Abstract
Background
S. miltiorrhiza root rot is a soil-borne disease mainly caused by Fusarium solani and Fusarium oxysporum, which has spread rapidly in China in recent years. To reduce the amount of pesticides to control this plant fungal disease, biological control using endophytic bacteria is a promising method. Many endophytic bacteria show good biocontrol potential against various plant fungal diseases. The aims of this study were to isolate and identify endophytic bacteria with antifungal activity from Salvia miltiorrhiza plant tissue. In order to increase antifungal substances production, the culture conditions of the isolated DS-R5 strain were optimized through response surface methodology.
Results
Thirteen endophytic bacteria with antifungal activity against the target pathogenic fungus were successfully screened. The DS-R5 strain that had the strongest antifungal activity was identified based on morphological, physiological and biochemical characteristics, 16S rRNA and gyrB sequence analysis.The results of response surface methodology experiments showed that the optimal values of the three significant factors were as follows: medium volume, 51.0 ml; initial pH, 6.7; fermentation temperature, 33.1 °C. Under these optimal culture conditions, the titer of antifungal substances produced by the DS-R5 strain was 77.6% higher than that under the initial culture conditions.
Conclusions
The antifungal activity of endophytic bacteria from Salvia miltiorrhiza has been demonstrated for the first time, which may benefit future crop quality and production. In addition, response surface methodology can be well applied the optimization of culture conditions for antifungal substance, which lays the foundation for further research on strain DS-R5.
Keywords: Salvia miltiorrhiza root rot, Biocontrol bacteria, Isolation and identification, Antifungal substance, Optimization, Response surface methodology
Introduction
Salvia miltiorrhiza is a perennial herbal medicinal plant whose root and rhizome is used as medicine to remove blood stasis and relieve pain, promote blood circulation and regulate menstruation, nourish the heart and reduce anxiety. It is also widely used in the treatment of cardiovascular and cerebrovascular diseases [1–3]. Under artificial cultivation conditions, due to poor diversity and low richness of the biological communities in the planting environment, serious diseases and pests often occur, which affect the growth and development of S. miltiorrhiza and reduce yield and quality of the medicinal material [4]. S. miltiorrhiza root rot is a soil-borne disease, mainly caused by Fusarium solani and Fusarium oxysporum, that has spread rapidly in China in recent years [5]. After becoming infected, S. miltiorrhiza plants grow weak, and the root xylem completely rots to become black and brown, which affects the appearance, character and quality to the point that they do not meet the requirements for medicinal use. At present, the main control method for fungal diseases of medicinal plants is chemical control. Although chemical control can effectively inhibit the occurrence of fungal diseases in the short term, the application of chemical agents greatly increases pesticide residues in the soil, pollutes the environment and enhances the disease resistance of pathogenic microorganisms [6]. In recent years, more and more studies have shown that biological control using endophytes is an important alternative to chemical control [7–9].
Endophytic bacteria live in healthy plant tissues without causing significant damage to the plant and can improve the resistance of plants to biotic or abiotic stresses, making them a good choice as potential biocontrol bacteria [10]. People have been looking for endophytic bacteria for biocontrol strains against many plant pathogens for decades, and many endophytic bacteria show good biocontrol potential against various plant fungal diseases [11]. Paenibacillus polymyxa is a gram-positive spore-producing bacteria formerly known as Bacillus polymyxa that is a good target for screening biocontrol bacteria of plant diseases [12]. To control many different plant diseases, P. polymyxa releases many different antimicrobial components, including proteins, phenols, peptides, pyrazines and nucleosides [13]. As P. polymyxa can be used as a biological pesticide to control a variety of plant diseases and it is safe to use without environmental pollution, P. polymyxa has been designated as one of the commercially available microorganisms by the U.S. Environmental Protection Agency [14].
In the process of microbial fermentation, the environmental conditions need to be strictly controlled in order to improve the yield of fermentation products [15]. To date, optimization of culture conditions is still the most important method to improve fermentation efficiency. Therefore, the optimization of culture conditions for the production of antimicrobial substances is of great significance. At present, one of the main problems to be solved in the application of biological control agents is that biocontrol bacteria secrete less effective antifungal substances during the fermentation process [16]. In order to maximize antifungal activity, the fermentation medium and culture conditions for antifungal substances production by the DS-R5 strain must be optimized. In our previous study, the fermentation medium for antifungal substances production by the DS-R5 strain was optimized using one-factor-at-a-time, Plackett–Burman (PB) design and Box-Behnken design experiments. Increasingly, researchers have shown that environmental conditions such as culture time, shaker speed, temperature, pH and inoculum percentage play a significant role in inducing antimicrobial substance synthesis [17, 18].
In this study, the endophytic strain DS-R5 with strong antifungal activity against F. solani was isolated from S. miltiorrhiza root tissue and identified as P. polymyxa using morphology and molecular biology. The results of pot experiments showed that the control efficacy of the DS-R5 strain on S. miltiorrhiza root rot was 61.4%. Therefore, the DS-R5 strain has good biocontrol potential for preventing and controlling S. miltiorrhiza root rot and offers the prospect of further development. In addition, response surface methodology (RSM) was used to optimize the culture conditions for antifungal substances synthesis in order to maximize their production. Our objectives in studying the specific components of active antifungal substances of the DS-R5 strain are to clarify its antifungal mechanism and to assess its potential as a biocontrol agent for preventing and controlling S. miltiorrhiza root rot.
To the best of our knowledge, we have isolated and identified a biocontrol bacterium from the medicinal plant S. miltiorrhiza for the first time in this study and demonstrated its antagonistic effects on F. solani, the pathogen of S. miltiorrhiza root rot. The results of this research demonstrate the potential of the DS-R5 strain as a biocontrol agent for preventing and controlling S. miltiorrhiza root rot. In addition, this study provides a reference for the optimization of culture conditions and lay a theoretical foundation for the preparation of biocontrol agents by the DS-R5 strain.
Materials and methods
Isolation of endophytic antagonistic bacteria and growth conditions
Healthy S. miltiorrhiza plants with good growth were collected and cleaned with water. The material surface was disinfected by soaking in 75% ethanol for 1 min and rinsing with sterile water three times, then soaking in 0.1% mercuric chloride for 3 min and rinsing with sterile water three times. The treated S. miltiorrhiza tissues, including the root, stem and leaf, were ground into a pulp with a mortar on a sterile operating table, diluted 10 times in sterile water, coated on LB medium and cultured at 37 °C for 3 days. When visible colonies appeared on the culture medium, the colonies were picked and pure cultures were obtained by repeated streak-line separation. Sterile water was used to take the last rinse of S. miltiorrhiza tissues and coat it on LB medium. No colonies grew after incubation at 37 °C for 3 days, indicating that the sample surface was thoroughly disinfected.
The fermentation medium for antifungal substances contained the following components: 15.0 g/l sucrose, 30 g/l soluble starch, 7.0 g/l corn steep liquor, 10.0 g/l (NH4)2SO4, and 0.7 g/l KH2PO4.
The DS-R5 strain was inoculated into Luria–Bertani (LB) medium and cultured at 37 °C in a rotary shaker (KS4000i, IKA, German) at 200 rpm for 16 h to prepare the inoculum. A 2% (v/v) inoculum was inoculated into a flask (250 ml) containing 100 ml fermentation medium and incubated at 37 °C in a rotary shaker at 200 rpm for 72 h to produce antifungal substances.
Sample collection and pathogenic fungi
Healthy 3-year-old S. miltiorrhiza plants were collected from an S. miltiorrhiza plantation at the foot of Mount Tai, China. During sampling, the entire plant was pulled out with its roots, placed in a sterile bag and brought back to the laboratory for endophytic bacteria isolation.
The tested pathogenic fungi used in this study, F. solani, Rhizoctonia solani, Alternaria alternate, Fusarium oxysporum, Colletotrichum orbiculare, Fusarium graminearum, Botryosphaeria ribis and Fusarium pseudograminearum, are all preserved in our laboratory.
Crude extract preparation of antifungal substances produced by the DS-R5 strain
After fermentation, the supernatant of the fermentation broth was adjusted to pH 2.0 with 6 mol/l hydrochloric acid, then centrifuged at 6000 rpm for 10 min after overnight storage at 4 °C. The precipitate was then lyophilized to obtain a light yellow powder, which was the crude extract of antifungal substances used to make the standard curve for titer determination.
Titer determination of the DS-R5 strain fermentation broth
A crude extract solution of antifungal substances with a concentration of 10,000 mg/l was prepared with 70% methanol solution, then diluted into 5000, 2500, 1250, 625 and 312.5 mg/l solutions. After titer determination, the medium was melted and cooled to 50–55 °C, and the spore suspension of F. oxysporum at a concentration of approximately 1 × 105 colony forming units/ml was added at a ratio of 0.5% (v/v) to prepare mixed spore plates. Sterilized Oxford disks were placed on the mixed spore plates, and 200 µl of antifungal substance solution was added at various concentrations. After culture at 30 °C for 48 h, the diameter of the inhibition zone were measured using the cross method to create a standard curve for the titer bioassay. The titer of the fermentation broth was then calculated according to the following validated standard equation: y = 8.7619x-6.4762 (R2 = 0.9911), where y is the diameter of the inhibition zone (mm) and x is the logarithmic value of the concentration of the antifungal substance solution.
Screening of antagonistic bacteria by plate confrontation assay
According to the method reported by Khalil et al. [19], the endophytic antagonistic bacteria isolated from S. miltiorrhiza were screened for their antagonistic activity against the target pathogen F. oxysporum using the dual plate confrontation assay method. A 5-mm puncher was used to punch holes on F. oxysporum plates cultured for 7 days. The 5-mm fungal cake was inoculated in the center of a potato dextrose agar (PDA) plate, and the isolated endophytic bacteria were inoculated in parallel lines 2.5 cm away from the fungal cake. A fungal plate without antagonistic bacteria was used as the control and placed in a 30 °C incubator for constant temperature culture. When the pathogen colonies in the control group were overgrown on the bottom of the dish, the width of the inhibition zone of the treatment group was measured. Each treatment was repeated three times.
Identification of endophytic antagonistic bacteria
Morphological, physiological and biochemical identification
The DS-R5 strain was inoculated onto LB plates and incubated in a 37 °C incubator (CT-260R, BOLV, China) for 24 h. The morphological characteristics of colonies were observed and conventional Gram staining was carried out. The morphology of bacterial cells was observed using an optical microscope (CX-31, Olympus, Japan) and a scanning electron microscope (S-3400 N, Hitachi, Japan). Physiological and biochemical identification of the DS-R5 strain was carried out according to Bergey’s Manual of Systematic Bacteriology (second edition, 2004).
Molecular identification
Single bacterial colonies were selected and cultured in liquid LB medium at 30 °C with shaking at 160 rpm for 14–18 h to obtain a bacterial suspension. Genomic DNA was extracted using a bacterial genomic DNA extraction kit.
Amplification of the 16S rRNA gene: The 16S rRNA gene sequence was amplified by polymerase chain reaction (PCR) according to the protocol of Aravind et al. [20]. Using total DNA as the template, the 16S rRNA sequence of the DS-R5 strain was amplified with universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACT-3′). The PCR reaction contained the following: 12.5 μl 2 × Taq PCR Master Mix, 1.0 μl upstream and downstream primers (10 μmol/l), 0.5 μl DNA template and 10.0 μl ddH2O. The PCR reaction conditions were as follows: 95 °C for 5 min; 30 cycles of 94 °C for 1 min, 55–58 °C for 1 min and 72 °C for 90 s; and 72 °C for 10 min.
Amplification of the gyrB gene: The gyrB gene exists in most bacteria and, because it does not undergo frequent horizontal transfer, can be used to effectively distinguish between closely related species [21]. In order to clarify the species of the DS-R5 strain, gyrB gene cloning and sequencing were used for its identification. The primer pair gyrB-F (5′-GAAGTCATCATGACCGTTCTGCAYGCNGGNGGNAARTTYGA-3′) and gyrB-R (5′-AGCAGGGTACGGATGTGCGAGCCRTCNACRTCNGCRTCNGTCAT-3′) was used for amplification of the gyrB gene [22]. The PCR reaction contained the following: 12.5 μl 2 × Taq PCR Master Mix, 0.5 μl upstream and downstream primers (10 μmol/l), 1.0 μl DNA template and 10.5 μl ddH2O. The PCR reaction conditions were as follows: 94 °C for 5 min; 35 cycles at 94 °C for 30 s, 55–58 °C for 30 s and 72 °C for 1 min; and 72 °C for 10 min.
PCR products of the 16S rRNA and gyrB genes were sequenced by Qingdao Yixin Testing Technology Co., Ltd (Qingdao, China). BLAST comparisons were performed on the National Center for Biotechnology Information (NCBI) to select strains with high similarity. The neighbor-joining method was used to construct a phylogenetic tree in MEGA 7.0 software.
Determination of the antimicrobial spectrum of the DS-R5 strain
Antagonistic experiments were conducted with the DS-R5 strain against eight common pathogenic plant fungi. The tested fungi were prepared into a cake with a diameter of 5 mm and placed in the center of a PDA plate. Then, the DS-R5 strain was spread 2.5 cm away from the test pathogen cake. After 7 days of constant temperature culture at 30 °C, the width of the inhibition zone was determined.
Pot experiments of the DS-R5 strain against S. miltiorrhiza root rot
Sixty potted S. miltiorrhiza seedlings with good growth were selected and divided into three groups: a healthy control group (no inoculation), a pathogenic control group (inoculation with pathogenic fungi) and a treatment group (inoculation with pathogenic fungi and antagonistic bacteria), with 20 plants in each group. The pathogenic control group and the treatment group were irrigated with a 20-ml F. solani spore suspension with a concentration of 1 × 106 spores/ml. After 10 days, the roots of the pathogenic control group and the treatment group were irrigated with 20 ml DS-R5 strain fermentation broth. The group inoculated with 20 ml uninoculated sterile medium was used as a healthy control group. After 60 days, the control effect of the DS-R5 strain on S. miltiorrhiza root rot was analyzed statistically. According to the lesion area on the root tissue of S. miltiorrhiza seedlings, the disease was divided into five grades: grade 1, no lesions; grade 2, lesion area < 5%; grade 3, lesion area of 5–20%; grade 4, lesion area of 21 repeated streaked separation 50%; and grade 5, lesion area > 50%. The disease index was calculated as follows: disease index = Σ (disease grade × number of plants of this disease grade) / (maximum disease grade × total number of plants) × 100. The control efficacy was calculated as follows: control efficacy (%) = (disease index of control group—disease index of treatment group) / control disease index × 100.
Analytical methods for optimizing culture conditions
One‑factor‑at‑a‑time experimental design
The main factors affecting antifungal substance synthesis are medium volume, initial pH, inoculum size, fermentation time, rotary speed and fermentation temperature. Considering the culture conditions reported in previous studies, the ranges of values of the main factors in this study were set as follows: medium volume of 30, 60, 90, 120, 150 and 180 ml in 250 ml shaking flasks; initial pH of 4.0, 5.0, 6.0, 7.0, 8.0 and 9.0; inoculum size of 0.5, 1.0, 1.5, 2.0, 2.5 and 3.0%; fermentation time of 3, 4, 5, 6, 7 and 8 days; rotary speed of 0, 50, 100, 150, 200 and 250 rpm; and fermentation temperature of 20, 25, 30, 35, 40 and 45 °C.
RSM experimental design
PB design
The PB design is a two-level experimental design method that screens for important factors using the least number of experiments to accurately estimate the effect values of different factors [23]. Based on the data from one‑factor‑at‑a‑time experiments, a PB design with a set of 12 experiments was used to identify the significant factors affecting antifungal substances production, with the response value as the titer of the antifungal substances. The six parameters, A, B, C, D, E and F, represent medium volume, initial pH, inoculum size, fermentation time, rotary speed and fermentation temperature, respectively. The high (+ 1) and low (-1) levels of these factors are shown in Table 1, with the high level being approximately 1.5 times that of the low level. Regression analysis was carried out using Minitab 17.0 software (Minitab, LLC, State College, PA, USA) to assess statistical significance.
Table 1.
Factors and levels of PB design
| Factors | Level | |
|---|---|---|
| -1 | + 1 | |
| A: medium volume (ml) | 50 | 70 |
| B: initial pH | 5.0 | 7.0 |
| C: inoculum size (%) | 1.5 | 2.5 |
| D: fermentation time (d) | 6.0 | 8.0 |
| E: rotary speed (rpm) | 120 | 180 |
| F: fermentation temperature (°C) | 25 | 35 |
Steepest ascent design
After significant influencing factors were screened from PB design results, the gradient direction of the response value was taken as the climbing direction. According to the effect value of each factor, the change step size was determined to design steepest ascent experiments so that the response value quickly approached the maximum response region.
Central composite design (CCD)
CCD experiments were carried out using the three most important factors identified in PB experiments while fixing other non-critical factors. The factors and levels of CCD are shown in Table 2. The experimental data were fitted by quadratic regression to obtain a quadratic equation with interaction terms and square terms, and the main effect and interaction effect of each factor were analyzed. Finally, the optimal value was obtained within a certain level range. The commonly used second-order equation model of response surface analysis is as follows:
| 1 |
Table 2.
Factors and levels of CCD
| Factors | Level | ||||
|---|---|---|---|---|---|
| -1.682 | -1 | 0 | + 1 | + 1.682 | |
| A: medium volume (ml) | 43 | 50 | 60 | 70 | 77 |
| B: initial pH | 4.3 | 5.0 | 6.0 | 7.0 | 7.7 |
| F: fermentation temperature (°C) | 21.6 | 25 | 30 | 35 | 38.4 |
where Y refers to the response value, X1, X2 and X3 are the three most significant parameters identified by the PB design; α0 refers to the coefficient of the model; α1, α2 and α3 refer to the linear correlation coefficient; α12, α13 and α23 refer to the interaction coefficients of factors and α11; and α22 and α33 refer to the square coefficients of each factor.
Verification experiments
By fitting the experimental results of the optimization experiments with the RSM, each coefficient in the model was obtained by regression, and the extreme point and the value of the corresponding independent variable was determined by mathematical analysis of the multivariate function. Experiments were carried out according to the calculated parameters to verify the reliability of the model and determine the final optimization results.
Statistics and data analysis
The data obtained from experiments were processed using SPSS 19.0 statistical software (IBM Corp., Armonk, NY, USA), and Duncan’s new multiple range method was used to analyze the significance of differences between treatments.
Results
Isolation and screening of endophytic antagonistic bacteria from S. miltiorrhiza
A total of 72 strains of bacteria were isolated from the roots, stems and leaves of S. miltiorrhiza. Among them, 39 strains were isolated from the root tissue, accounting for 54.2% of the total isolated bacteria, and 24 and 9 strains were isolated from the stems and leaves, accounting for 33.3% and 12.5% of the total isolated bacteria, respectively. Confrontation tests showed that 13 of the 72 endophytic bacteria had varying degrees of antagonistic effects against the target pathogenic fungus F. solani (Table 3). Among them, the DS-R5 strain had the strongest inhibitory effect, with an inhibitory zone width of 20.5 mm. Therefore, the DS-R5 strain was chosen for further research.
Table 3.
Antagonistic effect of endophytic bacteria from S. miltiorrhiza against F. solani
| Source of strain | Strain code | Width of inhibitory zone (mm) |
|---|---|---|
| DS-R5 | 20.5 ± 1.9a | |
| DS-R12 | 13.4 ± 1.4d | |
| Root | DS-R18 | 16.8 ± 1.1b |
| DS-R25 | 9.2 ± 0.7e | |
| DS-R29 | 9.5 ± 0.9e | |
| DS-R31 | 15.9 ± 2.5c | |
|
DS-S4 DS-S6 DS-S7 DS-S11 DS-S14 |
14.7 ± 1.2b 18.1 ± 2.0b 7.4 ± 1.1j 7.9 ± 1.3i 12.6 ± 1.7 g |
|
|
DS-S6 DS-S7 |
18.1 ± 2.0a 7.4 ± 1.1j |
|
| Stem | DS-S7 | 7.4 ± 1.1d |
| DS-S11 |
7.9 ± 1.3d 12.6 ± 1.7 g |
|
| DS-S14 | 12.6 ± 1.7c | |
| Leaf |
DS-L3 DS-L5 |
14.8 ± 2.3a |
| DS-L5 | 12.1 ± 1.9b |
Note: Data are mean ± standard deviation. Different lowercase letters in the same column indicate significant differences at the P < 0.05 level by Duncan’s new multiple range test
Identification of the DS-R5 strain
Morphological characteristics
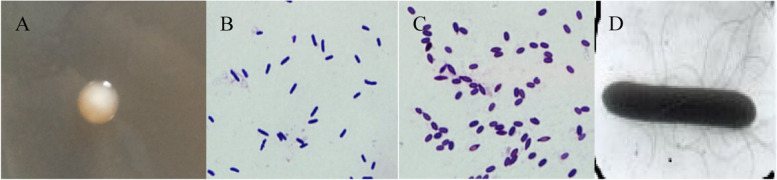
After the DS-R5 strain was cultured on LB medium for 36 h, the colonies were smooth, wet, round in shape and not easily agitated (Fig. 1A). Under the light microscope, the bacteria cells were rod-shaped and gram-positive with oval spores growing in the middle (Fig. 1B, C). Scanning electron microscopy showed that the size of the bacterial cells was about 0.8 µm × 2.8 µm and that there were peripheral flagella (Fig. 1D). Some physiological and biochemical characteristics of the DS-R5 strain are shown in Table 4. The DS-R5 strain can use glucose and glycerol to produce acid. Starch hydrolysis, gelatin hydrolysis, anaerobic growth, contact enzyme, nitrate, Voges-Proskauer (V-P) and hydrolyzed casein reactions were positive. Oxidase, citrate, succinate and H2S production reactions were negative. The strain did not grow on medium containing 5% NaCl. Overall, the experimental physiological and biochemical parameters of the DS-R5 strain were consistent with P. polymyxa [24].
Fig. 1.
Morphological characteristics of the DS-R5 strain
Table 4.
Physiological and biochemical properties of the DS-R5 strain
| Property | Result | Property | Result |
|---|---|---|---|
| Glucose | + | Amylo hydrolysis | + |
| Glycerol | + | Catalase | + |
| Oxidase | - | Casein hydrolysis | + |
| Succinate | - | V-P reaction | + |
| Anaerobic | + | Citrate utilization uuuutilization | - |
| Nitrate-reducing | + | H2S production | - |
| Isinglass Hydrolysis | + | Tolerance to NaCl | < 5% |
Note: “ + ” positive; “-” negative
Molecular identification of the DS-R5 strain
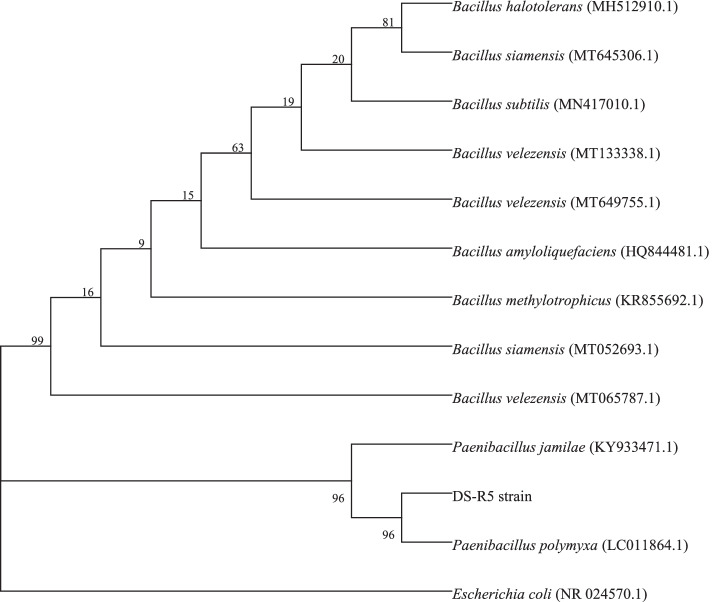
The 16S rRNA gene sequence of the DS-R5 strain was 1517 bp in length. After the sequence was submitted to NCBI, it was found by BLAST comparison that the 16S rRNA gene sequence of the DS-R5 strain was more than 99% similar to the gene sequences of P. polymyxa, Paenibacillus mucilaginosus, and B. subtilis. The phylogenetic tree showed that the DS-R5 strain and P. polymyxa belonged to the same branch with a support rate as high as 96% (Fig. 2). The phylogenetic tree based on the gyrB gene sequence showed that the DS-R5 strain and P. polymyxa belonged to the same branch and that the support rate was high (Fig. 3). Therefore, the DS-R5 strain was preliminarily identified as P. polymyxa. The combined morphological, physiological and biochemical characteristics and the results of gene sequence analysis of the 16S rRNA and gyrB genes confirmed that the DS-R5 strain was P. polymyxa.
Fig. 2.
The phylogenetic tree of the DS-R5 strain based on 16S rRNA gene sequencing
Fig. 3.
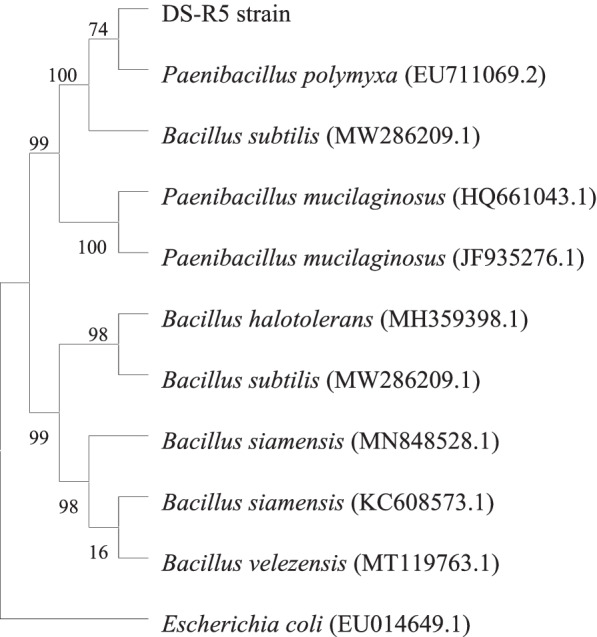
The phylogenetic tree of the DS-R5 strain based on gyrB gene sequencing
Antimicrobial spectrum of the DS-R5 strain
The DS-R5 strain had different degrees of inhibitory effects on eight common pathogenic plant fungi (Table 5). Its antifungal activity against R. solani and F. solani was strongest, with inhibition zone widths of 21.2 mm and 20.5 mm, respectively. Its antifungal activity against F. graminearum and F. pseudograminearum was weak, with inhibition zone widths of 7.9 and 6.5 mm, respectively. Tests showed that the DS-R5 strain had a broad antimicrobial spectrum.
Table 5.
Inhibitory effects of the DS-R5 strain on eight plant pathogens
| Pathogenic fungi | Width of inhibitory zone (mm) | Pathogenic fungi | Width of inhibitory zone (mm) |
|---|---|---|---|
| F. solani | 20.5 ± 1.9 b | F. oxysporum | 17.8 ± 2.6 c |
| R. solani | 21.2 ± 3.1 a | F. pseudograminearum | 6.5 ± 0.8 g |
| A. alternata | 15.9 ± 2.1 d | C. orbiculare | 13.1 ± 1.7 e |
| F. graminearum | 7.9 ± 1.1 f | B. ribis | 12.8 ± 1.3 e |
Note: Data are mean ± standard deviation. Different lowercase letters indicate significant differences at the P < 0.05 level by Duncan’s new multiple range test
Control efficacy of the DS-R5 strain against S. miltiorrhiza root rot
The results of pot experiments showed that the disease index of the healthy control group without inoculation was 6.7, the disease index of the pathogenic control group inoculated with pathogenic fungi was 60.5 and the disease index of the treatment group inoculated with pathogenic fungi and the DS-R5 strain was 26.3. These results show that inoculation with the antagonistic strain DS-R5 had a significant effect on S. miltiorrhiza root rot, with a control efficacy of 61.4% in the treatment group (Table 6).
Table 6.
Control efficacy of the DS-R5 strain against S. miltiorrhiza root rot
| Treatment | Disease index | Control efficacy (%) |
|---|---|---|
| Healthy control group | 6.7 ± 1.2 c | - |
| Pathogenic control group | 60.5 ± 7.2 a | - |
| Treatment group | 26.3 ± 5. 9b | 61.4 ± 8.1 |
Note: Different lowercase letters in the same column indicate significant differences at the P < 0.05 level by Duncan’s new multiple range test
Analytical results for fermentation condition optimization
Standard curve for antifungal substance bioassays
Bioassays were performed on a standard solution at five concentrations (10,000, 5000, 2500, 1250, 625, and 312.5 mg/l/), and the diameter of the zone of inhibition was recorded. Then, a standard curve was made with the logarithm of the concentration as the horizontal coordinate and the diameter of the zone of inhibition as the vertical coordinate (Fig. 4). It can be seen from the figure that the logarithm of the concentration had a linear relationship with the diameter of the inhibition zone, with a determination coefficient R2 = 0.9911, which meets the determination requirements.
Fig. 4.
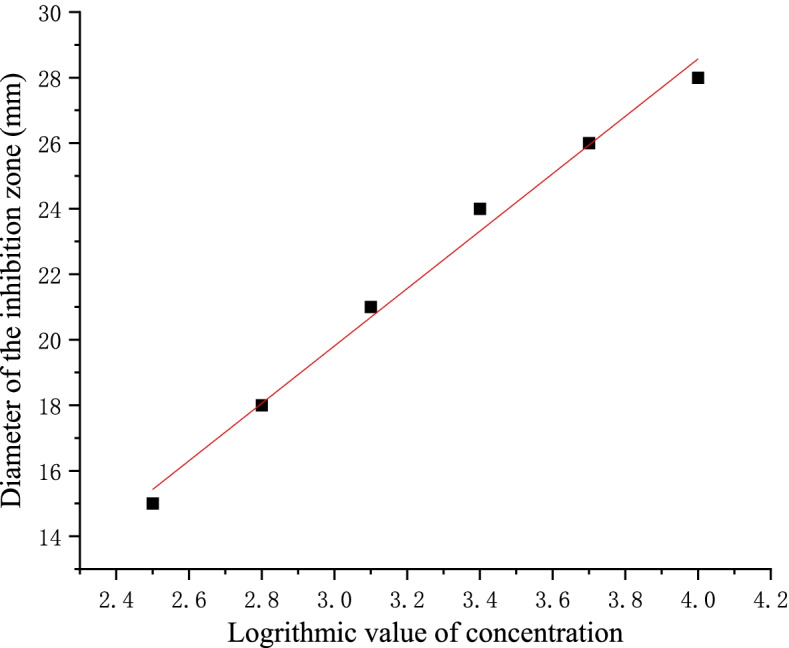
Standard curve for measuring antifungal substance titer
One‑factor‑at‑a‑time experimental design
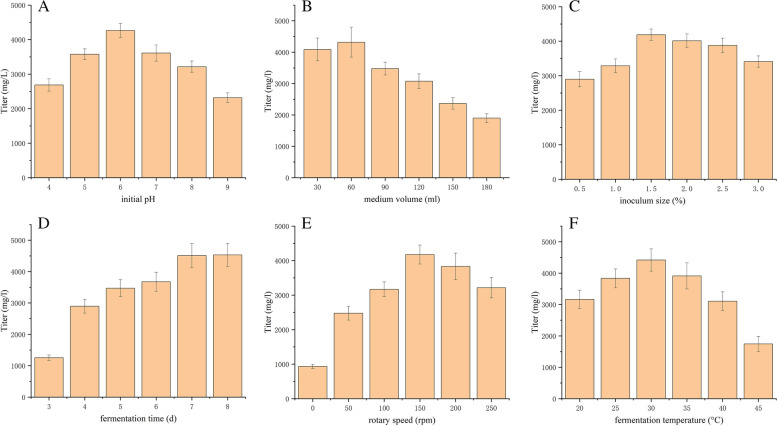
One‑factor‑at‑a‑time experiments were conducted to investigate the effect of different initial pH values, medium volumes, fermentation times, inoculum sizes, rotary speeds and fermentation temperatures on antifungal substance titers of the DS-R5 strain. Figure 5A shows the effect of medium volume in 250-ml shaking flasks on antifungal substance titers of the DS-R5 strain. The antifungal substance titer was highest when the medium volume was 50 ml, but the antifungal substance titer decreased rapidly when the medium volume was higher than 50 ml. It can be seen from Fig. 5B that the initial pH of the fermentation medium had great influence on the antifungal substance titer. When the initial pH was 4.0–9.0, the antifungal substance titer first increased and then decreased as the pH value increased. When the initial pH was 6.0, the antifungal substance titer increased significantly and reached its maximum (4322 mg/l). This indicated that the antifungal substance titer could be inhibited under conditions of excessive acidity or alkalinity. Therefore, pH 6.0 was selected as the optimal initial pH of the medium. The effect of inoculum size on the antifungal substance titer of the DS-R5 strain is shown in Fig. 5C. As inoculum size increased, the antifungal substance titer increased. At 2.0%, the antifungal substance titer reached its highest value at 4261 mg/ml. When the inoculum size was greater than 2.0%, the titer gradually decreased. Therefore, the optimal inoculum size was determined to be 2.0%. As fermentation time increased, the antifungal substance titer first increased and then tended to be stable (Fig. 5D). The titer was highest at a fermentation time of 7 days (4317 mg/ml). Therefore, the optimal fermentation time was determined to be 7 days. When the rotary speed was increased from 0 to 150 rpm, the antifungal substance titer increased. However, when the rotary speed exceeded 150 rpm, the titer began to decrease (Fig. 5E). Therefore, 150 rpm was selected as the optimal rotary speed. The effect of fermentation temperature on the antifungal substance titer of the DS-R5 strain is shown in Fig. 5F. In the range of 20–30 °C, the titer increased with increasing temperature. The titer was highest at 30 °C, then decreased rapidly at fermentation temperatures higher than 30 °C. Therefore, the fermentation temperature was set at 30 °C.
Fig. 5.
Results of one‑factor‑at‑a‑time experiments. Effect of (A) initial pH, (B) medium volume, (C) fermentation time, (D) inoculum size, (E) rotary speed and (F) fermentation temperature on antifungal substance titer
RSM experimental design results
Optimization of culture conditions by PB design
PB experimental design and response values are shown in Table 7. Regression analysis of each response value in Table 6 was carried out using Minitab 17.0 software to obtain the partial regression coefficient of each factor and analyze its significance. The effect size and significance of each factor after Minitab software analysis are shown in Table 8. The results of model analysis showed that the P value of the model was less than 0.0001, indicating that the regression equation was extremely significant. The model fit well in the entire regression region under study; the multiple correlation coefficient R2 = 0.9553 indicated good correlation, and the corrected coefficient of determination Rarj2 = 0.9016 indicated that 90.16% of the variability in the experimental data could be explained by this regression model. The significance order of the six culture condition factors on the antifungal substance titer was as follows: medium volume > fermentation temperature > initial pH > inoculum size > rotary speed > fermentation time, of which medium volume, initial pH and fermentation temperature were significant factors. The regression equation derived from the above PB design was as follows: . Initial pH, rotary speed and fermentation temperature had positive effects, whereas medium volume, inoculum size and fermentation time had negative effects. Based on the above analysis, medium volume (A), initial pH (B) and fermentation temperature (F) were selected for further optimization.
Table 7.
Design and results of PB experiments
| Runs | A | B | C | D | E | F | Titer (mg/l) | |
|---|---|---|---|---|---|---|---|---|
| Experimental | Predicted | |||||||
| 1 | 40(-1) | 7.0(+ 1) | 1.5(-1) | 6(-1) | 120(-1) | 35.0(+ 1) | 4200 | 4237 |
| 2 | 60(+ 1) | 5.0(-1) | 1.5(-1) | 6(-1) | 180(+ 1) | 35.0(+ 1) | 3048 | 2986 |
| 3 | 40(-1) | 5.0(-1) | 1.5(-1) | 6(-1) | 120(-1) | 25.0(-1) | 3133 | 3061 |
| 4 | 60 (+ 1) | 5.0(-1) | 2.5(+ 1) | 6(-1) | 120(-1) | 25.0(-1) | 2333 | 2380 |
| 5 | 60 (+ 1) | 5.0(-1) | 2.5(+ 1) | 8(+ 1) | 120(-1) | 35.0(+ 1) | 2667 | 2699 |
| 6 | 40 (-1) | 5.0(-1) | 2.5(+ 1) | 8(+ 1) | 180(+ 1) | 25.0(-1) | 3267 | 3188 |
| 7 | 60 (+ 1) | 7.0(+ 1) | 1.5(-1) | 8(+ 1) | 120(-1) | 25.0(-1) | 2600 | 2512 |
| 8 | 40 (-1) | 7.0(+ 1) | 2.5(+ 1) | 8(+ 1) | 120(-1) | 35.0(+ 1) | 3867 | 3926 |
| 9 | 60 (+ 1) | 7.0(+ 1) | 2.5(+ 1) | 6(-1) | 180(+ 1) | 35.0(+ 1) | 3200 | 3278 |
| 10 | 60 (+ 1) | 7.0(+ 1) | 1.5(-1) | 8(+ 1) | 180(+ 1) | 25.0(-1) | 3000 | 2943 |
| 11 | 40 (-1) | 5.0(-1) | 1.5(-1) | 8(+ 1) | 180(+ 1) | 35.0(+ 1) | 3667 | 3602 |
| 12 | 40 (-1) | 7.0(+ 1) | 2.5(+ 1) | 6(-1) | 180(+ 1) | 25.0(-1) | 3333 | 3302 |
Table 8.
Analysis of data generated using PB design
| Coded variable | Variable | Coefficient estimate | T-value | P-value | Significance |
|---|---|---|---|---|---|
| Intercept | Constant | 3192.9 | 65.93 | 0.000 | *** |
| A | medium volume | -384.9 | -7.85 | 0.001 | *** |
| B | initial pH | 173.8 | 3.59 | 0.016 | * |
| C | inoculum size | -81.8 | -1.69 | 0.152 | |
| D | fermentation time | -14.9 | -0.31 | 0.771 | |
| E | rotary speed | 59.6 | 1.23 | 0.273 | |
| F | fermentation temperature | 248.6 | 5.13 | 0.004 | ** |
Note:
*Significant at the P < 0.05 level
**Significant at the P < 0.01 level
*** Significant at the P < 0.001 level
Steepest ascent design
The coefficient estimates for medium volume, initial pH and fermentation temperature presented in Table 8 indicated that initial pH and fermentation temperature had positive effects, whereas medium volume had negative effects on the titer of antifungal substance. Based on the above results, the directions of the gradients for medium volume, initial pH and fermentation temperature were determined using steepest ascent design (Table 9). As shown in Table 9, the path of steepest ascent started from the center of the variables chosen from the PB design and moved along the path, with medium volume, initial pH and fermentation temperature moving by 5, 0.5 and 1.0, respectively. The maximum titer was obtained in test 3, when the point was nearest the maximum titer response. Therefore, this point (medium volume 50 ml, initial pH 7.0, fermentation temperature 32 °C) was chosen as the center point for further optimization by CCD.
Table 9.
Steepest ascent path experimental design and results
| Test | medium volume (ml) | initial pH | fermentation temperature (°C) | Titer (mg/l) |
|---|---|---|---|---|
| 1 | 60 | 6.0 | 30.0 | 6363 |
| 2 | 55 | 6.5 | 31.0 | 6889 |
| 3 | 50 | 7.0 | 32.0 | 7467 |
| 4 | 45 | 7.5 | 33.0 | 7148 |
| 5 | 40 | 8.0 | 34.0 | 6422 |
| 6 | 35 | 8.5 | 35.0 | 5012 |
| 7 | 30 | 9.0 | 36.0 | 4566 |
Optimization of significant variables using CCD
To find the optimal culture conditions for antifungal substance production, CCD with five coded levels was used to further optimize conditions with regard to medium volume, initial pH and fermentation temperature. The experimental design and response values from CCD are shown in Table 10. A quadratic regression equation with the titer of antifungal substance as the objective function was established, and variance analysis and significance tests were performed on the obtained regression equation. After regression analysis of the experimental data, the quadratic polynomial equation was obtained as follows:
Table 10.
Experimental design and response values from CCD
| Run | Factors | Titer (mg/l) | |||
|---|---|---|---|---|---|
| A:medium volume (ml) | B:initial pH | F:fermentation temperature (°C) | Experimantal | Predicted | |
| 1 | + 1.682(58) | 0(7.0) | 0(32) | 6199 | 6235 |
| 2 | 0(50) | 0(7.0) | 0(32) | 7470 | 7398 |
| 3 | + 1(55) | -1(6.5) | + 1(33) | 5990 | 6026 |
| 4 | 0(50) | 0(7.0) | 0(32) | 7691 | 7639 |
| 5 | + 1(55) | -1(6.5) | -1(31) | 5782 | 5814 |
| 6 | -1(45) | -1(6.5) | -1(31) | 5733 | 5688 |
| 7 | -1.682(42) | 0(7.0) | 0(32) | 5720 | 5673 |
| 8 | 0(50) | -1.682(6.2) | 0(32) | 6132 | 6185 |
| 9 | 0(50) | 0(7.0) | -1.682(30.3) | 5619 | 5725 |
| 10 | + 1(55) | + 1(7.5) | -1(31) | 6353 | 6279 |
| 11 | -1(45) | + 1(7.5) | -1(31) | 5561 | 5607 |
| 12 | + 1(55) | + 1(7.5) | + 1(33) | 6899 | 6823 |
| 13 | -1(45) | + 1(7.5) | + 1(33) | 5886 | 5936 |
| 14 | 0(50) | + 1.682(7.8) | 0(32) | 6341 | 6281 |
| 15 | 0(50) | 0(7.0) | + 1.682(33.7) | 6058 | 6090 |
| 16 | 0(50) | 0(7.0) | 0(32) | 7900 | 7983 |
| 17 | 0(50) | 0(7.0) | 0(32) | 7863 | 7793 |
| 18 | -1(45) | -1(6.5) | + 1(33) | 5659 | 5588 |
| 19 | 0(50) | 0(7.0) | 0(32) | 7893 | 7953 |
| 20 | 0(50) | 0(7.0) | 0(32) | 7937 | 8025 |
where Y is the titer of antifungal substances and A, B and F are the coded factors medium volume, initial pH and fermentation temperature, respectively. Standard variance analysis of the regression model is shown in Table 11. The regression model had a P value < 0.0001, revealing that the model was statistically significant. The lack of fit was not significant, indicating that any inability to fit caused by experimental error could be ignored. The model terms A, B, F, AB, A2, B2 and F2 were found to be significant, which revealed that medium volume, initial pH, fermentation temperature and the interaction of medium volume with initial pH were significant. The correlation coefficient of the regression equation was R2 = 0.9810, indicating that the obtained equation had good fit and that this equation could be used to predict test results.
Table 11.
ANOVA analysis for CCD regression equations
| Source | Sum of squares | Df | Mean square | F-value | P-value |
|---|---|---|---|---|---|
| Model | 15206097 | 9 | 1689566 | 57.29 | 0.000*** |
| A | 654877 | 1 | 654877 | 22.21 | 0.001** |
| B | 260592 | 1 | 260592 | 8.84 | 0.014* |
| F | 222534 | 1 | 222534 | 7.55 | 0.021* |
| A2 | 5742543 | 1 | 5742543 | 194.73 | 0.000*** |
| B2 | 4098925 | 1 | 4098925 | 139.00 | 0.000*** |
| F2 | 6547266 | 1 | 6547266 | 222.02 | 0.000*** |
| AB | 253828 | 1 | 253828 | 8.61 | 0.015* |
| AF | 31626 | 1 | 31626 | 1.07 | 0.325 |
| BF | 67896 | 1 | 67896 | 2.30 | 0.160 |
| Residual | 294893 | 10 | 29489 | ||
| Lack of Fit | 133078 | 5 | 26616 | 0.82 | 0.582 |
| Pure error | 161815 | 5 | 32363 | ||
| Cor total | 1915500990 | 19 |
R2 = 0.9810, adjusted R2 = 0.9639, predicted R2 = 0.9186
Note:
*Significant at the P < 0.05 level
**Significant at the P < 0.01 level
*** Significant at the P < 0.001 level
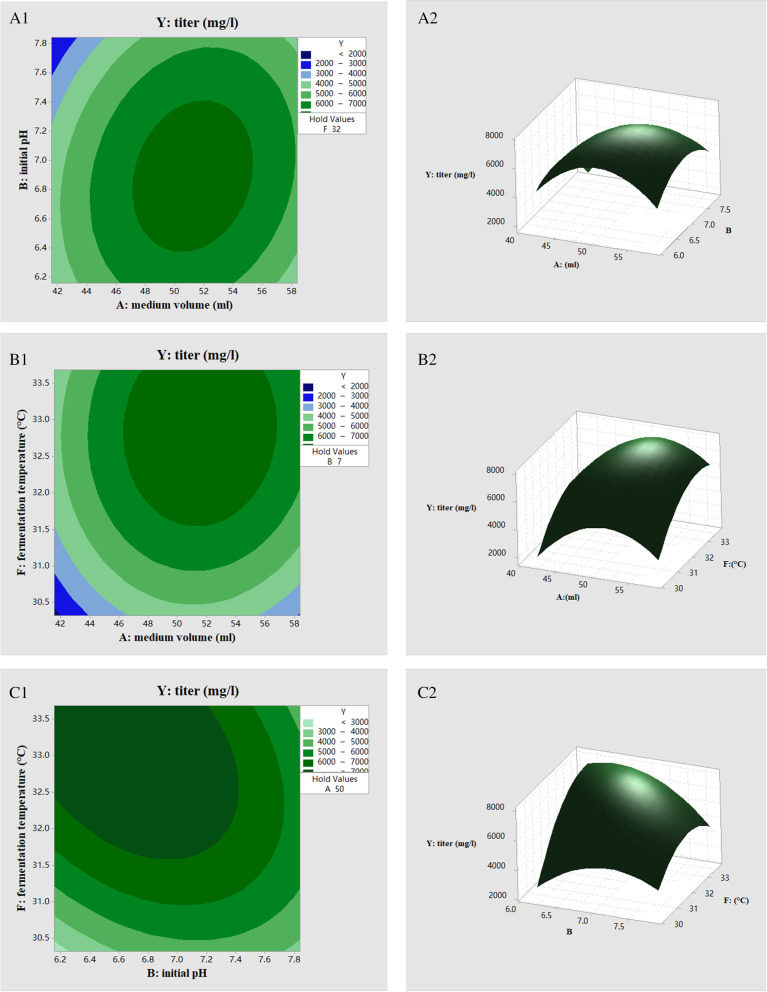
According to the obtained quadratic regression equation and variance analysis results, the contour plot and corresponding response surface curve was drawn using Minitab software to more intuitively observe the effect of the interactions between factors on the titer of antifungal substance (Fig. 6). When one factor was fixed at the optimal value, the titer of antifungal substance first increased then decreased as the other two factors increased until the vertex of the curved surface reached the maximum titer point. In the contour plot map, closer circles indicate less significant interactions between the two factors, whereas an ellipse represents a significant interaction [25]. The contour plot maps showed that the response surface curve of the interaction between medium volume and initial pH was the most curved, indicating that the interaction between these two factors was stronger than the interactions between other factors, which was also consistent with the results of variance analysis. In order to determine the optimal values of the main influencing culture condition factors, the derivation of three independent variables of the regression equation was obtained. With the derivative equal to 0, the extreme point of the model was obtained as follows: medium volume, 51.0 ml; initial pH, 6.7; fermentation temperature, 33.1 °C. According to PB design results, the optimal values of other culture conditions were as follows: inoculum size, 1.5%; fermentation time, 6.0 days; rotary speed, 180 rpm. Under these optimal conditions, the model predicted a maximum antifungal substance titer of 8036 mg/l, which was a 77.6% increase compared to the original culture conditions (4357 mg/l). In order to verify the feasibility of RSM, the optimal culture conditions obtained by regression analysis were verified. The validation test was repeated three times, and the average value was 4295 mg/l, which was not significantly different from the predicted value. The experimental results showing that the validation value was close to the predicted value indicated that the model reflects the actual fermentation situation and can be used to predict the relationship between the antifungal substance titer and fermentation conditions.
Fig. 6.
Response surface curve and corresponding contour plot of the effect of three variables on antifungal substance titer. A interaction of medium volume and initial pH; B interaction of medium volume and fermentation temperature; C interaction of initial pH and fermentation temperature
Discussion
Although 16S rRNA gene sequences are widely used to identify bacteria and construct phylogenetic relationships between bacteria, most 16S rRNA gene-based phylogenetic tree topologies are not reliable due to high similarity between sequences among taxa with close kinship [26]. In recent years, it has been found that many protein-coding genes can also be used as markers for phylogenetic identification. These include the gyrA, gyrB and ropD genes, and their use can compensate for the deficiencies of the 16S rRNA gene [27]. Among these protein-coding genes, the molecular evolution rate of the gyrB gene is higher than that of the 16S rRNA gene, so it is often used for the identification of closely related species [28]. In this study, the DS-R5 strain was identified as P. polymyxa by colony morphology and cellular, physiological and biochemical characteristics combined with 16S rRNA and gyrB gene sequence analysis.
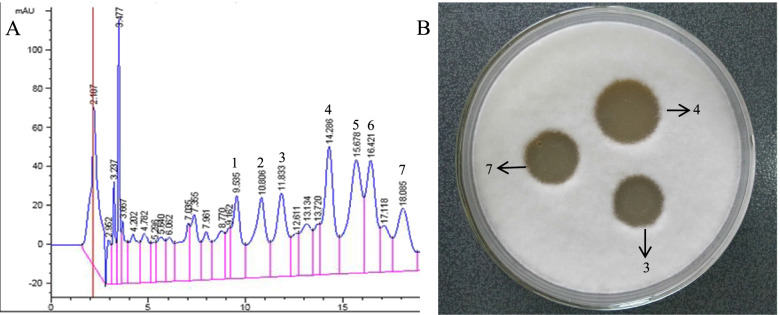
In our previous stuy, the antifungal substances produced by the DS-R5 strain were purified by using hydrochloric acid precipitation, methanol extraction and molecular sieve, and then freeze-dried to obtain the crude active substances. The crude is further separated by high performance liquid chromatography (HPLC) under the following conditions: Ultimate XB-C18 semi-preparative column (21.2 × 250 mm, 5 µm), flow rate 5.0 ml/min, column temperature 25 °C, injection volume 1 ml, mobile phase ratio: acetonitrile: water (55:45, adding 0.1% trifluoroacetic acid). The results of HPLC showed that seven main substances were isolated from the fermentation broth of the DS-R5 strain (Fig. 7A). Antifungal activity tests revealed that three of these seven substances had antifungal activity (Fig. 7B). These experiments also showed that the yields of the three antifungal substances were not all proportional to the results of culture condition optimization. Thus, we could not use the yield of one of the antifungal substances as the response variable for optimizing culture conditions. Therefore, when we carried out RSM to optimize the culture conditions, we selected the total antifungal effect of these three antifungal substances as the response variable. Generally speaking, the evaluation of the antimicrobial effect of biocontrol strains is based on the amount of a specific antimicrobial substance or the width of the inhibition zone. Zhao et al. optimized the fermentation conditions of the Bacillus sp. BH072 strain by RSM using the amount of antifungal substance iturin A as the optimization response variable [29]. Ju et al. optimized the nutritional requirements for antimicrobial activity of Streptomyces rimosus AG-P1441 using the width of the inhibition zone as optimization response variable [30]. In this study, we use the titer of the fermentation broth of the DS-R5 strain as the optimization response variable. Since the titer of the fermentation broth is proportional to the diameter of the inhibition zone, in essence, we also use the the diameter of inhibition zone as the response variable in the optimization process.
Fig. 7.
The HPLC and the antifungal activity result of the isolated substances
The production of antifungal substances by many biocontrol bacteria is generally believed to be an aerobic process [31], so dissolved oxygen is an important factor in the fermentation of P. polymyxa. In shake flasks, oxygen supply is related to the volume of media in the flask and the rotary speed. Decreasing the medium volume or increasing the rotary speed can improve dissolved oxygen levels in the medium. However, too little media or too high rotary speeds are also not conducive to the production of antifungal substances. If the amount of medium in the flask is too small, increased evaporation of the solution during the fermentation process can lead to large errors in the results. Similarly, once the rotary speed of the shaker reaches a certain value, the amount of dissolved oxygen will not increase further because of the upper limit of the dissolved oxygen concentration in water. In this study, medium volume was found to be the most important variable influencing the titer of antifungal substances of P. polymyxa DS-R5 at an individual level and showed significant influence at an interactive level. The titer of antifungal substances increased as medium volume in the flask decreased, indicating that higher dissolved oxygen levels are beneficial to antifungal substance production. The rotary speed of the shaker had a positive effect on the titer of antifungal substances, indicating that an appropriate increase in rotary speed would increase the titer of antifungal substances. However, the effect of rotary speed on the titer of antifungal substances was not significant, indicating that the range of rotary speeds we selected in PB design experiments had reached the upper limit and could not be further increased.
P. polymyxa can produce a variety of antifungal substances, such as peptides, proteins, nucleosides, pyrazines and phenols, and can prevent and control a variety of plant diseases [13]. The production of Bacillus secondary substances is affected by the composition of culture medium and fermentation conditions. Different medium components and fermentation conditions can significantly affect the growth of strains and the production of metabolites. There have been many previous RSM optimization studies on the antagonistic effect of antifungal substances. Zhao et al. demonstrated that the antifungal apecific activity of the Bacillus sp. BH072 strain was enhanced from 350.11 AU/mg to 513.92 AU/mg under the optimized medium and culture conditions, which was 46.8% higher than the previous result [29]. Wang et al. optimized the culture conditions of the P. polymyxa Cp-S316 strain by using ractional factorial design and central composite designs. Anfungal substances production could reach 4687.71 μg/ml, which was 3.05 times higher than that without optimization [32]. In this study, the antifungal substances production can reach 8036 mg/l after optimization by one‑factor‑at‑a‑time experimental design and CCD, which was a 77.6% increase compared to the original culture conditions. The optimization results indicate that RSM can be well used to optimize the culture conditions for antifungal substance and provides an important experimental basis for the industrial-scale production of the DS-R5 strain.
This study also found that both live DS-R5 bacteria and the antifungal substances produced in the fermentation broth could inhibit the growth of F. solani (Fig. 8), thereby allowing for biological control of S. miltiorrhiza root rot. Regarding the future application and development of S. miltiorrhiza root rot control measures, we recommend the use of live bacteria because the antifungal substances extraction process is cumbersome. In addition, the application of spores produced by the DS-R5 strain is also recommended because isolation of antifungal substances produced by the DS-R5 strain can be performed rapidly. This study isolating P. polymyxa DS-R5 from S. miltiorrhiza lays a strong foundation for the biological control of S. miltiorrhiza root rot and provides a basis for future studies clarifying the effect of P. polymyxa DS-R5 on S. miltiorrhiza root rot. This study provides theoretical guidance for the development and application of biocontrol strains for S. miltiorrhiza root rot and provides functional strains and effective metabolites for the development of new drugs.
Fig. 8.
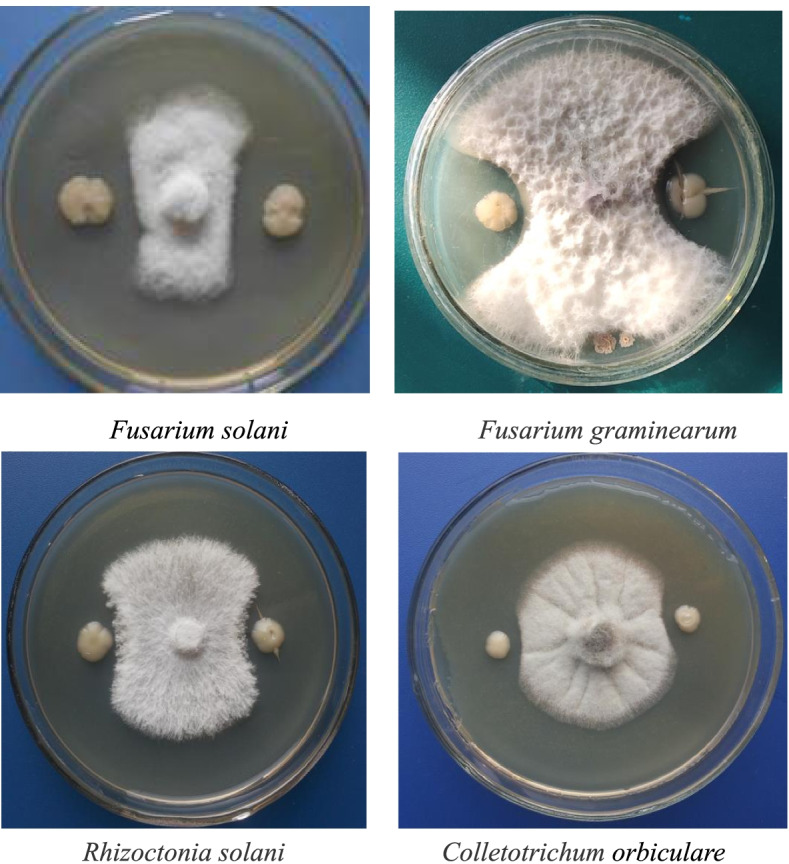
Antagonistic effect of DS-R5 strain on some pathogenic fungi
Conclusion
In this work we studied the isolation and screening of biocontrol bacteria strains that prevent and control S. miltiorrhiza root rot. A total of 72 strains of bacteria were isolated from the roots, stems and leaves of S. miltiorrhiza. Among the 72 isolates obtained from root, stem and leaf tissue, the DS-R5 strain had the strongest inhibitory effect against the target pathogenic fungus F. solani. The DS-R5 strain was identified as P. polymyxa by colony morphology and cellular, physiological and biochemical characteristics combined with 16S rRNA and gyrB gene sequence analysis. In addition, using one‑factor‑at‑a‑time experimental design, PB design and CCD, the fermentation conditions for production of antifungal substances by P. polymyxa DS-R5 in a shake flask system were optimized. The optimized fermentation conditions were as follows: medium volume, 51.0 ml; initial pH, 6.7; fermentation temperature, 33.1 °C; inoculum size, 1.5%; fermentation time, 6.0 days and rotary speed 180 rpm. Significantly higher antifungal substance titers (8036 mg/l) were obtained under the optimized fermentation conditions than under original fermentation conditions. To the best of our knowledge, this study is the first to isolate and purify biocontrol bacteria that prevent and control S. miltiorrhiza root rot and to optimize culture conditions for biocontrol bacteria to produce antifungal substances using PB design and CCD experiments.
Acknowledgements
We thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
Authors’ contributions
MS conceived the study. RSa designed and supervised all of the experimental works. SH, DH, ML, YY, and RSh carried out the experiments. MS analyzed the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by Shandong traditional Chinese medicine science and technology development plan project (2019–0347).
Availability of data and materials
All data and material are available upon request to correspondence author.
Declarations
Ethics approval and consent to participate
It is not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Guo Y, Li Y, Xue L, Severino RP, Gao S, Niu J, Qin LP, Zhang D, Bromme D. Salvia miltiorrhiza: an ancient Chinese herbal medicine as a source for anti-osteoporotic drugs. J Ethnopharmacol. 2014;155(3):1401–1416. doi: 10.1016/j.jep.2014.07.058. [DOI] [PubMed] [Google Scholar]
- 2.Hu J, Zhao M, Hou Z, Shang J. The complete chloroplast genome sequence of Salvia miltiorrhiza, a medicinal plant for preventing and treating vascular dementia. Mitochondrial DNA B Resour. 2020;5(3):2460–2462. doi: 10.1080/23802359.2020.1778574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L, Ma R, Liu C, Liu H, Zhu R, Guo S, Tang M, Li Y, Niu J, Fu M, et al. Salvia miltiorrhiza: A Potential Red Light to the Development of Cardiovascular Diseases. Curr Pharm Des. 2017;23(7):1077–1097. doi: 10.2174/1381612822666161010105242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang L, Miao ZQ, Yang G, Shao AJ, Huang LQ, Shen Y, Wang X, Chen ML. Research wilt disease of Salvia miltiorrhiza and its pathogen. Zhongguo Zhong Yao Za Zhi. 2013;38(23):4040–4043. [PubMed] [Google Scholar]
- 5.Wang TL, Guan W, Sun K, Wang S, Guo LP. Progress in researches on pathogens, epidemiology and integrated control of diseases on Salvia miltiorrhiza in China. Zhongguo Zhong Yao Za Zhi. 2018;43(11):2402–2406. doi: 10.19540/j.cnki.cjcmm.20180329.001. [DOI] [PubMed] [Google Scholar]
- 6.Ngo MT, Van Nguyen M, Han JW, Kim B, Kim YK, Park MS, Kim H, Choi GJ. Biocontrol Potential of Aspergillus Species Producing Antimicrobial Metabolites. Front Microbiol. 2021;12:804333. doi: 10.3389/fmicb.2021.804333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong F, Zhang XH, Li YH, Wang JF, Zhang SS, Hu XF, Chen JS. Characterization of the endophytic antagonist pY11T-3-1 against bacterial soft rot of Pinellia ternata. Lett Appl Microbiol. 2010;50(6):611–617. doi: 10.1111/j.1472-765X.2010.02841.x. [DOI] [PubMed] [Google Scholar]
- 8.Park YH, Chandra Mishra R, Yoon S, Kim H, Park C, Seo ST, Bae H. Endophytic Trichoderma citrinoviride isolated from mountain-cultivated ginseng (Panax ginseng) has great potential as a biocontrol agent against ginseng pathogens. J Ginseng Res. 2019;43(3):408–420. doi: 10.1016/j.jgr.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yue HM, Wang M, Gong WF, Zhang LQ. The screening and identification of the biological control fungi chaetomium spp. against wheat common root rot. FEMS Microbiol Lett. 2018;365(22):1–6. [DOI] [PubMed]
- 10.Etminani F, Harighi B. Isolation and Identification of Endophytic Bacteria with Plant Growth Promoting Activity and Biocontrol Potential from Wild Pistachio Trees. Plant Pathol J. 2018;34(3):208–217. doi: 10.5423/PPJ.OA.07.2017.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rana KL, Kour D, Kaur T, Devi R, Yadav AN, Yadav N, Dhaliwal HS, Saxena AK. Endophytic microbes: biodiversity, plant growth-promoting mechanisms and potential applications for agricultural sustainability. Antonie Van Leeuwenhoek. 2020;113(8):1075–1107. doi: 10.1007/s10482-020-01429-y. [DOI] [PubMed] [Google Scholar]
- 12.Khan MS, Gao J, Chen X, Zhang M, Yang F, Du Y, Moe TS, Munir I, Xue J, Zhang X. Isolation and Characterization of Plant Growth-Promoting Endophytic Bacteria Paenibacillus polymyxa SK1 from Lilium lancifolium. Biomed Res Int. 2020;2020:8650957. doi: 10.1155/2020/8650957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beatty PH, Jensen SE. Paenibacillus polymyxa produces fusaricidin-type antifungal antibiotics active against Leptosphaeria maculans, the causative agent of blackleg disease of canola. Can J Microbiol. 2002;48(2):159–169. doi: 10.1139/w02-002. [DOI] [PubMed] [Google Scholar]
- 14.Grady EN, MacDonald J, Liu L, Richman A, Yuan ZC. Current knowledge and perspectives of Paenibacillus: a review. Microb Cell Fact. 2016;15(1):203. doi: 10.1186/s12934-016-0603-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhai LX, Guo-Li LI, Leng Y, Shi-Weng LI, Chen XM, Chen T, Liu GX. High density fermentation of recombinant Kluyveromyces lactis producing antimicrobial peptide PSI and purification of its products. Food Ferment Industries 2018;4:211–8.
- 16.Yun TY, Feng RJ, Zhou DB, Pan YY, Chen YF, Wang F, Yin LY, Zhang YD, Xie JH. Optimization of fermentation conditions through response surface methodology for enhanced antibacterial metabolite production by Streptomyces sp. 1–14 from cassava rhizosphere. PLoS One. 2018;13(11):e0206497. doi: 10.1371/journal.pone.0206497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X, Guo P, Li M, Li H, Hu Z, Liu X, Zhang Q. Optimization of Culture Conditions for Amoxicillin Degrading Bacteria Screened from Pig Manure. Int J Environ Res Public Health. 2020;17(6):1973. doi: 10.3390/ijerph17061973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C, Xu H, Zhang Y, Wu S, Chen D, Qian G, Hu B, Fan J. Optimization of culture conditions for promoting heat-stable antifungal factor production level in Lysobacter enzymogenes. FEMS Microbiol Lett. 2019;366(17):fnz007. doi: 10.1093/femsle/fnz007. [DOI] [PubMed] [Google Scholar]
- 19.Khalil MMR, Fierro-Coronado RA, Penuelas-Rubio O, Villa-Lerma AG, Plascencia-Jatomea R, Felix-Gastelum R, Maldonado-Mendoza IE. Rhizospheric bacteria as potential biocontrol agents against Fusarium wilt and crown and root rot diseases in tomato. Saudi J Biol Sci. 2021;28(12):7460–7471. doi: 10.1016/j.sjbs.2021.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aravind R, Kumar A, Eapen SJ, Ramana KV. Endophytic bacterial flora in root and stem tissues of black pepper (Piper nigrum L.) genotype: isolation, identification and evaluation against Phytophthora capsici. Lett Appl Microbiol. 2009;48(1):58–64. doi: 10.1111/j.1472-765X.2008.02486.x. [DOI] [PubMed] [Google Scholar]
- 21.Jiang F, Huang H, Yang N, Feng H, Li Y, Han B. Isolation, identification, and biological control in vitro of tail rot pathogen strain from Hippocampus kuda. PLoS One. 2020;15(4):e0232162. doi: 10.1371/journal.pone.0232162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.La Duc MT, Satomi M, Agata N, Venkateswaran K. gyrB as a phylogenetic discriminator for members of the Bacillus anthracis-cereus-thuringiensis group. J Microbiol Methods. 2004;56(3):383–394. doi: 10.1016/j.mimet.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 23.Pal D, Patel G, Dobariya P, Nile SH, Pande AH, Banerjee UC. Optimization of medium composition to increase the expression of recombinant human interferon-beta using the Plackett-Burman and central composite design in E. coli SE1. 3 Biotech. 2021;11(5):226. doi: 10.1007/s13205-021-02772-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu F, Sun L, Lu Z, Bie X, Fang Y, Liu S. Isolation and identification of an endophytic strain EJS-3 producing novel fibrinolytic enzymes. Curr Microbiol. 2007;54(6):435–439. doi: 10.1007/s00284-006-0591-7. [DOI] [PubMed] [Google Scholar]
- 25.Jiapeng T, Yiting L, Li Z. Optimization of fermentation conditions and purification of cordycepin from Cordyceps militaris. Prep Biochem Biotechnol. 2014;44(1):90–106. doi: 10.1080/10826068.2013.833111. [DOI] [PubMed] [Google Scholar]
- 26.Savi DC, Aluizio R, Galli-Terasawa L, Kava V, Glienke C. 16S-gyrB-rpoB multilocus sequence analysis for species identification in the genus Microbispora. Antonie Van Leeuwenhoek. 2016;109(6):801–815. doi: 10.1007/s10482-016-0680-y. [DOI] [PubMed] [Google Scholar]
- 27.Liu G, Lin X, Xu S, Liu G, Liu F, Mu W. Screening, identification and application of soil bacteria with nematicidal activity against root-knot nematode (Meloidogyne incognita) on tomato. Pest Manag Sci. 2020;76(6):2217–2224. doi: 10.1002/ps.5759. [DOI] [PubMed] [Google Scholar]
- 28.Wang LT, Lee FL, Tai CJ, Kasai H. Comparison of gyrB gene sequences, 16S rRNA gene sequences and DNA-DNA hybridization in the Bacillus subtilis group. Int J Syst Evol Microbiol. 2007;57(Pt 8):1846–1850. doi: 10.1099/ijs.0.64685-0. [DOI] [PubMed] [Google Scholar]
- 29.Zhao X, Han Y, Tan XQ, Wang J, Zhou ZJ. Optimization of antifungal lipopeptide production from Bacillus sp. BH072 by response surface methodology. J Microbiol. 2014;52(4):324–332. doi: 10.1007/s12275-014-3354-3. [DOI] [PubMed] [Google Scholar]
- 30.Ju Y, Son KH, Jin C, Hwang BS, Park DJ, Kim CJ. Statistical optimization of culture medium for improved production of antimicrobial compound by Streptomyces rimosus AG-P1441. Food Sci Biotechnol. 2018;27(2):581–590. doi: 10.1007/s10068-017-0257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang YH, Feng JT, Zhang Q, Zhang X. Optimization of fermentation condition for antibiotic production by Xenorhabdus nematophila with response surface methodology. J Appl Microbiol. 2008;104(3):735–744. doi: 10.1111/j.1365-2672.2007.03599.x. [DOI] [PubMed] [Google Scholar]
- 32.Wang ZW, Liu XL. Medium optimization for antifungal active substances production from a newly isolated Paenibacillus sp. using response surface methodology. Bioresour Technol. 2008;99(17):8245–8251. doi: 10.1016/j.biortech.2008.03.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data and material are available upon request to correspondence author.