Abstract
Agrobacterium tumefaciens transfers oncogenic T-DNA and effector proteins to plant cells via a type IV secretion pathway. This transfer system, assembled from the products of the virB operon, is thought to consist of a transenvelope mating channel and the T pilus. When screened for the presence of VirB and VirE proteins, material sheared from the cell surface of octopine strain A348 was seen to possess detectable levels of VirB2 pilin, VirB5, and the VirB7 outer membrane lipoprotein. Material sheared from the cell surface of most virB gene deletion mutants also possessed VirB7, but not VirB2 or VirB5. During purification of the T pilus from wild-type cells, VirB2, VirB5, and VirB7 cofractionated through successive steps of gel filtration chromatography and sucrose density gradient centrifugation. A complex containing VirB2 and VirB7 was precipitated from a gel filtration fraction enriched for T pilus with both anti-VirB2 and anti-VirB7 antiserum. Both the exocellular and cellular forms of VirB7 migrated as disulfide-cross-linked dimers and monomers when samples were electrophoresed under nonreducing conditions. A mutant synthesizing VirB7 with a Ser substitution of the lipid-modified Cys15 residue failed to elaborate the T pilus, whereas a mutant synthesizing VirB7 with a Ser substitution for the disulfide-reactive Cys24 residue produced very low levels of T pilus. Together, these findings establish that the VirB7 lipoprotein localizes exocellularly, it associates with the T pilus, and both VirB7 lipid modification and disulfide cross-linking are important for T-pilus assembly. T-pilus-associated VirB2 migrated in nonreducing gels as a monomer and a disulfide-cross-linked homodimer, whereas cellular VirB2 migrated as a monomer. A strain synthesizing a VirB2 mutant with a Ser substitution for the reactive Cys64 residue elaborated T pilus but exhibited an attenuated virulence phenotype. Dithiothreitol-treated T pilus composed of native VirB2 pilin and untreated T pilus composed of the VirB2C64S mutant pilin distributed in sucrose gradients more predominantly in regions of lower sucrose density than untreated, native T pili. These findings indicate that intermolecular cross-linking of pilin monomers is not required for T-pilus production, but cross-linking does contribute to T-pilus stabilization.
Bacterial type IV secretion systems are of significant clinical concern. The type IV systems are composed of the well-known conjugation machines that are responsible for the rapid transmission of antibiotic resistance genes throughout bacterial populations under selective pressure (10, 27). Type IV systems also are composed of a more recently described group of secretion machines that mediate the delivery of effector molecules to the cytosols of eukaryotic cells during infection (8, 23). The list of medically important pathogens that utilize type IV systems for interkingdom macromolecular translocation now includes Helicobacter pylori, Bordetella pertussis, Legionella pneumophila, and Brucella and Bartonella species. All type IV systems share two common features: (i) these systems export macromolecules to other cells, usually via cell-to-cell contact, and (ii) these systems are ancestrally related to conjugation machines. These features distinguish the type IV systems from other bacterial secretion systems such as the ATP-binding cassette superfamily (type I) (18), the terminal branch of the general secretory pathway (type II) (31), the secretion systems ancestrally related to flagella (type III) (25), the immunoglobulin A autotransporters (type V) (31), and the recently described Tat export pathway (34).
Agrobacterium tumefaciens uses a type IV secretion system to export oncogenic T-DNA and proteins to susceptible plant cells during infection. The products of the virB operon and of the virD4 gene are required for type IV secretion. These proteins are proposed subunits of a gated channel through which substrates are translocated and of an extracellular pilus that mediates productive contacts with target cells. A general picture is emerging about the assembly pathway and structure of the T-DNA transfer system (8, 23). However, it is still not known where and how T-pilus polymerization initiates at the cell envelope or indeed whether the T pilus is physically joined to the mating channel. One model suggests that T-pilus polymerization begins at the inner membrane and proceeds outward through the periplasmic space and across the outer membrane, presumably through a gated structure. This model is reminiscent of the pathway by which Pseudomonas aeruginosa and many other pathogens assemble the type IV family of pili (which show no common ancestry with the pili elaborated by type IV secretion systems, i.e., the A. tumefaciens T pilus) (26). An alternative model suggests that the T-pilus subunits are delivered across the periplasm by the action of a chaperone to an outer membrane complex corresponding to the site of pilus assembly. This model is analogous to the chaperone-usher system used in type 1 pilus biogenesis (32).
VirB2 is the structural subunit of the A. tumefaciens T pilus (22). VirB2 undergoes two novel processing reactions, cleavage of an unusually long ∼5-kDa signal sequence and cyclization via formation of a covalent bond between the N-terminal Asp and C-terminal Glu residues (12, 22). Recent evidence suggests that the VirB5 protein associates in small amounts with the T pilus. VirB5 localizes predominantly in the periplasm where it might participate in T-pilus assembly, although genetic findings have been interpreted as evidence for the association of VirB5 along the length or at the tip of the T pilus (29).
In this study, we initiated work on the T-pilus assembly pathway. The likely outer membrane components participating in assembly include the small (4.5-kDa) outer membrane lipoprotein VirB7, VirB3, and VirB9. In previous studies, it was reported that VirB7 assembles as a disulfide-cross-linked homodimer and a heterodimer with VirB9 (1, 2, 30). Further, VirB7-VirB9 heterodimer formation was shown to have a stabilizing effect on several VirB proteins, leading to the proposal that the heterodimer functions as a nucleation center during biogenesis of the transporter T pilus (3, 14). Here, we have assayed for the association of VirB proteins with T pili purified from the cell surface of octopine strain A348. We report that the VirB7 monomer and homodimer, but not the VirB7-VirB9 heterodimer, are readily removed from the cell surface upon shearing. Furthermore, the VirB7 monomer and homodimer copurify with the T pilus. Mutational studies established the importance of VirB7 lipid modification and disulfide cross-linking for elaboration of the T pilus and identified a possible stabilizing effect of intermolecular disulfide cross-linking among VirB2 pilin subunits. Our findings support a model whereby sorting of the VirB7 lipoprotein as homo- and heteromultimeric complexes to the outer face of the outer membrane is a critical intermediate step in T-pilus biogenesis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
A348 is A. tumefaciens strain A136 containing pTiA6NC (15). The A348 derivatives, PC1001 to PC1011, carry nonpolar null mutations of virB1 to virB11, respectively (4). In the A348 derivative PC1000, the entire 9.5-kb virB operon is deleted from pTiA6NC (13). Plasmids pXZ16 and pXZ14 express virB7 derivatives with Ser codon substitutions for the Cys15 and Cys24 codons, respectively (30). Plasmid pVS10, which expresses virB2 with a Cys64-to-Ser substitution mutation, was constructed by oligonucleotide-directed mutagenesis with uracil-containing pBB8 (4) template and the mutagenic oligonucleotide 5′-GGATAAACGTACTTATATTGTTAACC-3′ according to the method of Kunkel et al. (20). The mutation was confirmed by sequencing across the virB fragment using an ABI 373A DNA sequencer (Perkin-Elmer). Plasmid pVS8, which expresses a glutathione S-transferase (GST)–VirB2 fusion protein, was constructed by PCR amplification of codons 48 to 121 of VirB2 with oligonucleotides 5′-CGGGATCCCAATCTGCGGGTGGCGGCACC-3′ and 5′-GGAATTCTCAACTACCGCCAGTGAGCGTTTG (BamHI and EcoRI sites are underlined). The amplified 200-bp BamHI-EcoRI fragment was ligated to the GST expression vector, pGEX-2T (Pharmacia).
Bacterial media and growth conditions have been previously described (13). For induction of the vir genes, A. tumefaciens cells were grown in MG/L medium to an optical density at 600 nm (OD600) of 0.5, harvested by centrifugation, and inoculated at an initial OD600 of 0.2 into induction medium (IM) (AB mimimal medium [pH 5.5] containing 1 mM phosphate supplemented with 200 μM acetosyringone [AS]) (13). Cultures were incubated with shaking at 22°C for 18 h and then harvested for protein analysis. In A. tumefaciens and Escherichia coli, plasmids were maintained by the addition of carbenicillin (50 μg/ml), kanamycin (50 μg/ml), or tetracycline (5 μg/ml) to the growth medium.
Protein analysis and immunoblotting.
Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) or a Tricine-SDS-PAGE system as previously described (28). Vir proteins were visualized by SDS-PAGE, protein transfer to nitrocellulose membranes, and immunoblot development with goat anti-rabbit antibodies conjugated to alkaline phosphatase and histochemical substrates. Alternatively, blots were developed with anti-rabbit antibodies conjugated to horseradish peroxidase and antibody-antigen interactions were visualized by chemiluminescence (Amersham, Arlington Heights, Ill.). Molecular size markers were from GIBCO-BRL (Grand Island, N.Y.).
Antibody specificities were previously documented for the VirB1, VirB4, VirB5, VirB7 through VirB11, and VirE2 proteins (see reference 36). Anti-VirB2 antiserum was raised by overproduction and purification of a GST-VirB2 fusion protein from E. coli BL21(DE3, pVS8). Briefly, a 100-ml culture was grown to an OD600 of 0.3, IPTG (isopropyl-β-d-thiogalactopyranoside) (1 mM final concentration) was added to induce gst-′virB2 expression, and cells were incubated with shaking for 4 h at 37°C. GST-′VirB2 present in the soluble fraction of cell lysates was purified by passage through a glutathione column and elution according to the manufacturer's instructions (Pharmacia). Purified GST-′VirB2 was sent to Cocalico Biologicals, Inc. (Reamstown, Pa.), for injection into New Zealand White rabbits.
Purification of T pili.
T pili were isolated according to the protocol of Lai and Kado (22). Briefly, A. tumefaciens strains were grown to an OD600 of 0.5 in MG/L medium at 28°C. Cells were pelleted, diluted fivefold in IM, and incubated for 6 h at 22°C. Two hundred microliters of AS-induced culture was spread on IM agar plates, and the plates were incubated for 3 days at 18°C. Cells were then gently scraped off the plates in 50 mM KPO4 buffer, pH 5.5, and pelleted by centrifugation at 14,000 × g for 15 min at room temperature. The supernatant was removed and the cell pellet was resuspended in 50 mM phosphate buffer. This suspension was passed through a 25-gauge needle 10 times to collect flagella, pili, and surface proteins. The sheared bacterial cells were pelleted by centrifugation at 14,000 × g for 30 min at 4°C. The remaining supernatant was filtered through a 0.22-μm-pore-size cellulose acetate membrane to remove unpelleted cells. When necessary, culture supernatants and sheared materials were concentrated with trichloroacetic acid or acetone as described previously (21).
T pili were purified by successive fractionation of exocellular material with gel filtration and sucrose density gradient centrifugation. The exocellular material suspended in 50 mM Tris (pH 8)–50 mM MgCl2 (buffer A) was applied at a total protein concentration of 10 mg/ml to a gel filtration column (30 by 1.5 cm) prepared with Toyopearl HW55 resin (TosoHaas, Montgomeryville, Pa.) according to the manufacturer's instructions. Material was fractionated with a Pharmacia Gradi-frac chromatography system with a flow speed of 0.6 ml/min, and fractions of 1.0 ml were collected. Fractions were analyzed for the presence of Vir proteins, by SDS-PAGE and immunodevelopment of blots as described above, or for the presence of total proteins, by silver staining of polyacrylamide gels. Protein molecular size markers included apoferritin (443 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), carbonic anhydrase (29 kDa), and cytochrome c (12 kDa). Size markers were electrophoresed both together with and independently of the test sample. Gel filtration fractions containing VirB2 pilin were layered on top of a 20 to 70% linear sucrose density gradient (5 ml) prepared with buffer A and ultracentrifuged in an SW55 Beckman rotor at 80,000 × g for 20 h at 4°C. Fractions of 0.5 ml were collected from the bottoms of the centrifugation tube and analyzed for the presence of Vir proteins by immunoblotting and for the presence of other proteins by silver staining. Immunoprecipitation with anti-VirB antiserum was carried out as previously described (28).
Electron microscopy.
Five microliters of purified T pili suspended in 50 mM Tris-HCl (pH 8.0)–50 mM MgCl2 buffer was deposited on carbon-Formvar films on 300-mesh, 3-mm2 copper grids. Samples were placed on grids for 30 s, excess liquid was removed, and the grids were rinsed three times by successive additions of triple-distilled H2O for 30 s. Samples were then stained with 2% uranyl acetate for 45 s and rinsed again with H2O before air drying.
Virulence assays.
Virulence assays were performed by inoculating wound sites of Kalanchoe daigremontiana leaves with ∼108 CFU of the various bacterial strains. Controls for the tumorigenesis assays included coinoculating the same leaf with wild-type A348 (virulent), avirulent ΔvirB mutants (4), and the various test strains. Each experiment was repeated at least four times, and tumor formation was monitored over a 3- to 4-month period.
RESULTS
Cofractionation of VirB2 and VirB7 during T-pilus purification.
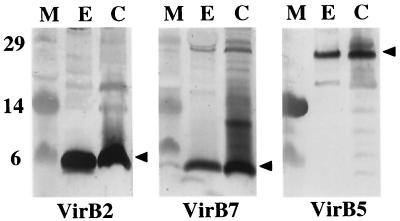
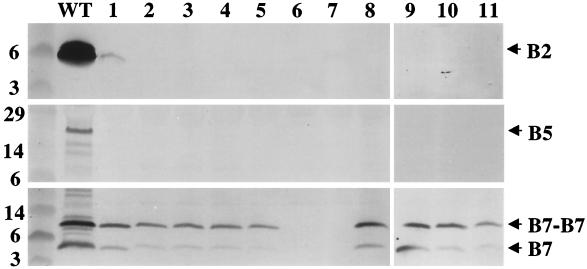
We isolated the T pilus from the octopine strain A348 according to a protocol developed by C. Kado and colleagues (21). Briefly, cells were induced for vir gene expression on agar plates, harvested, and sheared by successive passaging through a narrow-gauge needle. Cells were then removed from the exocellular fraction by low-speed centrifugation and filtration, and the surface proteins and organelles were concentrated by high-speed centrifugation. Silver staining of the concentrated exocellular material showed the presence of numerous protein species (data not shown), and immunoblot analysis showed the presence of VirB2, VirB5, and the outer membrane lipoprotein VirB7 (Fig. 1). Blots shown in Fig. 1 were developed with an alkaline phosphatase substrate. We were unable to detect VirB proteins other than VirB2, VirB5, and VirB7 in concentrated exocellular fractions either by development of blots with alkaline phosphatase or with chemiluminescence (data not shown). We also were unable to detect a species corresponding to VirB1∗, a proteolytic fragment of VirB1 encoded by the nopaline pTiC58 plasmid (24), in either the cellular or exocellular fractions of A348, which carries the octopine plasmid pTiA6NC. Finally, we did not detect the periplasmic protein ChvE or cytoplasmic VirE1 or the VirE2 single-stranded DNA-binding protein (SSB) in contrast to a previous finding (6). Thus, we conclude that under the growth and induction conditions of our experiments, VirB2, VirB5, and VirB7 are the only VirB or VirE proteins exocellularly localized or released at appreciable levels into the extracellular milieu.
FIG. 1.
Identification of VirB2, VirB5, and VirB7 in the exocellular fraction obtained by shearing of A348 cells. Blots were developed with antisera to the VirB proteins listed, and an arrowhead marks the position of the corresponding VirB protein. Abbreviations: E, exocellular fraction containing surface organelles and proteins; C, cell pellet recovered after removal of surface material by shearing as described in the text; M, molecular mass markers, with sizes in kilodaltons indicated at the left.
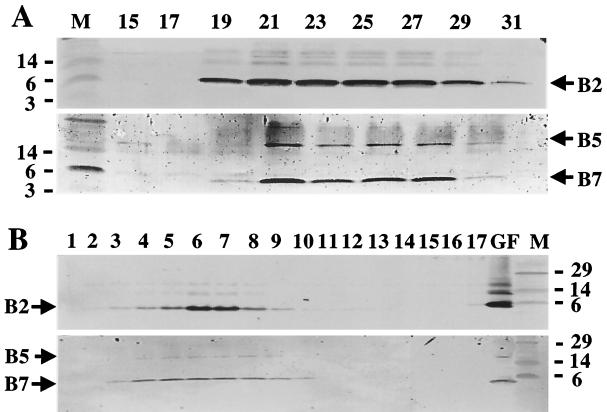
To purify the T pili, the sheared material was fractionated by gel filtration chromatography and then by sucrose density gradient centrifugation. In a representative experiment shown in Fig. 2A and B, fractions 21 to 27 eluting from the gel filtration columns contained abundant amounts of VirB2 and fractions 19, 29, and 31 contained lower but detectable levels (Fig. 2A). Of considerable interest, both VirB5 and VirB7 exhibited very similar elution profiles, with the most abundant levels of these proteins being present in fractions 21 to 27 (Fig. 2A). The molecular mass of the complex(es) composed of VirB2, VirB5, and VirB7 exceeded 440 kDa, as judged by elution of the 440-kDa ferritin size marker in fractions 31 to 35.
FIG. 2.
Purification of T pilus through sucessive steps of gel filtration chromatography and sucrose density gradient centrifugation. (A) Gel filtration fractions containing the VirB2, VirB5, and VirB7 proteins; the bottom blot is skewed slightly to the right relative to the top blot. (B) Sucrose density gradient fractions containing VirB2, VirB5, and VirB7. Top blots in each panel were developed with anti-VirB2 antiserum, and bottom blots were developed with anti-VirB5 and anti-VirB7 antisera. Positions of VirB proteins and sizes (in kilodaltons) of molecular mass markers (M) are denoted. Lane GF, gel filtration fraction 21 loaded onto the sucrose gradient.
Gel filtration fraction 21, which contained the most abundant amounts of VirB2, VirB5, and VirB7, was centrifuged through a 20 to 70% linear sucrose density gradient. As shown in Fig. 2B, these VirB proteins copartitioned in the sucrose gradients, providing further evidence for coassociation of these proteins in a high-molecular-weight structure. We further found that high-salt (1 M NaCl) treatment of gel filtration fractions containing VirB2, VirB5, and VirB7 did not eliminate comigration of these proteins in sucrose gradients, suggesting that formation of this putative VirB complex is mediated by hydrophobic rather than electrostatic interactions (data not shown). Of interest, VirB2 was present at appreciably greater levels in sucrose gradient fractions 6 and 7 than in adjacent fractions, whereas VirB5 and VirB7 were distributed at comparable levels across several fractions. This difference in fractionation behavior might be due to the presence of two subpopulations of T pilus in the pilus preparations, one devoid of and a second associated with a basal structure that we propose is composed of VirB5 and VirB7.
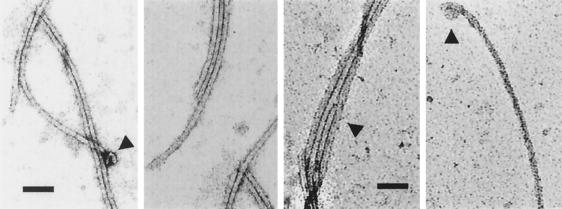
Next, we examined the content of the sucrose fractions containing the VirB proteins by transmission electron microscopy. Sucrose was removed by dialysis and pili were concentrated by centrifugation (see Materials and Methods). We detected T pili similar in appearance to those visualized previously (Fig. 3) (21). Typically, we saw long, flexible pili with an estimated diameter of ∼10 nm, as well as bundles of several T pili. Occasionally, we also saw structures at one end of the pilus resembling the terminal sacculi observed previously (29).
FIG. 3.
Transmission electron microscopy showing purified T pili obtained from sucrose density gradients. The first two images were from fraction 6 and second two were from fraction 8 of the sucrose gradient shown in Fig. 2. Arrowheads denote morphological features of interest, including single T pili (diameter, ∼10 nm), clumps of T pili, and occasional terminal sacculi as observed previously (20, 27). Pili were examined at a magnification of 40,000. Bar, 100 nm.
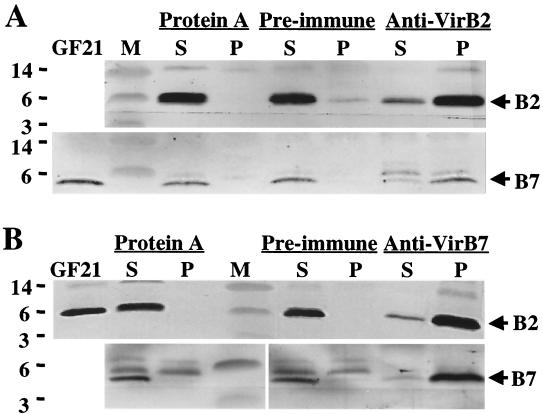
To further assay for VirB protein complexes, material from the relevant gel filtration fractions was subjected to immunoprecipitation analysis with anti-VirB antiserum. As represented by experiments shown in Fig. 4 in which gel filtration fraction 21 (Fig. 2A) was used as starting material, protein A-Sepharose and preimmune serum failed to precipitate these VirB proteins. However, both the anti-VirB2 antiserum and anti-VirB7 antiserum precipitated substantial amounts of VirB2 and comparatively less VirB7. This uneven ratio of VirB2 and VirB7 in the precipitated complexes and in the sucrose fractions might be due to the association of VirB7 in minor amounts at one position of the T pilus, possibly at the pilus base.
FIG. 4.
Coprecipitation of a VirB2 and VirB7 complex presumptively corresponding to the T pilus with anti-VirB2 and anti-VirB5 antiserum from gel filtration fraction 21 (GF21) (see Fig. 2). Blots showing VirB2 and VirB7 proteins in complexes precipitated with anti-VirB2 antiserum (A) and with anti-VirB7 antiserum (B) are shown. Positions of VirB proteins and sizes (in kilodaltons) of molecular mass markers (M) are denoted. Abbreviations: S, supernatant fraction following precipitation; P, pellet recovered upon precipitation with the reagents listed at the top of each panel.
Exocellular location of VirB7 in mutants defective in T-pilus production.
The above findings strongly suggest that VirB7 associates with the T pilus, as was shown previously for VirB5 (29). To determine whether the exocellular locations of VirB7 and VirB5 are dependent on T-pilus production, we assayed for the presence of these proteins in fractions obtained from each of the nonpolar ΔvirB mutants constructed in this laboratory (4). Previous work determined that the ΔvirB1 mutant, PC1001, as well as other nonpolar virB mutants, possesses no extracellular pilin (21, 29). We detected a very low level of VirB2 pilin in the exocellular fraction from the ΔvirB1 mutant and none in fractions from any of the other ΔvirB mutants (Fig. 5). We detected low levels of VirB5 in the exocellular fraction from wild-type A348 and none in fractions from the 11 ΔvirB mutants (Fig. 5).
FIG. 5.
Presence of the VirB7 homodimer in the exocellular fractions from the nonpolar ΔvirB mutants of A348 (4). Exocellular fractions were from wild-type strain A348 (lane WT) and PC1001 (ΔvirB1) through PC1011 (ΔvirB11) (lanes 1 through 11, respectively). Blots were developed with antisera to the VirB proteins denoted at the right. Samples analyzed with anti-VirB2 and anti-VirB5 antisera were electrophoresed under reducing conditions; samples analyzed with anti-VirB7 antisera were electrophoresed under nonreducing conditions to show the relative abundances of the VirB7 homodimer and monomer.
By contrast, VirB7 was present in the exocellular fractions of all mutants except for those from which virB6 or virB7 was deleted (Fig. 5). Results of studies describing the contribution of VirB6 to VirB7 dimer formation are reported elsewhere (17; S. Jakubowski, Z. Liu, and P. J. Christie, submitted for publication). Exocellular VirB7 migrated predominantly as a disulfide-cross-linked homodimer when samples were electrophoresed through gels under nonreducing conditions (Fig. 5). We show elsewhere that the cellular form of VirB7 accumulates at wild-type levels in all virB mutants except for ΔvirB6 and ΔvirB7 (Jakubowski et al., submitted); thus, the VirB7 homodimer apparently sorts to the outer face of the outer membrane independently of T-pilus production and also independently of most of the VirB proteins. However, it is also evident that exocellular VirB7 is present in reduced levels in all virB mutants compared to wild-type A348 (Fig. 5). This finding suggests that the VirB proteins are important for accumulation of wild-type levels of exocellular VirB7. Precisely how VirB7 interacts with the T pilus is unknown. Our inability to detect integral membrane VirB proteins—including VirB4, VirB6, VirB8, VirB10, and VirB11 (inner membrane), VirB9 (outer membrane), and ChvE (periplasm)—in exocellular fractions appears to exclude the possibility that VirB7 is associated with membrane vesicles. Interestingly, however, we determined that exocellular VirB7 from a ΔvirB2 mutant partitioned in sucrose gradients as a high-molecular-weight species. Although this species does not comigrate with VirB7 from T-pilus-producing cells, these findings raise the possibility that exocellular VirB7 is released from the surface of pilus-deficient cells in association with specialized vesicles (data not shown). Current studies in this laboratory are characterizing the composition and possible function(s) of the exocellular VirB7 complexes isolated from pilus-deficient cells.
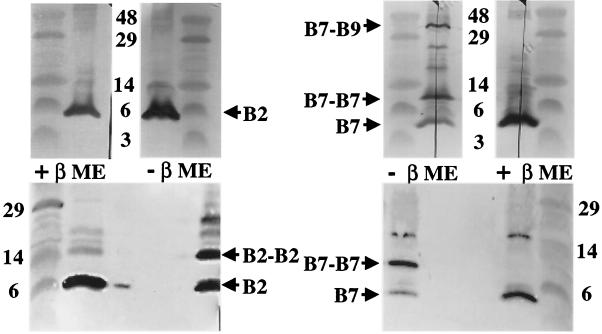
VirB7 and VirB2 homodimers associate with T pilus.
Cellular VirB7 accumulates as a monomer, homodimer, and VirB7-VirB9 heterodimer (Fig. 6, top right panel) (30). The exocellular form of VirB7 that copurifies with the T pilus is predominantly homodimeric (Fig. 6, bottom right panel). Some monomeric VirB7 was evident, but the 36-kDa VirB7-VirB9 heterodimer was not detected in pilus samples electrophoresed under nonreducing conditions (Fig. 6, bottom right panel).
FIG. 6.
Association of VirB2 and VirB7 homodimers with the T pilus. Top panels, VirB2 and VirB7 species present in extracts from A348 cells obtained after shearing to remove surface proteins and organelles. Extracts were electrophoresed under reducing (+βME) and nonreducing (−βME) conditions. The cellular form of VirB2 pilin migrates as a monomer, whereas the cellular form of VirB7 migrates predominantly as a disulfide-cross-linked dimer. A cross-reactive species of unknown composition is detected at a position corresponding to ∼18 kDa. Bottom panels, VirB2 and VirB7 species associated with T pili enriched from the exocellular fraction of A348 cells. The T-pilus-associated forms of both VirB2 and VirB7 migrated in nonreducing gels at positions corresponding to monomeric and disulfide-cross-linked homodimeric species. Positions of VirB2 and VirB7 species and sizes (in kilodaltons) of molecular mass markers are denoted.
Of further interest, although the cellular form of VirB2 is monomeric, T-pilus-associated VirB2 migrated both as an apparent homodimer and as a monomer (Fig. 6, left panels). The pilus preparations contained similar levels of these two forms of pilin. The material examined in Fig. 6 was derived from gel filtration fraction 21, which corresponded to the earliest-eluting, and presumably the largest, forms of T pilus (Fig. 2A). When we assayed for pilin dimer formation in the total exocellular fraction derived from T-pilus-producing cells or from the gel filtration fractions containing smaller, presumably more extensively broken forms of T pilus, we found that a smaller fraction, estimated at less than 20% of pilin, migrated as an apparent homodimer (Fig. 7). We propose that these and other experimental findings presented below might be due to disulfide cross-linking between a subset of pilin monomers that are located at a discrete position along the T pilus.
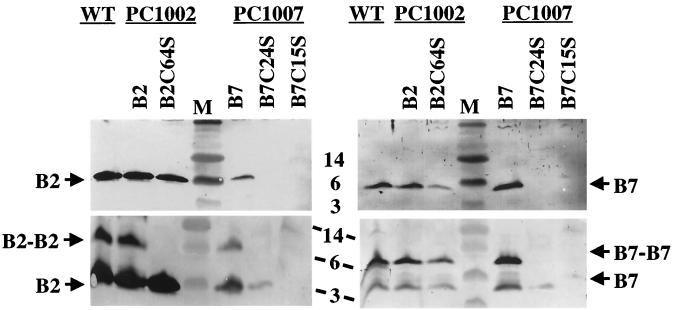
FIG. 7.
Effects of Cys-to-Ser substitution mutations on accumulation of exocellular VirB2 and VirB7. Exocellular fractions were from A348 (WT), strains PC1002 carrying pBB8 (B2) or pVS10 (B2C64S), and strains PC1007 carrying pPC974 (B7), pXZ14 (B7C24S), or pXZ16 (B7C15S). Top panels, protein samples were electrophoresed under reducing conditions. Bottom panels, protein samples were electrophoresed under nonreducing conditions. Blots on the left were developed with anti-VirB2 antiserum, and blots on the right were developed with anti-VirB7 antiserum. Sizes (in kilodaltons) of molecular mass markers (M) are denoted in the center.
The blots shown in the lower panels of Fig. 6 were generated by electrophoresing the pilus-containing samples through the same gel to clearly show the distinct migration of the VirB2 and VirB7 homodimers with predicted sizes of ∼14 and 9 kDa, respectively. In several repetititions of these experiments we have been unable to detect a cross-reactive species migrating at ∼11 kDa, the expected size of a VirB2-VirB7 heterodimer.
Role of VirB2 and VirB7 disulfide cross-linking and VirB7 lipid modification for T-pilus assembly.
The mature form of VirB2 possesses one internal Cys residue at position 64. To determine if Cys64 is essential for T-pilus production, we examined the effect of a Cys64Ser substitution mutation. As shown in Fig. 7, PC1002 cells expressing the alleles for native VirB2 and for VirB2C64S accumulated both proteins at abundant levels in the exocellular fraction. In contrast to native VirB2, the VirB2C64S mutant protein failed to migrate as an apparent homodimer when electrophoresed under nonreducing conditions (Fig. 7). Thus, disulfide cross-linking of VirB2 homodimers appears not to be essential for T-pilus production.
Mature VirB7 has two Cys residues, N-terminal Cys15 that undergoes lipid modification and internal Cys24 that reacts to form the disulfide-cross-linked dimers (13, 14). VirB7C15S is unstable and nonfunctional, as shown by a failure of the corresponding allele to complement a ΔvirB7 mutation (14). To examine the importance of VirB7 lipid modification for T-pilus assembly, we assayed for the presence of T pili in exocellular fractions of strain PC1007(pXZ16) expressing virB7C15S. As shown in Fig. 7, PC1007(pXZ16) cells accumulated undetectable levels of exocellular VirB7C15S and VirB2 pilin, demonstrating the importance of VirB7 lipid modification for pilus formation. Our previous studies showed that Cys24 is important for stability and functionality of VirB7 (31), although another study reported that cooverexpression of virB7C24S, virB8, and virB9 permitted successful T-DNA transfer (9). To examine the importance of VirB7 disulfide cross-linking for T-pilus production, we assayed for pilus production by strain PC1007(pXZB14) expressing virB7C24S. As shown in Fig. 7, avirulent PC1007(pXZB14) cells accumulated very low levels of exocellular forms of VirB2 and VirB7C24S proteins. Thus, VirB7 disulfide cross-linking contributes to efficient T-pilus production.
We next examined whether disulfide-mediated dimerization of VirB2 affects T-pilus function. Virulence assays were carried out with isogenic PC1002 mutants expressing alleles for either native VirB2 or VirB2C64S. Interestingly, whereas strain PC1002(pBB8) exhibits wild-type virulence (4), the isogenic strain PC1002(pVS10) synthesizing the substitution mutant exhibited an attenuated virulence phenotype. Tumor induction was delayed by approximately 1 week, and tumors were appreciably smaller than were tumors incited by strain PC1002(pBB8) (Fig. 8). The extent of the attenuated function of the C64S mutant pilin was assessed by inoculation of wound sites with serial dilutions of PC1002(pBB8) and PC1002(pVS10) cultures. Results of these assays indicated that the VirB2Cys64S substitution mutant was approximately 10- to 100-fold less virulent than the isogenic wild-type strain (data not shown).
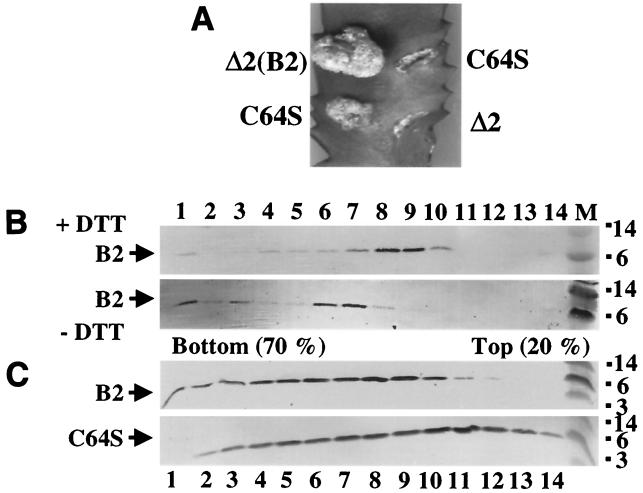
FIG. 8.
Effects of a Cys64Ser substitution mutation and DTT treatment of wild-type pilin on T-pilus assembly. (A) Virulence assays of PC1002 cells (Δ2) expressing wild-type VirB2 or VirB2C64S by inoculation of equivalent numbers of cells onto wounded Kalanchoe leaves. Cells synthesizing the C64S mutant incited tumors of variable sizes that were reproducibly smaller than those incited by cells synthesizing wild-type pilin. (B) T pilus composed of wild-type pilin enriched by gel filtration fraction chromatography was treated with 5 mM DTT, and both untreated and DTT-treated T-pilus samples were fractionated through identically prepared sucrose density gradients. (C) T pilus composed of wild-type or C64S mutant pilin present in the concentrated exocellular fractions from PC1002(pBB8) or PC1002(pVS10) cells, respectively, was fractionated through sucrose density gradients.
To assess the effect of the C64S mutation on T-pilus integrity, we centrifuged the concentrated, exocellular fractions obtained from PC1002(pBB8) and PC1002(pVS10) through sucrose gradients and examined fractions for T-pilus distribution. Care was taken to ensure that the gradients were identically prepared and fractionated. As shown in Fig. 8, the pili composed of wild-type and mutant pilin distributed differently in the gradients. Whereas T pili assembled from wild-type pilin were most abundant in sucrose fractions 6 to 8, the T pili assembled from the C64S mutant partitioned predominantly in fractions having a lower sucrose density. To further assess the importance of intermolecular disulfide cross-linking for T-pilus stability, we incubated a gel filtration fraction enriched for T pilus composed of wild-type pilin in the presence of 5 mM dithiothreitol (DTT) for 1 h at 30°C. Initial studies showed that DTT-treated pilin migrates exclusively as a monomer in nonreducing gels (data not shown). DTT-treated and untreated pili were again size-fractionated by centrifugation through identically prepared sucrose gradients. Interestingly, the DTT-treated wild-type T pili also fractionated at a position of lower sucrose density than did untreated T pili (Fig. 8). In the above-mentioned gradients, both VirB5 and VirB7 exhibited shifts in distribution similar to those of VirB2 pilin (data not shown). Thus, both a C64S mutation and reduction of the C64S disulfide cross-link of wild-type pilin had the same phenotypic effect, namely, a sucrose fractionation profile suggestive of the presence of shorter or less stable T pili than wild-type T pili not exposed to a reductant. It is noteworthy that neither the C64S mutation nor DTT treatment caused complete dissociation of the T pilus. On the basis of these findings, we suggest that VirB2 disulfide cross-linking, while not essential for T-pilus production, nevertheless does contribute to pilus integrity and function possibly by stabilizing subunit contacts at a specific position(s) along the pilus.
DISCUSSION
The assembly of pili associated with type IV secretion systems is a poorly understood process. Recent studies have established that T-pilus production at the A. tumefaciens cell surface requires all of the VirB proteins, including VirB1, a putative transglycosylase that is dispensable for T-DNA transfer (4, 21). These findings led to the proposal that the presumed virB-encoded mating apparatus provides the channel for translocation of VirB2 pilin from its inner membrane reservoir to its site of polymerization (20). If this scenario is correct, VirB2 pilin can be considered a bona fide substrate of the translocation channel, adding to the list of secretion substrates such as VirE2 SSB, VirF, and the T-strand-VirD2 and the RSF1010-MobA transfer intermediates. It is of interest, however, that the VirD4 putative ATPase is dispensable for T-pilus production but is required for intercellular substrate trafficking (21). VirD4 is a member of a family of coupling proteins that are proposed to function as docking sites for secretion substrates at the base of the transfer machine (16). Therefore, if VirB2 pilin is indeed translocated through the mating channel, it must enter this secretory machine via a different pathway from that used by the secretion substrates destined for intercellular transfer.
It is alternatively possible that the T-pilus biogenesis pathway does not depend on prior assembly of the translocation channel. According to this model, some of the VirB proteins might participate in the dynamics of pilus biogenesis, while the remaining VirB proteins provide a structure for anchoring the pilus to the cell envelope. Candidates that might actively promote the delivery of VirB2 pilin to its site of assembly include the VirB11 ATPase, whose multimerization as a double-ring, chaperone-like structure is inferred from genetic and biochemical findings (28) and a recent crystal structure of the H. pylori HP0525 homolog (35). In addition, VirB5 has been postulated to function as a periplasmic chaperone that delivers pilin from its inner membrane reservoir to the outer membrane by a route independent of the virB-encoded mating channel (23). Most of the remaining VirB proteins—i.e., VirB4, VirB6, VirB7, VirB8, VirB9, and VirB10—might assemble as a transenvelope structure that could alternatively (or dually) serve as a platform for pilus assembly or the mating channel for intercellular translocation of the VirE2, VirF, and DNA transfer intermediates.
To begin dissecting the functions of VirB proteins in the pilus assembly pathway, a reasonable starting point is the elucidation of the roles of the three VirB proteins, VirB3, VirB7, and VirB9, that localize at the outer membrane (13, 19; Jakubowski et al., submitted). These outer membrane proteins are the best candidates for assembling as a structure required for pilus production. Previous work determined that VirB7 is processed as a lipoprotein and that it assembles as a disulfide-cross-linked homodimer and a VirB7-VirB9 heterodimer (1, 13, 14, 30). On the basis of observed stabilizing activities, we postulated that the VirB7-VirB9 heterodimer functions as a nucleation center for recruitment of VirB proteins during assembly of the T-DNA transfer machine (14). However, our early studies did not provide any clues about the function of the VirB7 homodimer. Despite the capacity of a virB9 deletion mutant to assemble VirB7 homodimers, this mutant still accumulated low levels of several VirB proteins, suggesting that the homodimer is not a general stabilizing factor for the VirB proteins (4). We also assayed for isomerization of the homodimer disulfide cross-link with a strain expressing virB7 from the Plac promoter and virB9 from the PvirB promoter. In this experiment, cells were pretreated with IPTG to induce virB7 expression and assembly of the VirB7 homodimer, washed to remove IPTG, and incubated with AS to induce expression of virB9. Despite the accumulation of preassembled VirB7 homodimer, these cells did not accumulate detectable levels of the VirB7-VirB9 heterodimer, suggesting that the homodimer is not simply an intermediate in the VirB7-VirB9 assembly pathway (X. R. Zhou and P. J. Christie, unpublished data).
In the present study, we showed that the VirB7 homodimer localizes exocellularly independently of most of the other VirB proteins. Several lines of evidence suggest that VirB7 assembles at a discrete site(s) on the cell surface as a complex or structure that is relevant to pilus biogenesis. First, we have found that the VirB7 homodimer cofractionates with two pilus-associated proteins, VirB5 (29) and the VirB2 structural subunit (22), through successive steps of gel filtration and sucrose density gradient fractionation used for purification of the T pilus. VirB2, VirB5, and VirB7, but no other VirB proteins, were detected in exocellular fractions or purified T-pilus preparations from A348 cells. Second, the exocellular VirB7 homodimer tightly associates with the T pilus. This was shown by its cofractionation with a high-molecular-weight, VirB2-containing structure in the presence of high salt and by the demonstration that VirB7 and VirB2 coprecipitate from exocellular fractions. Third, exocellular VirB7 produced by a ΔvirB2 mutant and by wild-type cells displayed different distribution patterns in sucrose gradients. These last two findings appear to exclude the possibility that VirB7 complexes coincidentally cofractionate with VirB2 during the T-pilus purification. In addition, we note that the production of exocellular VirB7 homodimer requires VirB6, and the abundance of this species is influenced by production of each of the VirB proteins (Fig. 5) (19). These findings further support the notion that exocellular VirB7 lipoprotein forms a physiologically relevant complex with the T pilus.
We propose that the site of interaction between the VirB7 lipoprotein and the T pilus is the outer membrane; at this location, the covalently attached lipid groups most probably embed into the outer leaflet anchoring VirB7 to the membrane. It should be noted, however, that the lipid anchor is not tenacious; previous work has shown that lipoproteins bound to the membrane exclusively by fatty acylation are easily removed from the membranes during cell fractionation (see reference 13). This property probably explains why the monomeric and homodimeric forms of VirB7 were coextracted with the T pilus during shearing of the cells and pilus purification. A loose membrane association might also account for the presence of exocellular VirB7 in the various virB gene deletion mutants. We suspect that in these mutants VirB7 fails to establish stabilizing contacts with the T-pilus–translocation channel; therefore, VirB7 might be easily extracted from the membranes upon shearing. We acknowledge, however, that it might be premature to conclude that VirB7 functions exclusively in pilus biogenesis during the infection process. For example, previous work has established that T pili (and other related conjugative pili) are readily sloughed from the cell surface during growth (21, 22). Any molecules released from the surfaces of bacterial pathogens are potential signals for eukaryotic target cells; thus, exocellular VirB7 lipoprotein either in association with sloughed T pili or released to the milieu independently of the T pilus could activate host cellular processes required for the establishment of successful infection. Current studies in the laboratory are examining the possibility that VirB7 lipoprotein contributes to the A. tumefaciens infection process by a mechanism distinct from its role in T-pilus biogenesis at the bacterial cell surface.
Both VirB7 lipoprotein and outer membrane protein VirB9 clearly are essential for pilus formation on the basis of their demonstrated roles in stabilizing other VirB proteins and the failure of ΔvirB7 and ΔvirB9 mutants to elaborate T pili (3, 14, 21). The VirB7-VirB9 heterodimer was not detected in T-pilus preparations, but this likely is because VirB9 is an integral outer membrane protein and therefore is refractory to removal from the cell surface by shearing. On the basis of the available data, we therefore propose that both VirB7 dimers assemble at the outer membrane as a heteromultimeric complex or structure required for pilus assembly. The specific function of this putative structure remains to be elucidated, but two possibilities include the configuration as a platform corresponding to the site of pilus assembly or as a channel that permits passage of pilin subunits or the T pilus itself across the outer membrane.
With respect to the latter possibility, oligomeric ring-like complexes termed secretons are now known to be required for assembly and function of many secretion and organellar biogenesis systems of gram-negative bacteria (32). Relevant to pilus biogenesis, secretons mediate the delivery of structural subunits to the pilus assembly site at the outer membrane, e.g., E. coli PapC (33), or they serve as channels for pilus outgrowth, e.g., Pseudomonas aeruginosa PilQ (26). Relevant to VirB7-VirB9, in several cases the secreton structural subunit interacts with a cognate lipoprotein (31). Indeed, the outer membrane lipoprotein WzaK30 was itself recently shown to assemble as an oligomeric channel; this secreton promotes translocation of group 1 capsular polysaccharide to the surface of E. coli (11). An intriguing speculation that awaits further study is whether one or both of the VirB7 dimers assemble as an oligomeric ring at the outer membrane for T-pilus assembly and/or substrate translocation.
Finally, our studies have contributed insights about the T-pilus assembly pathway with respect to VirB2 processing requirements. Others have shown that VirB2 is processed most probably by signal peptidase I at a sequence that is conserved among other members of this pilin family (22). VirB2 then undergoes a novel cyclization reaction that results in a head-to-tail peptide bond formation (12). Both the signal sequence cleavage and cyclization reactions are thought to occur immediately upon localization of the pilin at the inner membrane. We have shown that this inner membrane reservoir of VirB2 does not form intermolecular cross-links. Conceivably, the pilin is embedded into the membrane in such a way as to render the unique Cys64 residue unavailable for disulfide cross-linking. At some point, an unknown signal activates pilus polymerization; this reaction involves a dynamic restructuring of pilin monomers from their inner membrane location to a site of pilus assembly. We suggest that it is during this recruitment step that a subset of pilin monomers form intermolecular cross-links. It is formally possible that these cross-links form spontaneously, because the Cys64 residue marks a site of close contact between two pilin monomers in the assembled T pilus. However, two lines of evidence suggest that pilin cross-linking is a physiologically relevant event. First, mutants synthesizing VirB2C64S reproducibly displayed an attenuated virulence phenotype compared to an isogenic strain synthesizing wild-type pilin. Second, both T pili assembled from VirB2C64S and DTT-treated, wild-type pili displayed fractionation patterns in sucrose gradients suggestive of a reduction in pilus length compared to untreated, wild-type T pili. On the basis of these findings, we suggest that disulfide cross-linking might occur at a discrete site along the pilus, possibly at its base, in order to stabilize specific contacts or regions of the T pilus. These cross-links are dispensable for pilus production but might be important for pilus retention at the plant wound site.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
This work was supported by NIH grant GM48746.
We thank members of the laboratory for helpful discussions. We thank Yasunori Machida for providing the ChvE-specific antiserum.
REFERENCES
- 1.Anderson L B, Hertzel A V, Das A. Agrobacterium tumefaciens VirB7 and VirB9 form a disulfide-linked protein complex. Proc Natl Acad Sci USA. 1996;93:8889–8894. doi: 10.1073/pnas.93.17.8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baron C, Thorstenson Y R, Zambryski P C. The lipoprotein VirB7 interacts with VirB9 in the membranes of Agrobacterium tumefaciens. J Bacteriol. 1997;179:1211–1218. doi: 10.1128/jb.179.4.1211-1218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaupré C E, Bohne J, Dale E M, Binns A N. Interactions between VirB9 and VirB10 membrane proteins involved in movement of DNA from Agrobacterium tumefaciens into plant cells. J Bacteriol. 1997;179:78–89. doi: 10.1128/jb.179.1.78-89.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger B R, Christie P J. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J Bacteriol. 1994;176:3646–3660. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C-Y, Winans S C. Controlled expression of the transcriptional activator gene virG in Agrobacterium tumefaciens by using the Escherichia coli lac promoter. J Bacteriol. 1991;173:1139–1144. doi: 10.1128/jb.173.3.1139-1144.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L, Li C M, Nester E W. Transferred DNA (T-DNA)-associated proteins of Agrobacterium tumefaciens are exported independently of virB. Proc Natl Acad Sci USA. 2000;97:7545–7550. doi: 10.1073/pnas.120156997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chilcott G S, Hughes K T. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol Mol Biol Rev. 2000;64:694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christie P J. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally-related to conjugation machines. Mol Microbiol. 2001;40:294–305. doi: 10.1046/j.1365-2958.2001.02302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das A, Anderson L B, Xie Y H. Delineation of the interaction domains of Agrobacterium tumefaciens VirB7 and VirB9 by use of the yeast two-hybrid assay. J Bacteriol. 1997;179:3404–3409. doi: 10.1128/jb.179.11.3404-3409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de la Cruz I, Davies I. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol. 2000;8:128–133. doi: 10.1016/s0966-842x(00)01703-0. [DOI] [PubMed] [Google Scholar]
- 11.Drummelsmith J, Whitfield C. Translocation of group 1 capsular polysaccharide to the surface of Escherichia coli requires a multimeric complex in the outer membrane. EMBO J. 2000;19:57–66. doi: 10.1093/emboj/19.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenbrandt R, Kalkum M, Lai E M, Lurz R, Kado C I, Lanka E. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J Biol Chem. 1999;274:22548–22555. doi: 10.1074/jbc.274.32.22548. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez D, Dang T A T, Spudich G M, Zhou X-R, Berger B R, Christie P J. The Agrobacterium tumefaciens virB7 gene product, a proposed component of the T-complex transport apparatus, is a membrane-associated lipoprotein exposed at the periplasmic surface. J Bacteriol. 1996;178:3156–3167. doi: 10.1128/jb.178.11.3156-3167.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandez D, Spudich G M, Zhou X-R, Christie P J. The Agrobacterium tumefaciens VirB7 lipoprotein is required for stabilization of VirB proteins during assembly of the T-complex transport apparatus. J Bacteriol. 1996;178:3168–3176. doi: 10.1128/jb.178.11.3168-3176.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garfinkel D J, Simpson R B, Ream L W, White F F, Gordon M P, Nester E W. Genetic analysis of crown gall: fine structure map of the T-DNA by site-directed mutagenesis. Cell. 1981;27:143–153. doi: 10.1016/0092-8674(81)90368-8. [DOI] [PubMed] [Google Scholar]
- 16.Hamilton C M, Lee H, Li P-L, Cook D M, Piper K R, Beck von Bodman S, Lanka E, Ream W, Farrand S K. TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugal transfer system of pTiC58. J Bacteriol. 2000;182:1541–1548. doi: 10.1128/jb.182.6.1541-1548.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hapfelmeier S, Domke N, Zambryski P C, Baron C. VirB6 is required for stabilization of VirB5 and VirB3 and formation of VirB7 homodimers in Agrobacterium tumefaciens. J Bacteriol. 2000;182:4505–4511. doi: 10.1128/jb.182.16.4505-4511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holland I B, Blight M A. ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J Mol Biol. 1999;293:381–399. doi: 10.1006/jmbi.1999.2993. [DOI] [PubMed] [Google Scholar]
- 19.Jones A L, Shirasu K, Kado C I. The product of the virB4 gene of Agrobacterium tumefaciens promotes accumulation of VirB3 protein. J Bacteriol. 1994;176:5255–5261. doi: 10.1128/jb.176.17.5255-5261.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunkel T A, Bebenek K, McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 21.Lai E M, Chesnokova O, Banta L M, Kado C I. Genetic and environmental factors affecting T-pilin export and T-pilus biogenesis in relation to flagellation of Agrobacterium tumefaciens. J Bacteriol. 2000;182:3705–3716. doi: 10.1128/jb.182.13.3705-3716.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai E M, Kado C I. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J Bacteriol. 1998;180:2711–2717. doi: 10.1128/jb.180.10.2711-2717.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai E M, Kado C I. The T-pilus of Agrobacterium tumefaciens. Trends Microbiol. 2000;8:361–369. doi: 10.1016/s0966-842x(00)01802-3. [DOI] [PubMed] [Google Scholar]
- 24.Llosa M, Zupan J, Baron C, Zambryski P. The N- and C-terminal portions of the Agrobacterium VirB1 protein independently enhance tumorigenesis. J Bacteriol. 2000;182:3437–3445. doi: 10.1128/jb.182.12.3437-3445.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macnab R M. The bacterial flagellum: reversible rotary propellor and type III export apparatus. J Bacteriol. 1999;181:7149–7153. doi: 10.1128/jb.181.23.7149-7153.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nunn D. Bacterial type II protein export and pilus biogenesis: more than just homologies? Trends Cell Biol. 1999;9:402–408. doi: 10.1016/s0962-8924(99)01634-7. [DOI] [PubMed] [Google Scholar]
- 27.Ochman H, Lawrence J G, Groisman E A. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 28.Rashkova S, Zhou X-R, Christie P J. Self-assembly of the Agrobacterium tumefaciens VirB11 traffic ATPase. J Bacteriol. 2000;182:4137–4145. doi: 10.1128/jb.182.15.4137-4145.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt-Eisenlohr H, Domke N, Angerer C, Wanner G, Zambryski P C, Baron C. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J Bacteriol. 1999;181:7485–7492. doi: 10.1128/jb.181.24.7485-7492.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spudich G M, Fernandez D, Zhou X-R, Christie P J. Intermolecular disulfide bonds stabilize VirB7 homodimers and VirB7/VirB9 heterodimers during biogenesis of the Agrobacterium tumefaciens T-complex transport apparatus. Proc Natl Acad Sci USA. 1996;93:7512–7517. doi: 10.1073/pnas.93.15.7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stathopoulos C, Hendrixson D R, Thanassi D G, Hultgren S J, St. Geme J W, III, Curtiss R., III Secretion of virulence determinants by the general secretory pathway in gram-negative pathogens: an evolving story. Microbes Infect. 2000;2:1061–1072. doi: 10.1016/s1286-4579(00)01260-0. [DOI] [PubMed] [Google Scholar]
- 32.Thanassi D G, Hultgren S J. Assembly of complex organelles: pilus biogenesis in gram-negative bacteria as a model system. Methods. 2000;20:111–126. doi: 10.1006/meth.1999.0910. [DOI] [PubMed] [Google Scholar]
- 33.Thanassi D G, Saulino E T, Lombardo M J, Roth R, Heuser J, Hultgren S J. The PapC usher forms an oligomeric channel: implications for pilus biogenesis across the outer membrane. Proc Natl Acad Sci USA. 1998;95:3146–3151. doi: 10.1073/pnas.95.6.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas J D, Daniel R A, Errington J, Robinson C. Export of active green fluorescent protein to the periplasm by the twin-arginine translocase (Tat) pathway in Escherichia coli. Mol Microbiol. 2001;39:47–53. doi: 10.1046/j.1365-2958.2001.02253.x. [DOI] [PubMed] [Google Scholar]
- 35.Yeo H-J, Savvides S N, Herr A B, Lanka E, Waksman G. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV system. Mol Cell. 2000;6:1461–1472. doi: 10.1016/s1097-2765(00)00142-8. [DOI] [PubMed] [Google Scholar]
- 36.Zhou X-R, Christie P J. Mutagenesis of Agrobacterium VirE2 single-stranded DNA-binding protein identifies regions required for self-association and interaction with VirE1 and a permissive site for hybrid protein construction. J Bacteriol. 1999;181:4342–4352. doi: 10.1128/jb.181.14.4342-4352.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]