Abstract
Purpose
Talaromyces marneffei (TM) is an opportunistic fungus leading to multi-organ damages and poor prognosis in immunocompromised individuals. TM infections in children are rare and our knowledge to TM infection is insufficient. To investigate the clinical characteristics of TM-infected children and to explore the underlying mechanisms for host against TM, we analysed TM-infected patients diagnosed in our hospital.
Methods
Eight patients with TM infections have been identified in Shenzhen Children’s Hospital during 2017–2021. Clinical data were collected from medical records. Immunological features were evaluated by flow cytometry. Literatures were also reviewed to summarize the reported inborn errors of immunity (IEIs) with TM infections.
Results
All 8 children were HIV-negative. The most common symptom of TM infections was fever (8/8), followed by weight loss (7/8), pneumonia (7/8), hepatomegaly (7/8), splenomegaly (6/8), anemia (6/8), lymphadenopathy (5/8), thrombocytopenia (3/8), diarrhea (3/8), rashes or skin lesions (3/8), and osteolytic lesions (1/8). Five children died during the follow-ups. CD3+ T cells were decreased in 6 patients. Eight patients had reduced natural killer cells. All patients went gene sequencing and were finally diagnosed as IEIs, including STAT1 gain-of-function, IL-2 receptor common gamma chain deficiency, adenosine deaminase deficiency, CD40 ligand deficiency, and STAT3 deficiency. Another 4 types of IEIs (CARD9, IFN-γ receptor 1, RelB, and NFKB2 deficiency), have been reported with TM infections based on literature review.
Conclusion
TM infections resulted in systemic injuries and high mortality. The spectrum of IEIs underlying TM infections indicated that T cell-mediated immunity, IFN-γ, IL-17 signalings and NF-κB pathways were important for host responses against TM infection. In reverse, for HIV-negative children without other secondary immunodeficiencies, IEIs should be considered in TM-infected children.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11046-022-00659-0.
Keywords: Talaromycesmarneffei, Inborn errors of immunity, Primary immunodeficiency diseases, Gene mutations, Children
Introduction
Inborn errors of immunity (IEIs), also known as primary immunodeficiency diseases (PIDs), are sorts of diseases caused by monogenic mutations, resulting in dysfunctions in immunity, and present vulnerable to infectious disease, autoimmune diseases, autoinflammatory diseases, and malignancies. Till now, more than 430 IEIs have been reported and classified into 10 categories [1]. For each category, they have unique clinical and immune characteristics, and are prone to different pathogen infections, indicating distinct underlying immunity defects [2, 3]. It’s a great challenge to diagnose and treat these rare diseases. Characteristic clinical manifestations, including typical infectious spectrum, will provide clues for disease recognition and diagnosis.
Talaromyces marneffei (TM, formerly known as Penicillium marneffei), is an important opportunistic pathogen and can result in local or disseminated talaromycosis in immunocompromised individuals. This is a dimorphic fungus, growing as a mold at 25 °C, and as a yeast at 37 °C [4]. The morphological conversion is associated with its virulence [5]. It was first isolated from bamboo rat in Vietnam in 1956 [6], and is mainly endemic in Southeast Asia, Northeast India, and South China [7–10]. Typical symptoms of disseminated talaromycosis include fever, anemia, weight loss, hepatosplenomegaly, lymphadenopathy, and multiply organ involvements [11, 12]. In spite of antifungal treatments, the mortality of talaromycosis remains high [13].
TM infections are widely reported in human immunodeficiency virus (HIV)-infected adults, especially in those with severely decreased CD4+ T cells (less than 100 cells/μL) [14], implying the role of cellular immunity against this fungus. Less commonly, TM infections are also described in people with secondary immunodeficiency conditions, such as malignancies [15], post-transplants [16], autoimmune diseases [17], and diabetes mellitus [18]. Talaromycosis is uncommon in otherwise healthy persons. Thus, TM infections in persons who are HIV negative and without secondary immunodeficiencies may suggest underlying primary immunodeficiencies, that is, IEIs. Actually, some HIV-negative adults with TM infections were detected with anti-IFN-γ autoantibodies, belonging to a kind of adult-onset IEIs [19, 20]. Most recently-reported children patients are also HIV-negative, and more infected children were revealed to have IEIs, such as hyperimmunoglobulin M syndrome (HIGM) [21], hyper-IgE syndrome (HIES) [22, 23], and STAT1 gain-of-function (GOF) disorder [24]. However, the reported spectrum of IEIs with TM infections is very limited. The clinical and immunological data on HIV-negative children with TM infections are even scarcer.
In this study, we retrospectively reviewed 8 children with TM infections and all of them were finally diagnosed as IEIs. Analysis of their clinical, immunological and genetic features may increase pediatricians’ awareness of this fungus infection. TM infections in HIV-negative children could help to diagnose different categories of IEIs. Besides, the spectrum of IEIs with TM infections may in reverse shed light on the underlying mechanisms for anti-TM infections.
Materials and Methods
Patients and Clinical Data
A total of 8 patients with TM infections were identified in Shenzhen Children’s Hospital during 2017–2021. All of the patients were HIV-negative. Clinical data were collected from patients’ medical records, including demographic information, initial clinical symptoms, onset age, family histories of IEIs, recurrent infections, complications, physical examinations, routine laboratory examinations, and follow-ups. Informed consents were obtained from each patients’ parents or guardians. This retrospective study was approved by the Ethics Committee of Shenzhen Children’s Hospital (2019050).
TM Identification
Microbiological Culture
Specimens including blood, bone marrow (BM), sputum, bronchoalveolar lavage fluid (BALF), ascites, cerebrospinal fluid (CSF), lung biopsies, or lymph nodes biopsies, were cultured on Sabouraud dextrose agar at 25 °C. Typical TM colonies present flat green-yellowish surfaces and produce diffusible red pigments to the surroundings. Multinucleate hyphal can be observed under microscope and cells divide by septation. When incubated at 35 °C, the conidia convert to yeasts and divide by medial fission.
Histopathological Examination
Wrights-stained bone marrow smears were observed under microscope. In positive infections, numerous yeast-like cells were observed inside and outside host cells. The yeast-likes cells were 2–4 μm in diameter and had clear transverse septa. Tissue sections from lymph node biopsy were stained with hematoxylin and eosin (H&E), Grocott methenamine silver (GMS), and periodic acid–Schiff (PAS) stain. Positive sections showed that small yeast-like cells distributed within macrophages and histiocytes, and divided by fission.
Megagenomic Next-Generation Sequencing (mNGS)
DNA in BALF was extracted using TIANamp Micro DNA Kit (TIANGEN, Beijing, China). DNA libraries were constructed through DNA-fragmentation, end-repair, adapter-ligation and PCR amplification. Library qualities were analysed using Agilent 2100 and then qualified libraries were sequenced using MGISEQ-2000 platform by BGI (Shenzhen, China). Then low-quality reads were trimmed off. Human host sequences which were mapped to human reference genome (hg19) using Burrows-Wheeler Alignment (BWA) were computational subtracted [25]. The remaining data were classified by simultaneously aligning to four Microbial Genome Databases, consisting of bacteria, fungi, viruses and parasites. The classification reference databases were downloaded from NCBI (ftp://ftp.ncbi.nlm.nih.gov/genomes/).
Genetic Analysis
Genomic DNA was extracted from peripheral blood mononuclear cells (PBMCs) of patients and their parents using QIAamp DNA Mini Kit (Qiagen, Germany). Whole exome sequencing or targeted panel sequencing were performed using Illumina HiSeq 2000 by MyGenostics (Beijing, China). Targeted panel was able to capture 423 IEI-related genes using the GenCap custom enrichment kit (MyGenostics). Then the qualified paired-end clean reads were mapped to the human reference genome (hg19). Duplicated reads were removed using Picard (http://broadinstitute.github.io/picard/). Variants were called by genome analysis toolkit (GATK) pipeline [26]. The identified variants were annotated using ANNOVAR [27], associated with 1000 genomes, Exome Aggregation Consortium, and the Human Gene Mutation Database. REVEL (rare exome variant ensemble learner) was used for functional prediction [28]. Splice site was predicted by Human Splicing Finder [29]. The pathogenicity of mutations was assessed following the American College of Medical Genetics and Genomics guideline [30]. All putative pathogenic variants were confirmed by Sanger sequencing.
Flow Cytometry Analysis
Proportion of IL-17A-Producing Cells
PBMCs were stimulated by Cell Stimulation and Protein Transport Inhibitor Cocktail (2 μl/ml, Biogems, USA) for 4 h at 37 °C. Then, PBMCs were washed twice with phosphate buffer saline (PBS) and incubated with CD3-PerCP-Cy5.5 and CD8-BV510 (BD Biosciences, USA) for 20 min at room temperature. Cells were then washed twice with PBS, and fixed with Intracellular Fixation Buffer (eBioscience, USA) for 30 min at 4 °C. After fixation, cells were permeabilized with diluted Permeabilization Buffer (1 × , eBioscience, USA) for 20 min at room temperature. Cells were then washed twice and incubated with IL-17A-PE (eBioscience, USA) for 30 min at room temperature. Cells were then washed twice and resuspended and acquired on BD FACS Canto II flow cytometer.
Lymphocyte Subsets
Immunophenotyping of peripheral blood lymphocyte subsets were mainly based on Reference [31]. Briefly, EDTA-anticoagulated whole blood was split into 2 parts and stained with different panels of antibodies to explore T cells and B cells separately. For T cell subsets, the antibody panel contained CD3-PerCP-Cy5.5, CD4-FITC, CD8-BV510, CD45RA-PE-Cy7, CD27-APC, TCR aβ-PE, and TCR γδ-BV421. For B cell subsets, the antibody panel contained CD19-APC, CD27-V450, IgD-BV510, CD24-PE, and CD38-PerCP-Cy5.5 (All antibodies were purchased from BD Biosciences). After 20 min, erythrocytes in blood sample were lysed by red cell lysis buffer (TIANGEN, China). Then cells were centrifuged and washed twice with PBS. After resuspended, the cells were acquired on BD FACS Canto II, and the data were analyzed using FlowJo (V10.4, BD Biosciences).
CD40 Ligand (CD40L) Expression
PBMCs were stimulated by 50 ng/mL phorbol myristate acetate (PMA, Merck) and 500 ng/mL ionomycin (Merck) for 4 h at 37 °C. After washed twice with PBS, cells were stained with CD4-PerCP-Cy5.5 (BD Biosciences, USA) and CD40L-FITC (eBioscience, USA) for 30 min at room temperature. Then cells were washed twice with PBS and resuspended and acquired on BD FACS Canto II flow cytometer. The percentage of CD4 + CD40L + cells were determined.
Search Strategy for Literature Review
PubMed was systematically searched for English articles published from 1956 to 2022. China National Knowledge Infrastructure (CNKI) (http://cnki.net/) and Wanfang (http://www.wanfangdata.com.cn/) data were also searched for Chinese articles published during the same period. Search terms were (“Talaromyces marneffei” OR “Penicillium marneffei” OR “marneffei” OR “penicilliosis” OR “talaromycosis”) AND (“mutation” OR “immunodeficiency” OR “inborn errors of immunity”). Their Chinese equivalents were used for Chinese database search. Full-text version of the original articles were reviewed. Exclusion criteria were as follows: (1) The patients were HIV-positive; (2) No detailed mutation information was provided; (3) The reported mutations were unrelated to IEIs; (4) There was no convinced evidence for TM infections.
Statistical Analysis
Continuous data which were not in Gaussian distributions were described as median (range). Categorical data were presented as numbers and percentages.
Results
Identification of TM Infections
During 2017–2021, eight children with TM infections were identified in Shenzhen Children’s Hospital, and all of them had confirmed microbiological evidences. Seven patients had positive culture results, using specimens from blood (P2, P3, P5), sputum (P2, P4, P5), BALF (P2, P4), BM (P4, P5), CSF (P4), ascites (P6), lung biopsy (P1), and lymph node biopsy (P8), separately. Typical images of TM infection were observed in BM smears from P2 and P5. mNGS for BALF reinforced TM infections in P8. For P7, histopathological examination of the lymph node tissue revealed granulomatous inflammation. PAS and GMS staining showed that numerous yeast-like or sausage-like organisms distributed within macrophages, and had transverse septa, demonstrating TM infection.
Clinical Manifestations
This study included 2 females and 6 males, with a median onset age of TM infections at 23 months old (range: 4–102 m). All the 8 patients were born and lived in the southeast of China, including 5 patients in Guangdong, 1 patient in Hunan, 1 patient in Yunnan, and 1 patient in Hubei province (Table 1).
Table 1.
Clinical features of the 8 patients with TM infection
| Patients | Gender | TM Onset age | Medical history | Symptoms/signs | Concurrent infections | Genetic defect |
Outcome |
|---|---|---|---|---|---|---|---|
| P1 | F | 7y11m | 1y3m: thrush, diarrhea, severe pneumonia, disseminated tuberculosis |
Weight loss, fever, skin lesions, CMC, pneumonia, hepatosplenomegaly, lymphadenopathy, anemia |
M. catarrhalis, H .influenzae, S. pneumoniae, EBV, rotavirus |
STAT1 (WES) |
Improved |
| P2 | M | 1y5m | 1y3m: lymphatic tuberculosis | Weight loss, fever, pneumonia, ARDS, thrush, diarrhea, hepatomegaly, hepatic failure, pancytopenia |
CMV, K. pneumoniae, E. cloacae, B. cepacia |
ADA (IEI panel) |
Death |
| P3 | M | 1y1m | 5 m:axillary abscess; 6 m: severe pneumonia, ARDS, eosinophilia, CMV infections |
Weight loss, fever, pneumonia, abdominal distension, diarrhea, hematochezia, jaundice, hypothyroidism, hapatosplenomegaly, lymphadenopathy, portal hypertension, cavernous transformation of the portal vein anemia |
CMV, S. typhimurium |
CD40LG (IEI panel) |
Death |
| P4 | F | 2y5m | – | Weight loss, fever, disturbance of consciousness, intracranial infection, cerebral infarction, cerebral hernia, pneumonia, respiratory failure, lymphadenopathy, skin lesions, anemia | M. pneumoniae |
STAT3 (IEI panel) |
Death |
| P5 | M | 8 m |
3 m: PJP Family history: One brother died from TM infection at 6 m |
Fever, pneumonia, rash, edema, hematuresis, diarrhea, hepatosplenomegaly, anemia, thrombocytopenia | Rhinovirus |
IL2RG (IEI panel) |
Death |
| P6 | M | 4 m | alpha thalassaemia | Weight loss, fever, pneumonia, peritonitis, hepatosplenomegaly, pancytopenia, HLH, MODS, |
Disseminated tuberculosis, C. parapsilosis, rhinovirus |
IL2RG (WES) |
Death |
| P7 | M | 4y7m | – | Weight loss, fever, lymphadenopathy, hepatosplenomegaly | – |
STAT1 (WES) |
Improved |
| P8 | M | 8y6m | – | Weight loss, fever, pneumonia, osteolytic lesions, lymphadenopathy, hepatosplenomegaly, lymphopenia, CMC |
C. albicans, Rhinovirus, S. aureus, M. catarrhalis, H. influenzae |
STAT1 (WES) |
Improved |
TM T. marneffei, F female, M male, CMV cytomegalovirus, EBV Epstein-Barr virus, PJP pneumocystis jiroveci pneumonia, CMC chronic mucocutaneous candidiasis, ARDS acute respiratory distress syndrome, HLH hemophagocytic lymphohistiocytosis, MODS multiple organ dysfunction syndrome, WES whole exome sequencing, IEI inborn errors of immunity
All the 8 patients presented disseminated talaromycosis. The most common symptom of TM infections was fever (8/8), followed by weight loss (7/8), pneumonia (7/8), hepatomegaly (7/8), splenomegaly (6/8), anemia (6/8), lymphadenopathy (5/8), thrombocytopenia (3/8), diarrhea (3/8), rashes or skin lesions (3/8), and osteolytic lesions (1/8) (Table 1). Besides, eosinophilia was observed in P2 and P3 (Table S1).
There were life-threatening complications, including acute respiratory distress syndrome (ARDS) and hepatic failure (P2), cerebral infarction and hernia, and respiratory failure (P4), portal hypertension (P3), hemophagocytic lymphohistiocytosis (HLH) and multiple organ dysfunction syndrome (MODS) (P6). Six patients experienced episodes in intensive care unit.
Special pathogens need to be noticed. Three patients had previous or concurrent M.tuberculosis (TB) infections (P1, P2, and P6), two of whom with disseminated tuberculosis. P5 had Pneumocystis jiroveci pneumonia (PJP) at 3 months old. P1 and P8 had chronic mucocutaneous candidiasis, which caused by C. albicans. P3 had recurrent S. typhimurium enteritis, proven by positive stool culture. Three patients were co-infected by herpes virus. Epstein-Barr virus (EBV) and cytomegalovirus (CMV) DNAs were positively amplified in BALFs from P1 and P2, separately. CMV DNAs were detected in serum and urine in P3 and CMV-specific IgM antibodies were also positive. Bacteria including M.catarrhalis and H.influenzae were also found concurrent with TM infections.
For laboratory examinations (Table S1), most patients had elevated erythrocyte sedimentation rate (ESR) (5/6) and c-reactive protein (CRP) (7/8). 3/6 patients had increased procalcitonin. Serum 1,3-beta-D-glucan was elevated in 5/7 patients. Higher aspartate amino transferase (AST) was observed in 5/8 patients. Serum lactate dehydrogenase (LDH) was elevated in 5/8 patients. As for conventional immunological assessments, CD3+ T cells were decreased in 6 patients (except for P3 and P7), together with decreased CD4+ and CD8+ T cells. Reduced CD19+ B cells were only observed in P2 and P8. All the patients had decreased natural killer (NK) cells.
Autoimmunity was found in four patients. P1 presented positive antinuclear antibodies, perinuclear antineutrophil cytoplasmic antibodies (pANCA), and autoantibodies to thyroglobulin, thyroid peroxidase, SS-A, ribonucleoprotein, and platelet. Another three patients (P3, P4, P8) exhibited autoantibodies and positive Coombs test. Low complement C3 level was observed in P2.
Genetic Analysis and IEI Diagnosis
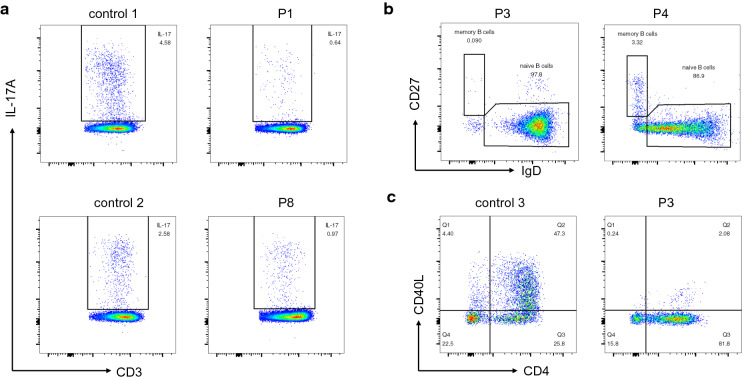
Based on the clinical symptoms and conventional immunological tests, all 8 patients further went gene sequencing (Table 1). Whole exome sequencing revealed heterozygous mutations in STAT1 in three patients: L351F in P1, D65N in P7, and M390I in P8. All these three missense mutations have been reported to cause STAT1 GOF mutations [24, 32, 33]. We further examined the proportion of IL-17A-producing T cells in P1 and P8, and found out that patients had decreased IL-17A-producing T cells (Fig. 1a).
Fig. 1.
Flow cytometric analysis of immunological functions in patients with T.marneffei infections. a Proportions of CD3 + CD8-IL-17A + T cells following stimulation with PMA and ionomycin were reduced in P1 and P8. b Percentages of CD19 + IgD-CD27 + memory B cells were decreased in P3 and P4. c The expression of CD40 ligand in CD4+ T cells stimulated by PMA and ionomycin was dramatically reduced in P3
The other five mutated genes found in patients were related to combined immunodeficiency (CID). Nonsense mutation W155X in IL2RG was found in P5 and P6, leading to X-linked severe combined immune deficiency (SCID). These two boys had lymphopenia, with very low CD3+ T cells and NK cells, and high proportion of B cells. The immunoglobulin levels were very low. Compound heterozygous mutations in ADA were detected in P2. He had profound lymphopenia, very low CD3+ T cells, CD19+ B cells and NK cells, consistent with T-B-NK- SCID phenotype. The immunoglobulin levels were not detected before intravenous immunoglobulin (IVIG). P3 had large fragment deletion in CD40LG. The IgG and IgA concentrations were low while the IgM level was within the normal range. Flow cytometry revealed that CD19 + IgD-CD27 + memory B cells were nearly absent (0.09%, normal range: 2.98–14.18%, Fig. 1b). After stimulated by PMA and ionomycin, CD40L expression was lower in P3 than that in control (2.08% vs. 47.3%, Fig. 1c). P4 had a heterozygous missense mutation in STAT3. The proportion of CD19 + IgD-CD27 + memory B cells was reduced (3.32%, normal range: 3.6–18.55%, Fig. 1b).
Treatments and Outcomes
Treatments were summarized in Table 2. All patients received anti-fungal drugs after TM suspicion or identification. Four of them were treated with additional anti-TB medications. One patient was diagnosed as HLH and treated with methylprednisolone and etoposide. Sulfamethoxazole and trimethoprim were added in 4 patients for PJP prophylaxis and IVIG were administered monthly in six patients. Besides medications, five patients ever had respiratory failure and were supported by mechanical ventilation. Four patients received fiberoptic bronchoscopy examination and bronchoalveolar lavage. Five patients had surgeries, including abscess excisions in P2 and P3, decompressive craniectomy in P4, and lymph nodes biopsies in P7 and P8.
Table 2.
Treatments of IEI patients with TM infections
| Patients | Antifungal | Anti-TB | Other treatments |
|---|---|---|---|
| P1 | VCZ, AmB, ITZ, FCZ | INH, RFP, PZA, LNZ | IVIG |
| P2 | VCZ, AmB | INH, RFP | CoSMZ, GCV, IVIG |
| P3 | VCZ, ITZ, AmB | – | CoSMZ, GCV, IVIG |
| P4 | VCZ, MCFG, AmB, ITZ | INH, RFP, PZA | ACV, DXM, IVIG |
| P5 | VCZ, AmB, ITZ | – | CoSMZ, IVIG |
| P6 | VCZ | INH, RFP, PZA, LNZ | MP, VP-16, CoSMZ, IVIG |
| P7 | AmB, ITZ | – | – |
| P8 | VCZ, ITZ | – | – |
IEI inborn errors of immunity, TM T. marneffei, AmB amphotericin B, VCZ voriconazole, ITZ itraconazole, FCZ fluconazole, MCFG micafungin, INH isoniazid, RFP rifampicin, PZA pyrazinamide, LNZ linezolid, CoSMZ compound sulfamethoxazole and trimethoprim, AZM azithromycin, GCV ganciclovir, ACV acyclovir, DXM dexamethasone, MP methylprednisolone, VP-16 etoposide, IVIG intravenous immunoglobulin
As for outcomes, 3 patients with STAT1 GOF mutations had improved. They were followed-up regularly and received itraconazole or fluconazole for long-term prophylaxis. The other 5 patients presenting combined immunodeficiencies, including SCID, HIGM, HIES, died although after comprehensive therapy.
Discussion
TM is an endemic opportunistic fungus, leading to multi-organ damages and poor prognosis in immunocompromised individuals. TM infections in children were rare. Most children patients were HIV-negative and more children patients were diagnosed as IEIs. In this study, we identified 8 children with TM infections, and finally all the patients were diagnosed as IEIs. We described TM infections in an adenosine deaminase (ADA) deficient child for the first time and enriched the spectrum of IEIs underlying TM infections.
Till now, 9 types of IEIs have been reported with TM infections, that is, CD40 ligand deficiency (12 cases), STAT1 GOF (11 cases), STAT3 deficiency (7 cases), X-SCID (3 cases), IFN-γ receptor 1 deficiency (2 cases), CARD9 deficiency (2 cases), ADA deficiency (1 case), RelB deficiency (1 case) and NFKB2 deficiency (1 case) (Table 3). There were more IEI patients reported with TM infections, however, the detailed mutations were not described. Thus, these cases were not included. For STAT1 GOF mutations, all the published cases were missense mutations. For CD40LG gene, deletions, especially large fragment deletions, and splice site mutations, were more frequently reported. Missense and in-frame deletions were common mutations in STAT3 deficiencies. The currently reported two cases with CARD9 mutations were both compound heterozygous mutations. All the three X-SCID patients had nonsense mutations in IL2RG, and died during the follow-ups.
Table 3.
Reported inborn errors of immunity in HIV-negative children with T. marneffei infection
| Patient number | Geneticdefect | Inheritance | Nucleotide change | Amino acid change | Mutation type | Outcome | References |
|---|---|---|---|---|---|---|---|
| P1 | STAT1 | AD GOF | c.1053G > T | p.L351F | Missense | Improved | This study |
| P2 | ADA | AR |
(1) c.730delG (2) c.202T > A |
(1) p.E244KfsX67 (2) p.Y68N |
Compound heterozygous mutations | Dead | This study |
| P3 | CD40LG | XL | – | – | Large fragment deletion including exon1-5 | Dead | This study |
| P4 | STAT3 | AD LOF | c.115G > A | p.E39K | Missense | Dead | This study |
| P5 | IL2RG | XL | c.464G > A | p.W155X | Nonsense | Dead | This study |
| P6 | IL2RG | XL | c.464G > A | p.W155X | Nonsense | Dead | This study |
| P7 | STAT1 | AD GOF | c.193G > A | p.D65N | Missense | Improved | This study |
| P8 | STAT1 | AD GOF | c.1170G > A | p.M390I | Missense | Improved | This study |
| P9 | STAT1 | AD GOF | c.800C > T | p.A267V | Missense | Improved | [32] |
| P10 | STAT1 | AD GOF | c.821G > A | p.R274Q | Missense | Improved | [42] |
|
P11 (adult) |
STAT1 | AD GOF | c.859 T > A | p.Y287N | Missense | Improved | [60] |
| P12 | STAT1 | AD GOF | c.863C > T | p.T288I | Missense | Improved | [32] |
| P13 | STAT1 | AD GOF | c.1053G > T | p.L351F | Missense | Improved | [24] |
| P14 | STAT1 | AD GOF | c.1074G > T | p.L358F | Missense | Dead | [32] |
| P15 | STAT1 | AD GOF | c.1170G > A | p.M390I | Missense | Improved | [32] |
| P16 | STAT1 | AD GOF | c.193G > A | p.D65N | Missense | Improved | [24] |
| P17 | STAT3 | AD LOF | c.1679_1681del | Not stated | Deletion | Improved | [21] |
| P18 | STAT3 | AD LOF | c.1593A > T | Not stated | Missense | Improved | [21] |
| P19 | STAT3 | AD LOF | c.1593A > T | p.K531N | Missense | Improved | [23] |
|
P20 (adult) |
STAT3 | AD LOF | c.92G > A | p.R31Q | Missense | Improved | [61] |
| P21 | STAT3 | AD LOF | c.1121A > G | p.D374G | Missense | Improved | [62] |
| P22 | STAT3 | AD LOF | c.1673G > A | p. G558D | Missense | Improved | [22] |
| P23 | CD40LG | XL | c.424_436del | Not stated | Deletion | Improved | [21] |
| P24 | CD40LG | XL | > 132 kb | Not stated | Large fragment deletion | Improved | [21] |
| P25 | CD40LG | XL | c.1978 + 1G > A | Not stated | Splicing error | Improved | [21] |
| P26 | CD40LG | XL | c.598A > T | Not stated | Missense | Improved | [21] |
| P27 | CD40LG | XL |
g.IVS1-3 T > G (c.157insAG) |
p.I53RfsX2 | Splicing error | Dead | [63] |
| P28 | CD40LG | XL | g.IVS1 + 1G > A (c.75-156del82bp) | p.M25IfsX26 | Splicing error | Improved | [63] |
| P29 | CD40LG | XL |
g.IVS3 + 1G > A (exon3 missing) |
– | Splicing error | Improved | [63] |
| P30 | CD40LG | XL |
g.IVS1-1G > A (c.158-161delTAGA) |
p.I53KfsX13 | Splicing error | Lost to follow-up | [63] |
| P31 | CD40LG | XL |
g.IVS4 + 1G > C (exon4 missing) |
p.L116-136del | Splicing error | Improved | [63] |
| P32 | CD40LG | XL | – | – | Large fragment deletion including exon4-5 | Improved | [63] |
| P33 | CD40LG | XL | g.IVS1 + 1G > A | p.M25IfsX26 | Splicing error | Improved | [32] |
| P34 | IL2RG | XL | c.185G > A | Not stated | Nonsense | Dead | [21] |
| P35 | CARD9 | AR |
(1) c.440 T > C (2) c.586A > G |
(1)p.L147P (2)p.K196E |
Compound heterozygous mutations | Dead | [40] |
| P36 | CARD9 | AR |
(1) c.1118G > C (2) c.610C > T |
(1)p.R373P (2)p.R204C |
Compound heterozygous mutations | Improved | [39] |
| P37 | IFNGR1 | AR | c.182dupT | p.V61fsX69 | Insertion | Dead | [32] |
| P38 | IFNGR1 | AR | c.182dupT | p.V61fsX69 | Insertion | Improved | [32] |
| P39 | RELB | AR | c.400_401insAGC | p.Q135_R136insQ | Insertion | Improved | [41] |
| P40 | NFKB2 | AD | c.2540dupT | – | Insertion | Improved | [42] |
HIV human immunodeficiency virus, AD autosomal dominant, AR autosomal recessive, XL X-linked, GOF gain-of-function
Although the invasion mechanisms were not clearly deciphered, it was widely believed that TM infections initiated from the inhalation of conidia [34, 35]. Then the conidia converted into yeast phase and were engulfed by macrophages [36]. The pathogenic yeasts could survive and replicate within macrophages, and even disseminated when the hosts were in immunocompromised situation [37]. Therefore, efficient macrophage activation and functions were key factors for anti-TM strategies. CD4+ T cells activated macrophages by offering CD40L-CD40 interactions and secreting IFN-γ [38]. Thus, CD4+ T lymphopenia, CD40 ligand absence, or interfered IFN-γ pathway could hamper macrophages to eliminate intracellular TM. HIV infections, IL2RG and ADA mutations all led to remarkable reduced CD4 + T cells. CD40 ligand deficiency impaired T cell-antigen presenting cell (APC) interactions. Autoantibodies against IFN-γ and IFN-γ receptor 1 deficiency blocked IFN-γ signaling. These defects were all reported to associate with TM infections.
On the other hand, patients with STAT1 GOF and STAT3 deficiency presented impaired Th17 responses [1], indicating that IL-17 signaling might be involved in anti-TM immunity. In addition, NF-κB (nuclear factor kappa light chain enhancer of activated B cells) pathways may also contribute to host strategies against TM infections. Patients with CARD9 [39, 40], RELB [41], or NFKB2 [42] mutations have been reported with TM infections. As an adaptor protein, CARD9 mediated signals from pattern recognition receptors (PRRs) which could recognize TM-related carbohydrates, to the downstream transcription factor NF-κB [43]. CARD9 deficiency impaired NF-κB activation, cytokines secretion, Th17 differentiation, and neutrophil killing [44, 45]. RelB and NF-κB2 are components of NF-κB family. RelB deficiency led to reduced T cell proliferations to mitogens and skewed T cell receptor (TCR) repertoire, together with impaired antibody responses, presenting combined immunodeficiency [46]. NFKB2 deficiency resulted in damaged B cell differentiation and hypogammaglobinemia. A series of lymphocyte subpopulations, including regulatory T cells, Th17 cells, and circulating T follicular helper cells were also decreased [47]. More cases and experiments are needed to demonstrate the roles of NF-κB pathway in TM infections.
We noticed reduced NK cells in all the eight patients in our study. The reported TM-infected children with CARD9, IFNGR1, or RELB mutations, also exhibited decreased NK cells [32, 39–41]. Zeng et al. [21] reported reduced NK cells in 62% of the TM-infected patients. In Guo’s study, 81.8% of the children with TM-infections exhibited decreased NK cells [48]. NK cells from patients with STAT1 GOF mutations displayed decreased IFN-γ production and reduced proliferation after stimulation of IL-15 [49]. NK cell was another important source of IFN-γ secretion, and participated in host responses against various pathogens, especially for herpes virus and intracellular bacteria [50, 51]. In our patients, EBV and CMV were observed in three TM-infected patients.
TM infection was a severe disease with high mortality. In our study, 5 patients died during the follow-ups (62.5%). Zeng et al. [21] reported 11 in 21 children (52.38%) died of TM infections. In Guo’s study, they enrolled 11 TM-infected HIV-negative children and four of them (36.36%) died during 1-year follow-ups [48]. Early recognition and comprehensive therapy may help to improve the prognosis.
Currently, guidelines recommended initial treatments for HIV-associated TM infections in adults with amphotericin B deoxycholate at a dose of 0.7–1 mg/kg/day for 2 weeks, followed by itraconazole at a dose of 400 mg per day for 10 weeks [52]. Considering the substantial side effects and limited availability of amphotericin B, other anti-fungal drugs were also used. The trial conducted by Le et al. proved that amphotericin B was superior to itraconazole as induction therapy for HIV-associated talaromycosis [53]. Huang’s trial indicated that voriconazole was noninferior to amphotericin B as an induction antifungal drug for HIV- associated disseminated talaromycosis [54]. There were also evidences supporting the use of amphotericin B, itraconazole, voriconazole, and posaconazole in management of TM infections, not fluconazole [55]. For TM infections in HIV-negative children patients, the treatments were still lacking consensus and remained a big challenge. Most of the infected children had underlying IEIs. They were susceptible to various pathogens except for TM and anti-bacteria and anti-virus therapy were necessary. Especially, intracellular bacteria, including mycobacteria and salmonella, were commonly co-infected with TM [19, 56], as hosts shared some core strategies to defend these intracellular pathogens. People also discussed the prophylaxis for TM infections [57–59]. For IEIs listed in Table 3, prophylactic anti-fungal drugs may be given to gain clinical benefits.
In conclusion, we retrospectively analysed TM infections in 8 Chinese children and all of them were diagnosed as IEIs, including STAT1 GOF, IL-2 receptor common gamma chain deficiency, ADA deficiency, CD40 ligand deficiency, and STAT3 deficiency. Based on our patients and literature review, the spectrum of IEIs underlying TM infections indicated that T cell-mediated immunity, IFN-γ, IL-17 signalings and NF-κB pathways were important for host responses against TM infection. In reverse, for children born or living in South China or other endemic areas, without HIV infection, without other secondary immunodeficiency, TM infections may be an indicator for IEIs and further immunological and genetical evaluations are needed.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We appreciate all the patients and their families for their kindness of permission and cooperation. We thank all their physicians and nurses for supporting this study.
Author Contributions
LLW, YL and XLL contributed equally to this work. LLW, YL, XLL and JY designed the project. LLW and YL performed the main experiments, analysed the data, and drafted the manuscript. XLL, YXL, YX, TYH, YYH, YBX, ZY, JYL, RHW, and XNZ collected clinical data and followed the patients. ZXQ helped with the experiments. JY and XLL reviewed the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by (1) Sanming Project of Medicine in Shenzhen (SZSM201812002); (2) National Natural Science Foundation of China (81900638).
Declarations
Conflict of interest
The authors declare no conflict of interest.
Ethics Approval
This study was conducted in line with the principles of the Declaration of Helsinki. Approval was given by the Ethics Committee of Shenzhen Children’s Hospital (2019050).
Consent to Participate
Informed consents were obtained from each patients’ parents.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Linlin Wang, Ying Luo, Xiaolin Li contributed equally to this work.
References
- 1.Tangye SG, Al-Herz W, Bousfiha A, Chatila T, Cunningham-Rundles C, Etzioni A, et al. Human inborn errors of immunity: 2019 update on the classification from the international union of immunological societies expert committee. J Clin Immunol. 2020;40(1):24–64. doi: 10.1007/s10875-019-00737-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Herz W, Essa S. Spectrum of viral infections among primary immunodeficient children: report from a national registry. Front Immunol. 2019;10:1231. doi: 10.3389/fimmu.2019.01231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bousfiha A, Jeddane L, Picard C, Ailal F, Bobby Gaspar H, Al-Herz W, et al. The 2017 IUIS phenotypic classification for primary immunodeficiencies. J Clin Immunol. 2018;38(1):129–143. doi: 10.1007/s10875-017-0465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guarner J, Brandt ME. Histopathologic diagnosis of fungal infections in the 21st century. Clin Microbiol Rev. 2011;24(2):247–280. doi: 10.1128/CMR.00053-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyce KJ, Andrianopoulos A. Fungal dimorphism: the switch from hyphae to yeast is a specialized morphogenetic adaptation allowing colonization of a host. FEMS Microbiol Rev. 2015;39(6):797–811. doi: 10.1093/femsre/fuv035. [DOI] [PubMed] [Google Scholar]
- 6.Capponi M, Segretain G, Sureau P. Penicillosis from Rhizomys sinensis. Bull Soc Pathol Exot Filiales. 1956;49(3):418–421. [PubMed] [Google Scholar]
- 7.Tsang CC, Lau SKP, Woo PCY. Sixty years from Segretain’s description: what have we learned and should learn about the basic mycology of Talaromyces marneffei? Mycopathologia. 2019;184(6):721–729. doi: 10.1007/s11046-019-00395-y. [DOI] [PubMed] [Google Scholar]
- 8.Hu Y, Zhang J, Li X, Yang Y, Zhang Y, Ma J, et al. Penicillium marneffei infection: an emerging disease in mainland China. Mycopathologia. 2013;175(1–2):57–67. doi: 10.1007/s11046-012-9577-0. [DOI] [PubMed] [Google Scholar]
- 9.Ranjana KH, Priyokumar K, Singh TJ, Gupta ChC, Sharmila L, Singh PN, et al. Disseminated Penicillium marneffei infection among HIV-infected patients in Manipur state. India J Infect. 2002;45(4):268–271. doi: 10.1053/jinf.2002.1062. [DOI] [PubMed] [Google Scholar]
- 10.Lee PP, Lau YL. Cellular and molecular defects underlying invasive fungal infections-revelations from endemic mycoses. Front Immunol. 2017;8:735. doi: 10.3389/fimmu.2017.00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larsson M, Nguyen LH, Wertheim HF, Dao TT, Taylor W, Horby P, et al. Clinical characteristics and outcome of Penicillium marneffei infection among HIV-infected patients in northern Vietnam. AIDS Res Ther. 2012;9(1):24. doi: 10.1186/1742-6405-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duong TA. Infection due to Penicillium marneffei, an emerging pathogen: review of 155 reported cases. Clin Infect Dis. 1996;23(1):125–130. doi: 10.1093/clinids/23.1.125. [DOI] [PubMed] [Google Scholar]
- 13.Sheng L, Shen Q, Zhou J. Efficacy of different antifungal drugs as initial treatment for patients with talaromycosis: a systematic review and meta-analysis. J Mycol Med. 2021;31(1):101108. doi: 10.1016/j.mycmed.2020.101108. [DOI] [PubMed] [Google Scholar]
- 14.Limper AH, Adenis A, Le T, Harrison TS. Fungal infections in HIV/AIDS. Lancet Infect Dis. 2017;17(11):e334–e343. doi: 10.1016/S1473-3099(17)30303-1. [DOI] [PubMed] [Google Scholar]
- 15.Liu W, Xu J, Lin B, Zhang Y, Xie F, Zhou C, et al. Pneumonia caused by Talaromyces marneffei in an epidermal growth factor receptor (EGFR) mutation-positive advanced lung adenocarcinoma patient: a case report. Ann Palliat Med. 2021;10(1):759–766. doi: 10.21037/apm-20-2137. [DOI] [PubMed] [Google Scholar]
- 16.Peng J, Chen Z, Cai R, Huang X, Lin L, Liang W, et al. Recovery from Talaromyces marneffei involving the kidney in a renal transplant recipient: a case report and literature review. Transpl Infect Dis. 2017;19(4):e12710. doi: 10.1111/tid.12710. [DOI] [PubMed] [Google Scholar]
- 17.Wei J, Qiu Y, Zeng W, Pan M, Zhang J. Talaromyces marneffei infection in systemic lupus erythematosus patients: report of two cases and review of the literature. Infect Drug Resist. 2020;13:3811–3816. doi: 10.2147/IDR.S265479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu Y, Zhang J, Liu G, Zhong X, Deng J, He Z, et al. Retrospective analysis of 14 cases of disseminated Penicillium marneffei infection with osteolytic lesions. BMC Infect Dis. 2015;15:47. doi: 10.1186/s12879-015-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin W, Liu J, Chen K, Shen L, Zhou Y, Wang L. Coinfection by Talaromyces marneffei and Mycobacterium abscessus in a human immunodeficiency virus-negative patient with anti-interferon-gamma autoantibody: a case report. J Int Med Res. 2021;49(1):300060520976471. doi: 10.1177/0300060520976471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo J, Ning XQ, Ding JY, Zheng YQ, Shi NN, Wu FY, et al. Anti-IFN-gamma autoantibodies underlie disseminated Talaromycesmarneffei infections. J Exp Med. 2020;217(12). [DOI] [PMC free article] [PubMed]
- 21.Zeng Q, Jin Y, Yin G, Yang D, Li W, Shi T, et al. Peripheral immune profile of children with Talaromyces marneffei infections: a retrospective analysis of 21 cases. BMC Infect Dis. 2021;21(1):287. doi: 10.1186/s12879-021-05978-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan M, Qiu Y, Zeng W, Tang S, Wei X, Zhang J. Disseminated Talaromyces marneffei infection presenting as multiple intestinal perforations and diffuse hepatic granulomatous inflammation in an infant with STAT3 mutation: a case report. BMC Infect Dis. 2020;20(1):394. doi: 10.1186/s12879-020-05113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan H, Huang L, Yang D, Lin Y, Lu G, Xie Y, et al. Pediatric hyperimmunoglobulin E syndrome: a case series of 4 children in China. Medicine (Baltimore) 2018;97(14):e0215. doi: 10.1097/MD.0000000000010215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X, Xu Q, Li X, Wang L, Yang L, Chen Z, et al. Molecular and phenotypic characterization of nine patients with STAT1 GOF mutations in China. J Clin Immunol. 2020;40(1):82–95. doi: 10.1007/s10875-019-00688-3. [DOI] [PubMed] [Google Scholar]
- 25.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016;99(4):877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human splicing finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37(9):e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding Y, Zhou L, Xia Y, Wang W, Wang Y, Li L, et al. Reference values for peripheral blood lymphocyte subsets of healthy children in China. J Allergy Clin Immunol. 2018;142(3):970–3e8. doi: 10.1016/j.jaci.2018.04.022. [DOI] [PubMed] [Google Scholar]
- 32.Lee PP, Lao-Araya M, Yang J, Chan KW, Ma H, Pei LC, et al. Application of flow cytometry in the diagnostics pipeline of primary immunodeficiencies underlying disseminated Talaromyces marneffei Infection in HIV-negative children. Front Immunol. 2019;10:2189. doi: 10.3389/fimmu.2019.02189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dotta L, Scomodon O, Padoan R, Timpano S, Plebani A, Soresina A, et al. Clinical heterogeneity of dominant chronic mucocutaneous candidiasis disease: presenting as treatment-resistant candidiasis and chronic lung disease. Clin Immunol. 2016;164:1–9. doi: 10.1016/j.clim.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 34.Hamilton AJ, Jeavons L, Youngchim S, Vanittanakom N. Recognition of fibronectin by Penicillium marneffei conidia via a sialic acid-dependent process and its relationship to the interaction between conidia and laminin. Infect Immun. 1999;67(10):5200–5205. doi: 10.1128/IAI.67.10.5200-5205.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weerasinghe H, Payne M, Beard S, Andrianopoulos A. Organism-wide studies into pathogenicity and morphogenesis in Talaromyces marneffei. Future Microbiol. 2016;11(4):511–526. doi: 10.2217/fmb.16.9. [DOI] [PubMed] [Google Scholar]
- 36.Chan YF, Chow TC. Ultrastructural observations on Penicillium marneffei in natural human infection. Ultrastruct Pathol. 1990;14(5):439–452. doi: 10.3109/01913129009007223. [DOI] [PubMed] [Google Scholar]
- 37.Pongpom M, Vanittanakom P, Nimmanee P, Cooper CR, Jr, Vanittanakom N. Adaptation to macrophage killing by Talaromyces marneffei. Future Sci OA. 2017;3(3):FSO215. doi: 10.4155/fsoa-2017-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tubo NJ, Jenkins MK. CD4+ T Cells: guardians of the phagosome. Clin Microbiol Rev. 2014;27(2):200–213. doi: 10.1128/CMR.00097-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ba H, Peng H, Cheng L, Lin Y, Li X, He X, et al. Case report: Talaromyces marneffei infection in a Chinese child with a complex heterozygous CARD9 Mutation. Front Immunol. 2021;12:685546. doi: 10.3389/fimmu.2021.685546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.You C-Y, Hu F, Lu S-W, Pi D-D, Xu F, Liu C-J, et al. Talaromyces marneffei infection in an HIV-negative child with a CARD9 mutation in China: a case report and review of the literature. Mycopathologia. 2021;186(4):553–61. doi: 10.1007/s11046-021-00576-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding X, Huang H, Zhong L, Chen M, Peng F, Zhang B, et al. Disseminated Talaromyces marneffei infection in a Non-HIV infant with a homozygous private variant of RELB. Front Cell Infect Microbiol. 2021;11:605589. doi: 10.3389/fcimb.2021.605589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang C, Wang N, Feng J, Li Y, Fang W, Zhang X, et al. Two cases of congenital immunodeficiency with disseminated infection of cyanobacteria marneffei and literature review. Chin Pediatric Emerg Med. 2020;27(11):861–864. [Google Scholar]
- 43.Wang Y, Zhang D, Hou Y, Shen S, Wang T. The adaptor protein CARD9, from fungal immunity to tumorigenesis. Am J Cancer Res. 2020;10(8):2203–2225. [PMC free article] [PubMed] [Google Scholar]
- 44.Corvilain E, Casanova JL, Puel A. Inherited CARD9 deficiency: invasive disease caused by ascomycete fungi in previously healthy children and adults. J Clin Immunol. 2018;38(6):656–693. doi: 10.1007/s10875-018-0539-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drummond RA, Franco LM, Lionakis MS. Human CARD9: a critical molecule of fungal immune surveillance. Front Immunol. 2018;9:1836. doi: 10.3389/fimmu.2018.01836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharfe N, Merico D, Karanxha A, Macdonald C, Dadi H, Ngan B, et al. The effects of RelB deficiency on lymphocyte development and function. J Autoimmun. 2015;65:90–100. doi: 10.1016/j.jaut.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 47.Klemann C, Camacho-Ordonez N, Yang L, Eskandarian Z, Rojas-Restrepo JL, Frede N, et al. Clinical and immunological phenotype of patients with primary immunodeficiency due to damaging mutations in NFKB2. Front Immunol. 2019;10:297. doi: 10.3389/fimmu.2019.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo J, Li BK, Li TM, Wei FL, Fu YJ, Zheng YQ, et al. Characteristics and prognosis of Talaromyces marneffei Infection in Non-HIV-infected children in southern China. Mycopathologia. 2019;184(6):735–745. doi: 10.1007/s11046-019-00373-4. [DOI] [PubMed] [Google Scholar]
- 49.Tabellini G, Vairo D, Scomodon O, Tamassia N, Ferraro RM, Patrizi O, et al. Impaired natural killer cell functions in patients with signal transducer and activator of transcription 1 (STAT1) gain-of-function mutations. J Allergy Clin Immunol. 2017;140(2):553–64 e4. doi: 10.1016/j.jaci.2016.10.051. [DOI] [PubMed] [Google Scholar]
- 50.Hamerman JA, Ogasawara K, Lanier LL. NK cells in innate immunity. Curr Opin Immunol. 2005;17(1):29–35. doi: 10.1016/j.coi.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 51.Mace EM, Orange JS. Emerging insights into human health and NK cell biology from the study of NK cell deficiencies. Immunol Rev. 2019;287(1):202–225. doi: 10.1111/imr.12725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guidelines for the prevention and treatment of opportunistic infections in adults and adolescents with HIV: recommendations from the Centers for Disease Control and Prevention, the National Institutes of Health, and the HIV Medicine Association of the Infectious Diseases Society of America.
- 53.Le T, Kinh NV, Cuc NTK, Tung NLN, Lam NT, Thuy PTT, et al. A Trial of Itraconazole or amphotericin B for HIV-associated Talaromycosis. N Engl J Med. 2017;376(24):2329–2340. doi: 10.1056/NEJMoa1613306. [DOI] [PubMed] [Google Scholar]
- 54.Huang W, Li T, Zhou C, Wei F, Cao C, Jiang J. Voriconazole versus amphotericin B as induction therapy for Talaromycosis in HIV/AIDS patients: a retrospective study. Mycopathologia. 2021;186(2):269–276. doi: 10.1007/s11046-021-00533-5. [DOI] [PubMed] [Google Scholar]
- 55.Lei HL, Li LH, Chen WS, Song WN, He Y, Hu FY, et al. Susceptibility profile of echinocandins, azoles and amphotericin B against yeast phase of Talaromyces marneffei isolated from HIV-infected patients in Guangdong, China. Eur J Clin Microbiol Infect Dis. 2018;37(6):1099–1102. doi: 10.1007/s10096-018-3222-x. [DOI] [PubMed] [Google Scholar]
- 56.Qiu Y, Huang J, Li Y, Zeng W, Pan M, Cen J, et al. Talaromyces marneffei and nontuberculous mycobacteria co-infection in HIV-negative patients. Sci Rep. 2021;11(1):16177. doi: 10.1038/s41598-021-95686-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chaiwarith R, Charoenyos N, Sirisanthana T, Supparatpinyo K. Discontinuation of secondary prophylaxis against penicilliosis marneffei in AIDS patients after HAART. AIDS. 2007;21(3):365–367. doi: 10.1097/01.aids.0000253374.19966.f9. [DOI] [PubMed] [Google Scholar]
- 58.Chariyalertsak S, Supparatpinyo K, Sirisanthana T, Nelson KE. A controlled trial of itraconazole as primary prophylaxis for systemic fungal infections in patients with advanced human immunodeficiency virus infection in Thailand. Clin Infect Dis. 2002;34(2):277–284. doi: 10.1086/338154. [DOI] [PubMed] [Google Scholar]
- 59.Chaiwarith R, Fakthongyoo A, Praparattanapan J, Boonmee D, Sirisanthana T, Supparatpinyo K. Itraconazole vs fluconazole as a primary prophylaxis for fungal infections in HIV-infected patients in Thailand. Curr HIV Res. 2011;9(5):334–338. doi: 10.2174/157016211797635991. [DOI] [PubMed] [Google Scholar]
- 60.Chen K, Tan J, Qian S, Wu S, Chen Q. Case report: Disseminated Talaromyces marneffei infection in a patient with chronic mucocutaneous candidiasis and a novel STAT1 gain-of-function mutation. Front Immunol. 2021;12:682350. doi: 10.3389/fimmu.2021.682350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang W, Ye J, Qiu C, Wang L, Jin W, Jiang C, et al. Rapid and precise diagnosis of T. marneffei pulmonary infection in a HIV-negative patient with autosomal-dominant STAT3 mutation: a case report. Ther Adv Respir Dis. 2020;14:1753466620929225. doi: 10.1177/1753466620929225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee PPW, Chan K-W, Lee T-L, Ho MH-K, Chen X-Y, Li C-H, et al. Penicilliosis in children without HIV infection—are they immunodeficient. Clin Infect Dis. 2012;54(2):e8–e19. doi: 10.1093/cid/cir754. [DOI] [PubMed] [Google Scholar]
- 63.Du X, Tang W, Chen X, Zeng T, Wang Y, Chen Z, et al. Clinical, genetic and immunological characteristics of 40 Chinese patients with CD40 ligand deficiency. Scand J Immunol. 2019;90(4):e12798. doi: 10.1111/sji.12798. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.