Abstract
Objectives
Community-acquired pneumonia (CAP) is a global health condition that affects populations from all age groups. The laboratory identification of Mycoplasma pneumoniae as a causative agent of CAP is challenging because of its atypical and fastidious nature. Therefore, this study assessed the diagnostic potential of PneumoCLART bacteria® in identifying M. pneumoniae as a causative agent of pneumonia in hospitalized adults.
Methods
This prospective study used a cross-sectional approach to assess the diagnostic potential of PneumoCLART bacteria® for detecting M. pneumoniae in sputum samples procured from 27 patients with pneumonia who required hospitalization.
Results
The PneumoCLART bacteria® results illustrated that 7 of 27 patients with pneumonia were positive for M. pneumoniae (26%). However, the quality of sputum varied among the M. pneumoniae-positive and M. pneumoniae-negative samples. Fifty percent of the specimens obtained from patients positive for M. pneumoniae were saliva-contaminated and unsuitable for analysis.
Conclusions
Because the leukocyte count was low and sputum specimens were saliva-contaminated, these findings require further validation to prove the utility of CLART® microarray technology for the identification of M. pneumoniae in pneumonia-positive patients. Conclusively, this prospective study included a small number of clinical samples, which likely affected its outcomes.
Keywords: Community-acquired pneumonia, Mycoplasma pneumoniae, CLART® microarray technology, PneumoCLART bacteria®, sputum, hospitalization, saliva contamination
Introduction
Community-acquired pneumonia (CAP) is a global concern that has crippled the healthcare systems of both developed and developing economies.1–8 Reportedly, a variety of respiratory viruses and bacteria are associated with CAP, including human metapneumovirus, respiratory syncytial virus, influenza A, adenovirus, Mycoplasma pneumoniae, Streptococcus pneumoniae, Haemophilus influenza, and Staphylococcus aureus.4,9 Among these microbes, M. pneumoniae accounts for a fairly high percentage of CAP cases in adult and pediatric populations, and nearly half of the affected individuals require hospitalization.4,10–14 Although M. pneumoniae is an important etiological agent of CAP, there are challenges associated with its diagnosis mainly because of its fastidious nature, seroprevalence, and possible transient asymptomatic carriage. Therefore, it is important to explore and develop efficient techniques for the detection of M. pneumoniae as an atypical pathogen of CAP.15–17
Because of their high sensitivity and specificity, multiplex molecular diagnostic modalities such as PneumoCLART bacteria® (Genomica, Madrid, Spain) allow pathogen-specific treatment and predict pathophysiological complications, making them methods of choice for detecting various respiratory tract pathogens. PneumoCLART bacteria® is particularly useful for detecting M. pneumoniae, a fastidious, slow-growing bacterial pathogen of the respiratory tract (incubation period is 7–21 days) with special growth requirements.18–22 Noteworthy, PneumoCLART bacteria® permits the simultaneous detection and genotyping of multiple diagnostically challenging yet important respiratory tract bacterial pathogens, including M. pneumoniae, from uncultured clinical respiratory specimens (sputum, nasopharyngeal exudates/lavages/aspirates, bronchoalveolar lavage, and bronchial suction) in a single test. This in turn considerably reduces the turnaround time (up to 6 hours) and cost of the assay. Taken together, rapid bacterial detection enables the clinician to modify the antimicrobial therapy for M. pneumoniae, which is a crucial factor in improving patients’ health and recovery prospects. Consequently, prolonged hospitalization and the use of ineffective regimens can be avoided, thereby reducing treatment costs. Another important aspect worth mentioning is that antibiotic therapy in patients does not affect their test results for the presence of M. pneumoniae using PneumoCLART bacteria®, which is a nucleic acid-based PCR technique. 23 Although cell culture remains the gold standard for the laboratory confirmation of M. pneumoniae, its clinical utility is limited for the aforementioned reasons. In addition, culture techniques often fail to identify M. pneumoniae, which shares similar symptomologies with other crucial bacterial pathogens of atypical pneumonia, and it is mostly a co-infecting pathogen in CAP.18–20,22 As a potential diagnostic and epidemiological tool, it is therefore pertinent to establish the clinical utility of PneumoCLART bacteria® by undertaking prospective studies for M. pneumoniae detection. Because the prevalence rate of M. pneumoniae as an agent of CAP in Indonesia is yet to be ascertained, this study assessed the positivity rate of M. pneumoniae from clinical specimens among patients with pneumonia using the PneumoCLART bacteria® method.
Materials and methods
Study outline and specimens
This cross-sectional, prospective study and collaborative work was conducted at Atma Jaya Hospital (Jakarta, Indonesia). The sputum samples of patients requiring hospitalization for pneumonia were included in this study. Consecutive sampling was performed for patients admitted to the aforementioned hospital between February 2017 and July 2017. Patients aged 18 years and older diagnosed with pneumonia by an attending doctor who could expectorate sputum were included in the study. Patients who declined to participate, failed to provide an informed consent form, or contracted other known causes of pneumonia such as active tuberculosis were excluded from this study. The routine bacterial cultivation and molecular methods using PneumoCLART bacteria® were performed simultaneously. The reporting of this study conforms to STROBE guidelines. 24
Sample size calculation
The sample size was calculated using the formula .
Zα is the conversion of the area under the normal curve at a certain confidence level against the standard deviation of 1.96 (when the accuracy interval applied was equal to 95%), P is the prevalence rate of bacterial pneumonia in adults in Indonesia, which equals 4.5% (as revealed from the number of adults with pneumonia admitted to Atma Jaya Hospital), Q equals 1 − P, (0.955) and d is the degree of desired precision (±10%), 0.1.
Using the aforementioned formula and values,
Twenty-seven research subjects were included in this study, slightly exceeding the calculated amount to account for potential sample mishandling.
Research ethics and patient consent
The clinical specimens were obtained with the prior verbal informed consent of the patients. 25 This study was performed with the approval of the Departmental Ethical Committee of the School of Medicine and Health Science, Atma Jaya Catholic University of Indonesia (Ethical approval number: 11/05/KEP-FKUAJ/2017; approval date: 5 November 2017).
Microbiology work-up, bacterial cultivation, and isolation
The routine bacterial cultivation was performed in fully equipped microbiology laboratories in the Microbiology Department, Faculty of Medicine, Atma Jaya Catholic University (North Jakarta, Indonesia) following standard guidelines. 26 It is worth mentioning that bacterial cultivation was performed to identify pathogens other than M. pneumoniae (atypical pathogen) in the sputum samples and assess specimen quality opposed to being used as a comparative detection technique.
Post-culture, Gram staining of sputum samples was performed routinely before isolation to identify bacterial pathogens other than M. pneumoniae (which lacks a defined cell wall). Leukocytes and squamous epithelial cells (SECs) were identified and counted. The same bacterial growth media, namely chocolate agar, blood agar, and MacConkey agar (all from Sigma-Aldrich, Inc., St. Louis, MO, USA), were used in both hospitals. Although microaerophilic conditions were achieved in a candle jar for bacterial incubation on chocolate agar at 37 ± 2°C for 24 to 48 hours, blood agar and MacConkey agar were incubated in aerobic conditions at 37 ± 2°C for 18 to 24 hours.27,28 Oxoid™ Microbact™ 12A/12B (Oxoid Limited, Basingstoke, Hampshire, UK), a microplate-based biochemical test, was used to identify gram-negative bacteria, and the conventional method according to Bergey’s system of classification.28–30 An automated VITEK®2 system (BioMérieux, Marcy-l’Étoile, France) was used for bacterial identification at Atma Jaya Hospital. 31 Nucleic acid extraction was conducted for all specimens that underwent bacterial cultivation without waiting for the cultivation result.
Bacterial identification
PneumoCLART bacteria® is an emerging molecular diagnostic technique with immense potential for the detection of human respiratory pathogens. In brief, the PneumoCLART bacteria® detection system is based on the precipitation of an insoluble product in areas on the microarray where the hybridization of amplification products with specific probes occurs. During PCR, amplified products are labeled with biotin. After amplification, these biotin-labeled products are hybridized with their respective specific complementary probes, which are then immobilized on specific known microarrays. Next, these probes are incubated with a streptavidin–peroxidase conjugate, which is bound to their specific probes. The peroxidase activity induces the appearance of a non-soluble product in the presence of the substrate o-dianisidine, which precipitates in the areas on the microarray where hybridization occurs. Finally, the precipitation of the substrate and control is read by the CAR® clinical array reader, which generates an objective clinical report (www.genomica.com).
In this study, the molecular analysis was performed using PneumoCLART bacteria®, which facilitated the in vitro detection and characterization of bacteria in the respiratory samples. Total nucleic acids were extracted from each specimen using an automated NUCLISENS® easyMag® system (BioMérieux) according to the manufacturer’s instructions, followed by amplification reaction and visualization of the amplified product in the CLART® strip (www.genomica.com). The amplification tubes had primers specific for a range of bacteria with established roles in respiratory tract infections including M. pneumoniae, S. aureus, S. pneumoniae, Chlamydia pneumoniae, Moraxella catarrhalis, Haemophilus, and Bordetella spp. (B. pertussis, B. parapertusis, B. bronchiseptica, B. holmesii). Endogenous genomic DNA and internal controls were included to ensure the efficiency of the assay and to eliminate false negatives. This examination was performed at Sentra Diagnostica Dinamika, a private laboratory in Jakarta, Indonesia. The laboratory staff involved in this study was unaware of the independent test results.
Results
This study enrolled 27 adults, including 11 women and 16 men aged 35 to 90 years, with pneumonia requiring hospitalization at Atma Jaya Hospital. Specifically, we sought patients who could expel active sputum spontaneously, and bacterial cultivation was performed.
The quality of the collected sputum was assessed, and the saliva-contaminated specimens accounted for almost 30% of the total number of samples (8/27) with SEC counts >10/low-power field (LPF). Our laboratory defines the adequacy of any specimen for the lower respiratory tract as an area containing SEC < 10/LPF and polymorphonuclear neutrophil or leukocyte counts ≥25/LPF.32,33 Accordingly, 15 representative specimens (55%) fulfilled the aforementioned criteria.
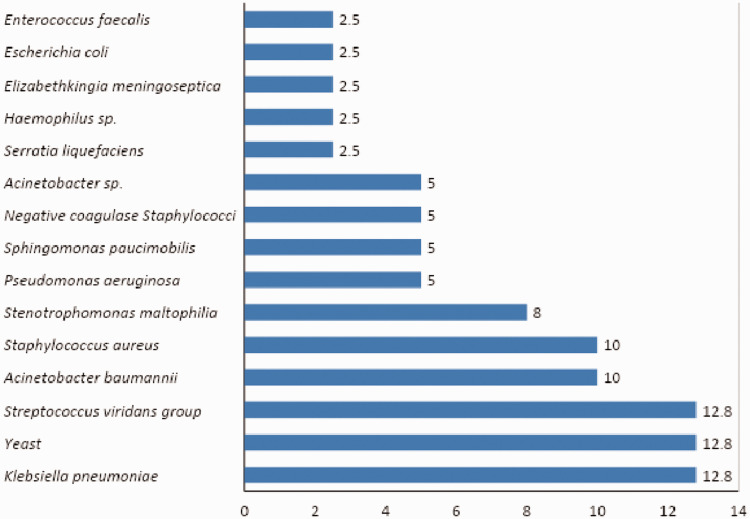
Gram staining performed before cultivation revealed the presence of various gram-negative and gram-positive organisms in the specimens. Accordingly, 39 organisms were isolated from 27 sputum specimens. Among these, Klebsiella pneumoniae (12.8%), yeast (12.8%), and viridans group Streptococci were present in abundance (12.8%, Figure 1).
Figure 1.
Percentage of each isolate cultivated from sputum specimens (n = 39).
According to the PneumoCLART bacteria® examination, 7 of 27 patients with pneumonia were positive for M. pneumoniae (26%). Among these, only two of the seven specimens positive for M. pneumoniae were adequate for analysis (Table 1). Notably, 50% of patients positive for M. pneumoniae required admission to the intensive care unit (ICU) at Atma Jaya Hospital (Figure 2).
Table 1.
Aerobic bacterial cultivation result and microscopic characteristics of the sputum from the Mycoplasma pneumoniae-positive group.
| Age (years) | Organism | Saliva contamination | Inflammation parameter | Location/origin |
|---|---|---|---|---|
| 42 | K. pneumoniae, P. aeruginosa | + | − | ICU |
| 67 | Haemophilus sp. | + | + | General ward |
| 37 | S. maltophilia, S. viridans | + | + | General ward |
| 48 | Yeast | − | + | General ward |
| 40 | K. pneumoniae | + | + | General ward |
| 74 | A. baumannii, S. maltophilia | − | + | ICU |
| 75 | Yeast | − | − | ICU |
ICU, intensive care unit.
Figure 2.
Result of Mycoplasma pneumoniae detection by PneumoCLART bacteria®.
Briefly, the time of disease onset in these patients ranged from less than 8 hours to 30 days. Among these, most of the patients received antibiotic treatment regimens comprising doripenem, levofloxacin, azithromycin, cefadroxil, levofloxacin, vancomycin, and meropenem. Further, the comorbidities in these patients included cardiovascular disease, diabetes mellitus (DM), hypertension, benign prostatic hyperplasia (BPH), chronic kidney disease (CKD), nucleus pulposus herniation, and scoliosis. Furthermore, the imaging features included pleural effusion, lung infiltrates, pleuropneumonia, bronchitis, scoliosis thoracalis, interposition colon, interstitial pneumonia, traction bronchiectasis, lower lobe atelectasis, bronchopneumonia, and pneumothorax. The clinical manifestations included dyspnea, hematemesis, necrotizing fasciitis, CKD, hyperkalemia, type II DM, melena, dehydration-associated hypotension, BPH, nucleus pulposus herniation, and suspected sepsis (attributable to elevated procalcitonin levels). Most importantly, normocytic normochromic anemia, anemia of chronic disease (ACD), ACD with infection anemia, thrombocytopenia, leukocytosis, infection or liver disease, and autoimmune disorder were also suspected in two patients who tested positive for infection by M. pneumoniae using PneumoCLART bacteria®.
Discussion
There are accumulating data on the high prevalence of M. pneumoniae as a major culprit causing CAP globally. In a prospective study, Liu et al. reported M. pneumoniae as the most prevalent pathogen (20.7%, 126/610) among urban Chinese adults with CAP. 34 Further, Wu et al. reported the highest detection rate of 56.9% (among 10,435 specimens) for M. pneumoniae among all pathogens tested in Chinese pediatric patients (age <16 years) with acute respiratory infections. 35 Similarly, Chen et al. identified M. pneumoniae as the predominant pathogen (positivity rate = 40.78%) among all tested pathogens in Chinese pediatric patients (aged 4–14 years). 36 Furthermore, Chen et al. reported a high M. pneumoniae prevalence (55%) in the pediatric population using an IgM antibody-based immune-gold labeling detection method. 37 In a retrospective study, M. pneumoniae was the most dominant causative agent in 14.5% of pediatric CAP cases (166 pneumonia-confirmed cases; aged 1–15 years) in Belgrade, Serbia. 38 Another prospective study identified M. pneumoniae infection among 27% of pediatric patients (140 children aged 2 months to 15 years) with CAP in Istanbul, Turkey. 39 In a cross-sectional study, Carcey et al. recorded a high positivity of 31.9% for M. pneumoniae in 20,020 serological samples from Chilean children using IgM serological testing (age <18 years). 40 In their consecutive cross-sectional study, Del et al. disclosed a high prevalence (25.19% or 170/675 pediatric patients) of M. pneumoniae-associated acute respiratory infections in Peruvian children (age <18 years). 41
Another retrospective study reported M. pneumoniae as a possible pathogen in Chinese patients (3852 adults and 3983 children) with respiratory tract infections admitted to the ICU during the epidemic (2011–2013). The positivity rate for M. pneumoniae was 21.2% with no statistically significant difference noted among different age groups during most of the epidemic. 42 In a 5-month study (from January 2017 to June 2017), Arfaatabar et al. observed a high frequency of M. pneumoniae among 520 patients with CAP in Tehran, Iran. 43 Su et al. determined that M. pneumoniae was highly prevalent in hospitalized children with community-acquired M. pneumoniae pneumonia (MPP) (66.4% [221/333 pediatric patients]). 44 A retrospective study by Cheng et al. investigated the epidemiology of M. pneumoniae in Chinese children with respiratory infections from June 2016 to May 2021. 45 Reportedly, the positivity rates did not differ significantly in relation to the season, age group, gender, or period (before or during the COVID-19 pandemic) as revealed from the M. pneumoniae specific-IgM antibody rapid immuno-chromatographic assay of the serum specimens of 569,887 pediatric patients. 45
Treatment strategies for CAP largely rely on the presented clinical symptoms (mild or severe) and infection type (bacterial- and/or viral-associated pneumonia; co-infection) suspected in patients with CAP.4,46–48 Over time, there has been a change in the CAP etiology concerning bacterial, viral, and fungal co-infections. It is therefore recommended to initially administer empiric antibiotic treatment to eradicate the major causative pathogens and resolve clinical symptoms.46–49 Precisely, the pre-hospitalization treatment regimen comprises oral macrolides (for example, azithromycin or clarithromycin and erythromycin), tetracycline (for example, doxycycline or vibramycin), and fluoroquinolones (for example, ciprofloxacin and levofloxacin) for managing CAP-associated clinical symptoms in patients. Alternatively, oral administration of amoxicillin or clavulanate and β-lactams (e.g., cefpodoxime, cefprozil, cefuroxime) can relieve mild symptoms in patients with no co-morbidities (e.g., renal failure, chronic obstructive pulmonary disease, diabetes, asplenia, congestive heart failure, chronic alcoholism, immunosuppressive conditions).46–48 However, clinical guidelines restrict the use of macrolides in patients with community-acquired MPP given the growing concerns for resistance in this pathogen.12,50–54 Moreover, β-lactam antibiotics are ineffective against M. pneumoniae because it lacks a rigid cell wall.47,48 Further, this study was undertaken to assess the diagnostic efficiency of PneumoCLART bacteria® regarding M. pneumoniae, but this study was not designed to recommend pre-hospitalization treatment for M. pneumoniae-associated CAP. The antibiotic treatment administered to the patients after admission to Atma Jaya Hospital was recorded.
M. pneumonia, as a fastidious pathogen, presents several challenges concerning its efficient and accurate diagnosis. 16 Of the seven samples positive for M. pneumoniae, two expectorated sputum specimens featured a leukocyte count of <25/LPF. In addition, 50% of the sputum specimens that failed to fulfill the criteria for adequacy were either contaminated with saliva or obtained from patients who were not admitted to the ICU. Further, one-fourth of specimens negative for M. pneumoniae featured a leukocyte count of <25/LPF. Among negative samples, fewer than 25% that failed to fulfill the criteria for adequacy were contaminated with saliva or obtained from patients not admitted to the ICU (Table 2). PneumoCLART bacteria® could be a useful diagnostic technique for detecting M. pneumoniae in patients with respiratory illnesses, especially community-acquired MPP. However, further studies including an appropriate number of specimens with stringent quality control measures for collection and analysis will be indispensable for strengthening the utility of this technique. The small sample size was a limitation of the present study.
Table 2.
Aerobic bacterial cultivation result and microscopic characteristics of sputum from the Mycoplasma pneumoniae-negative group.
| Age (years) | Organism | Saliva contamination* | Inflammation parameter# | Location/origin |
|---|---|---|---|---|
| 80 | S. liquefaciens | + | − | ICU |
| 72 | A. baumannii | + | − | General ward |
| 46 | Yeast | + | + | ICU |
| 65 | S. viridans | − | + | General ward |
| 45 | Acinetobacter sp. | + | + | General ward |
| 85 | S. gordonii, S. paucimobilis | − | + | IMC |
| 62 | A. lwoffii, S. haemolyticus, S. parasanguinis | − | + | IMC |
| 77 | S. aureus, P. aeruginosa | − | + | General ward |
| 52 | A. baumannii, E. meningoseptica | − | − | ICU |
| 67 | S. haemolyticus | − | + | ICU |
| 62 | A. baumannii | − | + | ICU |
| 88 | S. aureus | − | + | IMC |
| 54 | S. paucimobilis | − | + | ICU |
| 42 | K. pneumoniae | − | − | ICU |
| 72 | S. aureus | − | + | ICU |
| 76 | E. coli | − | + | IMC |
| 90 | K. pneumoniae | − | + | General ward |
| 86 | S. pseudoporcinus, E. faecalis, S. aureus | − | − | IMC |
| 52 | Yeast | − | + | ICU |
| 80 | Yeast | − | + | IMC |
*Squamous epithelial cell count >10/low power field (LPF); #polymorphonuclear neutrophil count ≥25/ LPF.
ICU, intensive care unit; IMC, intermediate care unit.
Conclusion
In our study, some samples featured a low leukocyte count and/or contamination by saliva; hence, the findings on the utility of CLART® microarray technology for the identification of M. pneumoniae in pneumonia-positive patients need further validation. Moreover, we speculate that the small sample size might have affected the outcomes of this prospective study. In conclusion, molecular diagnostic methods based on the amplification of nucleic acids (DNA/RNA) such as CLART® microarray technology might improve the sensitivity for M. pneumoniae identification provided a sufficient sample size is obtained and no samples are contaminated.
Acknowledgements
Experimental support from the Director and committee of Atma Jaya Hospital and Dr. Lely Saptawati of Dr. Moewardi Hospital (Surakarta, Central Jawa, Indonesia) is acknowledged.
Author contributions: ET, LHM, and SJ: Conceptualization, visualization, methodology, validation, data analysis and curation, original draft preparation, and review. LL and FP: Methodology, review, and editing. All authors contributed significantly to the study and approved the final manuscript.
Declaration of conflicting interests: The authors have no conflicts of interest to disclose.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: We thank the School of Medicine, Atma Jaya Catholic University of Indonesia (Jakarta, Indonesia) for providing financial support (funding number: 0831/III/D.FM-PM.10.01/04/2018) for this study.
Data availability
All relevant data are presented in the manuscript, and the data are accessible in the peer-review process.
ORCID iD
Enty Tjoa https://orcid.org/0000-0001-7187-4280
References
- 1.Chawla R. Epidemiology, etiology, and diagnosis of hospital-acquired pneumonia and ventilator-associated pneumonia in Asian countries. Am J Infect Control 2008; 36: S93–S100. [DOI] [PubMed] [Google Scholar]
- 2.Falade AG, Ayede AI. Epidemiology, aetiology and management of childhood acute community-acquired pneumonia in developing countries–a review. Afr J Med Med Sci 2011; 40: 293–308. [PubMed] [Google Scholar]
- 3.Song JH, Thamlikitkul V, Hsueh PR. Clinical and economic burden of community-acquired pneumonia amongst adults in the Asia-Pacific region. Int J Antimicrob Agents 2011; 38: 108–117. [DOI] [PubMed] [Google Scholar]
- 4.Jain S, Self WH, Wunderink RG, et al. Community-Acquired Pneumonia Requiring Hospitalization among U.S. Adults. N Engl J Med 2015; 373: 415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azmi S, Aljunid SM, Maimaiti N, et al. Assessing the burden of pneumonia using administrative data from Malaysia, Indonesia, and the Philippines. Int J Infect Dis 2016; 49: 87–93. [DOI] [PubMed] [Google Scholar]
- 6.Aston SJ. Pneumonia in the developing world: Characteristic features and approach to management. Respirology 2017; 22: 1276–1287. [DOI] [PubMed] [Google Scholar]
- 7.Bazie GW, Seid N, Admassu B. Determinants of community acquired pneumonia among 2 to 59 months of age children in Northeast Ethiopia: a case-control study. Pneumonia (Nathan) 2020; 12: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nasrin S, Tariqujjaman M, Sultana M, et al. Factors associated with community acquired severe pneumonia among under five children in Dhaka, Bangladesh: A case control analysis. PLoS One 2022; 17: e0265871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Musher DM, Thorner AR. Community-acquired pneumonia. N Engl J Med 2014; 371: 1619–1628. [DOI] [PubMed] [Google Scholar]
- 10.Ngeow YF, Suwanjutha S, Chantarojanasriri T, et al. An Asian study on the prevalence of atypical respiratory pathogens in community-acquired pneumonia. Int J Infect Dis 2005; 9: 144–153. [DOI] [PubMed] [Google Scholar]
- 11.Liu WK, Liu Q, Chen DH, et al. Epidemiology of acute respiratory infections in children in Guangzhou: a three-year study. PLoS One 2014; 9: e96674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao MC, Wang L, Qiu FZ, et al. Impact and clinical profiles of Mycoplasma pneumoniae co-detection in childhood community-acquired pneumonia. BMC Infect Dis 2019; 19: 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metsälä R, Ala-Korpi S, Rannikko J, et al. Mycoplasma pneumoniae may cause dyspnoea and hospitalisations in young healthy adults. Eur J Clin Microbiol Infect Dis 2021; 40: 1427–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qu J, Zhang J, Chen Y, et al. Aetiology of severe community acquired pneumonia in adults identified by combined detection methods: a multi-centre prospective study in China. Emerg Microbes Infect 2022; 11: 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daxboeck F, Krause R, Wenisch C. Laboratory diagnosis of Mycoplasma pneumoniae infection. Clin Microbiol Infect 2003; 9: 263–273. [DOI] [PubMed] [Google Scholar]
- 16.Kashyap S, Sarkar M. Mycoplasma pneumonia: Clinical features and management. Lung India 2010; 27: 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bajantri B, Venkatram S, Diaz-Fuentes G. Mycoplasma pneumoniae: A Potentially Severe Infection. J Clin Med Res 2018; 10: 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyashita N, Saito A, Kohno S, et al. ; CAP Study Group. Multiplex PCR for the simultaneous detection of Chlamydia pneumoniae, Mycoplasma pneumoniae and Legionella pneumophila in community-acquired pneumonia. Respir Med 2004; 98: 542–550. [DOI] [PubMed] [Google Scholar]
- 19.McDonough EA, Barrozo CP, Russell KL, et al. A multiplex PCR for detection of Mycoplasma pneumoniae, Chlamydophila pneumoniae, Legionella pneumophila, and Bordetella pertussis in clinical specimens. Mol Cell Probes 2005; 19: 314–322. [DOI] [PubMed] [Google Scholar]
- 20.Strålin K, Bäckman A, Holmberg H, et al. Design of a multiplex PCR for Streptococcus pneumoniae, Haemophilus influenzae, Mycoplasma pneumoniae and Chlamydophila pneumoniae to be used on sputum samples. APMIS 2005; 113: 99–111. [DOI] [PubMed] [Google Scholar]
- 21.Caliendo AM. Multiplex PCR and emerging technologies for the detection of respiratory pathogens. Clin Infect Dis 2011; 52: S326–S330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nummi M, Mannonen L, Puolakkainen M. Development of a multiplex real-time PCR assay for detection of Mycoplasma pneumoniae, Chlamydia pneumoniae and mutations associated with macrolide resistance in Mycoplasma pneumoniae from respiratory clinical specimens. Springerplus 2015; 4: 684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyashita N, Shimizu H, Ouchi K, et al. Assessment of the usefulness of sputum Gram stain and culture for diagnosis of community-acquired pneumonia requiring hospitalization. Med Sci Monit 2008; 14: CR171–CR176. [PubMed] [Google Scholar]
- 24.Von Elm E, Altman DG, Egger M, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Ann Intern Med 2007; 147: 573–577. [DOI] [PubMed] [Google Scholar]
- 25.Nijhawan LP, Janodia MD, Muddukrishna BS, et al. Informed consent: Issues and challenges. J Adv Pharm Technol Res 2013; 4: 134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vandepitte J, Engbaek K, Rohner P, et al. Basic Laboratory Procedures in Clinical Bacteriology 2nd Edition. Geneva: WHO 2003; 19–120. [Google Scholar]
- 27.Brooks GF, Carroll KC, Butel JS, et al. Medical Microbiology. Jawetz, Melnick and Adelbergs, Twenty-sixth Edition, McGraw-Hill Companies, New York. 2013; 67–72. [Google Scholar]
- 28.Linscott A. Collection, Transport, and Manipulation of Clinical Specimens and Initial Laboratory Concerns. In Leber A. (ed). Clinical Microbiology Procedures Handbook, Fourth Edition. ASM Press, Washington, DC. 2016; 2.1.1–2.1.30. [Google Scholar]
- 29.Burke V, Robinson J, Atkinson HM, et al. Biochemical characteristics of enterotoxigenic Aeromonas spp. J Clin Microbiol 1982; 15: 48–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farmer JJ, Davis BR, Hickman-Brenner FW, et al. Biochemical identification of new species and biogroups of Enterobacteriaceae isolated from clinical specimens. J Clin Microbiol 1985; 21: 46–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Hara CM. . Manual and automated instrumentation for identification of Enterobacteriaceae and other aerobic gram-negative bacilli. Clin Microbiol Rev 2005; 18: 147–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farida H, Gasem MH, Suryanto A, et al. Viruses and Gram-negative bacilli dominate the etiology of community-acquired pneumonia in Indonesia, a cohort study. Int J Infect Dis 2015; 38: 101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Musher DM, Montoya R, Wanahita A. Diagnostic value of microscopic examination of Gram-stained sputum and sputum cultures in patients with bacteremic pneumococcal pneumonia. Clin Infect Dis 2004; 39: 165–169. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y, Chen M, Zhao T, et al. Causative agent distribution and antibiotic therapy assessment among adult patients with community acquired pneumonia in Chinese urban population. BMC Infect Dis 2009; 9: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Z, Li Y, Gu J, et al. Detection of viruses and atypical bacteria associated with acute respiratory infection of children in Hubei, China. Respirology 2014; 19: 218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen K, Jia R, Li L, et al. The aetiology of community associated pneumonia in children in Nanjing, China and aetiological patterns associated with age and season. BMC Public Health 2015. a; 15: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen FQ, Yang YZ, Yu LL, et al. Prevalence of Mycoplasma pneumoniae: a cause for community-acquired infection among pediatric populaztion. Niger J Clin Pract 2015. b; 18: 354–358. [DOI] [PubMed] [Google Scholar]
- 38.Medjo B, Atanaskovic-Markovic M, Radic S, et al. Mycoplasma pneumoniae as a causative agent of community-acquired pneumonia in children: clinical features and laboratory diagnosis. Ital J Pediatr 2014; 40: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Somer A, Salman N, Yalçin I, et al. Role of Mycoplasma pneumoniae and Chlamydia pneumoniae in children with community-acquired pneumonia in Istanbul, Turkey. J Trop Pediatr 2006; 52: 173–178. [DOI] [PubMed] [Google Scholar]
- 40.Carcey J, Garcia P, Padilla O, et al. Increased prevalence of Mycoplasma pneumoniae serological positivity in Chilean young children. Allergol Immunopathol (Madr) 2016; 44: 467–471. [DOI] [PubMed] [Google Scholar]
- 41.Del Valle-Mendoza J, Orellana-Peralta F, Marcelo-Rodríguez A, et al. High Prevalence of Mycoplasma pneumoniae and Chlamydia pneumoniae in Children with Acute Respiratory Infections from Lima, Peru. PLoS One 2017; 12: e0170787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qu J, Yang C, Bao F, et al. Epidemiological characterization of respiratory tract infections caused by Mycoplasma pneumoniae during epidemic and post-epidemic periods in North China, from 2011 to 2016. BMC Infect Dis 2018; 18: 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arfaatabar M, Aminharati F, Azimi G, et al. High frequency of Mycoplasma pneumoniae among patients with atypical pneumonia in Tehran, Iran. Germs 2018; 8: 126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su M, Wang Q, Li D, et al. Prevalence and clinical characteristics of hospitalized children with community-acquired Mycoplasma pneumoniae pneumonia during 2017/2018, Chengde, China. Medicine (Baltimore) 2021; 100: e23786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng Y, Cheng Y, Dai S, et al. The Prevalence of Mycoplasma Pneumoniae Among Children in Beijing Before and During the COVID-19 Pandemic. Front Cell Infect Microbiol 2022; 12: 854505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lode HM. Managing community-acquired pneumonia: a European perspective. Respir Med 2007; 101: 1864–1873. [DOI] [PubMed] [Google Scholar]
- 47.Metlay JP, Waterer GW, Long AC, et al. Diagnosis and Treatment of Adults with Community-acquired Pneumonia. An Official Clinical Practice Guideline of the American Thoracic Society and Infectious Diseases Society of America. Am J Respir Crit Care Med 2019; 200: e45–e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Franco J. Community-acquired Pneumonia. Radiol Technol 2017; 88: 621–636. [PubMed] [Google Scholar]
- 49.Webb BJ, Sorensen J, Jephson A, et al. Broad-spectrum antibiotic use and poor outcomes in community-onset pneumonia: a cohort study. Eur Respir J 2019; 54: 1900057. [DOI] [PubMed] [Google Scholar]
- 50.Diaz MH, Benitez AJ, Winchell JM. Investigations of Mycoplasma pneumoniae infections in the United States: trends in molecular typing and macrolide resistance from 2006 to 2013. J Clin Microbiol 2015; 53: 124–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng X, Lee S, Selvarangan R, et al. Macrolide-Resistant Mycoplasma pneumoniae, United States. Emerg Infect Dis 2015; 21: 1470–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo DX, Hu WJ, Wei R, et al. Epidemiology and mechanism of drug resistance of Mycoplasma pneumoniae in Beijing, China: A multicenter study. Bosn J Basic Med Sci 2019; 19: 288–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao F, Liu J, Shi W, et al. Antimicrobial susceptibility and genotyping of Mycoplasma pneumoniae isolates in Beijing, China, from 2014 to 2016. Antimicrob Resist Infect Control 2019; 8: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waites KB, Ratliff A, Crabb DM, et al. Macrolide-Resistant Mycoplasma pneumoniae in the United States as Determined from a National Surveillance Program. J Clin Microbiol 2019; 57: e00968–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are presented in the manuscript, and the data are accessible in the peer-review process.