Abstract
The nucleotide sequences of the 27,939-bp-long upstream and 9,448-bp-long downstream regions of the carAaAaBaBbCAc(ORF7)Ad genes of carbazole-degrading Pseudomonas sp. strain CA10 were determined. Thirty-two open reading frames (ORFs) were identified, and the car gene cluster was consequently revealed to consist of 10 genes (carAaAaBaBbCAcAdDFE) encoding the enzymes for the three-step conversion of carbazole to anthranilate and the degradation of 2-hydroxypenta-2,4-dienoate. The high identities (68 to 83%) with the enzymes involved in 3-(3-hydroxyphenyl)propionic acid degradation were observed only for CarFE. This observation, together with the fact that two ORFs are inserted between carD and carFE, makes it quite likely that the carFE genes were recruited from another locus. In the 21-kb region upstream from carAa, aromatic-ring-hydroxylating dioxygenase genes (ORF26, ORF27, and ORF28) were found. Inductive expression in carbazole-grown cells and the results of homology searching indicate that these genes encode the anthranilate 1,2-dioxygenase involved in carbazole degradation. Therefore, these ORFs were designated antABC. Four homologous insertion sequences, IS5car1 to IS5car4, were identified in the neighboring regions of car and ant genes. IS5car2 and IS5car3 constituted the putative composite transposon containing antABC. One-ended transposition of IS5car2 together with the 5′ portion of antA into the region immediately upstream of carAa had resulted in the formation of IS5car1 and ORF9. In addition to the insertion sequence-dependent recombination, gene duplications and presumed gene fusion were observed. In conclusion, through the above gene rearrangement, the novel genetic structure of the car gene cluster has been constructed. In addition, it was also revealed that the car and ant gene clusters are located on the megaplasmid pCAR1.
Microorganisms exposed to xenobiotics often show adaptive activities. As the molecular mechanism for adaptation, gene recruitment is the key to establishing several simultaneous degradation activities and the entire catabolic capacity for the xenobiotics to which microorganisms are exposed. In fact, catabolic gene clusters which have similarities in the levels of gene organization and nucleotide sequences have been reported for phylogenetically unrelated bacterial strains isolated from geographically distinct areas (15, 19, 46, 58, 61). Mobile genetic elements, such as plasmids and transposons, have been considered to play an important role in distributing catabolic capacities by gene recruitment. Previously, many xenobiotic-degradative gene clusters have been reported to be located on mobile genetic elements (55, 57, 62). The recruited gene clusters can also undergo various types of rearrangements including deletion, duplication, inversion, and insertion into other replicons, especially when the bacteria adapt in order to acquire novel catabolic activities (18, 62).
The well-clustered genetic organization of entire pathways or independently functioning pathway segments has been reported for bacteria which degrade easily degradable compounds, such as naphthalene, toluene, and biphenyl (15, 23, 63). It can be assumed that these genetic structures developed through long exposure to corresponding compounds. On the other hand, the degradative genes involved in the degradation of highly recalcitrant compounds seem to be disordered or separated. It can be speculated that the different levels of maturation in genetic structures resulted from the difference in the length of exposure to xenobiotics or rarity of the gene(s) indispensable for degradation. That is, the degradative genes for a highly recalcitrant molecule are useful in revealing the maturation (evolution) stage of xenobiotic-degrading gene clusters.
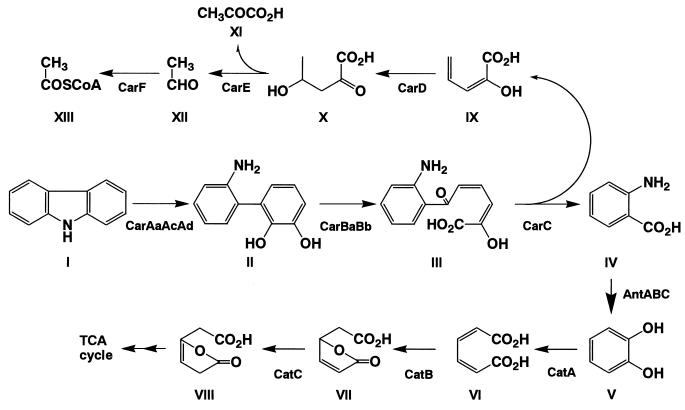
Carbazole (CAR) is a heterocyclic aromatic compound containing a dibenzopyrrole system derived from coal tar and shale oil (41). CAR is known to possess mutagenic and toxic activities (3) and also to be a recalcitrant molecule (12). Pseudomonas sp. strain CA10 is a microorganism having the ability to utilize CAR as a sole source of carbon, nitrogen, and energy (45). As shown in Fig. 1, strain CA10 degrades CAR to anthranilate and 2-hydroxypenta-2,4-dienoate through angular dioxygenation, meta-cleavage, and hydrolysis (45). The anthranilate produced is attacked by dioxygenase at the 1,2 positions to yield catechol, which is degraded by the β-ketoadipate pathway. On the other hand, 2-hydroxypenta-2,4-dienoate is converted to a tricarboxylic acid (TCA) cycle intermediate by the three additional conversions called the meta-cleavage pathway.
FIG. 1.
Degradation pathway of CAR in Pseudomonas sp. strain CA10. Enzyme designations: CarAaAcAd, carbazole 1,9a-dioxygenase; CarBaBb, 2′-aminobiphenyl-2,3-diol 1,2-dioxygenase; CarC, 2-hydroxy-6-oxo-6-(2′-aminophenyl)-hexa-2,4-dienoic acid (meta-cleavage compound) hydrolase; CarD, 2-hydroxypenta-2,4-dienoate hydratase; CarE, 4-hydroxy-2-oxovalerate aldolase; CarF, acetaldehyde dehydrogenase (acylating); AntABC, anthranilate 1,2-dioxygenase; CatA, catechol 1,2-dioxygenase; CatB, cis,cis-muconate lactonizing enzyme; CatC, muconolactone δ-isomerase. Compounds: I, CAR; II, 2′-aminobiphenyl-2,3-diol; III, 2-hydroxy-6-oxo-6-(2′-aminophenyl)-hexa-2,4-dienoic acid (meta-cleavage compound); IV, anthranilic acid; V, catechol; VI, cis,cis-muconic acid; VII, muconolactone; VIII, β-ketoadipic acid enol-lactone; IX, 2-hydroxypenta-2,4-dienoic acid; X, 4-hydroxy-2-oxovaleric acid; XI, pyruvic acid; XII, acetaldehyde; XIII, acetyl coenzyme A.
Using Tn5 mutagenesis, we have previously shown the presence of car and cat gene clusters encoding the enzymes involved in the initial degradation of CAR and the β-ketoadipate pathway, respectively (30). By shotgun cloning, we have isolated the car gene cluster encoding carbazole 1,9a-dioxygenase (CarAaAcAd), the meta-cleavage enzyme (CarBaBb), and the meta-cleavage compound hydrolase (CarC), which catalyze the first three steps in CAR degradation (49, 50). Genetic analysis of the car gene cluster revealed that the 1,263-bp DNA region containing the carAa gene is tandemly duplicated except for one base (49). Also, two copies of carAa genes are separated from the carAc and carAd genes by the 2-kb DNA fragment containing carBaBb and carC. So far, such a gene structure has not been observed in other degradative gene clusters, except for the different alleles of the car gene cluster. The genetic organization of the car gene cluster of strain CA10 implies the possibility that gene rearrangements occurred through evolutionary divergence in the natural environment. Because of the fact that the G+C content of the identified car gene cluster is lower than that of total DNA of strain CA10 (45, 50), the car gene cluster was also hypothesized to have been recruited from other microorganisms.
We herein describe in detail the genetic analysis of the car gene cluster and its flanking region. We also discuss the evolutionary events that occurred during the development of a novel genetic structure in this DNA region. In addition, it is reported that the car gene cluster is located on the megaplasmid, although the cat gene cluster is located on the chromosome of strain CA10.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
Bacterial strains, plasmids, and cosmids used in this study are listed in Table 1. A physical map of the plasmids and cosmids constructed in this study is also shown in Fig. 2. For extractions of total DNA and the megaplasmid, strain CA10 was grown on 2xYT medium (48) and nutrient broth (Eiken Chemical Co. Ltd., Tokyo, Japan), respectively, at 30°C with reciprocal shaking (300 strokes/min) for 16 h. To prepare the total RNA of strain CA10, carbon- and nitrogen-free mineral medium (CNFMM) supplemented with CAR as the sole source of carbon, nitrogen, and energy was used. CNFMM had the following composition (grams per liter): Na2HPO4, 2.2; KH2PO4, 0.8; MgSO4 · 7H2O, 0.2; FeSO4 · 7H2O, 0.01; CaCl2 · 2H2O, 0.01; and yeast extract, 0.05. CAR was dissolved in dimethyl sulfoxide (10 mg/ml) and sterilized by filtration with a 0.2-μm-pore-size membrane filter. Then, 1% (vol/vol) of this dimethyl sulfoxide solution was added to CNFMM.
TABLE 1.
Bacterial strains, plasmids, and cosmids used in this study
| Strain, plasmid, or cosmid | Relevant characteristics | Source or reference |
|---|---|---|
| Bacterial strains | ||
| Pseudomonas sp. strain CA10 | Car+a | 45 |
| Pseudomonas sp. strain TE1 | Car−a, AN(C− N+)b | 30 |
| Escherichia coli | ||
| JM109 | recA1 Δ(lac-proAB) endA1 gyrA96 thi-1 hsdR17 relA1 supE44 [F′traD36 proAB lacIqZΔM15] | 48 |
| DH5α | F− φ80d lacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17(rK− mK+) deoR thi-1 supE44 λ gyrA96 relA1 | Toyobo |
| XL1-Blue MR | Δ(mcrA)183, Δ(mcrCB-hsdSMR-mrr) 173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac | Stratagene |
| Plasmids | ||
| pUC18/19 | AprlacZ, pMB9 replicon | 48 |
| pBluescript II KS(+/−) | AprlacZ, pMB9 replicon | Stratagene |
| pBluescript II SK(−) | AprlacZ, pMB9 replicon | Stratagene |
| SuperCos1 | Apr Kmr cos | Stratagene |
| pT7Blue(R) | AprlacZ | Novagen |
| pUCA1 | Apr pUC119 with 6.9-kb EcoRI insert of strain CA10 DNA | 50 |
| pUCA601 | Apr pUC19 with 5.8-kb HindIII insert of strain CA10 DNA | This study |
| pUCA610 | Apr pUC19 with 4.0-kb SalI insert of strain CA10 DNA | This study |
| pBCA616 | Apr pBluescript II SK(−) with 1.0-kb SalI-XbaI insert of strain CA10 DNA | This study |
| pUCA617 | Apr pUC18 with 1.3-kb HincII-SphI insert of strain CA10 DNA | This study |
| pUCA618 | Apr pUC19 with 1.7-kb EcoRV insert of strain CA10 DNA | This study |
| pBCA620 | Apr pBluescript II SK(−) with 9.5-kb EcoRI insert of strain CA10 DNA | This study |
| pBCA711 | Apr pBluescript II SK(−) with 4.4-kb EcoRI insert of strain CA10 DNA | This study |
| pBCA721 | Apr pBluescript II SK(−) with 11.3-kb XhoI insert of strain CA10 DNA | This study |
| pBCA722 | Apr pBluescript II SK(−) with 7.2-kb XhoI-SalI insert of strain CA10 DNA | This study |
| pBCA731 | Apr pBluescript II SK(−) with 7.5-kb EcoRV-ClaI insert of strain CA10 DNA | This study |
| pUCA741 | Apr pUC19 with 8.9-kb SalI insert of strain CA10 DNA | This study |
| pBCA751 | Apr pBluescript II SK(−) with 5.3-kb PstI insert of strain CA10 DNA | This study |
| pTE11 | Apr pUC19 with 10.7-kb EcoRI insert of strain TE1 DNA containing the 7.7-kb Tn5-Mob region | 30 |
| pTCA16S | Apr pT7Blue(R) with 1.5-kb PCR-amplified DNA fragment containing partial 16S rRNA gene of strain CA10 | This study |
| Cosmids | ||
| pSCos701 to pSCos709 | Apr Kmr SuperCos1 with EcoRI insert of strain CA10 DNA | This study |
Car+ indicates the ability to grow on CAR as a sole source of carbon, nitrogen, and energy.
AN(C− N+) indicates that strain TE1 can grow on anthranilate as a sole nitrogen source but not as a sole carbon source.
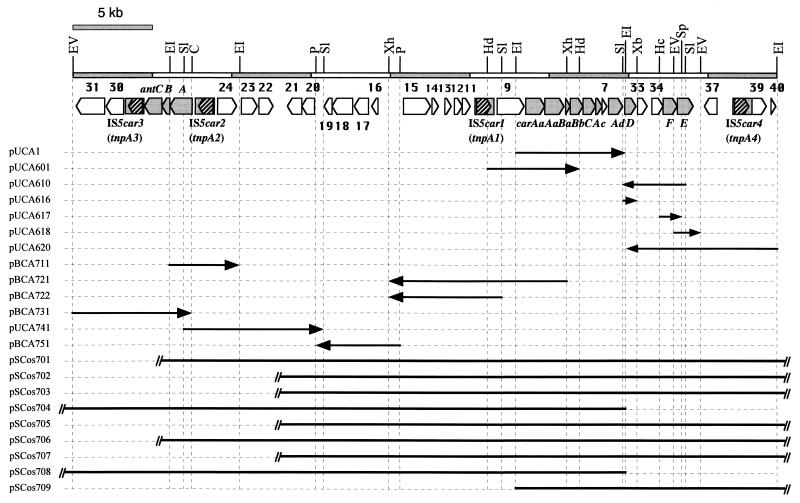
FIG. 2.
Physical map of the 44.3-kb DNA region containing the CAR-degrading car gene cluster. The pentagons and triangles in the physical map indicate the sizes, locations, and directions of transcription of the ORFs derived from the DNA sequence analysis. The shaded pentagons and triangles represent the car and ant gene clusters. The shaded boxes containing hatched pentagons represent the ISs, and the hatched pentagons indicate the sizes, locations, and directions of transcription of the transposase genes. The location of the subcloned DNA fragments and orientation of the lac promoter of the cloning vector are indicated by the lengths and the directions of the arrows, respectively. As for the cosmid clones, the DNA region which was revealed to be contained in the respective cosmids by Southern hybridization analysis is shown by the solid line. The double slash attached to the end of the solid line indicates that the DNA region outside the EcoRI site was contained in the respective cosmid insert. Only the restriction enzyme sites used for the construction of the plasmids are shown. Restriction enzyme site designations: C, ClaI; EI, EcoRI; EV, EcoRV; Hc, HincII; Hd, HindIII; P, PstI; Sl, SalI; Sp, SphI; Xb, XbaI; and Xh, XhoI.
Escherichia coli strains JM109 (48) and DH5α (Toyobo Co., Ltd., Tokyo, Japan) were used as host strains of the plasmid pUC19 (48); pBluescript II SK(−), KS(+), or KS(−) (Stratagene, La Jolla, Calif.); and their derivatives. For construction of the cosmid library of strain CA10 genomic DNA, cosmid vector SuperCos1 (Stratagene) and E. coli host strain XL1-Blue MR were used. E. coli strains were grown on 2xYT medium at 37°C. Ampicillin (50 μg/ml) or kanamycin (50 μg/ml) was added to selective media. For plate cultures, the above media solidified with 1.6% (wt/vol) agar were used.
DNA manipulation.
Plasmid or cosmid DNA was prepared from the E. coli host strain by the alkaline lysis method. Total DNA of strain CA10 was prepared as described previously (50). Restriction endonuclease and DNA Ligation Kit version 2 (Takara Shuzo Co., Ltd., Kyoto, Japan) were used according to the manufacturer's instructions. DNA fragments were extracted from the agarose gel by using Concert Rapid Gel Extraction Systems (Life Technologies, Rockville, Md.) according to the manufacturer's instructions. Other DNA manipulation procedures were performed according to the standard methods (48).
Gene walking to identify the flanking region of the car gene cluster.
A cosmid vector, SuperCos1, was first digested with XbaI, and the digested end of the DNA fragment was dephosphorylated with calf intestinal alkaline phosphatase (CIAP; Roche Diagnostics GmbH, Mannheim, Germany). Next, the resultant XbaI-CIAP-treated DNA fragment was digested with EcoRI. Total DNA of strain CA10 was partially digested with EcoRI and directly ligated to XbaI-CIAP/EcoRI-treated SuperCos1 vector. After ligation, the linear DNA formed was packaged in vitro into phage by using Gigapack III Gold Packaging Extract (Stratagene), which was used to infect host E. coli cells prepared according to the manufacturer's instructions. After the infection, host cells were spread onto the 2xYT agar plate supplemented with ampicillin to screen the clones carrying the flanking region of the car gene cluster. Colony hybridization was carried out according to the methods reported by Sambrook et al. (48) using a Biodyne B nylon membrane (Poll BioSupport Co., East Hills, N.Y.). The probe for hybridization was prepared from the 6.9-kb EcoRI DNA fragment of pUCA1 using the Megaprime DNA labeling system (Amersham Pharmacia Biotech UK Ltd., Little Chalfont, Buckinghamshire, England) and [α-32P]dCTP (110 TBq/mmol; Amersham Pharmacia Biotech UK Ltd.).
Prehybridization was performed at 65°C for at least 3 h in Rapid Hybridization buffer (Amersham Pharmacia Biotech UK Ltd.). Hybridization was performed at the same temperature for at least 16 h in the same buffer supplemented with the 32P-labeled probe newly denatured by heating at 98°C. The blots were washed once with 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS) (30 min, 65°C) and twice with 0.1× SSC–0.1% SDS (30 min, 65°C). The blots were visualized by using an imaging plate and an FLA-3000G image analyzer (Fuji Photo Film Co. Ltd., Tokyo, Japan).
Southern hybridization.
Southern blotting was performed by using a Biodyne B nylon membrane according to the recommendations of the manufacturer. A nonradioactive digoxigenin DNA labeling and detection kit (Roche Diagnostics GmbH) was used according to the manufacturer's instructions. Prehybridization and hybridization were carried out at 68°C. After hybridization, the membranes were washed three times with 2× SSC–0.1% SDS (20 min, room temperature) and three times with 0.1× SSC–0.1% SDS (20 min, 68°C).
Nucleotide sequence determination, homology search, and alignment.
To generate a population of DNA sequencing templates with randomly interspersed primer-binding sites, the GPS-1 Genome Priming System (New England Biolabs, Inc., Beverly, Mass.) was used. Nucleotide sequence determination was carried out by the chain termination method using a Li-Cor Model 4200L-2 auto-DNA sequencer (Li-Cor Inc., Lincoln, Nebr.) according to the manufacturer's instructions, and the nucleotide sequences obtained were analyzed with DNASIS-Mac software (version 3.7; Hitachi Software Engineering Co. Ltd., Yokohama, Japan). Homology searching was performed using the SWISS-PROT amino acid sequence data bank or the DDBJ/EMBL/GenBank DNA databases with the BLAST program (version 2.0.10) (1). Alignment of the deduced amino acid sequences of observed open reading frames (ORFs) was performed by using the CLUSTAL W package (version 1.6) (56).
Northern hybridization.
After the cultivation of strain CA10 in 5 ml of nutrient broth, cells were gathered from 4 ml of culture by centrifugation at 7,200 × g and then washed twice using CNF buffer, which contained 2.2 g of Na2HPO4 and 0.8 g of KH2PO4 per liter. The washed cells were suspended in 4 ml of CNF buffer. The cells were starved by incubation of the resultant cell suspension at 30°C with reciprocal shaking at 300 strokes/min for 6 h. The starved cells were washed twice as described above and finally suspended in 4 ml of CNF buffer. One hundred milliliters of CNFMM supplemented with CAR was inoculated with 10 μl of the resultant cell suspension of strain CA10. After 12 h of incubation with reciprocal shaking (300 strokes/min) at 30°C, the growing cells were harvested and used for extraction of total RNA. For the comparative study, 100 ml of nutrient broth was inoculated with 10 μl of the starved-cell suspension, and the nutrient broth-grown cells were similarly harvested as described above. The strain CA10 cells just after the starvation and after the cultivation on nutrient broth were used for extraction of total RNA. Total RNA from harvested cells was extracted using an RNeasy Mini or Midi kit combined with a QIAshredder and RNase-Free DNase set from Qiagen (Santa Clarita, Calif.) according to the manufacturer's instructions.
Two micrograms of total RNA was run on a 1% agarose gel containing 2.2 M formaldehyde using 1× morpholinepropanesulfonic acid (MOPS) buffer (20 mM MOPS, 5 mM CH3CO2Na, 1 mM EDTA; pH 7.0). After electrophoresis, RNA was transferred to a Biodyne B nylon membrane by capillary blotting overnight. The 32P-labeled probes for hybridization were prepared from the 200-bp SmaI DNA fragment (3′ portion of antA gene) of pBCA711 as described above. Prehybridization, hybridization, and detection of probes were performed as described above for colony hybridization.
Preparation of cell extract.
E. coli strain JM109 harboring plasmid pBCA616, pUCA617, or pUCA618 was grown to an optical density at 550 nm of 0.8. After further incubation in the presence of 1 mM isopropyl-β-d(−)-thiogalactopyranoside for 5 h at 37°C, the cells were harvested, washed twice with 50 mM sodium phosphate buffer (pH 7.5), and resuspended in 10 ml of the same buffer. The cell suspensions were sonicated and centrifuged at 21,600 × g at 4°C for 60 min, and the supernatants were used as cell extracts. The protein concentration was determined by using the Protein Assay Kit II (Bio-Rad Laboratories, Richmond, Calif.) with bovine serum albumin as a standard.
SDS-PAGE.
SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was performed with a 13% polyacrylamide gel. Protein staining of the gels was performed with Coomassie brilliant blue R-250.
Determination of enzymatic activities.
The hydratase activity for 2-hydroxypenta-2,4-dienoate of the cell extract was assayed by monitoring the decrease in A265 with a Beckman DU-7400 spectrophotometer equipped with a thermojacketed cuvette holder and a circulating water bath, as described by Harayama et al. (22). The substrate was prepared enzymatically from dl-allylglycine by the method used for synthesis of 2-oxopent-4-enoate (13). The assay was performed at 25°C using 1 ml of 10 mM Tris-HCl buffer (pH 7.0) containing 3.3 mM MgSO4 and 50 μg of crude cell extract/ml. The reaction was initiated by the addition of 50 μM substrate. One unit of activity was defined as the amount of the enzyme required to degrade 1 μmol of 2-hydroxypenta-2,4-dienoate per min at 25°C. The molar extinction coefficient of 2-hydroxypenta-2,4-dienoate was taken to be 19,200 cm−1 M−1 (47).
The acetoaldehyde dehydrogenase (acylating) activity was determined by monitoring the coenzyme A-stimulated production of NADH (A340) at 20°C as described by Shingler et al. (52). Assay mixtures (1 ml) contained 50 mM phosphate buffer (pH 7.5), 285 μM NAD+, 10 mM acetaldehyde, and 0.4 to 0.5 mg of crude cell extract/ml. The A340 of this mixture was monitored for 1 min, and the reaction was then initiated by the addition of 100 μM coenzyme A. One unit of activity was defined as the amount of the enzyme required to produce 1 mM NADH per min in the presence of coenzyme A under these conditions. The molar extinction coefficient of NADH was taken to be 6,300 cm−1 M−1.
Isolation of 16S rRNA gene from strain CA10 and determination of the sequence.
The 16S rRNA gene was amplified from the total DNA of strain CA10 by PCR using two primers that attached to positions 8 to 27 in the E. coli numbering system (5′-AGAGTTTGATC[A/C]TGGCTCAG-3′) and positions 1513 to 1492 (TACGG[A/T/C]TACCTTGTTACGACTT). The amplified DNA fragment was ligated to pT7Blue(R) (Novagen, Madison, Wis.), and then the resultant pTCA16S was used as the template to determine the nucleotide sequence of the partial 16S rRNA gene of strain CA10 as described above.
Extraction of the megaplasmid from strain CA10 cells.
After the cultivation using nutrient broth, strain CA10 cells were harvested from 5 ml of culture by centrifugation (7,200 × g for 2 min), washed with 1 ml of sterilized distilled water, and repelleted. According to a previously reported procedure (27), the resultant cells suspended in 50 μl of distilled water were lysed by adding 200 μl of lysing solution (50 mM Tris, 3% SDS, pH 12.6). The solution was incubated at room temperature for 15 min followed by heating at 80°C for 1 min and extracted with 250 μl of a phenol-chloroform solution (1:1, vol/vol) saturated with Tris-EDTA (TE) buffer (48). The resultant lysate was incubated for 3 h. After centrifugation (7,200 × g 15 min), the aqueous phase was subjected to electrophoresis in 0.7% (wt/vol) agarose H (Nippon Gene Co., Tokyo, Japan) in Tris-acetate-EDTA buffer (48).
Chemicals.
CAR and anthranilic acid were purchased from Tokyo Kasei Kogyo Co., Ltd. (Tokyo, Japan). All other chemicals were of the highest purity commercially available.
Nucleotide sequence accession number.
The nucleotide sequence data of the car gene cluster flanking region and the 16S rRNA gene of strain CA10 reported in this study were registered in the DDBJ, EMBL, and GenBank nucleotide sequence databases with the accession no. AB047548 and AB047273, respectively.
RESULTS
Cloning of the genes containing the flanking region of the car gene cluster.
A cosmid library of the strain CA10 genome was constructed, and approximately 1,800 of the resultant colonies were screened for the car gene cluster by probing with the 6.9-kb EcoRI insert DNA fragment of pUCA1. Thus, nine positive clones (pSCos701 to pSCos709) were isolated, and restriction mapping combined with Southern hybridization analyses was performed. The resultant restriction maps of these clones are shown in Fig. 2.
Determination of ORFs by nucleotide sequence analysis.
To determine the nucleotide sequence of the upstream region of the carAa gene, we constructed the deletion plasmids pUCA601, pUCA610, pBCA620, pBCA711, pBCA722, pBCA731, pUCA741, and pBCA751 from the cosmid inserts. The series of Tn7 mutant derivatives of each plasmid were constructed by using the GPS system, and the flanking region of the Tn7 insertion was sequenced. Consequently, the nucleotide sequences of 27,937-bp-long upstream and 9,448-bp-long downstream regions of the already-known car gene cluster were determined. There existed 23 (ORF9 to ORF31) and 9 (ORF32 to ORF40) additional ORFs in the upstream and downstream regions of the car gene cluster as shown in Fig. 2 and Table 2. The putative Shine-Dalgarno sequences (51) were found in the upstream regions of the putative initiation codons of ORFs except for ORF9, ORF10, ORF14, ORF15, ORF16, ORF22, ORF23, ORF25, ORF29, ORF31, ORF38, and ORF39 (data not shown).
TABLE 2.
Proposed functions of the proteins encoded by the genes located in the car gene cluster and its flanking region
| ORF (gene name) | Positions in the sequence with accession no. AB047548 | Probable function | Calculated molecular mass (Da) | G+C content (%) | Amino acid identity (%) | Homology
|
Accession no. | |
|---|---|---|---|---|---|---|---|---|
| Name(s) of protein(s) | Source | |||||||
| ORF9 | 26,723–28,426 | Unknown (fusion gene) | 63,238 | 55.5 | 40 | AntA | Acinetobacter sp. strain ADP1 | AF071556 |
| 27 | Xy1X | pDK1 (P. putida strain PaW630) | AF134348 | |||||
| ORF10 (tnpA1) | 25,375–26,355 | Transposase | 36,828 | 58.9 | 100 | TnpA2, TnpA4 | Pseudomonas sp. strain CA10 | AB047548 |
| 96 | TnpA3 | Pseudomonas sp. strain CA10 | AB047548 | |||||
| 96 | TnpA1 | P. stutzeri strain AN10 | AF039533 | |||||
| 96 | Transposase | IS1384 (P. putida strain H) | AF052751 | |||||
| 94 | TnpA2 | P. stutzeri strain AN10 | AF039534 | |||||
| ORF11 | 24,489–25,001 | Unknown | 18,850 | 49.3 | ||||
| ORF12 | 23,972–24,463 | Unknown | 18,038 | 49.2 | ||||
| ORF13 | 23,447–23,779 | Unknown | 12,384 | 55.0 | ||||
| ORF14 | 22,567–22,971 | Unknown | 14,672 | 65.2 | ||||
| ORF15 | 20,851–22,485 | Unknown | 57,008 | 68.7 | 13a | VgrG | E. coli strain ECOR-50 | AF044506 |
| 13a | VgrG | E. coli strain ec-11 | AF044503 | |||||
| ORF16 | 19,244–18,846 | Unknown | 14,101 | 62.6 | ||||
| ORF17 | 18,644–17,697 | Reductase component | 33,670 | 67.9 | 40 | VanB | Pseudomonas sp. strain ATCC 19151 | M22077 |
| 34 | PobB | Pseudomonas pseudoalcaligenes strain POB310 | X78823 | |||||
| ORF18 | 17,646–16,315 | Large (α) subunit of terminal oxygenase component | 49,399 | 63.4 | 24 | TdnA1 | pTDNI (P. putida strain UCC22) | D85415 |
| 25 | AtdA3 | pYA1 (Acinetobacter sp. strain YAA) | D86080 | |||||
| ORF19 | 16,325–15,792 | Small (β) subunit of terminal oxygenase component | 20,934 | 64.8 | 29 | PhnY | Burkholderia sp. strain RP007 | AF112137 |
| 26 | BnzB | P. putida strain BE-81 | P08085 | |||||
| ORF20 | 15,203–14,394 | Short-chain alcohol dehydrogenase | 27,878 | 69.1 | 33 | Short-chain alcohol dehydrogenase | Bacillus thermoleovorans strain B23 | AB040809 |
| 33 | Hypothetical protein X | Pseudomonas aeruginosa | S39654 | |||||
| ORF21 | 14,347–13,526 | Dienelactone hydrolase | 29,901 | 65.4 | 36 | ORF2 | Azospirillum brasilense strain Sp7 | Q43914 |
| 33 | ClcD | P. aeruginosa strain JB2 | AF087482 | |||||
| ORF22 | 11,659–12,645 | Putative regulatory protein | 36,411 | 67.6 | 30 | ORF3 | P. aeruginosa strain JB2 | AF087482 |
| 17 | BphS | Pseudomonas sp. strain IC | AF077670 | |||||
| ORF23 | 10,639–11,655 | Putative regulatory protein | 38,853 | 60.9 | 27 | OxoS | P. putida strain 86 | Y12655 |
| 19 | ThcR | Rhodococcus sp. strain NI86/21 | P43462 | |||||
| ORF24 | 9,062–10,318 | Channel-forming protein | 47,299 | 61.7 | 39 | PhaK | P. putida strain U | AF029714 |
| 35 | PhaK-like protein | P. putida strain DOT-T1E | AF031417 | |||||
| ORF25 (tnpA2) | 8,809–7,829 | Transposase | 36,828 | 58.8 | 100 | TnpA1 | Pseudomonas sp. strain CA10 | AB047548 |
| ORF26 (antA) | 7,487–6,084 | Large (α) subunit of terminal oxygenase component of anthranilate 1,2-dioxygenase | 53,154 | 61.1 | 72 | AntA | Acinetobacter sp. strain ADP1 | AF071556 |
| 43 | XylX | pDK1 (P. putida strain PaW630) | AF134348 | |||||
| ORF27 (antB) | 6,081–5,590 | Small (β) subunit of terminal oxygenase component of anthranilate 1,2-dioxygenase | 19,394 | 62.8 | 53 | AntB | Acinetobacter sp. strain ADP1 | AF071556 |
| 43 | BenB | P. putida | AF218267 | |||||
| ORF28 (antC) | 5,571–4,543 | Reductase component of anthranilate 1,2-dioxygenase | 37,984 | 65.6 | 54 | AntC | Acinetobacter sp. strain ADP1 | AF071556 |
| 33 | XylZ | pWW0 (P. putida strain mt-2) | P23101 | |||||
| ORF29 (tnpA3) | 4,385–3,405 | Transposase | 36,916 | 58.8 | 99 | TnpA1 | P. stutzeri strain AN10 | AF039533 |
| 99 | Transposase | IS1384 (P. putida strain H) | AF052751 | |||||
| 96 | TnpA1 | Pseudomonas sp. strain CA10 | AB047548 | |||||
| 94 | TnpA2 | P. stutzeri strain AN10 | AF039534 | |||||
| ORF30 | 3,245–2,085 | Putative ABC transporter substrate binding protein | 41,687 | 63.3 | 24 | HbaE | Rhodopseudomonas palustris strain GCA009 | U75364 |
| 15 | BraC | P. aeruginosa strain PAO | P21175 | |||||
| ORF31 | 1,929–214 | Putative membrane-spanning protein | 60,651 | 65.6 | 30 | OhbE | P. aeruginosa strain JB2 | AF087482 |
| 18 | HbaG | R. palustris strain GCA009 | U75364 | |||||
| ORF32 (carD) | 34,668–35,450 | 2-Hydroxypenta-2,4-dienoate hydratase | 27,458 | 51.6 | 74 | CumE | Pseudomonas fluorescens strain IP01 | D63377 |
| 73 | IpbE | pRE4 (P. putida strain RE204) | AF006691 | |||||
| ORF33 | 35,562–36,059 | Unknown | 18,625 | 48.0 | ||||
| ORF34 | 36,389–37,132 | Unknown | 27,415 | 44.4 | ||||
| ORF35 (carF) | 37,086–38,033 | Acetaldehyde dehydrogenase (acylating) | 33,265 | 48.2 | 71 | DmpF | Pseudomonas sp. strain CF600 | S24419 |
| 68 | XylQ | pWW0 (P. putida strain mt-2) | S35222 | |||||
| 68 | MhpF | E. coli strain K-12 | P77580 | |||||
| 67 | MhpF | Comamonas testosteroni strain TA441 | AB024335 | |||||
| ORF36 (carE) | 38,030–39,052 | 4-Hydroxy-2-oxovalerate aldolase | 36,651 | 50.6 | 82 | MhpE | C. testosteroni strain TA441 | AB024335 |
| 83 | MhpE | E. coli strain K-12 | P51020 | |||||
| ORF37 | 40,520–39,723 | Unknown | 29,534 | 52.3 | ||||
| ORF38 (tnpA4) | 41,554–42,534 | Transposase | 36,828 | 58.9 | 100 | TnpA1 | Pseudomonas sp. strain CA10 | AB047548 |
| ORF39 | 42,747–43,577 | C terminus of YkoW-like protein (truncated ORF) | 31,201 | 44.6 | 38b | Protein slr1305 | Synechocystis sp. strain PCC 6803 | S76238 |
| 36b | YkoW | Bacillus cereus strain ATCC 14579 | AJ243712 | |||||
| ORF40 | 43,934–44,203 | Unknown | 10,057 | 38.5 | ||||
Homology was observed in only about a 100-amino-acid region in the N terminus of the deduced amino acid sequence of ORF15.
Homology was observed in only about a 300-amino-acid region in the C terminus of YkoW or slr1305.
Homology search analyses.
To determine the functions of identified ORFs, we performed homology search analyses, the results of which are summarized in Table 2.
(i) ORF9 to ORF15.
The deduced amino acid sequence of ORF9 showed 40% identity with the large subunit (AntA) of the oxygenase component in anthranilate dioxygenase from Acinetobacter sp. strain ADP1 (11). However, the similarity with AntA was observed only in the N-terminal region of a putative protein encoded by ORF9, and the C terminus showed no homology to previously reported proteins. In addition, while homologous proteins (AntA [11], XylX [21], and BenA [40]) have 450 to 470 amino acid residues, ORF9 encodes a polypeptide with 567 amino acids.
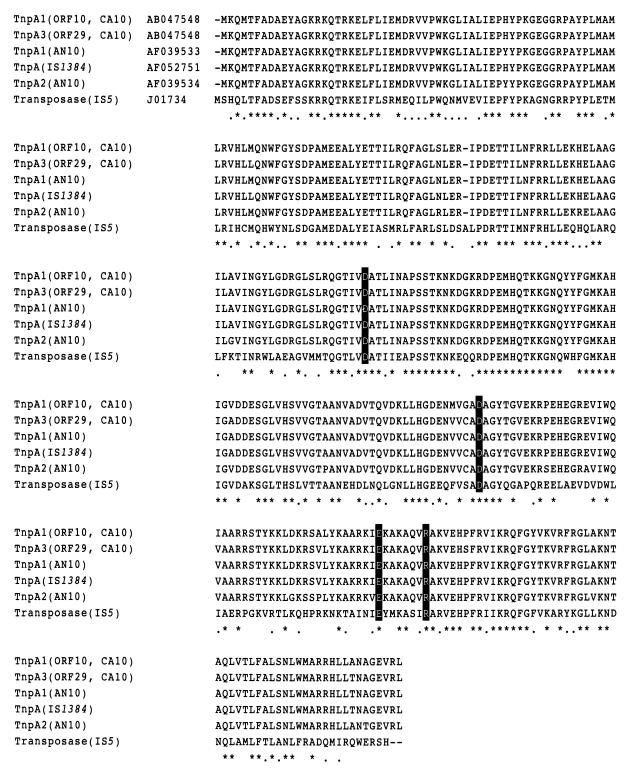
The deduced amino acid sequence of ORF10 showed 96, 96, and 94% identities with TnpA1 from Pseudomonas stutzeri AN10 (7), transposase in IS1384 from Pseudomonas putida strain H (AF052751), and TnpA2 from P. stutzeri AN10 (9), respectively. Figure 3 shows the amino acid sequence alignment with transposases contained in the insertion sequences (ISs) classified in the IS5 family. The critical DDE consensus motif and the R (or K) residue 7 amino acids downstream from the E residue in N2, N3, and C1 domains (32) were well conserved in the deduced amino acid sequence of ORF10, as shown in Fig. 3. Since these results suggest that the protein encoded by ORF10 functions as a transposase, ORF10 was designated tnpA1.
FIG. 3.
Alignments of TnpA1 and TnpA3 identified from strain CA10 with their homologous transposases. The aligned transposases are contained in ISs classified in the IS5 family (32). The identical and similar amino acid residues in all proteins are indicated by asterisks and dots, respectively. The gaps introduced into the alignment are indicated by hyphens. The critical DDE consensus motif and the well-conserved R (or K) residue 7 amino acids downstream from the E residue in N2, N3, and C1 domains (32) are indicated by the white type. The DDBJ, EMBL, and GenBank nucleotide sequence accession numbers are indicated after the names of transposases and the names of ISs or bacterial strains (in parentheses).
No significant homology between the putative proteins encoded by ORF11, ORF12, ORF13, and ORF14 and other proteins was observed. However, the 524-bp DNA region (24,440 to 24,963 in AB047548) containing almost all of ORF11 and the 3′ portion of ORF12 showed a high degree of identity (87%) to the 527-bp DNA region (23,923 to 24,449 in AB047548) containing almost all of ORF12 and its 5′-flanking region (data not shown). These two regions overlapped by 10 bp (AAACCTGCTG), and this sequence was found in both ends of each DNA region.
Although the deduced amino acid sequence of ORF15 showed little homology (13% identity) with the Rhs element (VgrG) in E. coli strain ECOR-11 (60), we could not speculate on the function of the protein encoded by ORF15, because the homologous region is limited to the N-terminal one-fifth of the deduced amino acid sequence of ORF15.
(ii) ORF16 to ORF21.
As summarized in Table 2, it is thought that the putative proteins encoded by three ORFs, ORF17 to ORF19, showed homology to the reductase component and the large (α) and small (β) subunits of the terminal oxygenase component of the multicomponent oxygenase system, respectively. Alignment analyses revealed that a putative plant-type iron-sulfur protein consensus sequence (CX4CX2CX28C) for the binding of a [2Fe-2S] cluster (44) and a possible NAD-ribose binding region were contained in the deduced amino acid sequence of ORF17 (data not shown). In the N terminus of the deduced amino acid sequence of ORF18, the consensus sequence of Rieske-type iron-sulfur proteins for the binding of a [2Fe-2S] cluster (CXHX16-17CX2H) (28) was found (Fig. 4). The consensus sequence for the binding of mononuclear iron (28) was also observed in ORF18 (Fig. 4). Based on these results, the proteins encoded by these ORFs constitute the multicomponent dioxygenase system classified in class IA by Batie et al. (6).
FIG. 4.
Alignments of the large (α) subunits of class I terminal oxygenase components with the postulated ORF18 and antA gene products. The identical and similar amino acid residues in all proteins are indicated by asterisks and dots, respectively. The gaps introduced into the alignment are indicated by hyphens. The amino acids conserved for the Rieske-type [2Fe-2S] cluster binding site and the mononuclear iron coordination site (28) are indicated by white type and shaded boxes, respectively. The DDBJ, EMBL, and GenBank nucleotide sequence accession numbers are indicated after the names of enzymes and the names of plasmids or bacterial strains (in parentheses).
Although no significant homology between the deduced amino acid sequence of ORF16 and those of other proteins was observed, the putative proteins encoded by ORF20 and ORF21 showed the short-chain alcohol dehydrogenase and dienelactone hydrolase, respectively (Table 2).
(iii) ORF22 to ORF31.
Homology search and alignment analyses revealed that ORF25 encodes a protein identical to TnpA1 and that the nucleotide sequence of ORF25 was identical to that of ORF10 except for one base. Because ORF25 was considered to encode transposase, we designated ORF25 as tnpA2. On the other hand, the deduced amino acid sequence of ORF29 showed 99% identity with those of both TnpA1 from P. stutzeri strain AN10 (7) and TnpA of IS1384 (AF052751) (Table 2). Ninety-six percent identity was observed between the deduced amino acid sequence of ORF29 and that of TnpA1 (TnpA2) of strain CA10. The critical DDE consensus motif and the R (or K) residue 7 amino acids downstream from the E residue (32) were also conserved in the deduced amino acid sequence of ORF29, as shown in Fig. 3. Based on these results, ORF29 was designated tnpA3.
Between two transposase genes (tnpA2 and tnpA3), there existed three ORFs. The deduced amino acid sequences of ORF26, ORF27, and ORF28 showed significant homologies with constituents of several class IB (6) multicomponent dioxygenases. Among them, the highest homology was observed with anthranilate 1,2-dioxygenase (AntABC) identified from Acinetobacter sp. strain ADP1 (11) as shown in Table 2. The gene organization of these three ORFs was quite similar to that of the antABC gene cluster of strain ADP1, although the genes located in the immediate vicinity of the antABC genes in the ant locus of strain ADP1 are not transposase genes. The consensus sequence of a Rieske-type iron-sulfur protein for the binding of a [2Fe-2S] cluster (28) was also found in the N terminus of the deduced amino acid sequence of ORF26 (Fig. 4). The consensus motif responsible for the coordination with mononuclear iron (28) was also conserved in ORF26 (Fig. 4). The conserved cysteines (CX4CX2CX31C) (44) which may coordinate a [2Fe-2S] cluster in plant-type iron-sulfur proteins and the potential cofactor binding domains (14, 34, 37, 40) were found in the deduced amino acid sequence of ORF28 (data not shown). The above results strongly indicate that ORF26 and ORF27 encode the large (α) and small (β) subunits of the terminal oxygenase component of anthranilate 1,2-dioxygenase, respectively. Also, ORF28 is considered to encode the reductase component of anthranilate 1,2-dioxygenase. Therefore, ORF26, ORF27, and ORF28 were designated antA, antB, and antC, respectively.
The deduced amino acid sequence of ORF22 and ORF23 showed homology to the regulatory proteins classified in the AraC/XylS family of positive transcriptional regulators (Table 2) (16). It was reported previously that 17 amino acid residues are well conserved among the AraC/XylS family of regulatory proteins and that this sequence was conserved in at least 60% of aligned proteins (16). Among these conserved amino acid residues, 10 and 13 amino acid residues were conserved in the deduced amino acid sequences of ORF22 and ORF23, respectively.
As a result of the homology search analyses (Table 2), it is predicted that ORF24, ORF30, and ORF31 encode the channel-forming protein on the outer membrane, the ATP binding cassette (ABC) transporter substrate binding protein, and the ABC transporter membrane-spanning protein, respectively. These predicted proteins might function in the transport of organic compounds into the cell.
(iv) ORF32 to ORF36.
The deduced amino acid sequences of ORF32, ORF35, and ORF36 showed significant homology to those of 2-hydroxypenta-2,4-dienoate hydratase, acetaldehyde dehydrogenase (acylating), and 4-hydroxy-2-oxovalerate aldorase, which were identified from various aromatic-compound-degrading bacteria (Table 2). Similar to other reported 2-hydroxypenta-2,4-dienoate hydratases, the pentapeptide (ADNAS), which has been suggested to have either a catalytic or a structural role in the respective enzymes (31), was observed near the central portion of the deduced amino acid sequence of ORF32 (data not shown). Based on the above results, ORF32, ORF35, and ORF36 were designated carD, carF, and carE, respectively.
On the other hand, the deduced amino acid sequences of ORF33 and ORF34 did not show significant homology with those of any other reported proteins.
(v) ORF37 to ORF40.
Pairwise comparison among the amino acid sequences of the transposases identified from a neighboring region of the car gene cluster revealed that the deduced amino acid sequence of ORF38 is identical to those of TnpA1 and TnpA2. Therefore, ORF38 was designated tnpA4. The deduced amino acid sequence of ORF39 showed homology (36 to 38% identity) with C-terminal regions (about 300 amino acids in length) of proteins of unknown function, YkoW in Bacillus cereus strain ATCC 14579 (43) and hypothetical protein slr1305 in Synechocystis sp. strain PCC6803 (S76238), although these two proteins have 840 and 892 amino acid residues, respectively. The deduced amino acid sequences of ORF37 and ORF40 did not show significant homology with those of any other reported proteins.
Analysis of ISs.
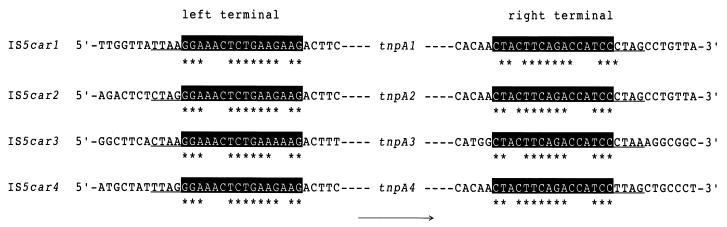
As shown above, four copies of homologous transposase genes were found in the flanking region of the car gene cluster of strain CA10. As a result of the alignment analysis of the neighboring region of transposase genes, inverted repeat sequences (IRs; 16 bp) were found as shown in Fig. 5. In addition, the transposases identified from strain CA10, TnpA1, TnpA2, TnpA3, and TnpA4, had significant homology with the IS5 family of transposases as shown in Table 2 and Fig. 3. Based on the above results, IS-like DNA regions containing tnpA1, tnpA2, tnpA3, and tnpA4 were designated IS5car1, IS5car2, IS5car3, and IS5car4, respectively, and considered to be classifiable in the IS5 subgroup of the IS5 family (32). The nucleotide sequence of IS5car1 is identical to that of IS5car4 and nearly identical to that of IS5car2 with one base pair mismatch. On the other hand, the nucleotide sequence of IS5car3 showed only 81.6% identity to that of IS5car1, although it showed 98% identity to the IS-like sequence containing tnpA1 from P. stutzeri strain AN10 (7). In the flanking region of three ISs, IS5car2, IS5car3, and IS5car4, a target site duplication consisting of 4-bp-long direct repeat sequences (DRs; CTAG, CTAA, and TTAG for IS5car2, IS5car3, and IS5car4, respectively) was observed, although the 4-bp-long DNA sequence located on the left border (TTAA) of IS5car1 was not identical with that of the right border (CTAG) (Fig. 5).
FIG. 5.
Terminal regions of ISs and their flanking sequences identified from strain CA10. The IRs and DRs are shown by white type and underlining, respectively. The complementary nucleotides between the left and right IRs are shown by asterisks. The direction of transposase genes is shown by the arrow.
Transposition of IS5car2 with its flanking sequence.
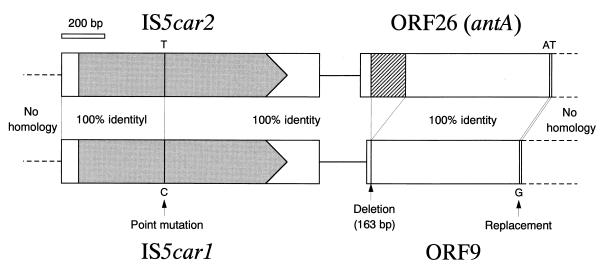
As shown in Fig. 6, the 2,279-bp-long DNA regions (8,878 to 6,600 in AB047548) containing IS5car2 and the 5′ portion of the antA gene were quite similar to the 2,115-bp-long DNA regions (25,306 to 27,420 in AB047548) containing IS5car1 and the 5′ portion of ORF9. The 888-bp-long 5′ portion of antA corresponds to the 698-bp-long 5′ portion of ORF9 and the 26-bp-long 5′ upstream sequence of ORF9. Deletion of a 163-bp-long DNA fragment and replacement of AT bases with a single G base were shown to occur in ORF9. The 3′ half of ORF9 following the transposed antA sequence showed no homology to that of the antA gene. These results strongly suggest that the transposition of IS5car2 and the 5′ portion of the antA gene resulted in the formation of IS5car1 and the fusion gene ORF9. However, immediately 3′ of the 698-bp-long ORF9 portion, we could not find the target site duplication which was considered to be formed by the transposition (data not shown).
FIG. 6.
Comparison of 2,279-bp-long DNA regions (8,878 to 6,600 in AB047548), containing IS5car2 and the 5′ portion of the antA gene, with 2,115-bp-long DNA regions (25,306 to 27,420 in AB047548), containing IS5car1 and the 5′ portion of ORF9. ISs and ORFs are shown by the boxes. Shaded pentagons within the boxes represent the locations, lengths, and directions of translation of transposase genes. The hatched box represents the deleted region of the antA gene in the nucleotide sequence of ORF9. Dashed lines represent the neighboring region of transposed DNA.
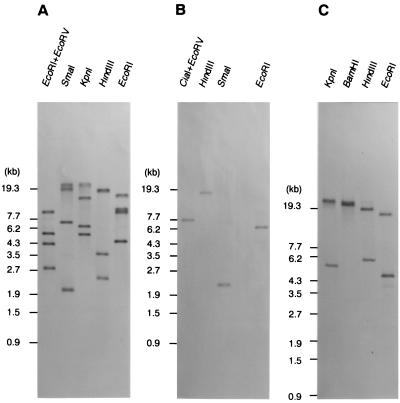
Genomic Southern hybridization of the identified gene.
The above-mentioned results indicate the possibility that the IS plays an important role in the development of the car and ant gene clusters. To reveal the copy numbers of IS5car2 and IS5car3 in the strain CA10 genome, Southern hybridization analyses were conducted. Using the probe made from the 332-bp PvuI-HindIII fragment in IS5car2, we detected four positive bands (Fig. 7A). Based on the determination of the hybridized band, three of the four bands were revealed to correspond to DNA fragments containing IS5car1, IS5car2, and IS5car4. Because the hybridization and washing were carried out under high-stringency conditions, IS5car3 could not be detected. Therefore, the additional bands indicate the presence of another copy of a nearly identical IS.
FIG. 7.
Southern blot analyses using the 332-bp PvuI-HindIII fragment (IS5car2) of pBCA711 (A), the 880-bp PstI-DraI fragment (IS5car3) of pBCA731 (B), and the 1,119-bp SmaI fragment (antA) of pBCA711 (C) as probes. Restriction endonucleases used for digestion of total DNA of strain CA10 are shown in the panels.
In Southern hybridization analysis using the 880-bp PstI-DraI fragment in IS5car3 as a specific probe, only a single band which corresponded to the DNA fragment carrying IS5car3 was detected (Fig. 7B). This indicates that strain CA10 has only one copy of IS5car3 on the genome.
It is quite likely that AntABC can catalyze the conversion of anthranilate to catechol and function as a critical enzyme in the whole CAR degradation by strain CA10. However, considering that the antABC gene is contained in the possible composite transposon, we could not exclude the possibility that another antA gene exists in the genome of strain CA10. Therefore, we performed Southern hybridization analysis of the total DNA of strain CA10 by using the 1,119-bp SmaI fragment of pBCA711 as a probe. The 1,119-bp SmaI fragment covers nearly all of the antA gene, and the probe made from this DNA fragment can hybridize to antA and ORF9. As a result of hybridization, only two positive bands were detected, and these bands corresponded to DNA fragments containing the antA gene and ORF9 (Fig. 7C). Therefore, we concluded that strain CA10 has only one copy of the intact antA gene in the genome.
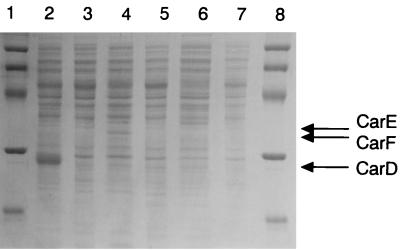
Detection of the products of carD, carE, and carF.
To confirm if ORF32, ORF35, and ORF36 could be translated to proteins with the predicted sizes in E. coli, the cellular proteins from the recombinant E. coli cells harboring plasmids encoding the respective genes were loaded onto an SDS–13% polyacrylamide gel (Fig. 8). Expression of ORF32, ORF35, and ORF36 yielded peptides having molecular masses of 27 (lane 2), 35 (lane 4) and 36 (lane 6) kDa, respectively. The estimated molecular masses were in accordance with the predicted molecular masses of CarD (27,458 Da), CarF (33,265 Da), and CarE (36,651 Da).
FIG. 8.
Detection of CarD, CarF, and CarE produced by E. coli cells. Total cellular proteins of E. coli cells were analyzed by SDS-PAGE. Lanes: 1 and 8, molecular mass standards of 94, 67, 43, 30, 20.1, and 14.4 kDa (top to bottom, respectively); 2, JM109(pBCA616); 3, JM109[pBluescript II SK(−)]; 4, JM109(pUCA617); 5, JM109(pUC18); 6, JM109(pUCA618); 7, JM109(pUC19).
Enzymatic activities of CarD and CarF.
CarD showed significant hydratase activity for 2-hydroxypenta-2,4-dienoate, and its enzymatic activity for 2-hydroxypenta-2,4-dienoate was 3.05 U/mg of protein in crude extracts from E. coli cells harboring pBCA616. The hydratase activity of negative-control E. coli cells harboring pBluescript II SK(−) was almost negligible (0.076 U/mg of protein).
Acetaldehyde dehydrogenase (acylating) activity (0.119 U/mg of protein) was detected in the crude cell extract of E. coli cells harboring pUCA617 expressing CarF, and the activity was 9.2 times higher than that of the crude cell extract of E. coli cells harboring pUC18.
Transcriptional analysis of the ant genes.
In order to determine whether the transcripts of the ant genes exist in the CAR-grown strain CA10 cells, Northern hybridization analyses were performed. As shown in Fig. 9, lane 3, a strong and clear hybridization, with the 200-bp SmaI fragment being specific for antA and not for ORF9, was observed, although the band detected was very blurry. A similar hybridization was observed when the antC gene was used as a probe (data not shown). In sharp contrast, no hybridization band was detected when equal amounts of total RNA extracted from the strain CA10 cells grown on nutrient medium were loaded in the lanes (as confirmed by ethidium bromide visualization of 16S and 23S rRNA bands before transfer [data not shown]) (Fig. 9, lane 2). These results indicate that the ant genes are strongly induced in the strain CA10 cells grown on CAR, although it is impossible to tell whether there is more than one band or whether the mRNA was degraded.
FIG. 9.

Northern hybridization analysis using the antA gene as a probe. Total RNA was extracted from the CA10 cells grown on nutrient medium (lane 2) and CAR (lane 3) to late log phase. As the control, total RNA was similarly extracted from strain CA10 cells just after starvation culture (lane 1).
Analysis of the 16S rRNA gene of strain CA10.
For use as the authentic gene encoded on the chromosome of strain CA10 in Southern hybridization analysis, we amplified the 16S rRNA gene by using PCR. After cloning the amplified DNA fragment into the plasmid vector, we determined the nucleotide sequence of the insert.
A partial nucleotide sequence of the 16S rRNA gene from strain CA10, corresponding to the region between positions 31 and 1,491 of the gene in E. coli, was determined. BLAST search analysis revealed that the nucleotide sequence of the 16S rRNA gene from strain CA10 showed 99.0, 97.9 to 98.0, 96.5, and 96.1 to 96.6% identity with those of Pseudomonas resinovorans (AB021373 and Z76668), Pseudomonas aeruginosa (AJ249451, AB037553, and U38445), Pseudomonas fragi (D84014), and P. stutzeri (U26420, AF152596, and U26262), respectively. These results suggest that strain CA10 is closely related to P. resinovorans.
Localization of the car gene cluster, the cat gene cluster, and transposase genes.
Electrophoresis of the cell lysate of strain CA10 revealed two bands (Fig. 10A). As shown in Fig. 10B, the probe made from the 16S rRNA gene hybridized to the lower band, suggesting that this lower band is a chromosome. This result indicated that strain CA10 cells have at least one megaplasmid. Clear and specific hybridization to the upper band (megaplasmid) was observed with the car gene (pUCA1 insert) used as the probe (Fig. 10C). These results indicate that the car and ant gene clusters which are responsible for CAR degradation by strain CA10 are encoded on this megaplasmid. Therefore, we designated this megaplasmid pCAR1.
FIG. 10.
Southern hybridization analysis of the chromosome and plasmid detected in strain CA10 cells using the 16S rRNA gene (EcoRI-PstI fragment of pTCA16S) (B), the car gene (EcoRI insert of pUCA1) (C), the cat gene (SalI fragment of pTE11) (D), and the tnpA2 gene (PvuI-HindIII fragment in IS5car2) (E) as probes. A photograph of the ethidium bromide-stained agarose gel is shown in panel A.
On the other hand, we have identified the Tn5 mutant of strain CA10, designated strain TE1, which is deficient in catechol metabolism in the CAR degradation pathway (30). The sequencing analysis of the Tn5 insertion-flanking DNA region revealed the presence of catechol-degradative catRBC genes (30). The catRB gene (1.1-kb SalI fragment of pTE11) hybridized to the lower band (chromosome) (Fig. 10D), suggesting that the cat gene cluster involved in CAR degradation by strain CA10 is located on the chromosome, as is the case with other catechol-degradative cat genes.
As shown in Fig. 7A and B, four copies of nearly identical ISs, including IS5car1, IS5car2, and IS5car4 and one copy of IS5car2, exist in the genome of strain CA10. To clarify whether the fourth IS5car1-like IS is located on pCAR1 or the chromosome, we performed Southern hybridization analyses using the PvuI-HindIII fragment in IS5car2 as a probe. As shown in Fig. 10E, clear hybridization to pCAR1 (upper band), not to the chromosome (lower band), was observed. This result indicated that all the related ISs are located on pCAR1.
DISCUSSION
Whole structure of the car gene cluster.
Homology searching, SDS-PAGE analyses, and the determination of enzymatic activity revealed that the genes encoding the meta-cleavage pathway enzymes involved in the conversion of 2-hydroxypenta-2,4-dienoate to a TCA cycle intermediate are located in the downstream region of the carAd gene (Fig. 2). Consequently, the car gene cluster consists of 10 genes (carAaAaBaBbCAcAdDFE) which are involved in the conversion of CAR to anthranilate and 2-hydroxypenta-2,4-dienoate and the degradation of 2-hydroxypenta-2,4-dienoate to a TCA cycle intermediate. In the case of bacterial gene clusters for the degradation of biphenyl, toluene, isopropylbenzene, phenol, and naphthalene (8, 17, 20, 25, 29, 31, 52), the genes for hydratase, acetaldehyde dehydrogenase (acylating), and aldolase which constitute the meta-cleavage pathway are assembled in this order and tandemly linked. Interestingly, the functionally unknown ORF33 and ORF34 are inserted between carD and carFE in the case of the car gene cluster of strain CA10. Such unusual organization of corresponding genes has been reported elsewhere for Rhodococcus sp. strain RHA1 (33) and Comamonas testosteroni strain TA441 (2). In the biphenyl-degrading bph operon of Rhodococcus sp. strain RHA1, the genes encoding the meta-cleavage enzyme (etbC), the meta-cleavage compound hydrolase (bphD), hydratase (bphE), and aldolase (bphF) are clustered, while the putative acetaldehyde dehydrogenase (acylating) (bphG) is not encoded in the neighboring region of the etbCbphDEF operon. In the 3-(3-hydroxyphenyl)propionic acid-degrading mhp operon of C. testosteroni strain TA441, two ORFs (ORF4 and ORF5) encoding proteins of unknown function are inserted between mhpD and mhpFE. Because genes similar to the two ORFs have been found in the gene clusters involved in the chlorocatechol degradation pathway and biosynthesis of poly(3-hydroxybutyrate-co-4-hydroxybutylate) (2), the products of ORF4 and ORF5 might have these functions, although disruption of the genes did not affect the 3-(3-hydroxyphenyl)propionic acid-degrading activity of strain TA441. As shown in Table 2, CarF and CarE showed significantly high homology with MhpF and MhpE, respectively, of strains TA441 and E. coli K-12. In contrast, CarD showed the highest identity with the isofunctional enzymes involved in isopropylbenzene degradation (Table 2), while the identities of CarD with MhpD proteins from C. testosteroni strain TA441 (2) and E. coli strain K-12 (AE000142) were only 45 and 40%, respectively. In addition, the nucleotide and deduced amino acid sequences of ORF33 and ORF34 showed no homology with those of mhp genes including ORF4 and ORF5 identified from strain TA441. On the basis of this observation and the fact that carAaBaBbCAcORF7Ad also showed no phylogenetic affiliation with mhp genes at both DNA and amino acid sequence levels (data not shown), we can speculate that carFE genes have been recruited from other loci, resulting in the present structure of the car gene cluster (Fig. 2). This hypothesis is in accordance with the fact that carEF is transcribed as an mRNA different from carAaAaBaBbCAcORF7AdD (M. Urata, H. Nojiri, T. Yoshida, H. Habe, and T. Omori, unpublished data).
Besides the above-mentioned carFE genes, there are several interesting features in the car gene cluster of strain CA10. The CarAa protein has the features of a class IA terminal oxygenase component according to the classification by Batie et al. (6), while the characteristics of the electron transport chain proteins, CarAc and CarAd, are related to those of class III (38). This discrepancy indicates the possibility that the origin of the carAa gene is different from those of carAc and/or carAd. In addition, the reading frames of carBb overlap with that of carC, indicating the strong possibility that carBaBbC genes originated from the same ancestral gene cluster. Three unknown ORFs (ORF7, ORF33, and ORF34) observed within the car gene cluster might be fusion or truncated genes formed by rearrangements, as is the case with ORF9. If we can isolate the ancestral genes of the car gene cluster, we might be able to obtain interesting information on the developmental process of a novel mosaic structure of the car gene cluster.
Involvement in CAR degradation by the proteins encoded by the genes located in the flanking region of the car gene cluster.
Southern hybridization analysis revealed that strain CA10 has only one copy of intact antABC genes on the genome (Fig. 7C). In addition, Northern hybridization analysis revealed that antABC genes were strongly induced in CAR-grown strain CA10 cells (Fig. 9). These results strongly suggest that AntABC functions as the critical enzyme catalyzing the conversion of anthranilate to catechol in CAR degradation by strain CA10. However, we have not succeeded in the expression of antABC in E. coli cells and detection of anthranilate 1,2-dioxygenase activity (data not shown). To support the evidence of the function of AntABC in CAR degradation, it is important to detect anthranilate 1,2-dioxygenase activity by using the expression system in other bacterial host strains, such as pseudomonads. In addition, it would also be interesting to determine the effects of gene disruption of antABC on the metabolic capacity of CAR and anthranilate in strain CA10. By identifying the ant gene cluster in addition to the car gene cluster, we have elucidated the genes encoding enzymes associated with the catabolic pathway from CAR to catechol. Several CAR-utilizing bacteria have already been isolated, and the degradation pathway, degradative enzymes, and corresponding genes have been studied (10). However, this is the first report on an entire gene set specific for CAR degradation in bacteria.
Genes encoding the two-component dioxygenase (ORF17, ORF18, and ORF19) were identified between the car and ant gene clusters. Because these dioxygenase genes are not expressed in CAR-grown strain CA10 cells (data not shown), this dioxygenase is not involved in CAR degradation by strain CA10. Homology search analysis of the terminal oxygenase component showed the possibility that the dioxygenase system is related to that involved in degradation of mononuclear aromatic compounds, such as benzoate and aniline. While benzoate is converted to catechol by benzoate 1,2-dioxygenase (40) and cis-diol dehydrogenase (39), aniline is converted to catechol by aniline 1,2-dioxygenase, glutamine synthetase-like protein, and glutamine amidotransferase-like protein (54). Considering that there is a putative dehydrogenase gene immediately downstream of the dioxygenase genes in the same direction, the substrate of this two-component dioxygenase system might be benzoate or a related compound. The detection of dioxygenase activity, the determination of substrate specificity, and gene disruption will reveal the function of this putative aromatic-compound-degradative enzyme.
No homologous proteins were found for ORF11 to ORF15. Because the insertion of IS5car2 and its flanking DNA region (IS5car1 and 5′ portion of ORF9) does not reduce the CAR-degrading capability of strain CA10, the DNA region of ORF11 to ORF15 is not considered to be involved in CAR degradation. This consideration is in accordance with the facts that the specific transcripts of ORF11 and ORF15 were not detected in both the CAR- and the nutrient broth-grown strain CA10 cells (data not shown), though carAaAaBaBbCAcORF7Ad genes were strongly induced in CAR-grown cells (M. Urata, H. Nojiri, T. Yoshida, H. Habe, and T. Omori, unpublished data).
Homology search analyses (Table 2) imply the possibility that the products of ORF24, ORF30, and ORF31 are involved in the transport of organic compounds into the cell. Also, ORF22 and ORF23 are predicted to encode the AraC/XylS family of positive regulatory proteins involved in degradation of aromatic compounds. To reveal whether these proteins are indispensable for CAR degradation by strain CA10, it is necessary to determine the effects of the disruption of these genes on CAR utilization by this strain.
Identification of ISs.
Sequencing analysis revealed the presence of four copies of homologous ISs in the flanking region of the car and ant gene clusters (Fig. 2 and Table 2). These ISs are considered to belong to the IS5 subgroup of the IS5 family (32) based on homology searching and alignment analyses. Among the four ISs, it is thought that IS5car2 and IS5car3 constitute the composite transposon containing the entire antABC genes (Fig. 2). The DR observed at either border of IS5car2 was not identical to that observed at IS5car3, although only the central TA was conserved between the two DRs. For the IS5 subgroup, a target sequence ([C/T]TA[G/A]) has been observed in which either all four base pairs or the central TA is duplicated on insertion (32). Therefore, this DNA region might be formed by the transposition of this composite transposon. These results also imply the possibility that antABC genes have been acquired in this locus during the evolution of the CAR degradation pathway in strain CA10. It is well known that many bacterial strains have the capability to convert catechol to a TCA cycle intermediate through meta or ortho cleavage (24). Therefore, the composite transposon containing only the anthranilate 1,2-dioxygenase gene is considered to be effective enough to distribute anthranilate metabolic capacity in the environment.
Considering the high identity among the ISs identified in this study, these four ISs could be considered four different alleles of the same gene. The nucleotide sequences of these ISs were highly homologous to those of IS-like sequences located in the adjacent locus of the naphthalene-degradative nah gene cluster of P. stutzeri strain AN10 (7, 8, 9) and IS1384 harbored by P. putida plasmid pPGH1 (AF052751). Bosch et al. also reported that a portion of the IS-like sequence in strain AN10 is homologous to the flanking sequences of several aromatic-compound-degradative genes in pseudomonads (7, 8). These results suggest that the alleles of IS5car1 and IS5car3 are widely distributed in pseudomonads or related genera and contribute to the gene rearrangement or distribution of aromatic-compound-degradative genes.
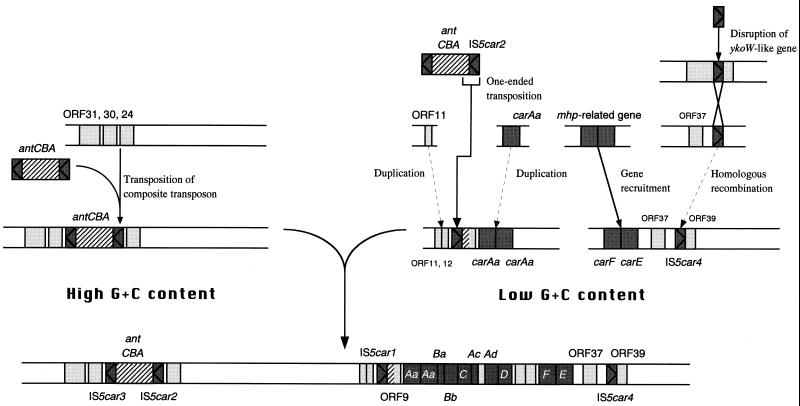
Novel structure of the car gene cluster and its flanking region.
A proposed model of formation of the present novel structure of the car gene cluster and its flanking region is shown in Fig. 11. The present study revealed that IS5car1 and the 5′ portion of ORF9 were formed by transposition of IS5car2 and the 5′ portion of antA. This phenomenon has been termed one-ended transposition and has been reported for Tn3, Tn21, and Tn1721, which lack one terminal IR (4, 5, 36). Also, van der Ploeg et al. reported that IS1247 was able to transpose itself together with the dhlB gene without the requirement for a second insertion element to be present downstream of dhlB (59). In contrast to the case with the transposition of IS1247 and the dhlB gene (59), we could not find the target site duplication or central TA duplication flanking the ends of the transposed DNA region containing IS5car1 and the 5′ portion of ORF9 (data not shown). In general, the one-ended transposition occurs at a much lower frequency than does a normal transposition. We must analyze the frequency of transposition (or one-ended transposition) of identified ISs and the putative composite transposon-containing antABC genes. Also, it is quite interesting to reveal the change of genetic structure in the flanking region of these putative transposable elements by their transposition. In addition, the deletion of the 163-bp DNA region and the replacement of AT bases with a G base occurred in the 5′ portion of ORF9, as shown in Fig. 6. In the 163-bp DNA regions deleted, we could not find DRs or IRs, which possibly promote the deletion by homologous recombination or conservative transposition. It is also interesting to reveal whether these deletions are linked to this one-ended transposition. On the other hand, IS5car1 and IS5car4 form the composite transposon structure containing the whole car gene cluster (Fig. 2). Although IS5car1 is formed by transposition of IS5car2 as shown in Fig. 6, the resultant composite transposon structure might function as a mobile genetic element. Considering the facts that the car gene cluster encodes the entire genes responsible for growth on CAR as a carbon and energy source and that carbazole 1,9a-dioxygenase can attack the dioxin skeleton (42, 49), it is quite interesting to reveal whether this putative composite transposon can transpose into other loci.
FIG. 11.
Proposed model of the developmental process of the novel structure in the car gene cluster and its flanking region. Shaded boxes represent the car genes and several neighboring ORFs. Shaded boxes with triangles indicate the ISs. Striped boxes represent the ant genes or the transposed DNA region from the antA gene within ORF9. The arrows with solid lines indicate the transposition (recombination). The arrows with dashed lines indicate the duplication or homologous recombination.
This study also revealed that the duplication of the gene had occurred around ORF11 and ORF12 (Fig. 11). There exist 10-bp-long DRs (AAACCTGCTG) in both ends of the duplicated DNA fragments, and a DR in the 3′ end of the 527-bp DNA region overlaps that in the 5′ end of the 524-bp DNA region. Although the DRs observed in the termini of these DNA regions were only 10 bp long, three copies of the DR might be a sequence of homologous recombinations to form this duplication. On the other hand, we have already reported that a 1,263-bp DNA region containing the entire carAa gene is tandemly duplicated and that there were only three bases (GGC) between these tandemly linked DNA regions (49) (Fig. 11). While the duplicated 1,263-bp DNA regions are identical except for one base, the nucleotide sequences of the 527- and 524-bp DNA regions showed 87% identity. This difference in the degree of conservation between the two duplicated DNA regions might come from the difference in time after the duplication occurred. In addition, it might be likely that the point mutation has been scarcely accumulated in carAa genes (duplicated 1,263-bp DNA regions) because two copies of carAa genes are advantageous for growth of strain CA10 on CAR as a sole carbon, nitrogen, and energy source.
While ORF39 possibly encodes a polypeptide with 276 amino acid residues, the proteins which showed similarity with the deduced amino acid sequence of ORF39 have 730 to 1,580 amino acids. Because the termination codon was not observed in the intergenic region between the right border of IS5car4 and the putative initiation codon of ORF39, we translated the intergenic region and then performed alignment analysis of the extended amino acid sequence with homologous proteins. Consequently, the amino acid sequence homologous to slr1305 and YkoW was found to extend to the immediate downstream region of IS5car4. These results strongly suggest that the intact ykoW-like gene of strain CA10 had been disrupted by the transposition of an IS or a composite transposon. In this study, the truncated ykoW-like gene (ORF39) was found in the immediate downstream region of the newly identified IS5car4. However, the 5′ half of the disrupted ykoW-like gene has not been found in the flanking regions of any ISs (data not shown). These results indicate that the homologous recombination between the IS inserting into the ykoW-like gene and the unidentified fourth IS5car1 homologue has occurred as shown in Fig. 11, though it remains possible that the large composite transposon consisted of IS5car4 and the fourth IS5car1 homologue transposed into the ykoW-like gene.
The G+C content of the total DNA of strain CA10 is 61.9% (45). While the G+C contents of carAaAaBaBbCAcORF7Ad genes are 48.0 to 53.3%, those of the 3′ portions (1,006 bp) of ORF9, ORF11, and ORF12 are 49.2 to 50.7% (Table 2). Similarly, the G+C contents of ORFs located in the downstream region of the car gene cluster except for the IS ranged from 38.5 to 52.3% (Table 2). On the other hand, the G+C contents of the 5′ portion of ORF9 and ORFs located upstream of ORF13 except for ISs are similar to or higher than that of total DNA of strain CA10 (Table 2). ORF13, located between the high- and low-G+C-content regions, showed a moderate G+C content (55.0%) (Table 2). These results might suggest that the gene fusion had occurred in or around ORF13 (Fig. 11), although the molecular mechanism has not been determined. To obtain information on the molecular mechanism of recombination, it is necessary to analyze the downstream region of ORF40 and to reveal the length of the low-G+C-content region containing the car gene cluster.
Although we can speculate on the proposed rearrangements of genes to form the novel present structure of the car gene cluster and its flanking region as shown in Fig. 11, we cannot determine the order in which recombination events occurred in this region. We have already isolated CAR-degrading bacteria having car gene cluster homologues (data not shown). Genetic analyses of such car gene cluster homologues and their flanking regions will provide us information useful in understanding the evolution of the car gene cluster and the molecular mechanism of recombination events.
Identification of CAR-degrading megaplasmid pCAR1.
As shown in Fig. 10C, the car gene cluster is located on the newly identified megaplasmid pCAR1. Considering that the catechol-degradative cat genes were located on the chromosome (Fig. 10D), it can be considered that strain CA10 had acquired the metabolic capacity for CAR by recruitment of pCAR1. In newly identified CAR-degrading bacteria, we have also detected the pCAR1-like plasmid containing the car gene cluster homologue, although chromosomally encoded car gene clusters were also found (J. Widada, H. Nojiri, S. Nakai, T. Yoshida, H. Habe, and T. Omori, unpublished data). This phenomenon suggests the possibility that pCAR1 plays an important role in the distribution of CAR-degrading activity in the environment.
The existence of a chromosomally encoded car gene cluster also implies the possibility that all or some of pCAR1 is able to integrate into the chromosome or another replicon as reported elsewhere for the 2,4-dichlorophenoxyacetic acid-degradative plasmid pKA2 (27) and the toluene-xylene-degradative plasmid pWW0 (26, 35, 53). Together with the fact that all ISs identified in strain CA10 are located on pCAR1 (Fig. 10), it might also be possible that the ISs specifically function in the integration of the car and ant gene clusters and their flanking regions into the other replicon. To confirm the above possibilities, the determination of the self-transmissibility of pCAR1 and the ability of pCAR1 to integrate into the chromosome is now under way.
ACKNOWLEDGMENTS
This work was supported by the Proposal-Based New Industry Creative Type Technology R&D Promotion Program (Project ID 98Z34017) from the New Energy and Industrial Technology Development Organization (NEDO) and the Program for Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN).
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arai H, Yamamoto T, Ohishi T, Shimizu T, Nakata T, Kudo T. Genetic organization and characteristics of the 3-(3-hydroxyphenyl)propionic acid degradation pathway of Comamonas testosteroni TA441. Microbiology. 1999;145:2813–2820. doi: 10.1099/00221287-145-10-2813. [DOI] [PubMed] [Google Scholar]
- 3.Arcos J C, Argus M F. Molecular geometry and carcinogenic activity of aromatic compounds. New perspectives. Adv Cancer Res. 1968;11:305–471. doi: 10.1016/s0065-230x(08)60390-5. [DOI] [PubMed] [Google Scholar]
- 4.Arthur A, Nimmo E, Hettle S, Sherratt D. Transposition and transposition immunity of transposon Tn3 derivatives having different ends. EMBO J. 1984;3:1723–1729. doi: 10.1002/j.1460-2075.1984.tb02038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avila P, de la Cruz F, Ward E, Grinsted J. Plasmids containing one inverted repeat of Tn21 can fuse with other plasmids in the presence of Tn21 transposase. Mol Gen Genet. 1984;195:288–293. doi: 10.1007/BF00332761. [DOI] [PubMed] [Google Scholar]
- 6.Batie C J, Ballow D P, Correll C C. Phthalate dioxygenase reductase and related flavin-iron-sulfur containing electron transferases. In: Muller F, editor. Chemistry and biochemistry of flavoenzymes. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 543–556. [Google Scholar]
- 7.Bosch R, García-Valdés E, Moore E R B. Genetic characterization and evolutionary implications of a chromosomally encoded naphthalene-degradation upper pathway from Pseudomonas stutzeri AN10. Gene. 1999;236:149–157. doi: 10.1016/s0378-1119(99)00241-3. [DOI] [PubMed] [Google Scholar]
- 8.Bosch R, García-Valdés E, Moore E R B. Complete nucleotide sequence and evolutionary significance of a chromosomally encoded naphthalene-degradation lower pathway from Pseudomonas stutzeri AN10. Gene. 2000;245:65–74. doi: 10.1016/s0378-1119(00)00038-x. [DOI] [PubMed] [Google Scholar]
- 9.Bosch R, Moore E R B, García-Valdés E, Pieper D H. NahW, a novel, inducible salicylate hydroxylase involved in mineralization of naphthalene by Pseudomonas stutzeri AN10. J Bacteriol. 1999;181:2315–2322. doi: 10.1128/jb.181.8.2315-2322.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bressler D C, Fedorak P M. Bacterial metabolism of fluorene, dibenzofuran, dibenzothiophene, and carbazole. Can J Microbiol. 2000;46:397–409. [PubMed] [Google Scholar]
- 11.Bundy B M, Campbell A L, Neidle E L. Similarities between the antABC-encoded anthranilate dioxygenase and the benABC-encoded benzoate dioxygenase of Acinetobacter sp. strain ADP1. J Bacteriol. 1998;180:4466–4474. doi: 10.1128/jb.180.17.4466-4474.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chemicals Inspection and Testing Institute Japan. Biodegradation and bioaccumulation: data of existing chemicals based on the CSCL, Japan. Tokyo, Japan: Japan Chemical Industry Ecology-Toxicology and Information Center; 1992. [Google Scholar]
- 13.Collinsworth W L, Chapman P J, Dagley S. Stereospecific enzymes in the degradation of aromatic compounds by Pseudomonas putida. J Bacteriol. 1973;113:922–931. doi: 10.1128/jb.113.2.922-931.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Correll C C, Batie C J, Ballou D P, Ludwig M L. Phthalate dioxygenase reductase: a modular structure for electron transfer from pyridine nucleotides to [2Fe-2S] Science. 1992;258:1604–1610. doi: 10.1126/science.1280857. [DOI] [PubMed] [Google Scholar]
- 15.Furukawa K. Molecular genetics and evolutionary relationship of PCB-degrading bacteria. Biodegradation. 1994;5:289–300. doi: 10.1007/BF00696466. [DOI] [PubMed] [Google Scholar]
- 16.Gallegos M-T, Schleif R, Bairoch A, Hofmann K, Ramos J L. AraC/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Habe H, Kimura T, Nojiri H, Yamane H, Omori T. Cloning and nucleotide sequence of the genes involved in the meta-cleavage pathway of cumene degradation in Pseudomonas fluorescens IP01. J Ferment Bioeng. 1996;81:247–254. [Google Scholar]
- 18.Hallet B, Sherratt D J. Transposition and site-specific recombination: adapting DNA cut-and-paste mechanisms to a variety of genetic rearrangements. FEMS Microbiol Rev. 1997;21:157–178. doi: 10.1111/j.1574-6976.1997.tb00349.x. [DOI] [PubMed] [Google Scholar]
- 19.Harayama S, Kok M, Niedle E L. Functional and evolutionary relationships among diverse oxygenases. Annu Rev Microbiol. 1992;46:565–601. doi: 10.1146/annurev.mi.46.100192.003025. [DOI] [PubMed] [Google Scholar]
- 20.Harayama S, Rekik M. Comparison of the nucleotide sequences of the meta-cleavage pathway genes of TOL plasmid pWW0 from Pseudomonas putida with other meta-cleavage genes suggests that both single and multiple nucleotide substitutions contribute to enzyme evolution. Mol Gen Genet. 1993;239:81–89. doi: 10.1007/BF00281605. [DOI] [PubMed] [Google Scholar]
- 21.Harayama S, Rekik M, Bairoch A, Neidle E L, Ornston L N. Potential DNA slippage structures acquired during evolutionary divergence of Acinetobacter calcoaceticus chromosomal benABC and Pseudomonas putida TOL pWWO plasmid xylXYZ, genes encoding benzoate dioxygenases. J Bacteriol. 1991;173:7540–7548. doi: 10.1128/jb.173.23.7540-7548.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harayama S, Rekik M, Ngai K-L, Ornston L N. Physically associated enzymes produce and metabolize 2-hydroxy-2,4-dienoate, a chemically unstable intermediate formed in catechol metabolism via meta cleavage in Pseudomonas putida. J Bacteriol. 1989;171:6251–6258. doi: 10.1128/jb.171.11.6251-6258.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harayama S, Rekik M, Wasserfallen A, Bairoch A. Evolutionary relationships between catabolic pathways for aromatics: conservation of gene order and nucleotide sequences of catechol oxidation genes of pWW0 and NAH7 plasmids. Mol Gen Genet. 1987;210:241–247. doi: 10.1007/BF00325689. [DOI] [PubMed] [Google Scholar]
- 24.Harwood C S, Parales R E. The β-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol. 1996;50:553–590. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- 25.Hofer B, Backhaus S, Timmis K N. The biphenyl/polychlorinated biphenyl-degradation locus (bph) of Pseudomonas sp. LB400 encodes four additional metabolic enzymes. Gene. 1994;144:9–16. doi: 10.1016/0378-1119(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 26.Jeenes D J, Williams P A. Excision and integration of degradative pathway genes from TOL plasmid pWW0. J Bacteriol. 1982;150:188–194. doi: 10.1128/jb.150.1.188-194.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ka J, Tiedje J M. Integration and excision of a 2,4-dichlorophenoxyacetic acid-degradative plasmid in Alcaligenes paradoxus and evidence of its natural intergeneric transfer. J Bacteriol. 1994;176:5284–5289. doi: 10.1128/jb.176.17.5284-5289.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kauppi B, Lee K, Carredano E, Parales R E, Gibson D T, Eklund H, Ramaswamy S. Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure. 1998;6:571–586. doi: 10.1016/s0969-2126(98)00059-8. [DOI] [PubMed] [Google Scholar]
- 29.Kikuchi Y, Yasukochi Y, Nagata Y, Fukuda M, Takagi M. Nucleotide sequence and functional analysis of the meta-cleavage pathway involved in biphenyl and polychlorinated biphenyl degradation in Pseudomonas sp. strain KKS102. J Bacteriol. 1994;176:4269–4276. doi: 10.1128/jb.176.14.4269-4276.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimura T, Zhang Y, Kodama T, Omori T. Isolation and characterization of Tn5-induced mutants deficient in carbazole catabolism. FEMS Microbiol Lett. 1996;135:65–70. [Google Scholar]
- 31.Lau P C K, Bergeron H, Labbé D, Wang Y, Brousseau R, Gibson D T. Sequence and expression of the todGIH genes involved in the last three steps of toluene degradation by Pseudomonas putida F1. Gene. 1994;146:7–13. doi: 10.1016/0378-1119(94)90827-3. [DOI] [PubMed] [Google Scholar]
- 32.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masai E, Sugiyama K, Iwashita N, Shimizu S, Hauschild J E, Hatta T, Kimbara K, Yano K, Fukuda M. The bphDEF meta-cleavage pathway genes involved in biphenyl/polychlorinated biphenyl degradation are located on a linear plasmid and separated from the initial bphACB genes in Rhodococcus sp. strain RHA1. Gene. 1997;187:141–149. doi: 10.1016/s0378-1119(96)00748-2. [DOI] [PubMed] [Google Scholar]
- 34.Mason J R, Commack R. The electron-transport proteins of hydroxylating bacterial dioxygenase. Annu Rev Microbiol. 1992;46:277–305. doi: 10.1146/annurev.mi.46.100192.001425. [DOI] [PubMed] [Google Scholar]
- 35.Meulien P, Broda P. Identification of chromosomally integrated TOL DNA in cured derivatives of Pseudomonas putida PAW1. J Bacteriol. 1982;152:911–914. doi: 10.1128/jb.152.2.911-914.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mötsch S, Schmitt R. Replicon fusion mediated by a single-ended derivative of transposon Tn1721. Mol Gen Genet. 1984;195:281–287. doi: 10.1007/BF00332760. [DOI] [PubMed] [Google Scholar]
- 37.Nakatsu C H, Straus N A, Wyndham R C. The nucleotide sequence of Tn5271 3-chlorobenzoate 3,4-dioxygenase gene (cbaAB) unites the class IA oxygenases in a single lineage. Microbiology. 1995;141:485–495. doi: 10.1099/13500872-141-2-485. [DOI] [PubMed] [Google Scholar]
- 38.Nam J-W, Nojiri H, Yoshida T, Habe H, Yamane H, Omori T. New classification system for oxygenase components involved in ring-hydroxylating oxygenations. Biosci Biotechnol Biochem. 2001;65:254–263. doi: 10.1271/bbb.65.254. [DOI] [PubMed] [Google Scholar]
- 39.Neidle E, Hartnett C, Ornston L N, Rekik M, Harayama S. Cis-diol dehydrogenases encoded by the TOL pWW0 plasmid xylL gene and Acinetobacter calcoaceticus chromosomal benD gene are members of the short-chain alcohol dehydrogenase superfamily. Eur J Biochem. 1992;204:113–120. doi: 10.1111/j.1432-1033.1992.tb16612.x. [DOI] [PubMed] [Google Scholar]
- 40.Neidle E L, Hartnett C, Ornston L N, Bairoch A, Rekik M, Harayama S. Nucleotide sequences of the Acinetobacter calcoaceticus benABC genes for benzoate 1,2-dioxygenase reveal evolutionary relationships among multicomponent oxygenases. J Bacteriol. 1991;173:5385–5395. doi: 10.1128/jb.173.17.5385-5395.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nestler F H M. Characterization of wood-preserving coal-tar creosote by gas-liquid chromatography. Anal Chem. 1974;46:46–53. [Google Scholar]
- 42.Nojiri H, Nam J-W, Kosaka M, Morii K, Takemura T, Furihata K, Yamane H, Omori T. Diverse oxygenations catalyzed by carbazole 1,9a-dioxygenase from Pseudomonas sp. strain CA10. J Bacteriol. 1999;181:3105–3113. doi: 10.1128/jb.181.10.3105-3113.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Økstad O A, Gominet M, Purnelle B, Rose M, Lereclus D, Kolstdø A B. Sequence analysis of three Bacillus cereus loci carrying PIcR-regulated genes encoding degradative enzymes and enterotoxin. Microbiology. 1999;145:3129–3138. doi: 10.1099/00221287-145-11-3129. [DOI] [PubMed] [Google Scholar]
- 44.Otaka E, Ooi T. Examination of protein sequence homologies. 5. New perspectives on evolution between bacterial and chloroplast-type ferredoxins inferred from sequence evidence. J Mol Evol. 1989;29:246–254. doi: 10.1007/BF02100208. [DOI] [PubMed] [Google Scholar]
- 45.Ouchiyama N, Zhang Y, Omori T, Kodama T. Biodegradation of carbazole by Pseudomonas spp. CA06 and CA10. Biosci Biotechnol Biochem. 1993;57:455–460. [Google Scholar]
- 46.Poelarends G J, Zandstra M, Bosma T, Kulakov L A, Larkin M J, Marchesi J R, Weightman A J, Janssen D B. Haloalkane-utilizing Rhodococcus strains isolated from geographically distinct locations possess a highly conserved gene cluster encoding haloalkane catabolism. J Bacteriol. 2000;182:2725–2731. doi: 10.1128/jb.182.10.2725-2731.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pollard J R, Bugg T D H. Purification, characterization and reaction mechanism of monofunctional 2-hydroxypentadienoic acid hydratase from Escherichia coli. Eur J Biochem. 1998;251:98–106. doi: 10.1046/j.1432-1327.1998.2510098.x. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 49.Sato S, Nam J-W, Kasuga K, Nojiri H, Yamane H, Omori T. Identification and characterization of genes encoding carbazole 1,9a-dioxygenase in Pseudomonas sp. strain CA10. J Bacteriol. 1997;179:4850–4858. doi: 10.1128/jb.179.15.4850-4858.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sato S, Ouchiyama N, Kimura T, Nojiri H, Yamane H, Omori T. Cloning of genes involved in carbazole degradation of Pseudomonas sp. strain CA10: nucleotide sequences of genes and characterization of meta-cleavage enzymes and hydrolase. J Bacteriol. 1997;179:4841–4849. doi: 10.1128/jb.179.15.4841-4849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shine J, Dalgarno L. Determination of cistron specificity in bacterial ribosomes. Nature. 1975;254:34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- 52.Shingler V, Powlowski J, Marklund U. Nucleotide sequence and functional analysis of the complete phenol/3,4-dimethylphenol catabolic pathway of Pseudomonas sp. strain CF600. J Bacteriol. 1992;174:711–724. doi: 10.1128/jb.174.3.711-724.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sinclair M I, Maxwell P C, Lyon B R, Holloway B W. Chromosomal location of TOL plasmid DNA in Pseudomonas putida. J Bacteriol. 1986;168:1302–1308. doi: 10.1128/jb.168.3.1302-1308.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takeo M, Fujii T, Maeda Y. Sequence analysis of the genes encoding a multicomponent dioxygenase involved in oxidation of aniline and o-toluidine in Acinetobacter sp. strain YAA. J Ferment Bioeng. 1998;85:17–24. [Google Scholar]
- 55.Tan H-M. Bacterial catabolic transposons. Appl Microbiol Biotechnol. 1999;51:1–12. doi: 10.1007/s002530051356. [DOI] [PubMed] [Google Scholar]
- 56.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsuda M, Tan H-M, Nishi A, Furukawa K. Mobile catabolic genes in bacteria. J Biosci Bioeng. 1999;87:401–410. doi: 10.1016/s1389-1723(99)80086-3. [DOI] [PubMed] [Google Scholar]
- 58.van der Meer J R, de Vos W M, Harayama S, Zehnder A J B. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol Rev. 1992;56:677–694. doi: 10.1128/mr.56.4.677-694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van der Ploeg J, Willemsen M, van Hall G, Janssen D B. Adaptation of Xanthobacter autotrophicus GJ10 to bromoacetate due to activation and mobilization of the haloacetate dehalogenase gene by insertion element IS1247. J Bacteriol. 1995;177:1348–1356. doi: 10.1128/jb.177.5.1348-1356.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Y D, Zhao S, Hill C W. Rhs elements comprise three subfamilies which diverged prior to acquisition by Escherichia coli. J Bacteriol. 1998;180:4102–4110. doi: 10.1128/jb.180.16.4102-4110.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams P A, Sayers J R. The evolution of pathways for aromatic hydrocarbon oxidation in Pseudomonas. Biodegradation. 1994;5:195–217. doi: 10.1007/BF00696460. [DOI] [PubMed] [Google Scholar]
- 62.Wyndham R C, Cashore A E, Nakatsu C H, Peel M C. Catabolic transposons. Biodegradation. 1994;5:323–342. doi: 10.1007/BF00696468. [DOI] [PubMed] [Google Scholar]
- 63.Yen K-M, Serdar C M. Genetics of naphthalene catabolism in pseudomonads. Crit Rev Microbiol. 1988;15:247–268. doi: 10.3109/10408418809104459. [DOI] [PubMed] [Google Scholar]