Abstract
RNA interference (RNAi) is the process by which double-stranded RNA (dsRNA) directs sequence-specific degradation of messenger RNA in animal and plant cells1,2. In mammalian cells, RNAi can be triggered by 21-nucleotide duplexes of small interfering RNA (siRNA)3. Here we describe inhibition of early and late steps of HIV-1 replication in human cell lines and primary lymphocytes by siRNAs targeted to various regions of the HIV-1 genome. We demonstrate that synthetic siRNA duplexes or plasmid-derived siRNAs inhibit HIV-1 infection by specifically degrading genomic HIV-1 RNA, thereby preventing formation of viral complementary-DNA intermediates. These results demonstrate the utility of RNAi for modulating the HIV replication cycle and provide evidence that genomic HIV-1 RNA, as it exists within a nucleoprotein reverse-transcription complex, is amenable to siRNA-mediated degradation.
RNAi is a ubiquitous mechanism of gene regulation in plants and animals4 in which target mRNAs are degraded in a sequence-specific manner5. RNAi is initiated by the dsRNA-specific endonuclease Dicer, which promotes processive cleavage of long dsRNA into double-stranded fragments between 21 and 25 nucleotides long, termed siRNAs5–8. Small interfering RNAs are incorporated into a protein complex that recognizes and cleaves target mRNAs9. Introduction of dsRNA into mammalian cells does not result in efficient Dicer-mediated generation of siRNA and therefore does not induce RNAi10,11. The requirement for Dicer in maturation of siRNAs can be bypassed by introducing synthetic 21-nucleotide siRNA duplexes, which inhibits expression of transfected and endogenous genes in a variety of mammalian cells3. HIV-1 uses RNA intermediates in its replication. Therefore, we examined whether siRNA duplexes specific for HIV-1 were capable of effecting the degradation of viral RNAs necessary for completion of early and late events in the viral replication cycle.
We directed 21-nucleotide siRNA duplexes against several regions of the HIV-1 genome, including the viral long terminal repeat (LTR) and the accessory genes vif and nef (Fig. 1a). Small interfering RNA duplexes were co-transfected with an HIV-1 molecular clone (HIVNL-GFP; ref. 12) into CD4-positive HeLa (Magi) cells13. Transfection of cells with an infectious molecular HIV-1 clone recapitulates late events in the viral life cycle, including production of viral RNAs, translation of viral proteins and release of virions. Compared with cells not transfected with siRNA duplexes, virus production, measured 24 h after transfection, was reduced 30-fold to 50-fold by homologous siRNAs (Fig. 1b). HIV production was inhibited to a lesser extent by single mismatch siRNAs (MTAR, M441), whereas a vif siRNA with four mismatches (M98) did not inhibit HIV production (Fig. 1b). Activation of the dsRNA-activated protein kinase PKR leads to an inhibition of protein translation in a sequence-non-specific manner relative to the inducing dsRNA. Activation with PKR was not involved in the inhibition of the negative-strand RNA virus RSV (respiratory syncytial virus) by siRNAs14. Similarly, there was no significant induction of activated PKR (phosphorylated on Thr 446) over levels in non-transfected cells by any of the siRNAs (Fig. 1c). To further exclude a PKR effect, Magi cells were co-transfected with two HIV-1 variants (HIV-1NL-GFP, HIV-1YU-2; ref. 15) and with siRNAs that are specifically targeted to either virus. Because of the presence of a green fluorescent protein (GFP) insertion in Nef, HIVNL-GFP should be targeted by the GFP-specific siRNA G388, whereas HIVYU-2, which lacks a GFP insert, should be insensitive to G388. In addition, we exploited sequence differences in the vif genes of these viruses. The M98 siRNA contains four mismatches relative to the HIVNL-GFP vif gene but is completely homologous to HIVYU-2 vif. Thus, M98 should direct the specific inhibition of HIVYU-2 RNA and not HIVNL-GFP RNA. Because of the GFP insertion in HIVNL-GFP, viral RNA produced in cells harbouring both viruses could be distinguished. In the absence of siRNAs, both HIVNL-GFP and HIVYU-2 RNAs were evident in co-transfected cells (Fig. 1d). However, co-transfection with the G388 siRNA resulted in a loss of HIVNL-GFP RNA but not HIVYU-2 RNA. Conversely, the M98 siRNA caused a loss in HIVYU-2 RNA without affecting HIVNL-GFP RNA (Fig. 1d). This sequence-specific inhibition is inconsistent with a sequence-non-specific PKR effect and indicates that siRNAs are inhibiting HIV production by causing the specific degradation of viral RNA. We next examined whether siRNAs could inhibit HIV gene expression (GFP fluorescence) in primary peripheral blood lymphocytes (PBLs), which are natural targets for HIV-1 infection. The frequency of GFP-expressing cells was markedly reduced in cells transfected with homologous siRNAs (T98, G388, nef) relative to cells transfected with mismatched siRNAs or non-transfected cells (Fig. 1e). The level of HIVNL-GFP RNA, as determined by polymerase chain reaction with reverse transcription (RT–PCR), was also markedly reduced in cells transfected with homologous siRNAs (results not shown). Therefore, the components of siRNA-activated RNAi are fully functional in cells naturally targeted by HIV-1 infection.
Figure 1.
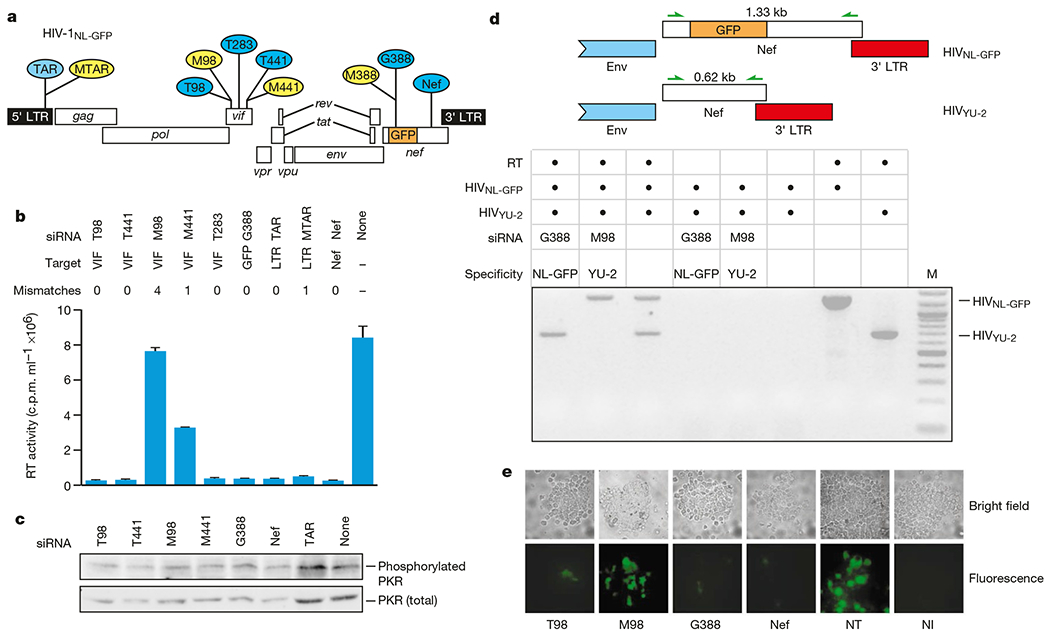
Small interfering RNAs inhibit late events in HIV replication by promoting degradation of HIV-1 RNA. a, HIV targets of siRNAs used in this study. Small interfering RNAs completely homologous to the target HIV sequence (HIVNL-GFP) are shown in blue and those harbouring nucleotide mismatches are shown in yellow. b, Effect of siRNAs on HIV-1 particle production. c, Total and active (phosphorylated) PKR levels in siRNA-transfected Magi cells. d, Small interfering RNAs mediate sequence-specific HIV RNA degradation. The presence of HIVNL-GFP or HIVYU-2 RNA was determined by RT–PCR using HIV Nef-specific primers. Because of the GFP insertion in HIVNL-GFP Nef, RNAs originating from HIVNL-GFP are 710 nucleotides larger than those originating from HIVYU-2. M, molecular weight marker (100 bp ladder, New England Biolabs). e, Effect of siRNAs on HIV expression in primary PBLs.
Upon HIV-1 infection, genomic viral RNA is introduced into the host cell cytoplasm in the form of a nucleoprotein complex, which comprises viral proteins in association with genomic viral RNA16. Within this complex, the viral reverse transcriptase enzyme directs the synthesis of viral cDNA intermediates from the genomic viral RNA template. Recent studies with RSV have indicated that genomic viral RNA, which is tightly associated with nucleocapsid protein, is resistant to siRNAs17. We investigated whether siRNAs were able to direct the specific degradation of genomic viral RNA of HIV-1. The experimental design is outlined in Fig. 2a. Magi cells were transfected with the various siRNAs and infected with HIVNL-GFP 20 h later. Transfection of cells with siRNAs did not significantly interfere with virus uptake per se, on the basis of levels of cell-associated p24 at 1 h after infection (Fig. 2b). The strategy for analysis of viral reverse-transcription intermediates in acutely infected cells is outlined in Fig. 2c. At 1 h after infection, genomic viral RNA was specifically detected in cells transfected with mismatched siRNAs and in non-transfected cells (M98, M441) but not in cells transfected with homologous siRNAs (Fig. 2d). Because genomic viral RNA is the template for the synthesis of viral cDNA intermediates, the synthesis of viral cDNAs, determined 36 h after infection, was dramatically inhibited in cells transfected with homologous siRNAs (T98, GFP, nef) (Fig. 2e). Small interfering RNAs bearing one-nucleotide mismatches (M441, M388) were partially inhibitory relative to the siRNA bearing four mismatches (Fig. 2e). Small interfering RNAs were quite stable in cells: HIV entry was suppressed to equal levels whether virus was added 20 h or 4 days after siRNA transfection (data not shown). Upon completion of viral cDNA synthesis, viral sequences integrate into cellular DNA to form a provirus. The level of provirus formation, as evidenced by the presence of junction sequences flanking viral and cellular DNA (Fig. 2e), was markedly reduced in cells transfected with homologous siRNAs (T98, G388, nef) relative to cells transfected with mismatched (M98) siRNAs or non-transfected cells (Fig. 2f). Collectively, these studies indicate that siRNAs interrupt early events in the HIV replication cycle by directing the specific degradation of genomic HIV-1 RNA, thereby preventing the subsequent synthesis of viral reverse-transcription intermediates and establishment of the provirus.
Figure 2.
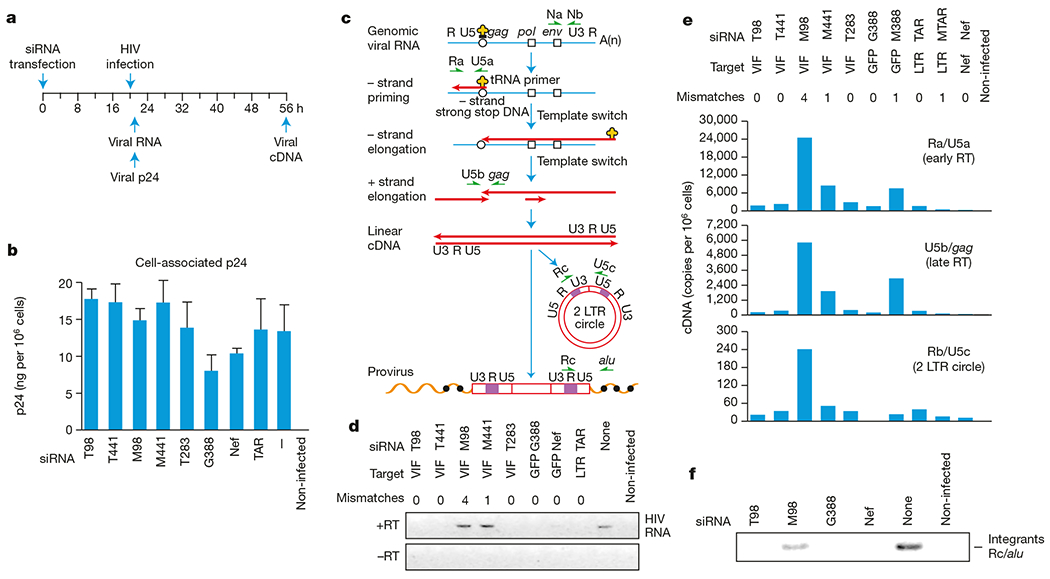
Small interfering RNAs block early events in HIV replication by promoting degradation of genomic HIV RNA. a, Experimental design. b, Levels of trypsin-resistant HIV gag p24 in siRNA-transfected cells. Dash indicates no siRNA transfected into the cells. c, Strategy for analysis of viral nucleic acid intermediates formed early after HIV infection. Major cDNA intermediates in viral reverse transcription are indicated. Blue line, viral RNA; red line, viral cDNA; open circles and squares, primer-binding sites for initiation of minus-strand synthesis and polypurine tracts for plus-strand synthesis, respectively. HIV-specific primers (green arrows) are shown next to the earliest cDNA intermediate they amplify. Integrated (proviral) HIV DNA was amplified using an HIV LTR-specific primer (Rc) and a primer directed to alu repeats (filled circles) within flanking cellular DNA. d, Effect of siRNAs on genomic viral RNA. e, Effect of siRNAs on formation of HIV-1 reverse transcription (RT) intermediates. f, Reduced levels of viral integration in siRNA-transfected cells.
Expression of siRNAs from plasmid templates offers several advantages over synthetic siRNAs, such as stable selection under selectable markers and inducible promoters, which are features that could be useful for genetic approaches to HIV therapy. Therefore, we examined whether expressed siRNAs could inhibit HIV. Modifying a strategy used previously in plants18,19, we constructed plasmids containing a 19-base pair (bp) region of the HIV-1 vif gene in 5′–3′ and 3′–5′ orientations under the control of a T7 promoter (Fig. 3a). Virus production was determined 24 h after a three-way transfection of Magi cells with an HIVNL-GFP molecular clone, the linearized Vif hairpin plasmid (Tl Vif) and a vector expressing T7 RNA polymerase (T7 Pol). In the presence of T7 RNA polymerase, T7 transcripts derived from BstBI-linearized expression plasmids would be predicted to comprise a GGUACC sequence from the T7 promoter, a 19-bp stem of self-complementary Vif sequences, a 3-, 5- or 7-nucleotide loop and a 3′ UU overhang. All three Vif hairpin plasmids containing 3-, 5- or 7- nucleotide loops potently suppressed virus production to 20–30-fold relative to non-transfected cells. By comparison, the presence of an identical plasmid lacking Vif sequences (TL ΔVif or a control plasmid pcDNA) had no effect on virus production in co-transfected cells (Fig. 3b). This inhibitory effect on virus production was reflected by a loss of viral RNA (Fig. 3c). The Vif hairpin plasmid (TL Vif7) also inhibited viral gene expression in primary lymphocytes, whereas there was no inhibitory effect of the plasmid lacking Vif sequences in these cells (Fig. 3d). These results indicate that a sequence-specific RNAi effect can be activated in established and primary cells by siRNAs derived from self-complementary hairpin-generating plasmids. This provides a rationale for gene-therapy approaches to HIV that complement existing post-transcriptional approaches for inhibiting HIV, including ribozymes and antisense RNA (reviewed in ref. 20).
Figure 3.
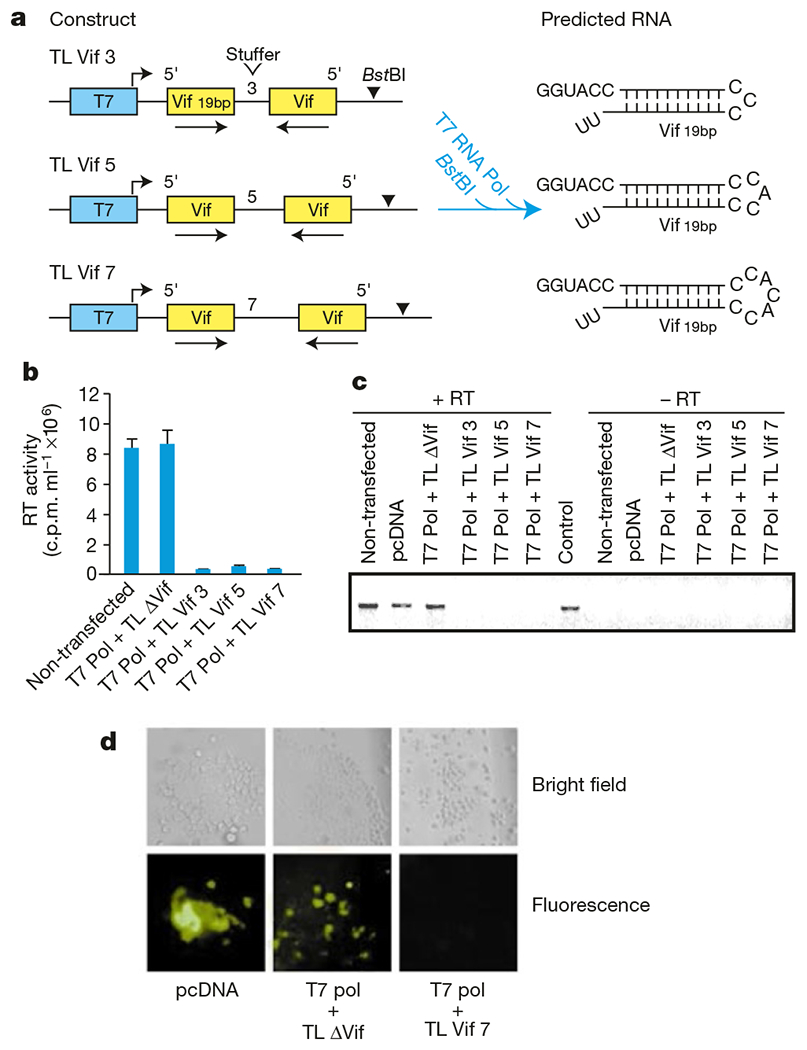
Inhibition of HIV replication by siRNAs derived from plasmid DNA templates. a, Strategy for production of hairpin siRNAs from plasmid vectors. Linearization of each construct with BstBI and transfection into cells with a plasmid expressing T7 RNA polymerase (Pol) predicts the expression of a hairpin RNA with a 19-bp self-complementary Vif stem and non-base-paired loops of 3, 5 and 7 nucleotides. b, Effect of Vif hairpin siRNAs on HIV particle production. Tl ΔVif is identical to plasmids that express Vif hairpin except that it lacks self-complementary Vif sequences. c, Vif hairpin siRNAs promote degradation of HIV RNA. PCR products amplified from HIVNL-GFP served as a control. d, Inhibition of HIV-1 expression by Vif hairpin siRNAs in primary PBLs.
The involvement of RNAi in transposon silencing21,22, suggests that RNAi is an ancient antiviral system that may have evolved as a defence mechanism to protect the host from invasion by mobile genetic elements including transposons and viruses. Several studies have indicated that it is difficult to induce RNAi in mammalian cells using long dsRNAs. Although long dsRNAs can modestly inhibit gene expression in mammalian cells, the effects are not sequence specific3,23 and are more consistent with inhibition by the interferon response. Intriguingly, it is now becoming apparent that underlying the non-specific dsRNA-activated interferon response in mammalian cells, there may indeed be a sequence-specific RNAi effect that can be activated by long dsRNA24–26. Silencing by long dsRNAs has now been observed in various cultured mammalian cells24,25. The mechanism of silencing is consistent with RNAi because there is evidence that the long dsRNAs are processed to siRNAs and target RNAs are specifically degraded. Our results indicate that 21-nucleotide siRNAs promote HIV RNA degradation in primary lymphocytes, suggesting that the major target cell for HIV replication possesses functional components of the siRNA-induced silencing complex that mediates specific cleavage of target RNA2. Future studies should determine whether sequence-specific RNAi that is independent of the interferon response can be activated against HIV by long dsRNAs.
Methods
Synthesis of siRNA
The following RNA oligonucleotides were purchased from Dharmacon: T98 (5′-GGAAAGCUAAGGACUGGUUdTdT-3′); T283 (5′ -AGCACACAAGUAGACCCUGdTdT-3′ ; T441:5′ -CUUGGCACUAGCAGCAUUAdTdT-3′); M98 (5′ GAAAGCUAGGGGAUGGUUdTdT-3′); M441 (5′-CUUGGCACUAACAGCAUUAdTdT-3′); G388 (5′-GACUUCAAGGAAGAUGGCAdTdT-3)′; M388 (5′-GACUUCAAGGGAGAUGGCAdTdT-3′); nef (5′ -GUGCCUGGCUAGAAGCACAdTdT-3′); TAR (5′ -AGACCAGAUCUGAGCCUGGdTdT-3′); and MTAR (5′ -AGACCAGAUAUGAG CCUGGdTdT-3′).
Plasmids
The T7 promoter was modified in the plasmid PCRscript (Stratagene) to form pCRT7. Oligonucleotides corresponding to nucleotides 5,323–5,342 of HIV-1 vif (Genbank accession number M19921) were inserted at the SrfI site of pCRT7. T7 Pol comprises T7 RNA polymerase from Escherichia coli BL21 (DE3) cloned into pcDNA 3.1 (Invitrogen).
Cells and transfections
Magi cells were grown in DMEM containing 10% fetal bovine serum (FBS). PHA-activated, elutriated PBLs were cultured in RPMI containing 10% FBS and 64 Uml−1 of interleukin-2 (ICN). Magi cells were transfected with oligofectamine (GIBCO) by the manufacturer’s protocol in the presence of 1 μg HIV plasmid and/or 60 pmol of siRNA oligonucleotides. Transfection efficiencies were 75–85%. For PHA-activated PBLs, 5 × 106 cells were electroporated using a Gene Pulser apparatus (Bio-Rad) at 250V, 960 μF, resistance R = ∞ with 5 μg plasmid and/or 200 pmol siRNA. Transfection efficiencies were 30–50% of viable cells. Three-way transfections with siRNA expression plasmids comprised 0.1 μg T7 Pol, 0.5 μg pTL Vif and 0.5 μg pNLGFP (Magi cells), or 0.5 μg T7 Pol, 2 μg TL Vif and 2 μg pNLGFP (for primary lymphocytes). Transfected cells were centrifuged (1,200g) on DAKO silanized slides and examined under bright-field illumination or fluorescence (wavelength 516 nm) on a Zeiss Axioplan 2 microscope.
PCR analysis
Real-time PCR was performed as previously reported27. Products were amplified from 5 to 20 μl of extrachromosomal DNA in 50-μl reactions containing 1 × HotStart Taq buffer (Qiagen), 200 nM dNTPs, 400 nM primers and 1.5 U HotStart Taq. Two-LTR junctions were amplified by the primers Rc (5′-TAGACCAGATCT GAGCCTGGGA -3′) and U5c (5′-GTAGTTCTGCCAATCAGGG AAG -3′). Early products were amplified by the primers Ra (5′-TCTCTGGTTAGACCAGATCTG-3′) and U5a (5′ -GTCTGAGGGATCTCTAGTTAC-3′), and late products were amplified with U5b (5′ -GGGAGCTCTCTGGCTAACT-3′) and gag (5′ -GGATTAA CTGCGAATCGTTC-3′) primers. The oligonucleotide probe for real-time PCR was as previously reported27.
Viral assays
For RT–PCR, 1–2 μg RNA was reverse transcribed and amplified by PCR using the Nef primers Na (5′-GACAGGGCTTGGAAAGG-3′) and Nb (5′ -TTAGCAGTTCTGAA GTACTC-3′) as described previously28. The integration assay was performed on DNAzol-extracted total DNA (Invitrogen) using the Alu primer SB704 (5′ -TGCTGGGATTACAG GCGTGAG-3′) and primer Rc for the first round of PCR (25 cycles). Nested PCR was performed under the same conditions using primers M667 (5′ -GGCTAACTAGGGAA CCCACTG-3′) and AA55 (5′-CTGCTAGAGATTTTCCACACTGAC-3′). For virus production, viral p24 (capsid) was measured by enzyme-linked immunosorbent assay according to the manufacturer’s protocol (Beckman-Coulter). Reverse transcription activity was measured as previously reported28.
PKR assay
We electrophoresed 20 μg of whole-cell lysates in triple detergent lysis buffer on a 10% SDS–polyacrylamide gel and electrotransferred to a nitrocellulose membrane (Amersham Hybond C + ). The membrane was probed with a phospho-Thr 446 PKR-specific antibody or a PKR-specific antibody (Upstate Biotechnology).
Acknowledgements
We thank A. Mann for research support, C. Mello and P. Zamore for discussions, B. Mellor for preparation of the figures, and T. Pinkos for manuscript preparation. We also acknowledge assay support provided by the University of Massachusetts Center for AIDS Research. HIVYU-2 was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH), from B. Hahn and G. Shaw. This study was supported by grants from the NIH and the Jenner Foundation to M.S.
Footnotes
Competing interests statement
The authors declare that they have no competing financial interests.
References
- 1.Sharp PA RNA interference–2001. Genes Dev. 15, 485–490 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Hutvagner G & Zamore PD RNAi: nature abhors a double-strand. Curr. Opin. Genet. Dev 12, 225–232 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Elbashir SM et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Fire A et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Zamore PD, Tuschl T, Sharp PA & Bartel DP RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101, 25–33 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Elbashir SM, Lendeckel W & Tuschl T RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15, 188–200 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammond SM, Bernstein E, Beach D & Hannon GJ An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404, 293–296 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Bernstein E, Caudy AA, Hammond SM & Hannon GJ Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Nykanen A, Haley B & Zamore PD ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107, 309–321 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Caplen NJ, Fleenor J, Fire A & Morgan RA dsRNA-mediated gene silencing in cultured Drosophila cells: a tissue culture model for the analysis of RNA interference. Gene 252, 95–105 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Ui-Tei K, Zenno S, Miyata Y & Saigo K Sensitive assay of RNA interference in Drosophila and Chinese hamster cultured cells using firefly luciferase gene as target. FEBS Lett. 479, 79–82 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Welker R, Harris M, Cardel B & Krausslich HG Virion incorporation of human immunodeficiency virus type 1 Nef is mediated by a bipartite membrane-targeting signal: analysis of its role in enhancement of viral infectivity. J. Virol 72, 8833–8840 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimpton J & Emerman M Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J. Virol 66, 2232–2239 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bitko V & Barik S Phenotypic silencing of cytoplasmic genes using sequence-specific double-stranded short interfering RNA and its application in the reverse genetics of wild type negative-strand RNA viruses. BMC Microbiol. 1, 34–45 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y et al. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: Identification of replication-competent and -defective viral genomes. J. Virol 65, 3973–3985 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore J & Stevenson M New targets for inhibitors of HIV-1 replication. Nature Rev. Mol. Cell Biol 1, 40–49 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Bitko V & Barik S An endoplasmic reticulum-specific stress-activated caspase (caspase-12) is implicated in the apoptosis of A549 epithelial cells by respiratory syncytial virus. J. Cell Biochem 80, 441–454 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Wang MB & Waterhouse PM High-efficiency silencing of a β-glucuronidase gene in rice is correlated with repetitive transgene structure but is independent of DNA methylation. Plant Mol. Biol 43, 67–82 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Varshawesley S et al. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27, 581–590 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Dornburg R & Pomerantz RJ HIV-1 gene therapy: promise for the future. Adv. Pharmacol 49, 229–261 (2000). [PubMed] [Google Scholar]
- 21.Ketting RF, Haverkamp TH,van Luenen HG & Plasterk RH Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99, 133–141 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Tabara H, Hill RJ, Mello CC, Priess JR & Kohara Y pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development 126, 1–11 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Caplen NJ, Parrish S, Imani F, Fire A & Morgan RA Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl Acad. Sci. USA 98, 9742–9747 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Billy E, Brondani V, Zhang H, Muller U & Filipowicz W Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl Acad. Sci. USA 98, 14428–14433 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paddison PJ, Caudy AA & Hannon GJ Stable suppression of gene expression by RNAi in mammalian cells. Proc. Natl Acad. Sci. USA 99, 1443–1448 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang S, Tutton S, Pierce E & Yoon K Specific double-stranded RNA interference in undifferentiated mouse embryonic stem cells. Mol. Cell. Biol 21, 7807–7816 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharkey M et al. Persistence of episomal HIV-1 infection intermediates in patients on highly active antiretroviral therapy. Nature Med. 6, 76–81 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brichacek B & Stevenson M Quantitative competitive RNA PCR for quantitation of virion associated HIV-1 RNA. Methods 12, 294–299 (1997). [DOI] [PubMed] [Google Scholar]


