Abstract
In August 2016, the Wisconsin Department of Health Services notified the U.S. Centers for Disease Control and Prevention of multidrug-resistant (MDR) Salmonella enterica serovar Heidelberg infections in people who reported contact with dairy calves. Federal and state partners investigated this to identify the source and scope of the outbreak and to prevent further illnesses. Cases were defined as human Salmonella Heidelberg infection caused by a strain that had one of seven pulsed-field gel electrophoresis (PFGE) patterns or was related by whole genome sequencing (WGS), with illness onset from January 1, 2015, through July 2, 2018. Patient exposure and calf purchase information was collected and analyzed; calves were traced back from the point of purchase. Isolates obtained from animal and environmental samples collected on-farm were supplied by veterinary diagnostic laboratories and compared with patient isolates using PFGE and WGS. Antimicrobial susceptibility testing by standardized broth microdilution was performed. Sixty-eight patients from 17 states were identified. Forty (63%) of 64 patients noted cattle contact before illness. Thirteen (33%) of 40 patients with exposure to calves reported that calves were sick or had died. Seven individuals purchased calves from a single Wisconsin livestock market. One hundred forty cattle from 14 states were infected with the outbreak strain. WGS indicated that human, cattle, and environmental isolates from the livestock market were genetically closely related. Most isolates (88%) had resistance or reduced susceptibility to antibiotics of ≥5 antibiotic classes. This resistance profile included first-line antibiotic treatments for patients with severe salmonellosis, including ampicillin, ceftriaxone, and ciprofloxacin. In this outbreak, MDR Salmonella Heidelberg likely spread from sick calves to humans, emphasizing the importance of illness surveillance in animal populations to prevent future spillover of this zoonotic disease.
Keywords: antimicrobial resistance, zoonotic disease, food safety, calves, Salmonella, One Health
Introduction
Nontyphoidal Salmonella causes an estimated 1.35 million infections and 26,500 hospitalizations in the United States annually (CDC, 2019). Infection often results from consumption of contaminated food or from contact with animals or their environments (Fey et al., 2000; Lynch et al., 2009; Gieraltowski et al., 2016). Livestock infected with Salmonella can shed the pathogen in feces and may not demonstrate clinical signs (McGuirk et al., 2003). Human symptoms include diarrhea (sometimes bloody), fever, and abdominal cramps and may depend on the immune status of the infected person (Molbak, 2005; CDC, 2015).
Some Salmonella infections can spread to the bloodstream, increase complications and disease severity, and require treatment with antimicrobials. Ciprofloxacin, ceftriaxone, or azithromycin is used as first-line treatment for severe Salmonella infections pending culture results (Shane et al., 2017; American Academy of Pediatrics, 2021). Clinically significant antimicrobial resistance in Salmonella can be predicted from the presence of certain genetic factors in the bacterial genome (McDermott et al., 2016). Multidrug-resistant (MDR) strains of Salmonella have been associated with increased risk of bloodstream infections and hospitalization; as a result, MDR Salmonella strains are a serious human health threat (Varma et al., 2005; CDC, 2019).
Salmonella enterica serovar Heidelberg was initially discovered in 1933 after a human outbreak in Heidelberg, Germany (Habbs, 1933). A Salmonella Heidelberg illness outbreak was reported in the United Kingdom in the 1960s, attributed to ingestion of contaminated milk from dairy cattle (Davies et al., 1962; Knox et al., 1963). More recently, Salmonella Heidelberg caused a multistate MDR illness outbreak linked to poultry consumption (Gieraltowski et al., 2016), and in 2017, Salmonella Heidelberg was among the Salmonella serotypes most commonly linked to human infection (Marder et al., 2018). Salmonella Heidelberg isolated from live animals and food products in the United States commonly demonstrates resistance to multiple antimicrobials, including tetracycline, streptomycin, kanamycin, and ampicillin (Lynne et al., 2009).
On August 3, 2016, the Wisconsin Veterinary Diagnostic Laboratory (WVDL) notified the Wisconsin Division of Public Health (WDPH) of an isolate of Salmonella Heidelberg cultured from an ill dairy bull calf submitted to the WVDL by a farm experiencing a recurrent Salmonella outbreak among calves, which was associated with unusually high calf mortality. Initial query of PulseNet, the national molecular subtyping network for foodborne disease surveillance, identified isolates from ill people matching those of the index calf.
Antimicrobial susceptibility testing (AST) of the calf’s isolate revealed resistance, as defined by the Clinical and Laboratory Standards Institute (CLSI), to multiple antimicrobials (CLSI, 2021). Consultation with the CDC National Antimicrobial Resistance Monitoring System (NARMS) team indicated that the AST resistance phenotype had not been previously reported to NARMS among human Salmonella Heidelberg isolates and included antimicrobials used for first-line treatment of salmonellosis (CDC, 2020a). Whole genome sequencing (WGS) demonstrated close genetic relatedness of a Salmonella isolate from an ill worker from the index farm and the calf’s isolate.
An investigation involving human and animal health and laboratory entities was initiated to identify additional illnesses and determine relatedness of the human and animal Salmonella Heidelberg isolates. This article describes the actions taken to investigate this outbreak of Salmonella Heidelberg with a novel MDR pattern in humans and animals and ongoing activities to disseminate information to prevent Salmonella Heidelberg illnesses.
Materials and Methods
This activity was reviewed by the CDC and was conducted consistent with applicable federal law and CDC policy.*
The initial case definition was a laboratory-confirmed Salmonella Heidelberg human infection with one of six pulsed-field gel electrophoresis (PFGE) patterns reported through PulseNet with illness onset on or after January 1, 2016. These PFGE patterns were found in cattle isolates and in isolates from ill people who reported contact with cattle. The case definition was subsequently expanded to encompass cases with illness onset on or after January 1, 2015, after case finding conducted using PulseNet identified seven additional clinical Salmonella Heidelberg isolates from 2015.
Case identification continued throughout 2018, and an additional PFGE pattern was identified with close genetic similarity (based on WGS analysis) to the outbreak strain causing illness in humans with reported calf exposure. The final case definition was a laboratory-confirmed Salmonella Heidelberg human infection with an isolate with one of seven implicated PFGE patterns, or related by WGS, with illness onset from January 1, 2015, through July 2, 2018.
To identify potential sources of infection and other epidemiologic linkages between cases, state and local public health officials interviewed ill people about food and environmental exposures occurring in the week before illness onset. Based on initial reports of exposure to dairy cattle and calves and identification of samples from dairy calves owned by an ill person with a related Salmonella Heidelberg isolate, ill people were asked more detailed questions on the type of contact with cattle, health status of cattle, date and location of purchase of cattle (purchase records), and cattle rearing and management practices.
Calf purchase records obtained from ill people were used by federal and state health and agriculture officials to examine and trace interstate movement of cattle from purchasers to livestock markets and transport companies or haulers. Trace back activities helped to inform environmental testing at a single livestock market. Health and agriculture departments in states that received cattle from these markets were informed of the outbreak.
The WVDL serotyped Salmonella isolates from cattle and calves submitted for necropsy and from fecal or environmental samples submitted for culture from farms experiencing animal morbidity or mortality consistent with Salmonella infection. Animal isolates identified as Salmonella Heidelberg were submitted to the Wisconsin State Laboratory of Hygiene (WSLH) for additional characterization, including PFGE and WGS; these data were submitted to PulseNet to determine genetic relatedness of human and cattle isolates. Additionally, some ill people who owned cattle granted investigators permission to collect samples from cattle.
Environmental sampling was conducted at a livestock market that voluntarily permitted investigation to determine if Salmonella Heidelberg could be isolated from locations where commingled calves were housed. Animal samples from 14 states were submitted to USDA-APHIS National Veterinary Services Laboratories (NVSL), WVDL, state veterinary diagnostic laboratories, or public health laboratories where culture for Salmonella, PFGE, and WGS analysis were performed. During this investigation, PulseNet was transitioning from using PFGE to WGS to monitor and track disease outbreaks; thus, both methods were utilized during this investigation (Kubota et al., 2019).
PFGE was performed following the PulseNet protocol for Salmonella (Ribot et al., 2006). Patterns were analyzed using BioNumerics 6.6 (Applied Maths, Sint-Martens-Latem, Belgium) and uploaded to the national database at CDC for comparison and pattern naming. WGS was performed using the Nextera XT library preparation kit (Illumina, San Diego, CA), followed by sequencing on the Illumina MiSeq platform. Sequences were shared with CDC for high-quality single-nucleotide polymorphism analysis and core genome multilocus sequence typing (Katz et al., 2017). All sequences have been submitted to the National Center for Biotechnology Information (NCBI) bioproject PRJNA230403.
The WSLH and NARMS laboratory performed AST by broth microdilution on select human clinical isolates using a Sensititre® panel (Trek Diagnostics, Westlake, OH) with 14 drugs: amoxicillin–clavulanic acid, ampicillin, azithromycin, cefoxitin, ceftriaxone, chloramphenicol, ciprofloxacin, gentamicin, meropenem, nalidixic acid, streptomycin, sulfisoxazole, tetracycline, and trimethoprim–sulfamethoxazole (CDC, 2018b). Breakpoints established by the CLSI were used to define susceptible, intermediate, and resistant ranges when available; otherwise, NARMS-established breakpoints were used (CDC, 2018b; CLSI, 2021). De novo assemblies from sequenced isolates were produced using shovill, v.1.0.4 (https://github.com/tseemann/shovill).
Screening of assemblies for resistance determinants was performed using staramr, v. 0.4.0, which employs the ResFinder database (updated July 30, 2020) using thresholds of 90% identity and 50% gene coverage and the PointFinder scheme for Salmonella spp. (updated August 30, 2019) (Tagg et al., 2020). A predicted resistance pattern was assigned based on the presence of resistance determinants in genome assemblies (McDermott et al., 2016). Multidrug resistance was defined as resistance to at least one antimicrobial in three or more drug classes (Magiorakos et al., 2012).
Results
Sixty-eight human cases of Salmonella Heidelberg infection were identified from 17 states; of these, 18 (26%) cases were reported from Wisconsin (Fig. 1). Estimated onset dates of infection in patients ranged from August 1, 2015, through July 2, 2018 (Fig. 2). The median patient age was 14 years, with a range of <1–89 years. Thirty-nine (57%) patients were 18 years old or younger (Fig. 3). Twenty-one (35%) patients were hospitalized. Median age of hospitalized patients was 24 years. No deaths were reported.
FIG. 1.
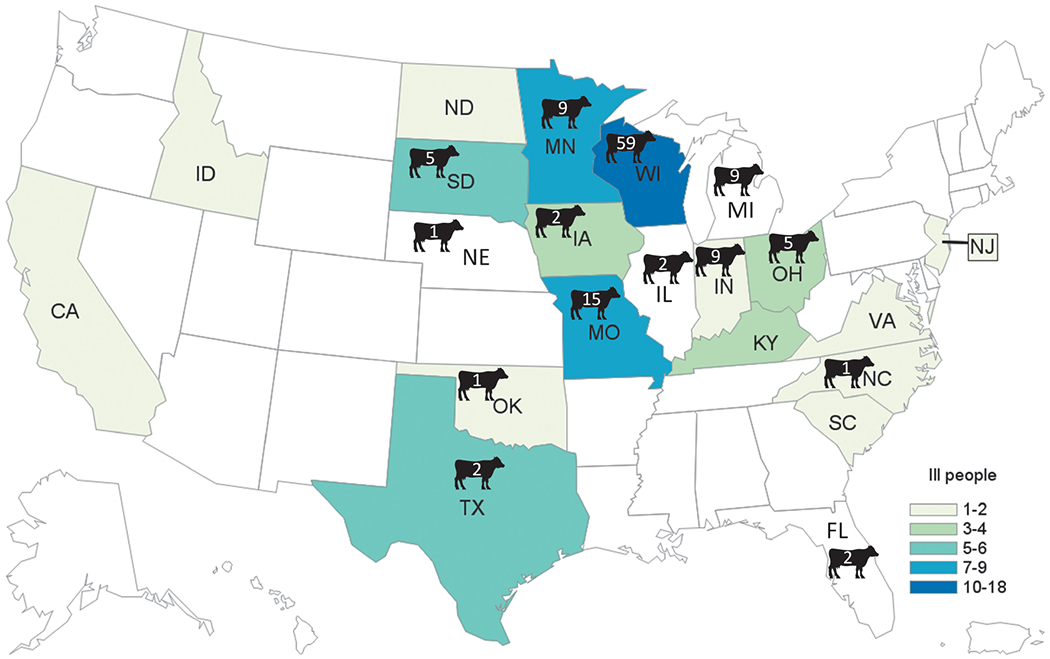
Human cases of Salmonella Heidelberg by state. A total of 17 states were associated with human cases in this outbreak. The highest number of cases (18) occurred in Wisconsin. Cattle icons denote states in which isolates positive for the Salmonella Heidelberg outbreak strain were detected in samples from cattle, and the corresponding number of positive isolates is indicated. Color images are available online.
FIG. 2.
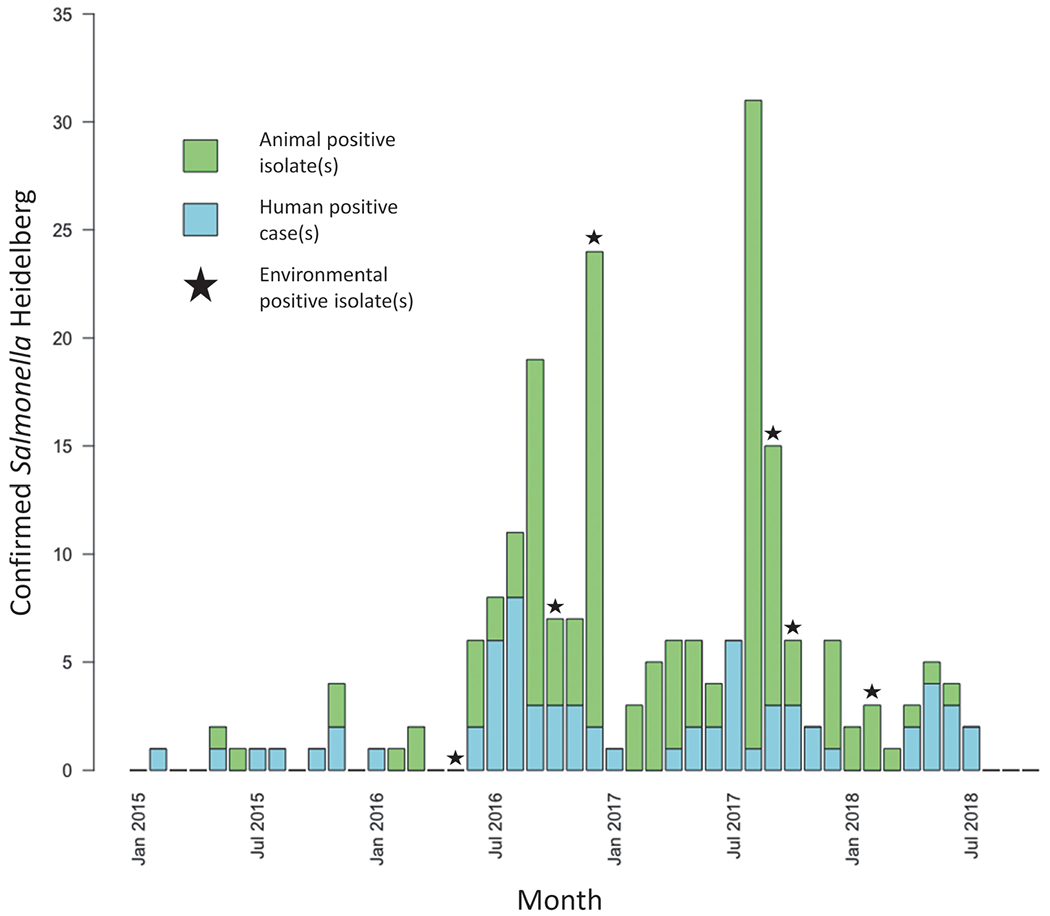
Epidemiologic curve of human Salmonella Heidelberg cases and animal isolates from January 1, 2015, to July 2, 2018. Seven human patients matching the outbreak case definition had estimated onset dates of Salmonella Heidelberg in 2015; 28 in 2016; 22 in 2017; and 11 in 2018. Animal isolates reflect dates when Salmonella Heidelberg was isolated through laboratory testing rather than animal illness onset date. Star icons indicate months in which Salmonella Heidelberg was isolated through environmental testing. Color images are available online.
FIG. 3.
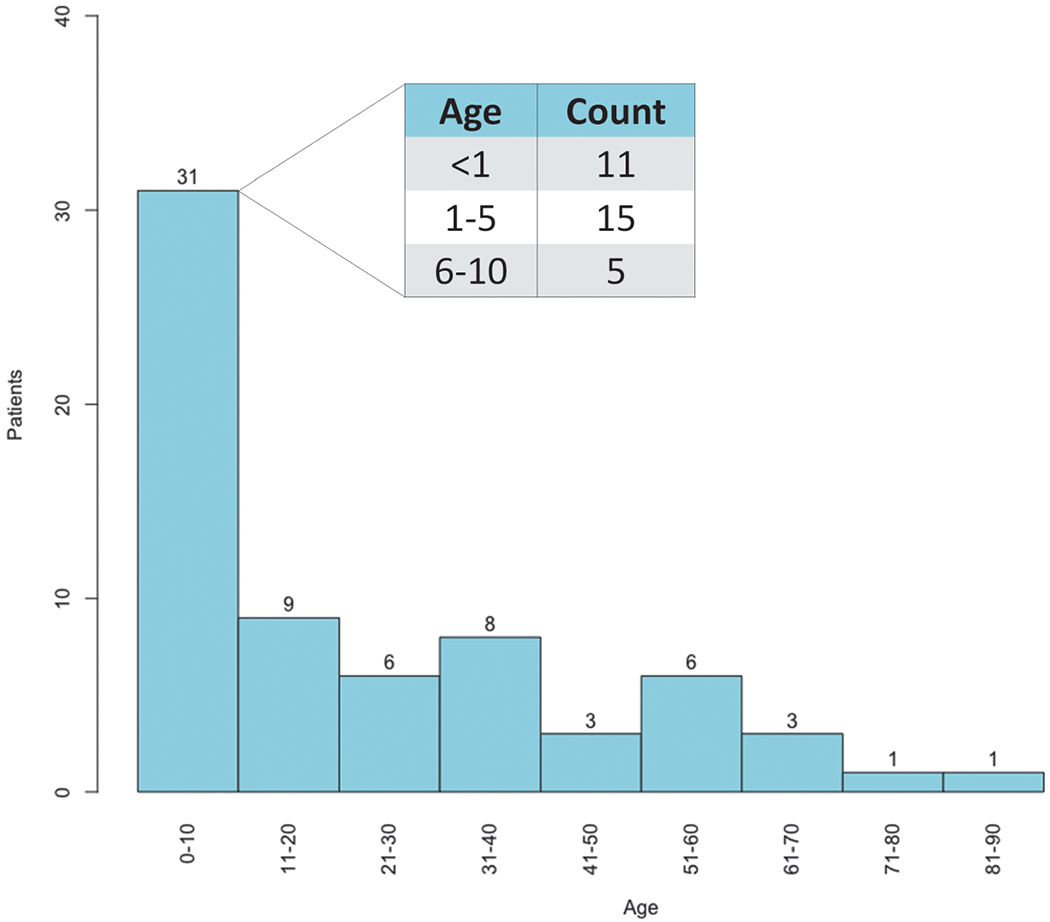
Age distribution of Salmonella Heidelberg human patients. The median age of human patients meeting the Salmonella Heidelberg outbreak case definition was 14 (range <1–89 years). Color images are available online.
Forty (63%) of 64 patients with available information reported exposure to cattle. Twenty-four (60%) of those 40 patients were 18 years old or younger. Eleven (28%) of the 40 patients reported contact with calves specifically, and 5 (13%) patients with cattle exposure lived on farms where calves were present. Two people indicated their reason for purchasing calves, and both reported purchasing for the purpose of youth agriculture club activities. Salmonella Heidelberg was also plausibly transmitted from person to person among family members in households with cattle; among five members of a single family infected with Salmonella Heidelberg with a single PFGE pattern, two were children below the age of 1 year who did not have direct cattle contact.
Thirteen (33%) of the 40 patients with cattle exposure reported that the cattle were sick. Eight (20%) specified that cattle or calves were sick with diarrhea, which is the most common illness affecting young calves (Lorenz et al., 2011). Ten (77%) of 13 patients who reported owning or caring for sick cattle specified that the sick animals were calves and reported calf mortality. Recent purchase of cattle or calves before illness onset was reported for 23 (58%) of 40 patients with cattle exposure. One patient reported owning a goat that was housed with sick cattle. This goat became ill and died, and the outbreak strain was isolated from samples collected from the goat (Fig. 4 and Supplementary Fig. S1).
FIG. 4.
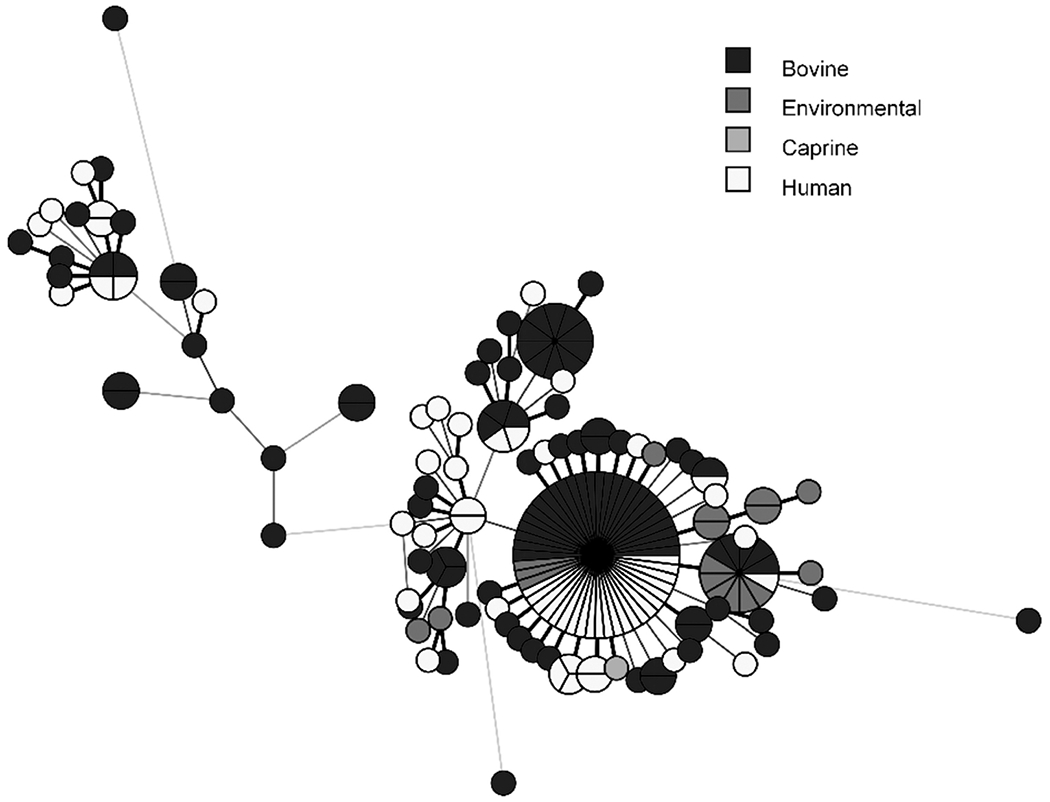
Minimum spanning tree diagraming genetic relatedness of human, animal, and environmental isolates. All sequences have an allele difference range of 0–35 alleles. The length of the lines indicates the allele difference between individual sequences. Line length is proportionally scaled, and the longer the line, the greater the allele difference is between the sequences. Isolates with no allelic variation are represented as slices of the same circle. The single caprine isolate was obtained from a goat that became infected with the outbreak strain, following housing with infected cattle. Color images are available online.
Calves linked to seven patients in six states (Wisconsin, Iowa, Minnesota, Missouri, Oklahoma, and South Dakota) had purchase records from a single livestock market (Market A) in Wisconsin. Purchase record and trace back information was more complete for calves sold from Wisconsin to other states than calves sold intrastate, as calves sold intrastate were not required to have the same level of identification as cattle moving between states (e.g., ear tags or brands). Market A was supplied with calves from multiple source farms in Wisconsin.
One hundred and seventy-five nonhuman isolates with PFGE patterns matching the human patients were identified either through sampling as part of the outbreak investigation or by review of PFGE results obtained during routine surveillance activities (Supplementary Fig. S1). Matching nonhuman isolates were collected from May 28, 2015, through June 19, 2018 (Fig. 2), in 14 states. Animal isolates accounted for 140 (80%) of 175 nonhuman isolates. Thirty-three (92%) of the 35 environmental isolates were obtained from sources in Wisconsin.
WGS analysis of isolates resulting from bacterial culture of Market A samples indicated that 10 environmental isolates were genetically closely related to human- and bovine-origin isolates identified in this investigation (Fig. 4). Animal transport vehicles for Market A were sampled and yielded isolates matching the outbreak strain of Salmonella Heidelberg.
Thirteen human isolates were submitted to CDC’s NARMS laboratory for phenotypic AST. All were MDR strains, with nonsusceptibility in up to 11 of the 14 antimicrobial agents tested (Table 1). Whole genome sequences from 59 human and 117 animal isolates revealed resistance determinants consistent with the phenotypic AST results; a majority of isolates from all sources contained the plasmid-mediated resistance genes, aadA1, aph(3′)-Ia, blaCMY-2, floR, fosA7, qnrB19, strA, strB, sull, sul2, tet(A), tet(B), and tet(O), and a novel gene dfrA34.
Table 1.
Antimicrobial Resistancea of Human and Bovine Salmonella Heidelberg Outbreak Isolates from January 1, 2015, Through July 2, 2018.
| Drug b | Human N = 59 n (%) | Bovine N = 117 n (%) | Total N = 176 n (%) |
|---|---|---|---|
| Ampicillin | 55 (93) | 101(86) | 156 (88) |
| Amoxicillin–clavulanic acid | 55 (93) | 101 (86) | 156 (88) |
| Azithromycin | 0 (0) | 0 (0) | 0 (0) |
| Cefoxitin | 55 (93) | 101 (86) | 156 (88) |
| Ceftriaxone | 55 (93) | 101 (86) | 156 (88) |
| Chloramphenicolc | 25 (42) | 44 (38) | 69 (39) |
| Ciprofloxacind | 59 (100) | 111 (95) | 170 (97) |
| Gentamicin | 0 (0) | 2 (1.7) | 2 (1.1) |
| Meropenem | 0 (0) | 0 (0) | 0 (0) |
| Streptomycin | 57 (97) | 101 (86) | 158 (90) |
| Sulfisoxazole | 57 (97) | 97 (83) | 154 (88) |
| Tetracycline | 57 (97) | 97 (83) | 154 (88) |
| Trimethoprim–sulfamethoxazole | 56 (95) | 96 (82) | 152 (86) |
| Kanamycine | 45 (76) | 90 (77) | 135 (77) |
| Fosfomycine | 59 (100) | 117 (100) | 176 (100) |
| Resistance to ≥5 antimicrobial classes | 55 (93) | 101 (86) | 156 (88) |
Resistance was determined by AST for 13 isolates; resistance was predicted based on whole genome sequencing for the remaining 163 isolates.
Resistance to nalidixic acid is not shown. Most (150/176) isolates contained the qnrB19 gene; 9/12 qnrB19(+) isolates tested phenotypically showed resistance to nalidixic acid.
Eighty-four out of 153 floR(+) isolates had an interruption in the floR gene, resulting in predicted loss of chloramphenicol resistance.
Includes intermediate susceptibility.
AST was not performed for these agents.
AST, antimicrobial susceptibility testing.
Following isolation of Salmonella Heidelberg from environmental sampling at Market A, education and cleaning and disinfection information were provided to the livestock market management and staff by investigators from Wisconsin and USDA. A recommendation was made to discontinue the use of a power washer that had been used to remove visible debris before disinfection to mitigate dissemination of bacteria throughout the facility. Records from the veterinary diagnostic laboratories indicated that ill calves originated from numerous farms and moved through multiple different livestock markets other than Market A.
Information on the Salmonella Heidelberg outbreak and preventive biosecurity measures was mailed to all licensed truckers, dealers, and livestock markets in Wisconsin. Additionally, this information was also provided to licensed veterinarians and producer groups. The CDC issued a public warning through a web posting in November 2016, with multiple updates through 2018 (CDC, 2018a). The outbreak notice included advice for livestock handlers and veterinarians and a section for health care providers regarding the MDR strain.
Discussion
The present study describes the first documented multistate outbreak of MDR Salmonella Heidelberg in humans linked to cattle exposure. This human illness outbreak likely occurred as the result of a spillover of an epidemic of MDR Salmonella Heidelberg infections in calves and cattle; this is supported by the detection of ill calves in veterinary diagnostic laboratories before detection of human illness and reports from patients, indicating that calves became sick before the onset of human illness. Initially, calves were traced back to a single livestock market (Market A); however, further investigation demonstrated that additional livestock markets sold calves that were linked to human illness.
Calves came into livestock markets, including Market A, from multiple source farms. However, trace back of calves to source farms was limited as records were not available for all cattle purchases from livestock markets or from farms. Isolates resulting from sampling at Market A were genetically closely related to isolates obtained from ill people reporting contact with this market through purchase of calves (Fig. 4). It is possible that additional livestock markets could have also yielded the outbreak strain had they been sampled.
The MDR Salmonella Heidelberg strain causing this outbreak might have been disseminated to many different farms, sale locations, and livestock markets through movement and commingling of calves and conveyance equipment, including livestock trailers. Trace back through calf purchase records collected from ill people, laboratory sampling, and WGS results indicated that interstate movement of cattle likely contributed to the geographic spread of the outbreak.
During the process of transportation over long distances, cattle commingle and share pathogens through direct contact with one another or contaminated conveyance equipment such as livestock trailers. Thus, infectious agents might spread over a wide geographic area if the same equipment is used to move animals to multiple locations without cleaning and disinfection between shipments (Barham et al., 2002; Arthur et al., 2008; Dewell et al., 2008). Stress of transportation has not been definitively proven to increase the rate at which Salmonella spp. are shed in feces of cattle, although studies have reported this finding (Corrier et al., 1990; Barham et al., 2002).
Increased shedding during shipment might increase the rate of contamination of animals themselves, which poses an additional risk of exposing animal handlers at the final destination, especially if trailers are not regularly cleaned (Barham et al., 2002; Arthur et al., 2008; Dewell et al., 2008). The USDA Title 9 Code of Federal Regulations (9 CFR 91.6) specifies requirements for cleaning and disinfection of vehicles used for interstate movement of animals (USDA, 2019). The responsibility falls on individual livestock haulers to institute proper routine biosecurity measures for transportation equipment to mitigate the risk of disease transmission and on veterinarians and other accredited personnel certifying animal transportation to ensure transportation equipment is cleaned and disinfected between animal shipments.
The resistance profile in this outbreak had not previously been identified among Salmonella strains reported to NARMS. This phenotype is conferred by the presence of 14 plasmid-borne genes, 12 of which are carried on an IncC plasmid (NCBI accession MH760469.1) (Folster et al., 2018; Tagg et al., 2019). The prevalence of MDR in Salmonella has increased over time among humans in the United States, which poses the challenge of severe salmonellosis steadily becoming more difficult to treat as empirical antimicrobial selection becomes more limited (Molbak, 2005; CDC, 2018b).
Recent Salmonella Heidelberg isolates specifically have demonstrated a broadened resistance profile compared with isolates dating back to the 1980s (Antony et al., 2018). In this outbreak, a high proportion of patients were hospitalized (35%), similar to previous outbreaks associated with MDR Salmonella Heidelberg (Folster et al., 2012; Grinnel et al., 2013; Gieraltowski et al., 2016). Resistance has been previously associated with elevated virulence, possibly secondary to coselection of virulence traits with resistance mechanisms (Fluit, 2005; Molbak, 2005).
Genomic analysis of isolates from calves and humans involved in this outbreak identified a specific safABCD genetic operon encoding adhesive fimbriae that confer enhanced pathogenesis. These have been identified with some frequency in Salmonella Typhimurium, but are generally not detected in Salmonella Heidelberg (Łaniewski et al., 2017; Antony et al., 2018).
Resistant Salmonella bacteria can be acquired from numerous sources along the food production chain, and evidence indicates that new MDR variants of Salmonella are continuing to emerge in food-producing animals just as in this outbreak (CDC, 2015; Cohen et al., 2020). Young age is a known risk factor for cattle carrying MDR Salmonella (Davidson et al., 2018; Springer et al., 2018). This might be from lack of competitive exclusion of resistant or pathogenic bacteria by the undeveloped intestinal microflora; however, more data are needed to substantiate this hypothesis (Davidson et al., 2018).
Exposure to antimicrobial drugs can also elevate the risk of developing antimicrobial resistance. Waste milk (milk that cannot be sold for human consumption) collected from cattle that have been administered systemic antibiotics can contain drug residues that might interact with intestinal flora and preferentially select for resistant bacteria (Van Vleck Pereira et al., 2016) when ingested by calves. Management practices that might have resulted in selection for and transmission of MDR Salmonella Heidelberg were not examined in this investigation, but were subsequently studied by state and federal partners (Lombard, 2017).
Treatment failures in humans and cattle were not assessed in this investigation. Case histories provided by practicing veterinarians to veterinary diagnostic laboratories mentioned the absence of any effective antimicrobial drug for treating calves infected with Salmonella Heidelberg and that sudden death in high-stressed calves occurred.
Of note, most infected patients in this outbreak were children (Fig. 3). Outbreaks of Salmonella Heidelberg in the United States have similarly reported a younger median patient age ranging from 14 to 23 years (CDC, 2012, 2014; Folster et al., 2012; Grinnel et al., 2013; Gieraltowski et al., 2016). Young age is a known risk factor for human salmonellosis, likely linked to decreased immune function and hygiene practices (Graham, 2002). This raises the possibility that children living on farms or participating in agricultural activities might be at a greater risk of zoonotic salmonellosis and therefore caregivers must be diligent in monitoring direct contact with animals and enforcing proper hand washing practices (CDC, 2015; Conrad et al., 2017).
During the active investigation, case frequency demonstrated a cyclical pattern in which the highest numbers of human cases were reported during the late spring and summer months (Fig. 2). It is well established that Salmonella excretion by cattle increases in the spring and summer months, which could contribute to farm contamination and human exposure (Davidson et al., 2006; Lombard et al., 2012; Abu Aboud et al., 2016; Likavec et al., 2016).
In this outbreak, this could also merely be an indicator of the time of year when humans were most likely to come in contact with cattle (e.g., calving season, petting zoos, and animal exhibits) (Conrad et al., 2017; Daly et al., 2017). Salmonella Heidelberg infections with this MDR strain continue to be monitored by CDC and state and local health departments.
Conclusions
This investigation highlights the importance of surveillance to identify disease outbreaks among animals, which can spillover or result in human illnesses. Understanding the complexity of animal movement within the industry was important to ensure broad dissemination of biosecurity information and infection prevention.
Early detection of emergence of MDR Salmonella strains, which are resistant to first-line antimicrobials used to treat severe human and animal infections, might help to prevent zoonotic and foodborne disease transmission through early implementation of disease prevention and control measures.
Supplementary Material
Acknowledgments
The authors would like to thank Amelia Bicknese, Christy Bennett, and Meseret Birhane for their contributions to antimicrobial resistance testing and data collection; the staff in the WVDL bacteriology department for the culture, serotyping, antimicrobial susceptibility testing, and stocking of Salmonella isolates; and Beth Tolar and Darlene Wagner for their assistance in bioinformatics. The authors would also like to thank Jeffrey P. Davis for his contributions in the epidemiologic investigation and trace back processes in Wisconsin.
Funding Information
The authors have no funding information to declare.
Footnotes
Disclosure Statement
The findings and conclusions of this article are those of the authors and do not necessarily represent the official position of the U.S. Centers for Disease Control and Prevention (CDC).
See, for example, 45 C.F.R. part 46, 21 C.F.R. part 56; 42 U.S.C. §241(d); 5 U.S.C. §552a; 44 U.S.C. §3501 et seq.
References
- Abu Aboud OA, Adaska JM, Williams DR, Rossitto PV, Champagne JD, Lehenbauer TW, Atwill R, Li X, Aly SS. Epidemiology of Salmonella sp. in California cull dairy cattle: Prevalence of fecal shedding and diagnostic accuracy of pooled enriched broth culture of fecal samples. PeerJ 2016;4:e2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Academy of Pediatrics. Red Book: Report of the Committee on Infectious Diseases, 32nd edition. Itasca, IL: American Academy of Pediatrics, 2021. [Google Scholar]
- Antony L, Behr M, Sockett D, Miskimins D, Aulik N, Christopher-Hennings J, Nelson E, Allard MW, Scaria J. Genome divergence and increased virulence of outbreak associated Salmonella enterica subspecies enterica serovar Heidelberg. Gut Pathogens 2018;10:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur TM, Bosileva JM, Brichta-Harhay DM, Kalchayanand N, King DA, Shackelford SD, Wheeler TL, Koohmaraie M. Source track of Escherichia coli O157:H7 and Salmonella contamination in the lairage environment at commercial U.S. beef processing plants and identification of an effective intervention. J Food Prot 2008;71:1752–1760. [DOI] [PubMed] [Google Scholar]
- Barham AR, Barham BL, Johnson AK, Allen DM, Blanton JR, Miller MF. Effects of the transportation of beef cattle from the feedyard to the packing plant on prevalence levels of Escherichia coli O157 and Salmonella spp. J Food Prot 2002;65:280–283. [DOI] [PubMed] [Google Scholar]
- CDC. Multistate outbreak of human Salmonella Heidelberg infections linked to “kosher broiled chicken livers” from Schreiber Processing Corportation (final update). Volume 2020. Centers for Disease Control and Prevention, National Center for Emerging and Zoonotic Infectious Diseases (NCEZID), Division of Foodborne, Waterborne, and Environmental Diseases (DFWED), 2012. Available at: https://www.cdc.gov/salmonella/2011/chicken-liver-1-11-2012.html, accessed October 14, 2021. [Google Scholar]
- CDC. Multistate outbreak of multidrug-resistant Salmonella Heidelberg infections linked to Foster Farms Brand chicken (final update). Volume 2020. Centers for Disease Control and Prevention, National Center for Emerging and Zoonotic Infectious Diseases (NCEZID), Division of Foodborne, Waterborne, and Environmental Diseases (DFWED), 2014. Available at: https://www.cdc.gov/salmonella/Heidelberg-10-13/index.html, accessed October 14, 2021. [Google Scholar]
- CDC. Salmonella infection. Volume 2020. Centers for Disease Control and Prevention DoF, Waterborne, and Environmental Diseases, 2015. Available at: https://www.cdc.gov/healthypets/diseases/salmonella.html, accessed October 14, 2021. [Google Scholar]
- CDC. Multistate outbreak of multidrug-resistant Salmonella Heidelberg infections linked to contact with dairy calves (final update). Volume 2020. Centers for Disease Control and Prevention, National Center for Emerging and Zoonotic Infectious Diseases (NCEZID), Division of Foodborne, Waterborne, and Environmental Diseases (DFWED), 2018a. Available at: https://www.cdc.gov/salmonella/heidelberg-11-16/index.html, accessed October 14, 2021. [Google Scholar]
- CDC. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): Human Isolates Surveillance Report for 2015 (Final Report). Atlanta, GA: U.S. Department of Health and Human Services, CDC, 2018b. [Google Scholar]
- CDC. Antibiotic Resistance Threats in the United States. Atlanta, GA: U.S. Department of Health and Human Services, CDC, 2019. [Google Scholar]
- CDC. National Antimicrobial Resistance Monitoring System (NARMS) Now: Human Data. Atlanta, GA: U.S. Department of Health and Human Services, CDC, 2020a. Available at: https://www.cdc.gov/narmsnow, accessed October 14, 2021. [Google Scholar]
- CDC. FoodNet Fast Population Survey Tool. Atlanta, GA: Centers for Disease Control and Prevention Foodborne Diseases Active Surveillance Network, 2021. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing CLSI Supplement M100. Malvern, PA: Clinical and Laboratory Standards Institute, 2021. [Google Scholar]
- Cohen E, Davidovich M, Rokney A, Valinsky L, Rahav G, Gal-Mor O. Emergence of new variants of antibiotic resistance genomic islands among multidrug-resistant Salmonella enterica in poultry. Environ Microbiol 2020;22:413–432. [DOI] [PubMed] [Google Scholar]
- Conrad C, Stanford K, Narvaez-Bravo C, Callaway T, McAllister T. Farm fairs and petting zoos: A review of animal contact as a source of zoonotic enteric disease. Foodborne Pathog Dis 2017;14:59–73. [DOI] [PubMed] [Google Scholar]
- Corrier D, Purdy C, DeLoach J. Effects of marketing stress on fecal excretion of Salmonella spp. in feeder calves. Am J Vet Res 1990;51:866–869. [PubMed] [Google Scholar]
- Daly RF, House J, Stanek D, Stobierski MG. Compendium of Measures to Prevent Disease Associated with Animals in Public Settings, 2017. J Am Vet Med Assoc 2017;251:1268–1292. [DOI] [PubMed] [Google Scholar]
- Davidson H, Sayers A, Smith R, Evans S, Weaver J, Pascoe S, Davies R. Risk factors associated with the salmonella status of dairy farms in England and Wales. Vet Rec 2006;159:871–880. [PubMed] [Google Scholar]
- Davidson K, Byrne B, Pires A, Magdesian K, Pereira R. Anti-microbial resistance trends in fecal Salmonella isolates from northern California dairy cattle admitted to a veterinary teaching hospital, 2002–2016. PLoS One 2018;13:e0199928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies E, Venn J. The detection of a bovine carrier of Salmonella Heidelberg. J Hyg (Lond) 1962;60:495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewell G, Simpson C, Dewell R, Hyatt D, Belk K, Scanga J, Morley P, Grandin T, Smith G, Dargatz D, Wagner B, Salman M. Risk associated with transportation and lairage on hide contamination with Salmonella enterica in finished beef cattle at slaughter. J Food Prot 2008;71:2228–2232. [DOI] [PubMed] [Google Scholar]
- Fey PD, Safranek TJ, Rupp ME, Dunne EF, Ribot E, Iwen PC, Bradfor PA, Angulo FJ, Hinrichs SH. Ceftriazone-resistant Salmonella infection acquired by a child from cattle. N Engl J Med 2000;342:1242–1429. [DOI] [PubMed] [Google Scholar]
- Fluit AC. Towards more virulent and antibiotic-resistant Salmonella? FEMS Immunol Med Microbiol 2005;43:1–11. [DOI] [PubMed] [Google Scholar]
- Folster J, Chen J, Tagg K, Bennett C, Watkins LF, Schlater L, Morningstar-Shaw B, Lantz K, Aulik N, Sockett D, Elbadawi L, Gundlach K, Valley A, Klos R, Stevenson L, Nichols M. Identification and characterization of a multidrug-resistant Salmonella enterica serotype Heidelberg outbreak associated with dairy cattle in the United States. In Intranational Association for Food Protection Annual Meeting. Salt Lake City, UT, 2018. Available at: https://iafp.confex.com/iafp/2018/meetingapp.cgi/Paper/17253, accessed October 14, 2021. [Google Scholar]
- Folster JP, Pecic G, Rickert R, Taylor J, Zhao S, Fedorka-Cray PJ, Whichard J, McDermott P. Characterization of multidrug-resistant Salmonella enterica serovar Heidelberg from a ground turkey-associated outbreak in the United States in 2011. Antimicrob Agents Chemother 2012;56:3465–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieraltowski L, Higa J, Peralta V, Green A, Schwensohn C, Rosen H, Libby T, Kissler B, Marsden-Haug N, Booth H, Kimura A, Grass J, Bicknese A, Tolar B, Defibaugh-Chavez S, Williams I, Wise M, Salmonella Heidelberg Investigation Team. National outbreak of multidrug resistant Salmonella Heidelberg infections linked to a single poultry company. PLoS One 2016;11:e0162369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham SM. Salmonellosis in children in developing and developed countries and populations. Curr Opin Infect Dis 2002;15:507–512. [DOI] [PubMed] [Google Scholar]
- Grinnel M, Provo G, Marsden-Haug N, Stigi KA, DeBess E, Kissler B, Crarey E, Tate H, Pringle J, Grass J, Folster JP, Williams I, Gieraltowski L, Laufer AS. Outbreak of Salmonella Heidelberg infections linked to single poultry producer—13 state, 2012–2013. MMWR Morb Mortal Wkly Rep 2013;62:553–556. [PMC free article] [PubMed] [Google Scholar]
- Habbs VH. About a new type of bacteria form the paratyphoid enteritis group. J Bacteriol 1933:367–374. [Google Scholar]
- Katz LS, Griswold T, Williams-Newkirk AJ, Wagner D, Petkau A, Sieffert C, Van Domselaar G, Deng X, Carleton HA. A comparative analysis of the Lyve-SET phylogenomics pipeline for genomic epidemiology of foodborne pathogens. Front Microbiol 2017;8:375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox WA, Galbraith NS, Lewis MJ, Hickie GC, Johnston HH. A milk-borne outbreak of food poisoning due to Salmonella Heidelberg. J Hyg (Lond) 1963;61:175–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota KA, Wolfgang WJ, Baker DJ, Boxrud D, Turner L, Trees E, Carleton HA, Gerner-Smidt P. PulseNet and the changing paradigm of laboratory-based surveillance for foodborne diseases. Public Health Rep 2019;134:22S–28S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łaniewski P, Baek C-H, Roland KL, Curtiss R, Hultgren SJ. Analysis of spleen-induced fimbria production in recombinant attenuated Salmonella enterica Serovar Typhimurium vaccine strains. mBio 2017;8:e01189–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Likavec T, Pires AFA, Funk JA. Association between thermal environment and Salmonella in fecal samples from dairy cattle in midwestern United States. Can J Vet Res 2016;80:183–188. [PMC free article] [PubMed] [Google Scholar]
- Lombard J Salmonella Heidelberg: An on-farm study of dairy operations. In InFORM, 2017. Available at: https://www.aphl.org/conferences/proceedings/Documents/2017/InFORM/20-Lombard.pdf, accessed October 14, 2021.
- Lombard JE, Beam AL, Nifong EM, Fossler CP, Kopral CA, Dargatz DA, Wagner BA, Erdman MM, Fedorka-Cray PJ. Comparison of individual, pooled, and composite fecal sampling methods for detection of Salmonella on U.S. dairy operations. J Food Prot 2012;75:1562–1571. [DOI] [PubMed] [Google Scholar]
- Lorenz I, Fagan J, More SJ. Calf health from birth to weaning. II. Management of diarrhoea in pre-weaned calves. Ir Vet J 2011;64:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch MF, Tauxe RV, Hedberg CW. The growing burden of foodborne outbreaks due to contaminated fresh produce: Risks and opportunities. Epidemiol Infect 2009;137:307–315. [DOI] [PubMed] [Google Scholar]
- Lynne AM, Kaldhone P, David DE, White DG, Foley SL. Characterization of antimicrobial resistance in Salmonella enterica serotype Heidelberg isolated from food animals. Foodborne Pathog Dis 2009;6:207–208. [DOI] [PubMed] [Google Scholar]
- Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012;18:268–281. [DOI] [PubMed] [Google Scholar]
- Marder EP, Griffin PM, Cieslak PR, Dunn J, Hurd S, Jervis R, Lathrop S, Muse A, Ryan P, Smith K, Tobin-D’Angelo M, Vugia DJ, Holt KG, Wolpert BJ, Tauxe R, Geissler AL. Preliminary incidence and trends of infections with pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 2006–2017. MMWR Morb Mortal Wkly Rep 2018;67:324–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott PF, Tyson GH, Kabera C, Chen Y, Li C, Folster JP, Ayers SL, Lam C, Tate HP, Zhao S. Whole-genome sequencing for detecting antimicrobial resistance in non-typhoidal Salmonella. Antimicrob Agents Chemother 2016;60:5515–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuirk SM, Peek S. Salmonellosis in cattle: A review. In 36th Annual Conference of the American Association of Bovine Practioners. Columbus, OH: University of Wisconsin, School of Veterinary Medicine, 2003. [Google Scholar]
- Molbak K Human health consequences of antimicrobial drug-resistant Salmonella and other foodborne pathogens. Clin Infec Dis 2005;41:1613–1620. [DOI] [PubMed] [Google Scholar]
- Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 2006;3:59–67. [DOI] [PubMed] [Google Scholar]
- Shane AL, Mody RK, Crump JA, Tarr PI, Steiner TS, Kotloff K, Langley JM, Wanke C, Warren CA, Cheng AC, Cantey J, Pickering LK. Infectious diseases society of america clinical practice guidelines for the diagnosis and management of infectious diarrhea. Clin Infect Dis 2017;65:e45–e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer HR, Denagamage TN, Fenton GD, Haley BJ, Van Kessel JAS, Hovingh EP. Antimicrobial resistance in fecal Escherichia coli and Salmonella enterica from dairy calves: A systematic review. Foodborne Pathog Dis 2018;16:23–34. [DOI] [PubMed] [Google Scholar]
- Tagg KA, Amir A, Ikram A, Chen JC, Meservey JYKE, Joung YJ, Batra JLHD, Leeper MM, Katz LS, Saeed A, Freeman M, Watkins LF, Salman M, Folster JP. Sequencing and characterization of five extensively drug resistant Salmonella enterica serotype Typhi isolates implicated in human infections from Punjab, Pakistan. Microbiol Resour Announc 2020;9: e01466–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagg KA, Francois Watkins L, Moore MD, Bennett C, Joung YJ, Chen JC, Folster JP. Novel trimethoprim resistance gene dfrA34 identified in Salmonella Heidelberg in the USA. J Antimicrob Chemother 2019;74:38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA. Code of Federal Regulations Title 9—Animal and Animal Products Section 91.6. Washington, DC: Office of the Federal Register, National Archives and Records Administration, 2019. [Google Scholar]
- Van Vleck Pereira R, Lima S, Siler JD, Foditsch C, Warnick LD, Bicalho RC. Ingestion of milk containing very low concentration of antimicrobials: Longitudinal effect on fecal microbiota composition in preweaned calves. PLoS One 2016;11:e0147525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma JK, Mølbak K, Barrett TJ, Beebe JL, Jones TF, Rabatsky-Ehr T, Smith KE, Vugia DJ, Chang H-GH, Angulo FJ. Antimicrobial-resistant nontyphoidal Salmonella is associated with excess bloodstream infections and hospitalizations. J Infect Dis 2005;191:554–561. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


