) repressed the transcription of the prtP-gusA fusion (36). Finally, the activities of PepN and PepX and the expression of three transport systems are greater when cells grow in chemically defined medium (CDM) compared to media containing complex peptide sources (8, 12, 18, 39). Similarly to PrtP, PepN and PepX activities are repressed in the presence of the dipeptide PL (39).
Here we describe a systematic study of the transcription of 16 genes involved in the proteolytic system of L. lactis. mRNA analysis showed that the transcription of several genes was regulated by the peptide supply. The activities of 15 promoter regions were measured during growth in several media by using luciferase fusion, enabling direct comparison of promoter strengths. We report that the transcription of eight promoters is regulated by the peptide content of the medium; of these, five promoters are repressed by specific dipeptides. On the other hand, pepP transcription is regulated by the carbon source.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains used in this study are listed in Table 1. E. coli TG1 was used for plasmid propagation (14). E. coli was grown at 37°C in Luria-Bertani medium (35). L. lactis strains were grown at 30°C on M17 glucose medium (M17) (53) or on CDM (51). GalCDM is CDM with glucose replaced by galactose (0.5% [wt/vol]). CDM was supplemented with Casitone (CDM Casitone; Sigma-Aldrich) at 1.5% (wt/vol) or at different concentrations (0.1 to 2% [wt/vol]) where specified, with Casamino Acids (CDM CAA; Difco Laboratories, Detroit, Mich.) at 1.5% (wt/vol) or the dipeptide PL (Sigma) or LP (Sigma) at 1 mM. When needed, erythromycin (5 μg/ml for L. lactis; 100 μg/ml for E. coli), tetracycline (5 μg/ml for L. lactis), or ampicillin (100 μg/ml for E. coli) was added to the culture medium.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain, plasmid, or oligonucleotide | Relevant markers and characteristics | Reference or origin |
|---|---|---|
| Strains | ||
| E. coli TG1 | supE Δthi(lac-proAB) hsdD5 (F′+ traD36 proAB lacIqZDM15) | 14 |
| L. lactis | ||
| MG1363 | Plasmid-free derivative of L. lactis subsp. cremoris NCDO712, Lac− Prt− | 13 |
| Wg2 | Wild-type L. lactis subsp. cremoris strain harboring pWV05, prtP1 | 27 |
| SK11 | Wild-type L. lactis subsp. cremoris strain harboring pSK111, prtP3 | 9 |
| Plasmids | ||
| pBSSK+ | Ampr, M13ori pBR322ori | Stratagene |
| pGEM-T Easy | Ampr, M13ori pBR322ori, linear T-overhang vector | Promega |
| pGhost 8 | Tetr, thermosensitive replicative plasmid in L. lactis | 34 |
| pJIM2366 | Emr, promoter probe vector containing luxAB genes | 47 |
| pJIM2374 | Emr, integrative promoter probe vector containing luxAB genes | 7 |
| Integrative plasmids carrying lux fusion | ||
| pJIM3029 | PpepC-lux, XbaI fusion of pJIM2374 and pBSSK+ containing PpepC on a 654-bp fragment | This work |
| pJIM3030 | PpepN-lux, XbaI fusion of pJIM2374 and pBSSK+ containing PpepN on a 767-bp fragment | This work |
| pJIM3031 | PpepX-lux, XbaI fusion of pJIM2374 and pBSSK+ containing PpepX on a 687-bp fragment | This work |
| pJIM3032 | PpepF22-lux, XbaI fusion of pJIM2374 and pBSSK+ containing PpepF22 on a 654-bp fragment | This work |
| pJIM3063 | PpepF21-lux, SpeI fusion of pJIM2374 and pGEM-T Easy containing PpepF21 on a 772-bp fragment | This work |
| pJIM3066 | PpepA-lux, BamHI fusion of pJIM2374 and pGEM-T Easy containing PpepA on a 575-bp fragment | This work |
| pJIM3064 | PoppD-lux, SpeI fusion of pJIM2374 and pGEM-T Easy containing PoppD on a 587-bp fragment | This work |
| pJIM3065 | PoppA-lux, SpeI fusion of pJIM2374 and pGEM-T Easy containing PoppA on a 688-bp fragment | This work |
| pJIM3080 | PpepDA1-lux, SpeI fusion of pJIM2374 and pGEM-T Easy containing PpepDA1 on a 763-bp fragment | This work |
| pJIM3081 | PpepM-lux, SpeI fusion of pJIM2374 and pGEM-T Easy containing PpepM on a 913-bp fragment | This work |
| pJIM3082 | PpepP-lux, SpeI fusion of pJIM2374 and pGEM-T Easy containing PpepP on a 778-bp fragment | This work |
| pJIM3106 | PpepQ-lux, SpeI fusion of pJIM2374 and pGEM-T Easy containing PpepQ on a 760-bp fragment | This work |
| pJIM3118 | PpepT-lux, SpeI fusion of pJIM2374 and pGEM-T Easy containing PpepT on a 866-bp fragment | This work |
| Replicative plasmids carrying lux fusion | ||
| PJIM3120 | PprtP1-lux, pJIM2366 carrying PprtP1 on a 1,051-bp SpeI-NotI fragment from strain Wg2 | This work |
| pJIM3119 | PprtP3-lux, pJIM2366 carrying PprtP3 on a 1,056-bp SpeI-NotI fragment from strain SK11 | This work |
| Oligonucleotide pairs used for amplification or promoter and probe fragments | ||
| PpepA | GGGAATTCTAAAGTTTTGGCATT/CGGGATCCATAAATCCGACCTCAT | X81089 |
| PpepC | GGGAATTCCTTTGATTATTCATC/CGGGATCCAGATTGTTTTTGATTC | M86245 |
| PpepDA1 | TGTAGCAGACCTTGTATCAGC/GACATCGCAACATTAGCACTA | AE005176 |
| PpepF21 | GGGAATTCCAATTGAGCGAACTA/CGGGATCCTTGAGACGCTTAATA | X99710 |
| PpepF22 | GGGAATTCTAGTTTTATTCGTCA/CGGGATCCTTGATACTTTGCTTCTC | X99710 |
| PpepM | ACCTTTCCAGATGACCCGTTA/GTGCCTACTTTGGCTTGTTCA | AE005176 |
| PpepN | GGGAATTCTTGTTTTAAATGATGG/CGGGATCCTTCTTCTTCATCAAAT | M87840 |
| PpepP | TGGTTCGTCGTGAAGCACCAT/TCATCAGCAATTGCACAGGCT | Y08842 |
| PpepT | GCTATTGAGCCTTTTACT/GTTGTTCCATCAGTATGA | L27596 |
| PpepQ | TCTCCATCGATTGTTCCGACT/TCATCTGCTGACTTAATCAAA | AE005176 |
| PpepX | GGGAATTCCAAACATCGCACCTA/CGGGATCCCAATCTAATTCTATA | M35865 |
| PoppD | GGGAATTCTGCTTTTATTATTTCCT/CGGGATCCTACTTGTTCTAAAA | L18760 |
| PoppA | GGGAATTCTTTGGGAACAATGATAA/CGGGATCCGTTACTTCTGAACCA | L18760 |
| PprtP | CGGAATTCCTTGTACAAGCTACT/CGGAATTCAGCTTATTGTAATCA | M26310 |
| pepQ | ATCCGACAACTCTAAATTATCT/TAGCAGCGCGAGCTCCTGATA | AE005176 |
| oppA | AGCAGCTACACTCCTAAGTGCT/CGGGATCCGTTACTTCTGAACCA | L18760 |
| oppD | AGTCTTAAACGTGGTGAAGT/CGGGATCCTACTTGTTCTAAAA | L18760 |
| PpepDA2 | TGAGGTTTCTGTCGCAGTCAT/CGGTCAGCAATGGTTACCTTA | AE005176 |
| dtpT | TGACTCACGTCGTGACACTGGAT/TCCGTTCAAGAGACCTGGAAGT | U05215 |
| dtpP | ACAGCACTCTATGCAAGGGTCATA/AGTCGCTGGATCAACCGCACGTT | AE005176 |
DNA manipulation procedures.
Plasmids and total DNA were prepared as previously described (32, 35, 50). Procedures for DNA manipulations, transformation of E. coli cells, and cloning were essentially as described by Maniatis et al. (35). Electrotransformation of L. lactis was performed as described by Holo and Nes (20). All enzymes for DNA technology were used according to the manufacturer's specifications. Oligonucleotides were synthesized on an Oligo 1000M DNA synthesizer system (Beckman). Standard procedures were used for Southern hybridization analysis (48). Digested chromosomal DNA (2 μg) was transferred to a nylon membrane (Hybond N+) and hybridized with DNA probes that were labeled with the ECL (enhanced chemiluminescence) direct nucleic acid labeling and detection system. Hybridization and detection were performed according to the Amersham ECL protocol. Sequence analysis of double-stranded DNA was carried out according to the Applied Biosystems protocol accompanying the 370A DNA sequencer. DNA was used in dideoxynucleotide chain termination sequencing reactions with a Big Dye terminator kit (Applied Biosystems) and was sequenced on both strands.
Construction of lux transcriptional fusions.
PCR fragment products containing different promoters were cloned in E. coli, and their sequences verified. For this purpose, chromosomal DNA from L. lactis MG1363 was used as the template to generate 500- to 900-bp fragments containing the promoters and the putative start codons of pepA, pepC, pepDA1, pepF2, pepM, pepN, pepP, pepT, pepQ, pepX, and the opp-pepO1 operon. The two pepF2 promoters were designated PpepF21 and PpepF22, and the two opp-pepO1 promoters were designated PoppD and PoppA (Fig. 2). PoppD is located upstream of the opp-pepO1 operon, and PoppA is between the oppC and oppA genes (46, 54). Plasmid DNA from L. lactis SK11 and Wg2 was used to amplify fragments carrying promoter regions of prtP1 and prtP3, encoding the PI and PIII proteinases, respectively. The specific oligonucleotide pairs used in this work are described in Table 1. PCR fragments containing promoter regions of pepC, pepN, pepX, and pepF2 (PpepF22) were cut by EcoRI and BamHI present in the primers and cloned in pBluescript KS(+) (pBSSK+) cut by the same enzymes. PCR fragments containing promoter regions of prtp1, prtP3, pepA, pepDA1, pepF2 (PpepF21), pepM, pepP, pepQ, pepT, and the opp-pepO1 operon (PoppD and PoppA) were cloned directly in pGEM-T Easy vectors (Promega).
FIG. 2.
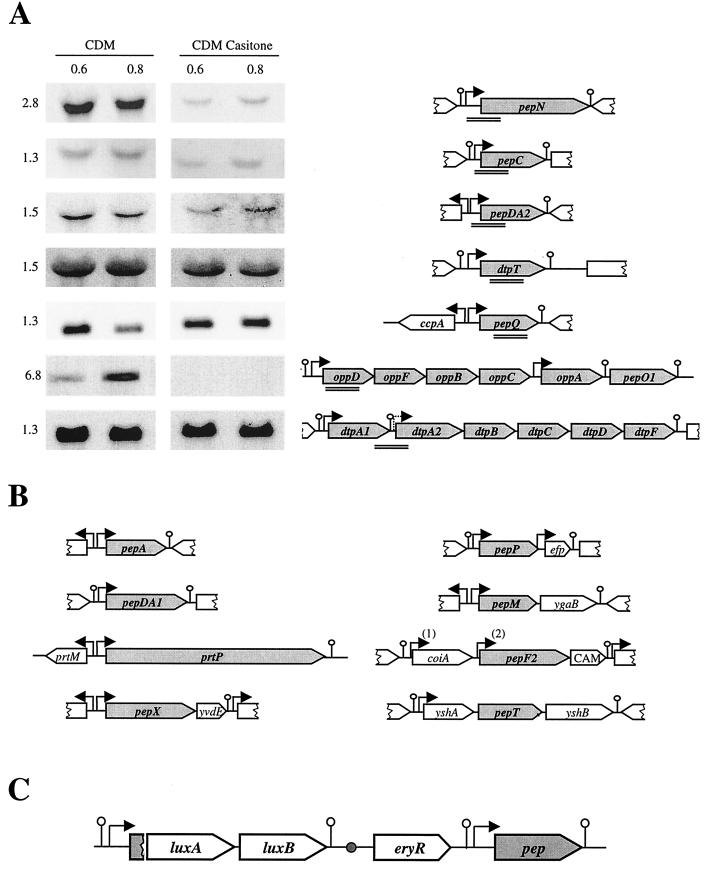
Genetic organization of genes encoding the proteolytic system of L. lactis. Schematic representation of the genetic organization of genes was deduced from published papers and completed by data of the L. lactis IL1403 genome sequence (AE005176) as mentioned in the text. (A) Northern blot analysis of the pepN, pepC, pepDA2, pepQ, and dtpT genes and the opp-pepOI and dtpP operons. Total RNA was extracted from L. lactis MG1363 at an OD600 of 0.6 and 0.8 in CDM and CDM Casitone. Total RNA was hybridized with PCR fragments used as probes denoted by double lines. The size of the messenger in kilobases is indicated on the left. Arrows and lollipops present the putative promoters and terminators, respectively. (B) Schematic representation of the genetic organization of prtP, pepA, pepDA1, pepM, pepX, pepP, pepF2, and pepT. Promoters were deduced only from DNA sequence analysis, except for the prtP promoter. The two pepF2 promoters, PpepF21 and PpepF22, are designated by “(1)” and “(2)”, respectively. prtM, efp, and cam (CAM on the diagram) encode the proteinase maturase, elongation factor P, and a potential methyltransferase, respectively. coiA is the counterpart of a gene whose product is involved in competence development in Streptococcus pneumoniae. ygaB, yshA, and yshB encode proteins sharing homology with membrane proteins and transporters, and yvdE encodes a protein similar to proteins of unknown function. Terminators localized downstream of yvdE, efp, and yshB and upstream of yshA and putative promoters localized upstream efp and yshA were deduced, in this work, from the sequence of the IL1403 genome (AE005176). (C) General scheme of the luciferase fusion integrated on the chromosome by single crossover. Upon insertion, the promoter is duplicated and drives expression of the luxAB genes and of the functional gene denoted in grey. The small circle represents the origin of replication of the integrative plasmid that is inactive after the helper plasmid is removed.
The integrative plasmids carrying the lux transcriptional fusions were constructed by fusing pGEM-T Easy or pBSSK+ containing promoter regions with the L. lactis intergrative vector pJIM2374 as described in Table 1. These plasmids were integrated at the promoter locus in the chromosome of L. lactis MG1363 by single crossover with pGhost 8 as a helper as described by Godon et al. (16). Strains were screened by PCR amplification with specific primers for monocopy integration of plasmid and further verified by Southern blotting. The resulting strains contained the lux genes downstream of the cloned promoter region followed by a copy of the intact gene (Fig. 2C). The lux transcriptional fusions with prtP1 and prtP3 promoters were obtained after cloning fragments carrying promoters from pGEM-T Easy into pJIM2366, an L. lactis replicative vector, as described in Table 1. These plasmids were transformed into L. lactis MG1363.
Determination of luciferase activity in L. lactis and growth rate of culture.
Luciferase assays were carried out on a Bertold Lumat LB9501 apparatus. One milliliter of L. lactis culture was mixed with 5 μl of nonaldehyde, and the light emission was immediately measured. The value of the peak obtained was standardized to the optical density at 600 nm (OD600) of the culture. Luciferase activity was measured throughout the growth of the culture. Values reported in Fig. 3 and Table 2 were measured at OD600 of 0.4. Luciferase assays were determined on L. lactis culture grown in several media where the growth rate differs significantly depending on the nitrogen and carbon sources (generation times are 190 min in GalCDM, and 60 min in CDM, with or without dipeptides and CAA, and 50 min in M17 or CDM Casitone). In this study, we considered that a 2- to 3.5-fold modulation in luciferase measurement was not significant if the growth rate was different. Indeed, variation in the growth rate might significantly affect the balance between synthesis, degradation, and dilution of the reporter gene during growth.
FIG. 3.
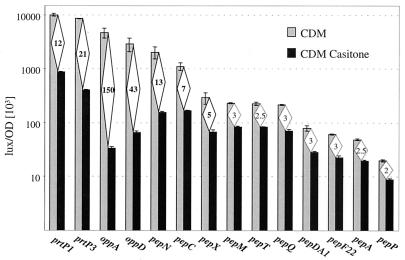
Histogram of luciferase activities obtained from lux fusions with 14 promoters. L. lactis MG1363 strains carrying the fusions were grown in CDM and CDM Casitone. The values reported correspond to those obtained at an OD600 of 0.4. Error bars indicate standard deviations. Diamonds contain the strength of repression corresponding to the ratio of luciferase activities obtained in CDM and in CDM Casitone.
TABLE 2.
Expression of proteolytic system gene promoter-lux fusions in CDM supplemented with dipeptides LP and PL at 1 mM
| Promoter | CDM (lux/OD unit [103]; mean ± SD) | CDM + LP (lux/OD unit [103]; mean ± SD) | Strength of repression by LPa | CDM + PL (lux/OD unit [103]; mean ± SD) | Strength of repression by PLb |
|---|---|---|---|---|---|
| PprtP3c | 8,540 ± 95 | 3,800 ± 100 | 2 | 4,300 ± 300 | 2 |
| PoppA | 5,040 ± 300 | 760 ± 30 | 6.5 | 635 ± 1 | 8 |
| PoppD | 2,820 ± 160 | 750 ± 50 | 4 | 620 ± 65 | 4.5 |
| PpepN | 2,020 ± 140 | 830 ± 50 | 2.5 | 900 ± 70 | 2 |
| PpepC | 1,180 ± 130 | 630 ± 20 | 2 | 790 ± 10 | 1.5 |
| PpepX | 320 ± 30 | NDd | ND | 305 ± 20 | 1 |
| PpepQ | 215 ± 4 | ND | ND | 210 ± 10 | 1 |
Ratio between the values obtained in CDM and CDM + LP.
Ratio between the values obtained in CDM and CDM + PL.
PprtP3-lux fusion is carried on a high-copy-number plasmid.
ND, not determined.
Northern blot analysis.
RNA was isolated from L. lactis MG1363 grown in CDM and in CDM Casitone (1.5% [wt/vol]; Sigma) at different times of growth corresponding to an OD600 of 0.6 and 0.8. Total RNA was prepared as previously described for Bacillus subtilis (15). After extraction and treatment with phenol-chloroform, RNA was precipitated with ethanol. Then 25 μg of glyoxalated RNA was subjected to electrophoresis through a 1% agarose gel. Transfers and hybridizations were performed as described by Maniatis et al. (35). Hybridization was performed with PCR fragments generated with oligonucleotides presented in Table 1 and summarized in Fig. 2A. Hybridization data were collected on a Storm instrument and quantified by the ImageQuant image analysis software package (Molecular Dynamics).
RESULTS
Transcriptional organization of the genes involved in peptide utilization in L. lactis.
The genetic organization of proteolytic genes studied here is presented Fig. 2A and B. Potential promoters were deduced from DNA sequence analysis, except for the transcription initiation site of prtP (37, 56). Only sizes of pepN, pepV, and pepF1 transcripts have been determined (19, 46, 52). To experimentally confirm the functionality of the promoters presented here, we checked that PCR fragments containing the promoter regions of oppD, oppA, pepN, pepC, pepX, pepM, pepT, pepP, pepQ, pepDA1, pepF21, pepF22, and pepA were able to drive luciferase activity when cloned in pJIM2374 maintained under its replicative form in L. lactis (not shown). To confirm the structural organization of several genes of the proteolytic system and assess the effect of adding a rich peptide source to CDM, we carried out Northern blotting on total RNA extracted during the exponential growth phase from MG1363 cells grown in CDM with and without Casitone. RNA was hybridized with fragments covering part of pepQ, oppD, dtpT, pepN, pepC, pepDA2, dtpPA1, and dtpA2 as shown in Fig. 2A.
In CDM, pepN, pepC, pepDA2, dtpT, and pepQ produced single transcripts of 2.8, 1.3, 1.5, 1.5, and 1.3 kb, respectively (Fig. 2A). These transcriptional patterns were in agreement with the sizes of the genes and confirmed their monocistronic organization. The oppD probe revealed a single 6.8-kb band, which should end within pepO1 and thus be a 3′-end degradation of the expected 8-kb transcript, confirming the polycistronic organization of the opp genes (Fig. 2A). The full-size 8-kb transcripts covering dtpP is barely detectable (data not shown) compared to the clear short transcript covering the 3′ end of dtpPA1, suggesting that expression of the second part of the dtpP operon is weak. Except for opp-pepO1 and pepQ transcripts, all transcripts had constant relative amounts during exponential growth.
pepDA2, pepC, pepN, and opp-pepO1 transcripts were 3-, 3-, 15-, and 20-fold respectively, more abundant in CDM than in CDM Casitone, while the transcription of dtpT, pepQ, and dtpPA1 was not affected more than 1.5-fold by Casitone (Fig. 2A). These results suggested that transcription of the pepDA2, pepC, and pepN genes and the opp-pepO1 operon is negatively controlled by Casitone.
Effect of peptide supply on transcription of the proteolytic system.
To verify the results obtained by RNA analysis, we constructed 15 transcriptional luxAB gene fusions with promoters PprtP1, PprtP3, PoppD, PoppA, PpepN, PpepC, PpepX, PpepM, PpepT, PpepP, PpepQ, PpepDA1, PpepF21, PpepF22, and PpepA. The resulting fusions were inserted in single copy by homologous recombination at their loci in L. lactis MG1363 (Fig. 2C). The fusions with PprtP1 and PprtP3 were carried on multicopy plasmids, as prtP genes are in multicopy in natural strains. To test the effects of peptide sources on promoter expression, the activities of the lux fusions were compared in CDM, CDM Casitone (Fig. 3), and M17 (data not shown). CDM contains all amino acids necessary for L. lactis growth (51), Casitone is an enzymatic casein hydrolysate that contains 80% peptides and 20% amino acids (36), and M17 contains a complex nitrogen source, including rapidly assimilated peptides (53). Luciferase activities were reduced in CDM Casitone and M17 compared to CDM for all promoters except PpepF21, for which no significant activity was detected (0.2 lux/OD unit [103]). However, the difference depended markedly on the promoters. The strength of PpepP, PpepA, PpepF22, PpepDA1, PpepQ, PpepT, and PpepM was only 2- to 3-fold lower in Casitone, whereas that of PpepX, PpepC, PpepN, PprtP1, PprtP3, PoppD, and PoppA was 5- to 150-fold lower (Fig. 3). The decrease of transcription was similar in CDM Casitone compared to M17 for all promoters except PprtP1, which was repressed twofold more in M17 (480 ± 10 versus 875 ± 25 lux/OD unit [103]). These results suggested that Casitone and M17 contain components that significantly repress the transcription of at least seven promoters, PpepX, PpepC, PpepN, PprtP1, PprtP3, PoppD, and PoppA. In addition, prtP1 transcription might be repressed by specific components in M17. These results were in agreement with RNA analysis and indicated that repression by the regulatory components of Casitone and M17 occurs at the level of the initiation of transcription.
Repression depends on the Casitone concentration.
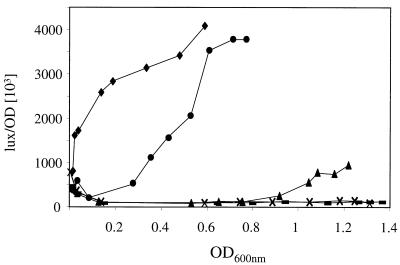
We studied the effect of the Casitone concentration on PoppA expression. Luciferase activities were more than 100-fold higher in CDM than in CDM supplemented with 1 or 2% Casitone (Fig. 4). At these concentrations, the repression of opp-pepO1 transcription was constant throughout cell growth. Interestingly, with 0.5 and 0.1% Casitone, expression was also repressed until the OD600 reached 0.8 and 0.3, respectively. The expression determined thereafter increased significantly (Fig. 4). These results indicated that the components which repressed opp-pepO1 transcription can be degraded or assimilated by L. lactis.
FIG. 4.
Luciferase activities of PoppA-lux fusion promoter during batch culture of L. lactis MG1363 in CDM (⧫) and in CDM containing 0.1% (●), 0.5% (▴), 1% (×), and 2% (—) Casitone.
Dipeptides are involved in the transcriptional repression of peptidases.
Since M17 and Casitone contain complex nitrogen sources, we decided to better define the elements affecting negatively peptidase expression. We tested the effects of different supplementary nitrogen sources on the activity of all lux fusions, such as CAA (acid casein hydrolysate containing 20% peptides and 80% amino acids) and dipeptides. All promoters repressed by Casitone except PpepX were also repressed by CAA. The rate of repression by CAA corresponding to the ratio of values obtained in CDM compared to those obtained in CDM with CAA is 2- to 12-fold for PprtP3, PprtP1, PpepC, PpepN, PoppD, and PoppA (data not shown). The rate of repression in CDM CAA is less than 1.5-fold for PpepX, PpepP, PpepA, PpepDA1, PpepQ, PpepT, and PpepM (data not shown). Although the nitrogen content of CAA is close to that of Casitone (two casein hydrolysates), the repression was approximately 5- to 10-fold lower in the medium with CAA, suggesting that the element-repressing peptidase genes are less abundant or less efficient in CAA. Effects of dipeptides LP and PL, known to regulate expression of the prtP3 gene (37), were tested on the promoters regulated by Casitone (Table 2). PL or LP repressed two- to eightfold the expression of luciferase genes under control of PprtP3, PpepC, PpepN, PoppD, and PoppA. These results indicated that at least five promoters out of seven previously shown regulated by Casitone were repressed by specific dipeptides, although at a lower level than by Casitone.
Effect of heat shock and catabolic repression on transcription of the proteolytic system.
It was reported earlier that a heat shock response might be involved in regulation of some peptidases (17). We therefore tested the effect of temperature on expression of the genes of the proteolytic system by monitoring the luciferase activity of lux fusions at 38°C. No significant difference of expression was observed at elevated temperature compared to 30°C, the usual temperature of growth for L. lactis (data not shown).
Since it was reported earlier that catabolic repression might be involved in the regulation of some peptidases (49), the activities of all lux fusions were compared in CDM containing as a carbon source glucose or galactose, in CDM, and in GalCDM. Galactose is assimilated slowly, which limits the growth rate (150 min, versus 60 min in the presence of glucose) and does not cause catabolic repression in L. lactis (33). The luciferase activity of the PpepP fusion was 8.5-fold higher in GalCDM than in CDM (170 ± 10 versus 20 ± 1 lux/OD unit [103]), whereas those of the other promoters did not increase significantly (<3.5-fold [data not shown; see Materials and Methods]). We searched for catabolite-responsive element (CRE) boxes, mediating catabolic repression in gram-positive bacteria (22), near the 15 promoters studied in lux fusions. Two mismatches from the consensus sequence TGNNANCGNTNNCA were allowed at any position except the central CG motif that is conserved in all known CRE boxes (Table 3) (22). Possible CRE boxes were found in the vicinity of pepM, pepP, pepX, and dtpT promoters (Table 3). None were detected within 250 bp from the −10 box of promoters of the other proteolytic genes. The absence of CRE boxes in most promoter regions was in agreement with the lack of significant variation of luciferase activities as a function of the carbon source. These results indicated that the expression of these genes were not under the control of catabolic repression. Absence of variation in luciferase activities of pepX and pepM fusions suggested that the potential CRE boxes found near their promoters were not active. On the other hand, the presence of four potential CRE sites and the differential expression of pepP depending on the carbon source strongly suggested that its transcription is controlled by catabolic repression.
TABLE 3.
Potential CRE boxes present in promoter regions of peptidase genes
| Gene | Distance (bp) toa:
|
No. of mismatches | Sequences of boxb | |
|---|---|---|---|---|
| Start codon | −10 box | |||
| pepM | +23 | +68 | 2 | TGAAATCGAGCAAA |
| pepP | −81 | −4 | 2 | AGGAAACGTTAACT |
| −65 | −20 | 2 | TGTTAGCGTTTTTG | |
| +99 | +154 | 1 | TGGAACCGCGGGCA | |
| +122 | +177 | 2 | TGACAGCGAAGCGA | |
| pepX | −265 | −221 | 2 | GTTCAACGTTAGCA |
| dtpT | +78 | +83 | 2 | TGGGAGCGTTTTTC |
| Consensus | TGNNANCGNTNNCA | |||
Relative position of potential CRE site upstream (−) or downstream (+) of the first codon and the −10 promoter box.
Mismatches to the CRE box consensus sequence are shown in bold.
DISCUSSION
L. lactis possesses a number of genes involved in the utilization of proteins present in the medium such as extracellular protease, peptide transport systems, and intracellular peptidases (Fig. 1). Here, we conducted a systematic study to determine some parameters regulating their expression. The sequences of many of these genes have been previously characterized (for recent reviews, see references 6 and 29), but promoter identification and transcriptional data were available only for prtP (37, 56) and for pepN, pepV, and pepF1 (19, 46, 52). We also included four genes and one operon revealed by the complete sequence of the L. lactis IL1403 genome (pepDA1, pepDA2, pepM, pepQ, and dtpP [2]). A schematic representation of the genetic organization of these genes, presented in Fig. 2, is deduced from published papers and completed by sequence analysis from the L. lactis IL1403 genome sequence (AE005176). By Northern blot analysis, we confirmed the sizes of pepN, pepC, pepDA2, dtpT, and pepQ predicted from nucleotide sequence analysis. We demonstrated that these genes had a monocistronic organization and that opp genes are in an operon. In this study, the use of lux as a gene reporter enabled us (i) to detect the presence of a promoter in cloned regions, (ii) to analyze the level of transcription as a function of growth in different environmental conditions, and (iii) to perform an initial comparison of the strengths of the 15 different promoters.
Catabolic repression might control pepP transcription only.
In Lactobacillus delbrueckii, CcpA, the regulator for catabolic repression in gram-positive bacteria, was proposed to regulate the transcription of pepQ and probably pepI and pepX (49). However, although pepQ is also divergently transcribed from ccpA in L. lactis (Fig. 2), we have shown that its transcription is not regulated by carbon source. This result is in agreement with the lack of CRE sites at less than 300 bp from the most probable transcriptional start of pepQ and strongly suggests that pepQ transcription is not under the control of catabolic repression. Analysis of the other lux fusions revealed that only the transcription of pepP was 8.5-fold more expressed in CDM with galactose. Analysis of the promoter region of pepP allowed us to find four potential CRE boxes present 4 and 20 bp upstream and 151 and 177 bp downstream of the −10 box of the promoter. Since these potential CRE boxes are located at a distance suitable to enable the repression of transcription of the PpepP, it is likely that pepP is effectively regulated by CcpA. This aminopeptidase cleaves off any N-terminal amino acid linked with proline, and it was proposed that PepP of E. coli is probably involved in the maturation of the N-terminal ends of proteins (38). Its coexpression with an elongation factor in many gram-positive bacteria argues for such a function (unpublished data). It is thus not surprising that pepP is regulated by factors other than those related to peptide supply such as carbon or energy metabolism.
Casitone-regulated genes encode key enzymes for proteolysis, while Casitone-independent genes would have another role.
The repression of transcription by nitrogen sources such as Casitone was particularly significant (5- to 150-fold) for prtP1, prtP3, pepX, pepN, pepC, and opp-pepO1 (Fig. 3). Transcriptional repression of pepN, pepC, and opp-pepO1 was confirmed by mRNA analysis. Furthermore, pepDA2, but not pepQ, dtpP, and dtpT, might also be regulated by Casitone (Fig. 2A). Interestingly, Casitone-regulated genes are those that have the highest expression level in CDM. Their promoter strength is similar to that of highly expressed lactococcal genes such as those encoding glycolytic enzymes (E. Jammet, personal communication). Moreover, functional studies have suggested that they play a significant role in protein utilization. The PrtP proteinase and Opp transport systems have been shown to be essential for growth in milk since they are involved in the first step of casein degradation and in the uptake of the resulting peptides, respectively (25, 26, 54). Moreover, although the inactivation of single peptidase genes does not generally lead to a drastic effect on growth in milk, the inactivation of pepN, pepC, and pepO1 leads to 25, 10, and 9% decreases, respectively, in growth rate in milk (6, 41). Combinations of these mutations have a drastic effect on growth, suggesting their crucial role in peptide or nutritional metabolism in the cell (30, 41). Last, the growth of the pepX mutant is clearly affected in medium containing casein as the sole peptide source (40 to 25% longer generation time) and in milk (15% longer generation time) (6, 30, 38). PepN, PepC, and PepO1 were showed to be the most important intracellular enzymes for the degradation of oligopeptides provided by the casein breakdown and PepX for peptides containing proline (6).
Most genes encoding these key enzymes in peptide utilization are either transcribed as single genes, such as the genes encoding PrtP, PepN, and PepC, or cotranscribed, such as the genes encoding oligopeptidase O and the oligopeptide transport system in the opp-pepO1 operon. By contrast, pepF2 (46), pepM (2), pepP (38), and pepT (40) appear to be linked to genes that are not involved in peptidolysis (Fig. 2). These genes as well as pepDA1, pepA, and pepQ are expressed at a low level and are not modulated by the peptide source. These results suggested they may not be involved in external peptide source utilization but have a role in other cellular processes. Indeed, as mentioned above, PepP might be involved in protein maturation (38). PepM, a methionine-specific aminopeptidase which removes N-terminal methionine residues from proteins, is essential and might also be involved in protein maturation in Salmonella enterica serovar Typhimurium (43). PepDA1 might result from the duplication of PepDA2 and have evolved to fulfill a particular role in the cell. The chromosomal pepF2 gene was also found duplicated on plasmids in several lactococcal strains. The loss of either copy results in a decrease in the growth rate in minimal media, suggesting a role of oligopeptidase PepF in protein turnover (46). Moreover, pepF2 is in an operon with a gene homologous to a gene induced during competence in Streptococcus pneumoniae, and PpepF21 displays a sequence signature similar to that present in the streptococcal regulated promoters involved in cellular competence (2).
Specific peptides control Casitone-regulated genes.
The transcription of prtP1, prtP3, pepN, pepC, pepX, pepDA2, and the opp-pepO1 operon, encoding the main components of the proteolytic system of L. lactis, is controlled by the complex nitrogen source contained in M17 and Casitone. Since Casitone is a proteolytic hydrolysate of casein, this result suggests that the signal for regulation is a peptide or a mixture of peptides. In addition to Casitone, CAA repressed the transcription of prtP1, prtP3, pepN, pepC, and opp-pepO1, although to a 10-fold-lower extent (data not shown). Moreover, we showed that addition of dipeptides LP and PL in the medium decreased the level of transcription of prtP3, pepN, pepC, and opp-pepO1 to the same extent as CAA (Table 3). These dipeptides have been previously reported to repress prtP3 transcription approximately 10-fold and decrease PepN and PepX enzymatic activities 1.7- and 1.5-fold, respectively (36, 39). These results suggest that PepX expression could be regulated at a level other than transcription since in our conditions, lux fusion showed that pepX expression was regulated by Casitone but not by dipeptides LP and PL.
The nature of the signal-repressing proteolytic gene is probably complex. Addition of CAA or specific dipeptides such as LP and PL has not the full repressing effect obtained with Casitone, and the efficiency of dipeptide repression is not increased when fivefold-higher concentrations are used (data not shown), suggesting that other peptides from Casitone might be more active or that a mix of specific peptides is required. However, the growth rate is higher in CDM Casitone than in CDM with CAA or dipeptides, indicating that peptides present in Casitone provide a better nitrogen source than CAA or dipeptides. Interestingly, the repressing factor(s) present in Casitone is used by L. lactis, since the duration of the repression is correlated with the amount of Casitone added in the medium (Fig. 4). A simple explanation would be that the repression requires transport and/or assimilation of certain peptides by the cell. Indeed, it has been shown that peptides are in competition to enter the cell (28) and that certain peptides are assimilated well whereas others accumulate in the medium during casein utilization (24).
Addition of peptide sources to CDM, a medium containing the 18 free amino acids (all amino acids except aspartic acid and glutamic acid [51]) required for rapid growth, causes repression of some proteolytic genes. However, even in an excess of free amino acids in CDM, their availability inside the cell could be limiting due to a low rate of uptake and lead to a partial starvation. This starvation could be overcome by addition of peptides efficiently taken up. Nevertheless, the major factor involved in this regulation is not due to a severe amino acid starvation, since addition of CAA or dipeptides LP and PL to CDM induces significant repression without improving the growth rate of L. lactis. Coordinate regulation of the proteolysis genes in L. lactis would thus depend on the content of the peptide source, presumably by a signal sensing the nutritional state of the cell for amino acid supply.
This study provided a set of data on the transcription of 16 genes potentially involved in protein utilization. Analysis of lux fusion data, including assessment of the relative level of transcription and regulation by environmental factors, provided new insight into the probable roles of the different components of the proteolytic system. Genes expressed at a low level are not regulated by the peptide source and probably encode enzymes involved in cellular functions other than peptide utilization. Moreover, the most highly expressed genes are repressed by the peptide source and encode the enzymes most important for proteolysis. Their transcription is repressed by dipeptides LP and PL, and more dipeptides should be tested to better define the signal-repressing proteolytic genes. Identification of the cellular factors that are involved in this repression mechanism will provide new understanding of the control of peptide or nutritional metabolism in L. lactis.
ACKNOWLEDGMENTS
This work was supported by contract BIO4-CT960016 in the Starlab project of the Commission of the European Communities.
We thank V. Monnet and M. Nardi for helpful discussions and P. Serror and D. Petranovic for critical reading of the manuscript.
REFERENCES
- 1.Baankreis R, Exterkate F A. Characterisation of a peptidase from Lactococcus lactis ssp cremoris HP that hydrolyses dipeptides and tripeptides containing proline or hydrophobic residues as the aminoterminal amino acid. Syst Appl Microbiol. 1991;14:317–323. [Google Scholar]
- 2.Bolotin, A., P. Winckler, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokine. The complete genome sequence of the lactic acid bacterium Lactococcus lactis. Genome Res., in press. [DOI] [PMC free article] [PubMed]
- 3.Booth M, Jennings P V, Ni Fhaolain I, O'Cuinn G. Prolidase activity of Lactococcus lactis subsp. cremoris AM2: partial purification and characterization. J Dairy Res. 1990;57:245–254. [Google Scholar]
- 4.Chapot-Chartier M P, Nardi M, Chopin M C, Chopin A, Gripon J C. Cloning and sequencing of pepC, a cysteine aminopeptidase gene from Lactococcus lactis subsp. cremoris AM2. Appl Environ Microbiol. 1993;59:330–333. doi: 10.1128/aem.59.1.330-333.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chopin A. Organization and regulation of genes for amino acid biosynthesis in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:21–38. doi: 10.1111/j.1574-6976.1993.tb00011.x. [DOI] [PubMed] [Google Scholar]
- 6.Christensen J E, Dudley E G, Pederson J A, Steele J L. Peptidases and amino acid catabolism in lactic acid bacteria. Antonie Leeuwenhoek. 1999;76:217–246. [PubMed] [Google Scholar]
- 7.Delorme C, Ehrlich S D, Renault P. Regulation of expression of the Lactococcus lactis histidine operon. J Bacteriol. 1999;181:2026–2037. doi: 10.1128/jb.181.7.2026-2037.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Detmers F J, Kunji E R, Lanfermeijer F C, Poolman B, Konings W N. Kinetics and specificity of peptide uptake by the oligopeptide transport system of Lactococcus lactis. Biochemistry. 1998;37:16671–16679. doi: 10.1021/bi981712t. [DOI] [PubMed] [Google Scholar]
- 9.de Vos W M, Vos P, de Haard H, Boerrigter I. Cloning and expression of the Lactococcus lactis subsp. cremoris SK11 gene encoding an extracellular serine proteinase. Gene. 1989;85:169–176. doi: 10.1016/0378-1119(89)90477-0. [DOI] [PubMed] [Google Scholar]
- 10.Exterkate F A. A dual-directed control of cell wall proteinase production in Streptococcus cremoris AM1: a possible mechanism of regulation during growth in milk. J Dairy Sci. 1985;68:562–571. [Google Scholar]
- 11.Exterkate F A. Pyrrolidone carboxylyl peptidase in Streptococcus cremoris: dependence on an interaction with membrane components. J Bacteriol. 1977;129:1281–1288. doi: 10.1128/jb.129.3.1281-1288.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foucaud C, Kunji E R, Hagting A, Richard J, Konings W N, Desmazeaud M, Poolman B. Specificity of peptide transport systems in Lactococcus lactis: evidence for a third system which transports hydrophobic di- and tripeptides. J Bacteriol. 1995;177:4652–4657. doi: 10.1128/jb.177.16.4652-4657.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilson T J. Ph.D. thesis. Cambridge, England: University of Cambridge; 1984. [Google Scholar]
- 15.Glatron M F, Rapoport G. Biosynthesis of the parasporal inclusion of Bacillus thuringiensis: half-life of its corresponding messenger RNA. Biochimie. 1972;54:1291–1301. doi: 10.1016/s0300-9084(72)80070-1. [DOI] [PubMed] [Google Scholar]
- 16.Godon J J, Pillidge C J, Jury K, Shearman C A, Gasson M J. Molecular analysis of the Lactococcus lactis sex factor. Dev Biol Stand. 1995;85:423–430. [PubMed] [Google Scholar]
- 17.Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 18.Hagting A, Knol J, Hasemeier B, Streutker M R, Fang G, Poolman B, Konings W N. Amplified expression, purification and functional reconstitution of the dipeptide and tripeptide transport protein of Lactococcus lactis. Eur J Biochem. 1997;247:581–587. doi: 10.1111/j.1432-1033.1997.00581.x. [DOI] [PubMed] [Google Scholar]
- 19.Hellendoorn M A, Franke-Fayard B M, Mierau I, Venema G, Kok J. Cloning and analysis of the pepV dipeptidase gene of Lactococcus lactis MG1363. J Bacteriol. 1997;179:3410–3415. doi: 10.1128/jb.179.11.3410-3415.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holo H, Nes I F. High-frequency transformation by electroporation of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hueck C J, Hillen W. Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the gram-positive bacteria? Mol Microbiol. 1995;15:395–401. doi: 10.1111/j.1365-2958.1995.tb02252.x. [DOI] [PubMed] [Google Scholar]
- 22.Hueck C J, Hillen W, Saier M H., Jr Analysis of a cis-active sequence mediating catabolite repression in gram-positive bacteria. Res Microbiol. 1994;145:503–518. doi: 10.1016/0923-2508(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 23.Hugenholz J, Exterkate F, Konings W N. The proteolytic systems of Streptococcus cremoris: an immunological analysis. Appl Environ Microbiol. 1984;48:1105–1110. doi: 10.1128/aem.48.6.1105-1110.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juillard V, Guillot A, LeBars D, Gripon J C. Specificity of milk peptide utilization by Lactococcus lactis. Appl Environ Microbiol. 1998;64:1230–1236. doi: 10.1128/aem.64.4.1230-1236.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Juillard V, Laan H, Kunji E R, Jeronimus-Stratingh C M, Bruins A P, Konings W N. The extracellular PI-type proteinase of Lactococcus lactis hydrolyzes beta-casein into more than one hundred different oligopeptides. J Bacteriol. 1995;177:3472–3478. doi: 10.1128/jb.177.12.3472-3478.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Juillard V, Le Bars D, Kunji E R, Konings W N, Gripon J C, Richard J. Oligopeptides are the main source of nitrogen for Lactococcus lactis during growth in milk. Appl Environ Microbiol. 1995;61:3024–3030. doi: 10.1128/aem.61.8.3024-3030.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kok J, Leenhouts K J, Haandrikman A J, Ledeboer A M, Venema G. Nucleotide sequence of the cell wall proteinase gene of Streptococcus cremoris Wg2. Appl Environ Microbiol. 1988;54:231–238. doi: 10.1128/aem.54.1.231-238.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunji E R, Fang G, Jeronimus-Stratingh C M, Bruins A P, Poolman B, Konings W N. Reconstruction of the proteolytic pathway for use of beta-casein by Lactococcus lactis. Mol Microbiol. 1998;27:1107–1118. doi: 10.1046/j.1365-2958.1998.00769.x. [DOI] [PubMed] [Google Scholar]
- 29.Kunji E R, Mierau I, Hagting A, Poolman B, Konings W N. The proteolytic systems of lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:187–221. doi: 10.1007/BF00395933. [DOI] [PubMed] [Google Scholar]
- 30.Kunji E R, Mierau I, Poolman B, Konings W N, Venema G, Kok J. Fate of peptides in peptidase mutants of Lactococcus lactis. Mol Microbiol. 1996;21:123–131. doi: 10.1046/j.1365-2958.1996.6231339.x. [DOI] [PubMed] [Google Scholar]
- 31.l'Anson K J, Movahedi S, Griffin H G, Gasson M J, Mulholland F. A non-essential glutamyl aminopeptidase is required for optimal growth of Lactococcus lactis MG1363 in milk. Microbiology. 1995;141:2873–2881. doi: 10.1099/13500872-141-11-2873. [DOI] [PubMed] [Google Scholar]
- 32.Loureiro Dos Santos A L, Chopin A. Shotgun cloning in Streptococcus lactis. FEMS Microbiol Lett. 1987;42:209–212. [Google Scholar]
- 33.Luesink E J, vanHerpen R E M A, Grossiord B P, Kuipers O P, de Vos W M. Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactococcus lactis are mediated by the catabolite control protein CcpA. Mol Microbiol. 1998;30:789–798. doi: 10.1046/j.1365-2958.1998.01111.x. [DOI] [PubMed] [Google Scholar]
- 34.Maguin E, Prevost H, Ehrlich S D, Gruss A. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol. 1996;178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 36.Marugg J D, Meijer W, van Kranenburg R, Laverman P, Bruinenberg P G, de Vos W M. Medium-dependent regulation of proteinase gene expression in Lactococcus lactis: control of transcription initiation by specific dipeptides. J Bacteriol. 1995;177:2982–2989. doi: 10.1128/jb.177.11.2982-2989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marugg J D, van Kranenburg R, Laverman P, Rutten G A, de Vos W M. Identical transcriptional control of the divergently transcribed prtP and prtM genes that are required for proteinase production in Lactococcus lactis SK11. J Bacteriol. 1996;178:1525–1531. doi: 10.1128/jb.178.6.1525-1531.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matos J, Nardi M, Kumura H, Monnet V. Genetic characterization of pepP, which encodes an aminopeptidase P whose deficiency does not affect Lactococcus lactis growth in milk, unlike deficiency of the X-prolyl dipeptidyl aminopeptidase. Appl Environ Microbiol. 1998;64:4591–4595. doi: 10.1128/aem.64.11.4591-4595.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meijer W C, Marugg J D, Hugenholtz J. Regulation of proteolytic enzyme activity in Lactococcus lactis. Appl Environ Microbiol. 1996;62:156–161. doi: 10.1128/aem.62.1.156-161.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mierau I, Haandrikman A J, Velterop O, Tan P S, Leenhouts K L, Konings W N, Venema G, Kok J. Tripeptidase gene (pepT) of Lactococcus lactis: molecular cloning and nucleotide sequencing of pepT and construction of a chromosomal deletion mutant. J Bacteriol. 1994;176:2854–2861. doi: 10.1128/jb.176.10.2854-2861.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mierau I, Kunji E R, Leenhouts K J, Hellendoorn M A, Haandrikman A J, Poolman B, Konings W N, Venema G, Kok J. Multiple-peptidase mutants of Lactococcus lactis are severely impaired in their ability to grow in milk. J Bacteriol. 1996;178:2794–2803. doi: 10.1128/jb.178.10.2794-2803.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mierau I, Tan P S T, Haandrikman A J, Kok J, Leenhouts K J, Konings W N, Venema G. Cloning and sequencing of the gene for a lactococcal endopeptidase, an enzyme with sequence similarity to mammalian enkephalinase. J Bacteriol. 1993;175:2087–2096. doi: 10.1128/jb.175.7.2087-2096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller C G, Kukral A M, Miller J L, Movva N R. pepM is an essential gene in Salmonella typhimurium. J Bacteriol. 1989;171:5215–5217. doi: 10.1128/jb.171.9.5215-5217.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mills O E, Thomas T D. Nitrogen sources for growth of lactic streptococci in milk. N Z J Dairy Sci Technol. 1981;15:43–55. [Google Scholar]
- 45.Nardi M, Chopin M C, Chopin A, Cals M M, Gripon J C. Cloning and DNA sequence analysis of an X-prolyl dipeptidyl aminopeptidase gene from Lactococcus lactis subsp. lactis NCDO 763. Appl Environ Microbiol. 1991;57:45–50. doi: 10.1128/aem.57.1.45-50.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nardi M, Renault P, Monnet V. Duplication of the pepF gene and shuffling of DNA fragments on the lactose plasmid of Lactococcus lactis. J Bacteriol. 1997;179:4164–4171. doi: 10.1128/jb.179.13.4164-4171.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Renault P, Corthier G, Goupil N, Delorme C, Ehrlich S D. Plasmid vectors for Gram-positive bacteria switching from high to low copy number. Gene. 1996;183:175–182. doi: 10.1016/s0378-1119(96)00554-9. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 49.Schick J, Weber B, Klein J R, Henrich B. PepR1, a CcpA-like transcription regulator of Lactobacillus delbrueckii subsp. lactis. Microbiology. 1999;145:3147–3154. doi: 10.1099/00221287-145-11-3147. [DOI] [PubMed] [Google Scholar]
- 50.Simon D, Chopin A. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie. 1988;70:559–566. doi: 10.1016/0300-9084(88)90093-4. [DOI] [PubMed] [Google Scholar]
- 51.Sissler M, Delorme C, Bond J, Ehrlich S D, Renault P, Francklyn C. An aminoacyl-tRNA synthetase paralog with a catalytic role in histidine biosynthesis. Proc Natl Acad Sci USA. 1999;96:8985–8990. doi: 10.1073/pnas.96.16.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stroman P. Sequence of a gene (lap) encoding a 95.3-kDa aminopeptidase from Lactococcus lactis ssp. cremoris Wg2. Gene. 1992;113:107–112. doi: 10.1016/0378-1119(92)90676-g. [DOI] [PubMed] [Google Scholar]
- 53.Terzaghi B, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tynkkynen S, Buist G, Kunji E R S, Kok J, Poolman B, Venema G, Haandrikman A J. Genetic and biochemical characterization of the oligopeptide transport system of Lactococcus lactis. J Bacteriol. 1993;175:7523–7532. doi: 10.1128/jb.175.23.7523-7532.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Alen-Boerrigter I J, Baankreis R, de Vos W M. Characterization and overexpression of the Lactococcus lactis pepN gene and localization of its product, aminopeptidase N. Appl Environ Microbiol. 1991;57:2555–2561. doi: 10.1128/aem.57.9.2555-2561.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van der Vossen J M, Kodde J, Haandrikman A J, Venema G, Kok J. Characterization of transcription initiation and termination signals of the proteinase genes of Lactococcus lactis Wg2 and enhancement of proteolysis in L. lactis. Appl Environ Microbiol. 1992;58:3142–3149. doi: 10.1128/aem.58.9.3142-3149.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Visser S, Exterkate F A, Slangen C J, de Veer G J C M. Comparative study of action of cell wall proteinases from various strains of Streptococcus cremoris on bovine αs1-, β-, and κ-casein. Appl Environ Microbiol. 1986;52:1162–1166. doi: 10.1128/aem.52.5.1162-1166.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]