Abstract
In eukaryotes, RNA polymerase (Pol) II transcribes chromatin and needs to move past nucleosomes, often resulting in nucleosome displacement. How Pol II unwraps the DNA from nucleosomes to allow transcription and how DNA rewraps to retain nucleosomes has been unclear. Here, we report the 3.0 Å cryo-electron microscopy structure of a mammalian Pol II-DSIF-SPT6-PAF1c-TFIIS-nucleosome complex stalled 54 base pairs within the nucleosome. The structure provides a mechanistic basis for nucleosome retention during transcription elongation where upstream DNA emerging from the Pol II cleft has rewrapped the proximal side of the nucleosome. Together, the structure uncovers a direct role for Pol II and transcription elongation factors in nucleosome retention and explains how nucleosomes are retained to prevent disruption of chromatin structure across actively transcribed genes.
One-Sentence Summary:
A 3.0 Å resolution cryo-EM structure explains nucleosome retention during transcription elongation.
Transcription requires the passage of RNA polymerase (Pol) II through chromatin. The nucleosome is a significant barrier to Pol II due to an extensive network of contacts between histone proteins and the DNA that is wrapped around them (1). In vitro, Pol II is either unable to move past these obstacles or needs to displace at least one H2A-H2B dimer to transcribe through a nucleosome (2–6). In contrast, in vivo experiments have demonstrated that most transcribed genes have intact nucleosomes and only very highly transcribed genes show nucleosomal loss (7, 8). Nucleosome maintenance during transcription is critical for cell survival and disruption of chromatin structure during transcription elongation has a major impact on transcription fidelity; even partial loss of nucleosomes results in aberrant transcription initiation events from non-promoter regions within gene bodies (9, 10). Thus, nucleosomes play a fundamental role in suppressing cryptic transcription events and restricting transcription initiation to gene promoters. Moreover, nucleosome displacement leads to loss of post-translational histone modifications that have a central regulatory role in demarcating transcriptionally active gene loci (11).
Understanding how nucleosomes are overcome and retained during transcription is a fundamental question. Recently, cryo-electron microscopy (cryo-EM) structures of Pol II-nucleosome complexes have begun to elucidate how transcription through nucleosomes is mediated (12–15). They have not, however, addressed the question of how nucleosomes are retained during transcription. Biochemical and in vivo data suggest that formation of an intra-nucleosomal Pol II containing template loop (“0-loop”) could play an important role in nucleosome retention (6, 16–19).
To investigate how Pol II traverses the nucleosome and concurrently maintains nucleosome integrity, we designed a nucleosomal substrate that allows for transcription of Pol II close to the nucleosomal dyad, a position previously described as a major stall site for Pol II (20). The nucleosomal DNA template contains a modified Widom 601 sequence that lacks deoxythymidine in the template strand from the transcription start site to a position 64 base pairs (bps) within the nucleosome (+64) (Fig. S1A). An RNA primer can be annealed to a 3’-DNA overhang at the entry site forming a 9 nt DNA·RNA hybrid to allow for primer extension by Pol II upon addition of NTPs. Mononucleosomes were assembled on this DNA template and purified S. scrofa Pol II and H. sapiens elongation factors PAF1c composed of subunits CTR9, CDC73, WDR61, PAF1, LEO1, and RTF1, DRB-sensitivity inducing factor (DSIF) composed of subunits SPT4 and SPT5, SPT6, TFIIS, the kinase positive transcription elongation factor b (P-TEFb), and 3’-dATP were then added (Fig. S1B). The transcription reaction was initiated by the addition of CTP, GTP, and UTP (Fig. S1C). 3’-dATP enabled robust stalling at the engineered stall site while serving as a P-TEFb substrate to allow for phosphorylation-dependent complex formation (Fig. S1C and D) (21). Subsequently, the transcription reaction mixture was subjected to size-exclusion chromatography, showing formation of a Pol II-DSIF-PAF1c-SPT6-TFIIS-nucleosome complex (Fig. 1A, Fig. S1D to F). Analysis of the transcription products by denaturing gel electrophoresis revealed that RNA fragments corresponding to nucleosomal stall sites +38, +54, and +64 were the predominant products (Fig. 1B) (20). Peak fractions containing the nucleosome-bound elongation complex were mildly crosslinked with glutaraldehyde and prepared for single-particle cryo-EM.
Fig. 1. Mammalian Pol II-DSIF-SPT6-PAF1c-TFIIS complex with rewrapped nucleosome.
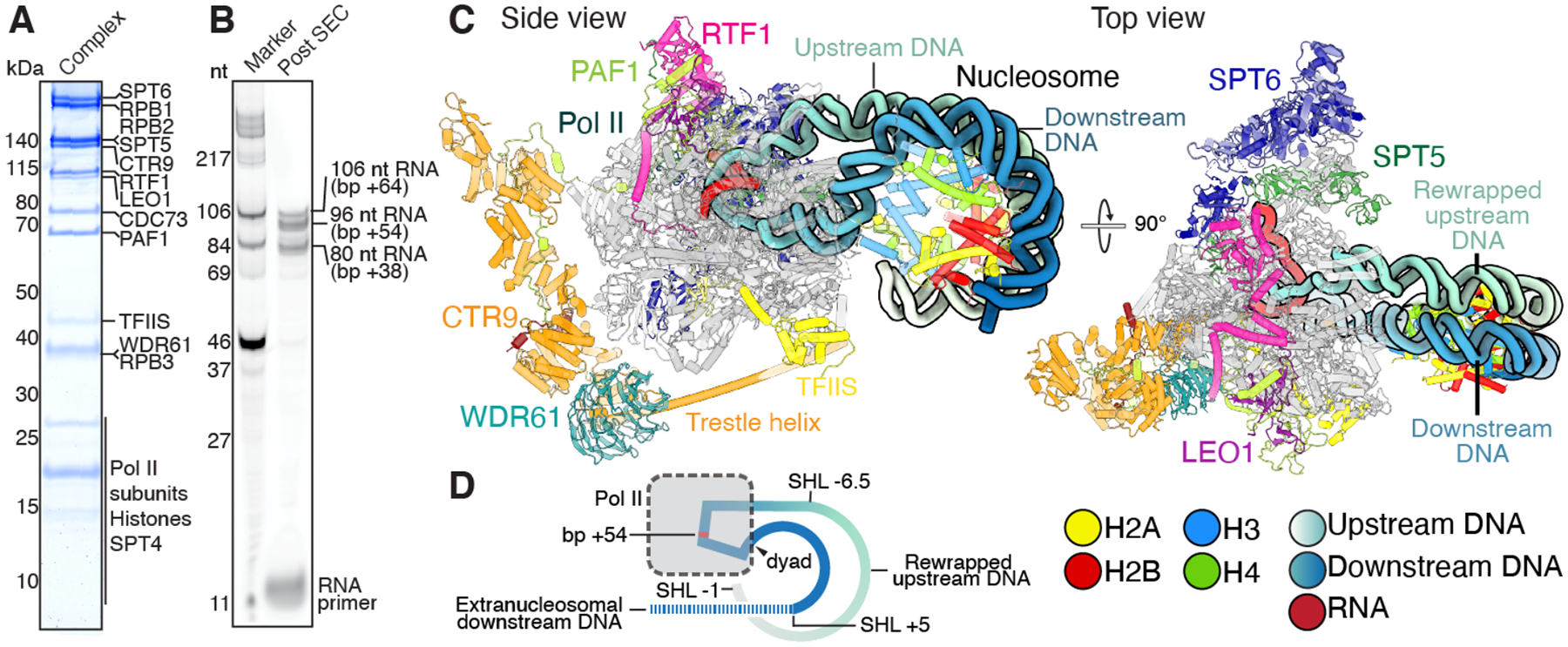
(A) SDS-PAGE of peak fraction used for cryo-EM analysis shows assembly of Pol II-DSIF-SPT6-PAF1c-TFIIS-nucleosome complex. (B) Denaturing gel of purified complex shows primer extension and accumulation of the elongation complex at nucleosomal bp +38, +54, and +64. Stall sites within the nucleosome and corresponding RNA lengths are indicated. (C) Two views of the mammalian Pol II-DSIF-PAF1c-TFIIS-nucleosome complex with rewrapped transcribed DNA. The views are related by a 90° rotation. Pol II, SPT5, SPT6, CTR9, WDR61, PAF1, LEO1, CDC73, RTF1, TFIIS, H2A, H2B, H3, and H4, are shown in gray, forest green, dark blue, orange, teal, limon, purple, firebrick red, pink, yellow, light yellow, red, light blue, and chartreuse, respectively. DNA template is colored upstream to downstream as a gradient from white to blue. Colors used throughout. (D) Schematic of the complex. Unresolved downstream DNA is shown as a dotted line.
We determined the structure of the activated elongation complex bound to a nucleosome at an overall resolution of 3.0 Å from 105,420 particles (Fig. 1C, Fig. S2 to S4, Movie S1, Table S1 and S3). A final composite map was assembled from masked refinements of the nucleosome, Pol II and the PAF1c complex with local resolutions ranging from 2.9 to 11.1 Å (Fig. S2 to S4). Known structures of the complete mammalian activated elongation complex, TFIIS, and the nucleosome were placed into this map and the structures were adjusted locally (21). The reconstruction shows DNA density extending from the upstream DNA towards the nucleosome. We observe a change in the location of the downstream DNA in the Pol II cleft compared to the DNA location of previously obtained elongation complexes (13, 15, 21, 22) (Fig. 1C and D, Fig. 2, Fig. S4F). We de-novo built the DNA to accommodate the observed changes in the DNA path. The model was real space refined and shows good stereochemistry (Table S3).
Fig. 2. Details of nucleosome rewrapping during transcription.
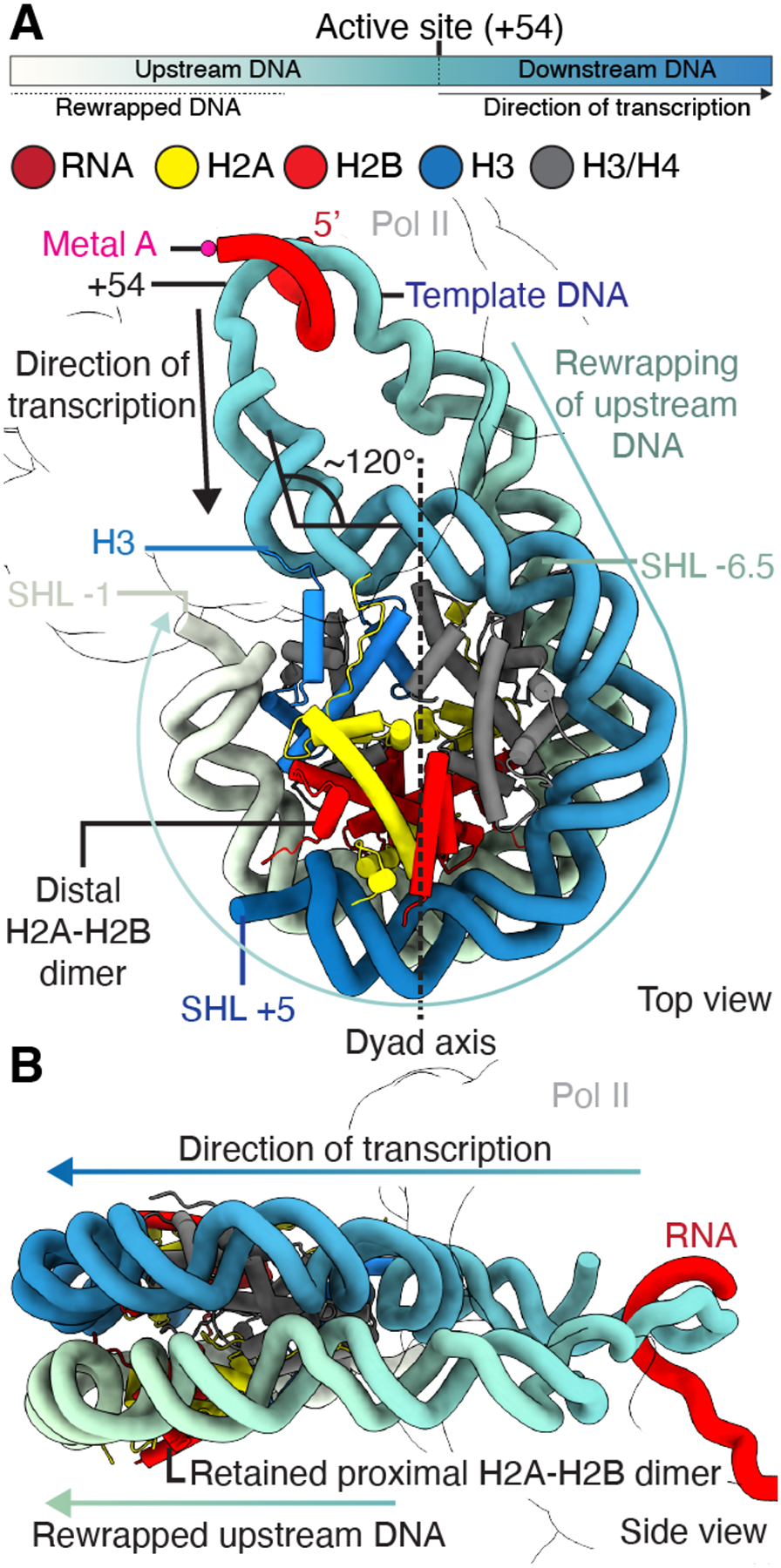
(A) Rewrapping of transcribed upstream DNA from SHL −6.5 to SHL −1 on the histone octamer and unwrapping of downstream DNA from SHL +5 to SHL +7. H3 inserts between rewrapped upstream and downstream DNA. (B) Nucleosome side view shows template looping with transcribed upstream DNA rewrapping the nucleosome. Directions of transcription and rewrapping are indicated.
High resolution features of the DNA·RNA hybrid and visualization of the complete transcription bubble allowed unambiguous assignment of the nucleic acid register (Fig. S4C, G and H). In our structure, Pol II has unraveled DNA up to nucleosomal super-helical location (SHL) −0.5 and the Pol II active site is located at nucleosomal bp +54 (Fig. 2A). It adopts a mixture of pre-translocated (nucleotide binding site occupied by newly extended RNA 3’ end) and post-translocated states (nucleotide binding site not occupied) (Fig. S4C). The +54 site that we observe has been selected by the transcribing Pol II since the engineered stall site is only at nucleosomal bp +64. This implies that stalling at +54 is a major barrier to Pol II.
The transcribed upstream DNA adopts a novel path when exiting the Pol II cleft. The upstream DNA projects away from Pol II towards the nucleosome and ~55 bps of transcribed DNA rewraps the proximal side of the nucleosome from SHL −6.5 to SHL −1 (Fig. 1D, Fig. 2A and B, Fig. S5 and S6). The path of the rewrapped DNA follows the DNA conformation of a canonical nucleosome. The upstream DNA stabilizes the proximal H2A-H2B dimer that plays a critical role in nucleosome retention during transcription (Fig. 2A and B) (2). The DNA rewrapping also has consequences for the path of the downstream DNA. Usually, the downstream DNA duplex and the DNA·RNA hybrid emanate at a 105° angle from the Pol II active site where the downstream DNA lies in the cleft between the clamp and RPB2 (Fig. S5) (22). Further downstream, the DNA contacts the Pol II jaw domain of RPB5. In our structure, the downstream DNA duplex is bent away from the jaw domain into the cleft by ~30°, losing contact with RPB5 (Fig. S5B) (22). After exiting the Pol II cleft, the downstream DNA is then contorted by ~120° and is bound by the histones from SHL −0.5 to SHL +5 (Fig. 2A and B, Fig. S5C). This DNA path differs from previous downstream DNA conformations of Pol II-nucleosome complexes where the downstream DNA projects in a straight B-DNA conformation towards the nucleosome (12–15). The small gap between downstream DNA at SHL −0.5 and rewrapped nucleosomal DNA at SHL −1 is filled by histone H3 N-terminal residues 39–43 (Fig. 2A). Additionally, we observe that downstream DNA starting at SHL +5 to SHL +7 is no longer bound by the nucleosome and projects away from the complex (Fig. S5D).
On the upstream side of Pol II, the rewrapped DNA displaces SPT4 and the SPT5 NGN and KOW1 domains of DSIF from their previously observed binding positions on Pol II (Fig. 3A) (21, 23). We, however, observe densities for SPT5 domains KOW2–3, KOWx-4, and KOW5 (Fig. S4K and L). This argues that the partial displacement of SPT4 and SPT5 from the tip of the Pol II clamp and from the RPB2 protrusion domain is required for rewrapping of nucleosomal DNA and supports nucleosome retention, as reported biochemically (24). Consistent with this observation, functional genomics data show changes in cross-linking patterns for Spt4/5 as Pol II transcribes through a nucleosome (25). We observe a second Pol II-DSIF-PAF1c-SPT6-TFIIS-nucleosome complex in our data stalled at nucleosomal bp +38 (Fig. S7–9, Table S2 and S3). This complex shows no rewrapping, but SPT4 and the SPT5 NGN and KOW1 domains are present and are positioned at the Pol II clamp as observed in prior studies (Fig. S9F and G).
Fig. 3. Interactions and changes in elongation complex conformation.
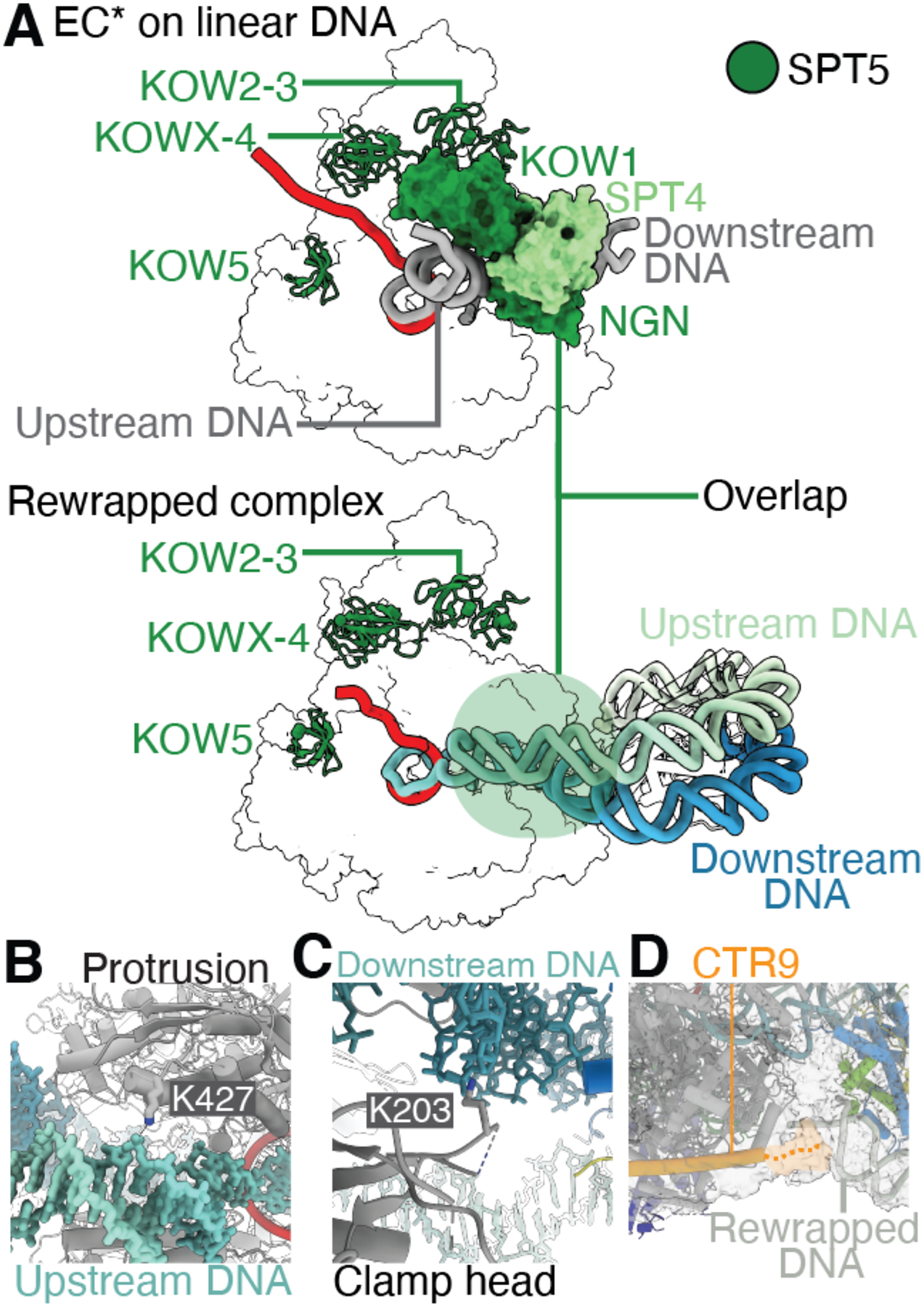
(A) Comparison of Pol II-DSIF-SPT6-PAF1c complex on linear DNA template (gray, PDB 6TED) with Pol II-DSIF-SPT6-PAF1c-TFIIS-nucleosome structure stalled at +54 reveals displacement of SPT4 and SPT5 NGN and KOW1 domains in the rewrapped complex. The trajectory of the upstream DNA duplex is altered compared to the Pol II-DSIF-SPT6-PAF1c complex on a linear DNA template. SPT5 KOW2–3, x-4, and 5 (forest green) are retained in the rewrapped complex. (B) Interaction of Pol II RPB2 protrusion domain residue K427 with nucleosomal DNA at the dyad. (C) Interaction of the Pol II clamp head (residue K203) with nucleosomal DNA. (D) The CTR9 C-terminus is positioned near the rewrapped nucleosomal DNA at SHL −1. Density in gray.
In the rewrapped nucleosome, we observe interactions between Pol II, the PAF1c subunit CTR9, and the nucleosome (Fig. 3B to D, Fig. S4I to L). These contacts actively stabilize the observed nucleosome conformation by contacting the rewrapped DNA. First, we observe an interaction between the RPB2 protrusion residue lysine 427 (K427) and the upstream DNA (Fig. 3B). RPB2 K427 is widely conserved among eukaryotes but is not found in the corresponding E. coli RNA polymerase subunit (Fig. S10). Thus, the protrusion domain may have evolved to support chromatin transcription by mediating DNA rewrapping. Second, like prior S. cerevisiae Pol II-nucleosome structures (13, 15), RPB1 clamp head residue lysine 203 (K203) contacts downstream nucleosomal DNA at SHL −0.5 (Fig. 3C). Third, we observe that the C-terminal end of the CTR9 trestle helix lies near the rewrapped DNA at SHL −1, potentially stabilizing the rewrapped DNA (Fig. 3D). These contacts formed by Pol II and transcription elongation factors with the retained nucleosome suggest that the transcription elongation machinery actively supports nucleosome retention by stabilizing the nucleosomal DNA through multiple contacts with the DNA phosphate backbone.
Together, our data show that Pol II can help retain nucleosomes during transcription by facilitating DNA rewrapping back onto the histone octamer surface (Fig. 4). This rewrapping and template looping supports the 0-loop hypothesis that was proposed based on biochemical data (6, 16). This model proposes that Pol II transcribes nucleosomes while DNA both upstream and downstream of Pol II is bound to the nucleosome. Our structure supports a loop size of ~90 bps (Fig. S6), comparable to atomic force microscopy measurements (6). Interestingly, unambiguous assignment of the DNA register and comparison with canonical nucleosomes show that 30 bps of previously linear entry side DNA rewrap the nucleosome from SHL −4 to SHL −1 (Fig. S6), resulting in an upstream nucleosomal shift after transcription (6, 26). Such a retrograde movement of nucleosomes has been observed both in vivo and in vitro (19, 26).
Fig. 4. Model of nucleosome retention.
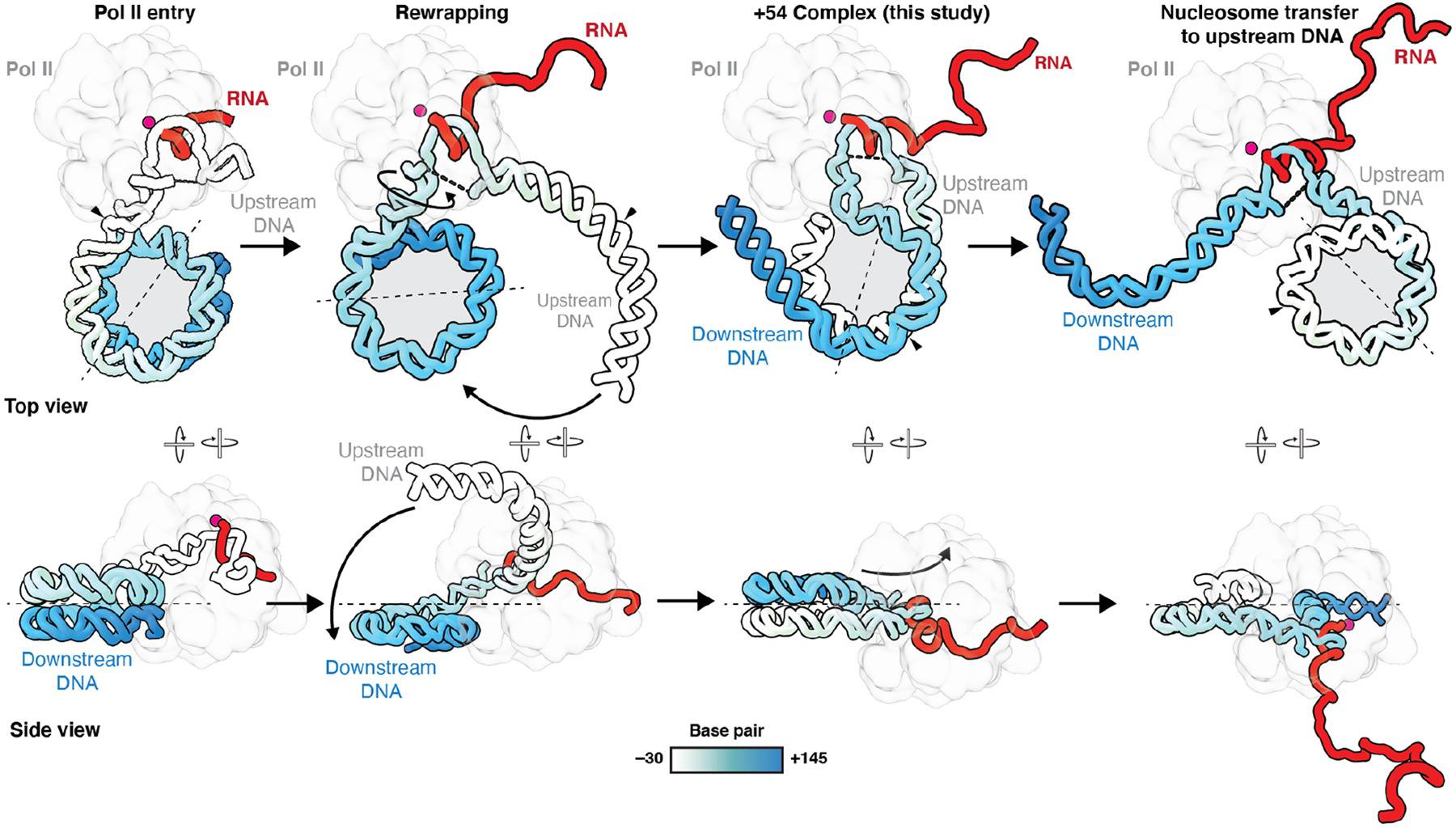
Model depicting Pol II transcription through a nucleosome. After Pol II has transcribed sufficiently into the nucleosome, the DNA rewraps. Further transcription may result in a complete transfer of the nucleosome to upstream DNA. The transfer may result in a positional shift of the nucleosome. Pol II is shown as an outline. Metal A is indicated as a magenta sphere. Histone octamer is shown as a gray circle. Nucleosomal entry base pair (bp +1) is indicated with a black arrow. Dyad axis is indicated as a dotted line.
Previous structural studies have shown that stabilization of the proximal H2A-H2B dimer can be achieved by the binding of a secondary, “foreign” DNA molecule in trans (14, 15). In contrast, our structure, wherein Pol II is accompanied by transcription factors DSIF, SPT6, and PAF1c, now reveals that rewrapping of upstream DNA can also occur in cis where the same DNA molecule rewraps the histone core (Fig. 4). This has wide implications for our understanding of the mechanism of nucleosome retention during transcription. Whereas binding of a secondary molecule in trans suggests a mechanism involving stabilization of the nucleosome to distal DNA, our structure provides evidence that nucleosomes are not fully disassembled during transcription (6, 17–19) (Supplementary Text).
We propose a model that explains nucleosome transfer to the upstream side of Pol II (Fig. 4). Transcription of Pol II past the dyad is likely to result in complete unravelling of the ~50 bp of downstream DNA that are still wrapped around the nucleosome from SHL −0.5 to SHL +5. Release of the downstream DNA from the octamer will result in a complete transfer of the nucleosome to the upstream DNA. At this point, Pol II only transcribes linear downstream DNA, which may explain why the distal part of the nucleosome does not pose any major transcriptional barrier (12, 20). Notably, the proximal and distal sides of the histone octamer switch during the transfer process.
Further rewrapping of upstream DNA may result in a reduced linker length between the transcribed nucleosome and the preceding nucleosome. Chromatin remodelers belonging to the CHD and ISWI families might be needed to reestablish nucleosome spacing (27). The chromatin remodeler CHD1, for example, could be readily recruited by the upstream positioned PAF1c subunit RTF1 to engage the retained nucleosome and correct nucleosome spacing (28).
Nucleosomes are critical for transcription fidelity by limiting transcription initiation to promoter regions and by maintaining transcriptional memory through epigenetic modifications. Our structure provides a structural foundation to explain how nucleosomes are retained during transcription elongation and provides a framework to understand the actions of the transcription machinery in a chromatin environment.
Supplementary Material
Acknowledgments:
We thank all members of the Farnung lab. We thank the Cramer lab for histone proteins. We thank Danesh Moazed, Karen Adelman, Steve Buratowski, Tom Rapoport, and Fred Winston for discussions and critical reading of the manuscript and valuable input. We thank The Harvard Cryo-EM Center for Structural Biology at Harvard Medical School for support with data collection.
Funding:
L.F. is supported by the Dorsett L. Spurgeon, MD (HMS 1929) Distinguished Research Award at Harvard Medical School, and by The Smith Family Awards Program for Excellence in Biomedical Research. S.M.V is supported by The Smith Family Awards Program for Excellence in Biomedical Research, the NIH Director’s New Innovator Award (DP2-GM146254), and Alex’s Lemonade Stand Foundation Crazy 8 Initiative (20071760).
Footnotes
Competing interests: The authors declare that they have no competing interests.
Supplementary Materials
References (29–44)
Data and materials availability:
Data are available in the main text or the supplementary materials. The cryo-EM reconstructions and final models for the Pol II–DSIF–nucleosome–complex stalled at +54 were deposited with the Electron Microscopy Data Base (EMD-26620) and the Protein Data Bank (PDB 7UNC). The cryo-EM reconstructions and final models for the Pol II–DSIF–nucleosome complex stalled at +38 were deposited with the Electron Microscopy Data Base (EMD-26621) and the Protein Data Bank (PDB 7UND).
References and Notes
- 1.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ, Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 389, 251 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Hsieh F-K, Kulaeva OI, Patel SS, Dyer PN, Luger K, Reinberg D, Studitsky VM, Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc National Acad Sci. 110, 7654–7659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole HA, Ocampo J, Iben JR, Chereji RV, Clark DJ, Heavy transcription of yeast genes correlates with differential loss of histone H2B relative to H4 and queued RNA polymerases. Nucleic Acids Res. 42, 12512–12522 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM, Nucleosome Remodeling Induced by RNA Polymerase II Loss of the H2A/H2B Dimer during Transcription. Mol Cell. 9, 541–552 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Bevington S, Boyes J, Transcription-coupled eviction of histones H2A/H2B governs V(D)J recombination. Embo J. 32, 1381–1392 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bintu L, Kopaczynska M, Hodges C, Lubkowska L, Kashlev M, Bustamante C, The elongation rate of RNA polymerase determines the fate of transcribed nucleosomes. Nat Struct Mol Biol. 18, 1394–1399 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee C-K, Shibata Y, Rao B, Strahl BD, Lieb JD, Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet. 36, 900–905 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Ramachandran S, Ahmad K, Henikoff S, Transcription and Remodeling Produce Asymmetrically Unwrapped Nucleosomal Intermediates. Mol Cell. 68, 1038–1053.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlissel G, Rine J, The nucleosome core particle remembers its position through DNA replication and RNA transcription. Proc National Acad Sci. 116, 20605–20611 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaplan CD, Laprade L, Winston F, Transcription Elongation Factors Repress Transcription Initiation from Cryptic Sites. Science. 301, 1096–1099 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Smolle M, Workman JL, Transcription-associated histone modifications and cryptic transcription. Biochimica Et Biophysica Acta Bba - Gene Regul Mech. 1829, 84–97 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farnung L, Ochmann M, Engeholm M, Cramer P, Structural basis of nucleosome transcription mediated by Chd1 and FACT. Nat Struct Mol Biol. 28, 382–387 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farnung L, Vos SM, Cramer P, Structure of transcribing RNA polymerase II-nucleosome complex. Nat Commun. 9, 5432 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehara H, Kujirai T, Fujino Y, Shirouzu M, Kurumizaka H, Sekine S, Structural insight into nucleosome transcription by RNA polymerase II with elongation factors. Science. 363, eaav8912 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Kujirai T, Ehara H, Fujino Y, Shirouzu M, Sekine S-I, Kurumizaka H, Structural basis of the nucleosome transition during RNA polymerase II passage. Sci New York N Y. 362, 595–598 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Kulaeva OI, Gaykalova DA, Pestov NA, Golovastov VV, Vassylyev DG, Artsimovitch I, Studitsky VM, Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat Struct Mol Biol. 16, 1272–1278 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Studitsky VM, Clark DJ, Felsenfeld G, A histone octamer can step around a transcribing polymerase without leaving the template. Cell. 76, 371–382 (1994). [DOI] [PubMed] [Google Scholar]
- 18.Bednar J, Studitsky VM, Grigoryev SA, Felsenfeld G, Woodcock CL, The Nature of the Nucleosomal Barrier to Transcription Direct Observation of Paused Intermediates by Electron Cryomicroscopy. Mol Cell. 4, 377–386 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C, Nucleosomal Fluctuations Govern the Transcription Dynamics of RNA Polymerase II. Science. 325, 626–628 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Z, Gabizon R, Brown AI, Lee A, Song A, Díaz-Celis C, Kaplan CD, Koslover EF, Yao T, Bustamante C, High-resolution and high-accuracy topographic and transcriptional maps of the nucleosome barrier. Elife. 8, e48281 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vos SM, Farnung L, Linden A, Urlaub H, Cramer P, Structure of complete Pol II–DSIF–PAF–SPT6 transcription complex reveals RTF1 allosteric activation. Nat Struct Mol Biol. 27, 668–677 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Bernecky C, Herzog F, Baumeister W, Plitzko JM, Cramer P, Structure of transcribing mammalian RNA polymerase II. Nature. 529, 551–554 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Bernecky C, Plitzko JM, Cramer P, Structure of a transcribing RNA polymerase II–DSIF complex reveals a multidentate DNA–RNA clamp. Nat Struct Mol Biol. 24, 809–815 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Crickard JB, Lee J, Lee T-H, Reese JC, The elongation factor Spt4/5 regulates RNA polymerase II transcription through the nucleosome. Nucleic Acids Res. 45, gkx220- (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uzun Ü, Brown T, Fischl H, Angel A, Mellor J, Spt4 facilitates the movement of RNA polymerase II through the +2 nucleosomal barrier. Cell Reports. 36, 109755 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Radman-Livaja M, Verzijlbergen KF, Weiner A, van Welsem T, Friedman N, Rando OJ, van Leeuwen F, Patterns and Mechanisms of Ancestral Histone Protein Inheritance in Budding Yeast. Plos Biol. 9, e1001075 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eriksson PR, Clark DJ, The yeast ISW1b ATP-dependent chromatin remodeler is critical for nucleosome spacing and dinucleosome resolution. Sci Rep-uk. 11, 4195 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM, Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. Embo J. 22, 1846–1856 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vos SM, Farnung L, Boehning M, Wigge C, Linden A, Urlaub H, Cramer P, Structure of activated transcription complex Pol II–DSIF–PAF–SPT6. Nature. 560, 607–612 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Vos SM, Farnung L, Urlaub H, Cramer P, Structure of paused transcription complex Pol II–DSIF–NELF. Nature. 560, 601–606 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farnung L, Vos SM, Wigge C, Cramer P, Nucleosome–Chd1 structure and implications for chromatin remodelling. Nature. 550, 539 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Punjani A, Zhang H, Fleet DJ, Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat Methods. 17, 1214–1221 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Tegunov D, Cramer P, Real-time cryo-electron microscopy data preprocessing with Warp. Nat Methods. 16, 1146–1152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr Sect D Biological Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Croll TI, ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr Sect D. 74, 519–530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE, UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, Adams PD, Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr Sect D Struct Biology. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kulaeva OI, Hsieh F-K, Studitsky VM, RNA polymerase complexes cooperate to relieve the nucleosomal barrier and evict histones. Proc National Acad Sci. 107, 11325–11330 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, Zhou K, Zhang N, Wei H, Tan YZ, Zhang Z, Carragher B, Potter CS, D’Arcy S, Luger K, FACT caught in the act of manipulating the nucleosome. Nature. 577, 426–431 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available in the main text or the supplementary materials. The cryo-EM reconstructions and final models for the Pol II–DSIF–nucleosome–complex stalled at +54 were deposited with the Electron Microscopy Data Base (EMD-26620) and the Protein Data Bank (PDB 7UNC). The cryo-EM reconstructions and final models for the Pol II–DSIF–nucleosome complex stalled at +38 were deposited with the Electron Microscopy Data Base (EMD-26621) and the Protein Data Bank (PDB 7UND).


