Abstract
Several 2-substituted benzoates (including 2-trifluoromethyl-, 2-chloro-, 2-bromo-, 2-iodo-, 2-nitro-, 2-methoxy-, and 2-acetyl-benzoates) were converted by phthalate-grown Arthrobacter keyseri (formerly Micrococcus sp.) 12B to the corresponding 2-substituted 3,4-dihydroxybenzoates (protocatechuates). Because these products lack a carboxyl group at the 2 position, they were not substrates for the next enzyme of the phthalate catabolic pathway, 3,4-dihydroxyphthalate 2-decarboxylase, and accumulated. When these incubations were carried out in iron-containing minimal medium, the products formed colored chelates. This chromogenic response was subsequently used to identify recombinant Escherichia coli strains carrying genes encoding the responsible enzymes, phthalate 3,4-dioxygenase and 3,4-dihydroxy-3,4-dihydrophthalate dehydrogenase, from the 130-kbp plasmid pRE1 of strain 12B. Beginning with the initially cloned 8.14-kbp PstI fragment of pRE824 as a probe to identify recombinant plasmids carrying overlapping fragments, a DNA segment of 33.5 kbp was cloned from pRE1 on several plasmids and mapped using restriction endonucleases. From these plasmids, the sequence of 26,274 contiguous bp was determined. Sequenced DNA included several genetic units: tnpR, pcm operon, ptr genes, pehA, norA fragment, and pht operon, encoding a transposon resolvase, catabolism of protocatechuate (3,4-dihydroxybenzoate), a putative ATP-binding cassette transporter, a possible phthalate ester hydrolase, a fragment of a norfloxacin resistance-like transporter, and the conversion of phthalate to protocatechuate, respectively. Activities of the eight enzymes involved in the catabolism of phthalate through protocatechuate to pyruvate and oxaloacetate were demonstrated in cells or cell extracts of recombinant E. coli strains.
Phthalate (benzene-1,2-dicarboxylate) is a central intermediate in the bacterial degradation of phthalate esters (75) as well as of certain fused-ring polycyclic aromatic hydrocarbons found in fossil fuels (72), including phenanthrene (3, 46), fluorene (29), and fluoranthene (80). Phthalate diesters are major industrial products, used primarily as plasticizers which are incorporated noncovalently into plastics such as polyvinyl chloride, polyvinyl acetate, and cellulose acetate to impart properties such as softness and flexibility to the polymer. Worldwide production of phthalate esters was estimated in 1993 to be 2.4 million metric tons per year (5). As a result of their common use as plasticizers, they are found at low levels throughout the environment (11). Extensive testing has led to some suggestions that certain phthalate esters may be teratogens or endocrine disruptors (40, 52); however, the effects of phthalate esters on human and environmental health remain unclear (62).
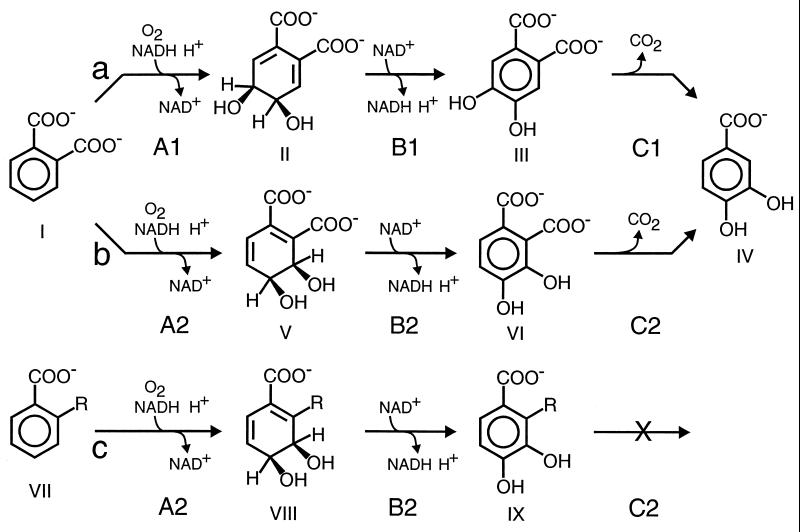
The metabolism of phthalate esters is initiated in bacteria by their hydrolysis to phthalate and two alcohols (75). Phthalate is further metabolized in aerobic bacteria by two different dioxygenase-initiated pathways through the common intermediate, protocatechuate (3,4-dihydroxybenzoate) (Fig. 1, compound IV). Gram-negative bacteria (Burkholderia cepacia, Comamonas testosteroni, and Pseudomonas sp. [4, 12, 64, 73, 75]) transform phthalate through cis-4,5-dihydroxy-4,5-dihydrophthalate and 4,5-dihydroxyphthalate to protocatechuate (Fig. 1, pathway a), while the gram-positive bacterium Arthrobacter keyseri (formerly Micrococcus sp.) 12B converts phthalate to protocatechuate through cis-3,4-dihydroxy-3,4-dihydrophthalate and 3,4-dihydroxyphthalate (Fig. 1, pathway b) (23, 24). Although the enzymes of the two pathways (reductive dioxygenases, dihydrodiol dehydrogenases, and decarboxylases) catalyze similar reactions, the work described here demonstrates that they are not closely related.
FIG. 1.
Early steps in the catabolism of phthalate by gram-negative bacteria and A. keyseri 12B and the transformation of 2-substituted benzoates by A. keyseri 12B. (a) Phthalate catabolic pathway in gram-negative bacteria; (b) phthalate catabolic pathway in A. keyseri 12B; (c) 2-substituted benzoate transformation in A. keyseri 12B. Chemicals; I, o-phthalate; II, cis-4,5-dihydroxy-4,5-dihydrophthalate; III, 4,5-dihydroxyphthalate; IV, protocatechuate; V, cis-3,4-dihydroxy-3,4-dihydrophthalate; VI, 3,4-dihydroxyphthalate; VII, 2-substituted benzoate; VIII, 2-substituted 3,4-dihydroxy-3,4-dihydrobenzoate; IX, 2-substituted protocatechuate. For dihydrodiols, cis but not absolute stereochemistry is intended. R = -CHO and -COOCH3 (prior to this study) and -CF3, -Cl, -Br, -I, -NO2, -COCH3, and -OCH3 (this study). Enzymes: A1, phthalate 4,5-dioxygenase; B1, cis−4,5-dihydroxy-4,5-dihydrophthalate dehydrogenase; Cl, 4,5-dihydroxyphthalate decarboxylase; A2, phthalate 3,4-dioxygenase; B2, cis-3,4-dihydroxy-3,4-dihydrophthalate dehydrogenase; C2, 3,4-dihydroxyphthalate 2-decarboxylase.
The enzymes of the plasmid-encoded A. keyseri 12B phthalate catabolic pathway have previously been shown to act on substrate analogs such as 2-formylbenzoate (23), the monomethyl ester of phthalate (25), and 3-methylphthalate (26). The transformations of 2-formylbenzoate and monomethylphthalate led to the accumulation of 2-substituted protocatechuates (Fig. 1, pathway c), presumably because these compounds lack a removable carboxyl group at the 2 position. In this study, several additional 2-substituted benzoates have been examined as substrates for phthalate-grown A. keyseri 12B. The ability of a product formed from one of these substrates, 2-trifluoromethylbenzoate, to form a colored chelate has been exploited in identifying recombinant bacteria containing cloned phthalate pathway genes. This has facilitated the cloning and characterization of the region of the A. keyseri 12B plasmid pRE1 which encodes the complete catabolism of phthalate.
(Part of this work has been presented previously in a preliminary form [R. W. Eaton, Abstr. Gen. Meet. Am. Soc. Microbiol., K-029, 1997].)
MATERIALS AND METHODS
Bacterial strains and plasmids.
Strains and plasmids used in this study are listed in Table 1. A. keyseri 12B was isolated by Paul Keyser from compost on a Pennsylvania farm, using dibutylphthalate as sole carbon and energy source (24, 43, 45).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference(s) or source |
|---|---|---|
| A. keyseri 12B | Grows with o-phthalate and dibutylphthalate | 24, 43, 45 |
| A. keyseri 12B-C14 | Derivative of A. keyseri 12B containing pRE1 (cured of pRE2 and pRE3) | This study |
| E. coli JM109 | recA endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB) (F′ traD36 proAB lacIqZΔM15) | 91 |
| E. coli BL21(DE3)(pLysS) | F−ompT hsdSB(rB mB−) gal dcm (DE3) pLysS. λDE3 prophage carries T7 RNA polymerase under lacUV5 control; Cmr; obtained from Novagen | 82 |
| pBBR1MCS2 | Kmr, multiple cloning site in lacZα | 48 |
| pBluescriptII KS | Apr, multiple cloning site in lacZα, between lac and T7 promoters; obtained from Stratagene Cloning Systems | 1 |
| pBluescriptII SK | Apr, multiple cloning site in lacZα, between lac and T7 promoters; obtained from Stratagene Cloning Systems | 1 |
| pLV59 | Encodes EcoRI restriction endonuclease and temperature-sensitive EcoRI methylase, Cmr, positive-selection cloning vector | 68 |
| pUCBM21 | Apr, derived from pUC18 with additional cloning sites inserted into lacZα; obtained from Boehringer Mannheim | 91 |
| pRE1 | 130-kbp plasmid from A. keyseri 12B and 12B-C14, encodes phthalate catabolism | This study |
| pRE752 | 14.1-kbp BglII fragment from pRE1 (map coordinates 19.4 to 33.5) inserted into pLV59 | This study |
| pRE754 | 7.79-kbp BglII fragment from pRE1 (map coordinates 10.5 to 18.3) inserted into pLV59 | This study |
| pRE755 | 8.07-kbp BglII fragment from pRE1 (map coordinates 2.4 to 10.5) inserted into pLV59, hybridized to pRE920 | This study |
| pRE761 | 1.15-kbp BglII fragment from pRE1 (map coordinates 18.3 to 19.4) inserted into pLV59 | This study |
| pRE790 | 2.4-kbp BglII fragment from pRE1 (map coordinates 0 to 2.4) inserted into pLV59, hybridized to pRE920 | This study |
| pRE824 | 8.14-kbp PstI fragment from pRE1 (from a mixture of plasmids from strain 12B) (map coordinates 17.2 to 25.4) inserted into pLV59, clone identified by screening on 2-trifluoromethylbenzoate-containing medium | This study |
| pRE826 | 8.14-kbp PstI fragment from pRE824 inserted into pUCBM21 | This study |
| pRE842 | 9.1-kbp HindIII fragment from pRE1 (from a mixture of plasmids from strain 12B) (map coordinates 16.3 to 25.8) inserted into pLV59; identified by hybridization to the PstI fragment of pRE824 | This study |
| pRE861 | 1.97-kbp HindIII-BglII fragment from pRE754 (map coordinates 16.3 to 18.3) inserted into HindIII-BamHI-digested pBluescriptII KS; carries pehA, lac orientation | This study |
| pRE871 | 9.1-kbp HindIII fragment from pRE842 inserted into pBluescriptII KS; lac orientation | This study |
| pRE899 | pRE871 with ClaI fragment (map coordinates 17.0 to 18.8) deleted; lacks phtB | This study |
| pRE920 | 16.5-kbp HindIII fragment from pRE1 (map coordinates-0.2 to 16.3) inserted into pLV59; identified by hybridization to a SmaI fragment (map coordinates 13.1 to 14.4) from pRE754 | This study |
| pRE995 | 5.4 kbp-ClaI-BglII fragment (map coordinates 5.1 to 10.5) from pRE755 inserted into ClaI-BamHI-digested pBluescriptII KS; carries most of the pcm operon, T7 orientation | This study |
| pRE1026 | 966-bp BspEI-XmaI fragment (map coordinates 23.0 to 24.0) from pRE824 inserted into XmaI-digested pBluescriptII SK; carries phtC, lac orientation | This study |
| pRE1043 | 1.86 kbp XhoI-BamHI fragment (map coordinates 6.1 to 8.0) from pRE920 inserted into pBluescriptII KS; carries pcmA, T7 orientation | This study |
| pRE1056 | 1.16-kbp PstI fragment (map coordinates 4.3 to 5.5) from pRE920 inserted into pBluescriptII SK; carries pcmF, possible dehydrogenase gene; T7 orientation | This study |
| pRE1058 | 1.19-kbp BssHI fragment (map coordinates 5.3 to 6.5) inserted into pBluescriptII SK; carries pcmB, lac orientation | This study |
| pRE1062 | 5.24-kbp SalI fragment (map coordinates 16.3 to 17.0 + 18.8 to 23.4) from pRE899 inserted into pBBR1MCS2; Kmr; carries phtAaAbAcAd downstream from lac promoter | This study |
| pRE1065 | 1.66-kbp NgoMIV fragment (map coordinates 7.4 to 9.0) from pRE920 inserted into XmaI-digested pBluescriptII SK; carries pcmC, T7 orientation | This study |
| pRE1066 | 9.1-kbp HindIII fragment from pRE842 inserted into pBBR1MCS2; Kmr; carries pht operon downstream from lac promoter | This study |
| pRE1089 | XbaI-digested 723-bp PCR product (map coordinates 16.4 to 17.1), made using Taq polymerase and primers ACG GTC TAG AAA GGA GGA AAG CAT GTC CGC G and TGC GTC TAG AGC GCT GGC ATG with pRE754 as template inserted into pBluescriptII SK; carries pehA, lac orientation | This study |
| pRE1096 | BspEI-XbaI-digested 3.8-kbp PCR product (map coordinates 13.2 to 17.1), made using Taq polymerase and primers TCA TTC CGG AGG AGA AGG GTA TGG ACG TAA and TGC GTC TAG AGC GCT GGC ATG with pRE754 as template, inserted into XmaI-XbaI-digested pBluescript II SK; carries ptrDABC pehA, T7 orientation | This study |
Chemicals and media.
Syntheses of 3,4-dihydroxyphthalic acid (25) and 2-pyrone-4,6-dicarboxylic acid (24) were described previously. Luria-Bertani (LB) medium (17) was used for the cultivation of bacteria except where noted. Minimal medium was R medium containing, per liter of H2O, 67 mM KH2PO4-NaOH buffer (pH 6.8), 1.2 g of (NH4)2SO4, 0.4 g of MgSO4 · 7H2O, 0.01 g of FeSO4 · 7H2O, and 0.02 ml concentrated HCl, supplemented with 0.5 mM biotin, 0.02% yeast extract, and phthalate or lactate (0.1%). Media were supplemented with antibiotics (100 μg of ampicillin ml−1, 30 μg of chloramphenicol ml−1, or 50 μg of kanamycin ml−1) and solidified by using 1.5% Bacto Agar (Difco Laboratories, Detroit, Mich.) as necessary.
Taxonomy.
Fatty acid methyl esters derived from strain 12B were analyzed using the Sherlock microbial identification system, version 1.06 (MIDI, Inc. Newark, N.J.), with the TSBA (revision 4.10) database.
The 16S ribosomal DNA (rDNA) from strain 12B was amplified using PCR with Taq DNA polymerase, primers rp2 and fd1 (88), and DNA template isolated from strain 12B (21). On an agarose gel, the product gave a single 1.5-kbp band, which was electroeluted essentially by the method of Dretzen et al. (19) but substituting an NA-45 membrane (Schleicher & Schuell) for DE81 paper. Both strands of the PCR product were sequenced (49).
Biotransformation of 2-substituted benzoates by strain 12B.
For biotransformations, 50 ml of an overnight culture of A. keyseri 12B in phthalate or lactate minimal medium was used to inoculate 1 liter of the same medium. The culture was incubated overnight at 30°C, then harvested by centrifugation, and washed with minimal medium (no supplements). Cells were resuspended in one-half to one-fifth volume of minimal medium containing 2-substituted benzoates (0.1 to 0.2% [wt/vol]), in some cases supplemented with lactate (0.1%). After overnight incubation at 30°C, cells were removed by centrifugation, and the supernatant was adjusted to pH 2 with HCl and extracted with ethyl acetate. The solvent was dried over anhydrous sodium sulfate and then removed under reduced pressure in a rotary evaporator. The residues, dissolved in 50% ethanol–50% water, were applied to a Sephadex LH-20 column (49 by 5 cm), from which products were eluted with the same solvent. This provided a means of separating the products from residual substrate and each other. The column effluent was monitored by recording UV-visible spectra of diluted effluent fractions (12.9 ml each). Peak fractions were pooled, and the solvent was removed by evaporation.
Products were analyzed by 1H and 13C nuclear magnetic resonance (NMR) spectroscopy in deuterated dimethyl sulfoxide (d6-DMSO) at 300 and 75 MHz, respectively, with a General Electric model QE plus spectrometer. Trimethylsilyl (TMS) derivatives generated by reaction with N,O-bis(trimethylsilyl)-trifluoroacetamide containing 1% trimethylchlorosilane were analyzed by gas chromatography-mass spectrometry (GC-MS) (22). Occasionally, metabolites were characterized by using thin-layer chromatography on Silica Gel 60F254 plates (EM Science).
Measurement of oxygen consumption with phthalate analogues.
For studies of oxygen consumption, A. keyseri 12B, grown with either phthalate or lactate as carbon source, was harvested by centrifugation, washed twice with at least 10 volumes of 50 mM KH2PO4-NaOH buffer (pH 7), resuspended in 2 volumes of the same buffer, and used immediately. Oxygen consumption by cell suspensions in the presence of substrates was measured polarographically by using a Clark-type oxygen electrode connected to an oxygen monitor (YSI model 5300; Yellow Springs Instruments, Yellow Springs, Ohio) as previously described (24).
Isolation of cured strains.
Plasmid-cured derivative 12B-C1, which is unable to grow with phthalate esters or phthalate as carbon source, was isolated previously following growth of 12B at 37°C, its maximum growth temperature (24). The phthalate catabolic genes in strain 12B are located on the largest of three plasmids (24). To facilitate cloning and analysis, it was useful to eliminate the two smaller plasmids, which, fortunately, are also sensitive to growth at elevated temperature. Phthalate-positive derivatives lacking one or both of the smaller plasmids were therefore isolated as follows. Strain 12B was inoculated from phthalate-minimal medium agar into 20 ml of LB medium in a 100-ml Erlenmeyer flask, which was then incubated overnight at 37°C with shaking. A loopful of this culture was used to inoculate another flask containing 20 ml of LB medium, which was then incubated as before; this was repeated for a total of six transfers. The culture was then streaked onto LB agar plates, which were incubated at 30°C overnight. Colonies that developed were transferred to two minimal medium plates containing either fumarate or phthalate as carbon source. Of 71 colonies growing on fumarate, only 31 grew on phthalate. Colonies growing on phthalate were analyzed for plasmids using the mini-plasmid isolation procedure of Birnboim and Doly (6) followed by electrophoresis of the uncut plasmids through agarose gels.
Preparation, analysis, and cloning of DNA.
Plasmid DNA was isolated from Arthrobacter strains by the method of Hansen and Olsen (31). Plasmids were isolated from Escherichia coli using either the boiling miniprep procedure of Holmes and Quigley (34) or the large-scale Brij lysis procedure (14). Cloning and analysis of clones were carried out as previously described (21, 27), with specific procedures described below.
Total plasmid DNA, isolated from strain 12B, was digested with the restriction endonuclease PstI and ligated to the positive-selection cloning vector, pLV59 (27, 68). Recombinant plasmids were used to transform E. coli JM109, which was then spread on chloramphenicol-LB agar plates. Following overnight incubation at 37°C, 192 colonies were picked to new agar plates and, from there, to wells of two 96-well microtiter plates containing 0.2 ml of minimal medium supplemented with 0.1% 2-trifluoromethylbenzoate, 0.05% phthalate, and 0.02% yeast extract. Microtiter plates and their contents were then incubated for several days at 30°C with occasional agitation and screening for color production.
The restriction endonuclease BglII cuts the largest of the three A. keyseri 12B plasmids, pRE1, into at least 13 fragments. To generate clones comprising most or all of pRE1, that plasmid, isolated from the cured strain 12B-C14, was digested with BglII, and fragments were ligated to BglII-digested pLV59. Recombinant E. coli transformants forming colonies on chloramphenicol-LB plates at 37°C were subsequently screened for inserted fragments. Some of the larger BglII fragments were purified by electrophoresis in low-melting-temperature agarose gels, from which they were recovered by using β-agarase (New England BioLabs) prior to ligation.
Recombinant bacteria carrying DNA fragments that overlap with previously cloned fragments were identified in colony hybridization experiments (30) using gel-purified 32P-labeled DNA fragments as probes.
Analysis of enzymes produced by recombinant bacteria.
For analysis of enzymes produced by recombinant bacteria, 5 ml of an overnight culture of a recombinant E. coli strain in LB-antibiotic medium was used to inoculate 250 ml of the same medium. After incubation at 30°C for 2 h, an inducer (1 mM isopropyl-B-d-thiogalactopyranoside [IPTG], 0.1% phthalate, or protocatechuate) was added and incubation was continued for 3 h. The culture was then harvested by centrifugation and washed with 50 mM KH2PO4-NaOH buffer (pH 6.8). Whole-cell biotransformations and product analyses were carried out as described above for A. keyseri 12B. For preparation of cell extracts (21), E. coli cells, resuspended in 3 ml of 50 mM KH2PO4-NaOH buffer (pH 6.8), were broken in a French pressure cell at 14,000 to 20,000 lb/in2, and particulate material was separated from soluble by centrifugation at 47,800 × g for 40 min at 4°C.
Some assays were carried out by recording enzyme-catalyzed changes in spectra or absorbance maxima of substrates and products over time using a Perkin-Elmer Lambda 6 double-beam spectrophotometer.
The ability of E. coli BL21(DE3)(pLysS)(pRE995) to transform protocatechuate to pyruvate and oxaloacetate was determined by incubating 2 ml of E. coli BL21(DE3)(pLysS)(pRE995) extract with 0.5 mmol of protocatechuate in 50 ml of 30 mM Tris-Cl buffer (pH 8.5) containing 20 μM MgCl2 and 120 μM NAD at room temperature for 20 h. At the end of the incubation, the pH of the mixture was adjusted to 2 with HCl, NaCl was added to 20%, and the reaction mixture was left at 4°C for several hours. Precipitated protein was removed by centrifugation, and the supernatant was extracted three times with 2 volumes ethyl acetate. Ethyl acetate was removed in the rotary evaporator, and the product was redissolved in 0.5 ml of water. It was then assayed for keto acids and used to prepare dinitrophenylhydrazones (DNPHs). Enzyme assays for pyruvate and oxaloacetate were carried out in spectrophotometer cuvettes containing the extracted product in 50 mM Tris-Cl (pH 8) with 150 μM NADH, and lactate dehydrogenase or malic dehydrogenase. The decrease in absorbance at 340 nm due to conversion of NADH to NAD+ was measured. DNPHs were prepared as described by Maruyama (55), extracted with ethyl acetate, and analyzed by thin-layer chromatography on silica gel plates, using ethyl acetate containing 1% acetic acid as the solvent.
DNA sequence determination.
Both strands of the DNA segments discussed here were sequenced by the dideoxy-chain termination method using double-stranded DNA as the template (91). The sequence of the pRE826 insert was determined by ACGT, Inc., Northbrook, Ill., and by the ICBR DNA Sequencing Core Laboratory, University of Florida, Gainesville. The latter completed the sequencing of pRE1-derived plasmids, including pRE920, pRE842, pRE752, and various subclones, as well the PCR-amplified A. keyseri 12B 16S rDNA. Primers were synthesized by the ICBR DNA Synthesis Core Laboratory, University of Florida, and by Gemini Biotech, Gainesville, Fla. Sequence data were aligned, edited, and compared by using DNASTAR programs (DNASTAR, Inc., Madison, Wis.). Searches in the GenBank database were carried out with the blastn or blastx program (2).
Nucleotide sequence accession numbers.
The DNA sequences obtained in this study are available from GenBank (accession number AF256196 for 16S rDNA; accession number AF331043 for the segment of pRE1).
RESULTS AND DISCUSSION
Taxonomy.
Major fatty acids from strain 12B identified as their methyl esters were 14:0 iso (2.32%), 15:0 iso (4.36%), 15:0 anteiso (72.94%), 16:0 iso (6.25%), 16:0, (1.94%), and 17:0 anteiso (12.19%). In the TSBA (revision 4.10) database, these showed strong correlation to Arthrobacter sp. and Arthrobacter histidinolovorans (similarity indices of 0.848). The similarity index with Arthrobacter ureafaciens was less significant, resulting from differences in the proportions of 14:0 iso, 16:0 iso, and 17:0 anteiso fatty acids (6.9, 12.13, and 6.27%, respectively, in A. ureafaciens).
Analysis of the 16S rDNA PCR product yielded a double-stranded DNA sequence of 1,452 bp with single-stranded ends of 10 and 22 bases (GenBank accession number [hereafter simply GenBank] AF256196). Comparison of the sequence to sequences in GenBank by using blastn showed a high degree of overall sequence homology to A. ureafaciens (GenBank X80744; 99%), A. nicotinovorans (GenBank X80743; 98.5%), and A. histidinolovorans (GenBank X83406; 98.2%). These are all members of the same branch within group I of the genus Arthrobacter (47). The region between bases 464 and 483 (E. coli numbering) in the 16S rDNA sequences of the four strains (strain 12B, GAAGCCCT–––TCGGGGTGAC; A. ureafaciens, GAAGCCCTCTTTGGGGGTGAC; A. nicotinovorans and A. histidinolovorans, GAAGCGTAA–––––––GTGAC) contains taxonomically significant insertions or deletions (the underlined bases are identical to those in strain 12B), which further suggest that the 16S rDNA of strain 12B is most closely related to that of A. ureafaciens. The combined analyses of fatty acid methyl esters and 16S rDNA sequences indicate that strain 12B is closely related to but different from A. ureafaciens and A. histidinolovorans. It has therefore been given the new species name Arthrobacter keyseri 12B after Paul Keyser, who not only isolated it (43) but also isolated another well-studied phthalate-degrading strain, Burkholderia cepacia DB01 (ATCC 29424; formerly Pseudomonas fluorescens PHK), from which he was the first to purify phthalate 4,5-dioxygenase (4, 12, 44, 45).
Biotransformation of phthalate analogues.
Media containing phthalate-grown A. keyseri 12B incubated with 2-trifluoromethylbenzoate, 2-nitrobenzoate, 2-iodobenzoate, 2-chlorobenzoate, 2-bromobenzoate, 2-acetylbenzoate, o-anisate (2-methoxybenzoate), monomethylphthalate (ester), 2,6-dichlorobenzoate, 2-chloro-6-fluorobenzoate, or 2-fluoro-6-iodobenzoate became red or purple. This color formation was accompanied by changes in the UV spectra of culture supernatants (not shown). Because the initial enzyme of the phthalate catabolic pathway, phthalate 3,4-dioxygenase (Fig. 1, enzyme A2), catalyzes insertion of a molecule of oxygen into its substrate, measurement of oxygen consumption is also a useful means of assaying the activities of this enzyme toward phthalate and potential phthalate analogues. Oxidation of phthalate and 2-substituted benzoates is inducible by growth with phthalate: phthalate-grown A. keyseri 12B consumed oxygen in the presence of phthalate and all of the 2-substituted benzoates listed above, while lactate-grown A. keyseri 12B did not consume oxygen in the presence of phthalate and its substrate analogs (data not shown). Other 2-substituted benzoates, including homophthalate (2-carboxyphenylacetate), 2-fluorobenzoate, 2-carboxycinnamate, N-acetylanthranilate, acetylsalicylate, salicylate, phthalide (2-hydroxymethylbenzoate), 2,5-dichlorobenzoate, and dicamba (3,6-dichloro-2-methoxybenzoate), were not transformed by phthalate-grown A. keyseri 12B.
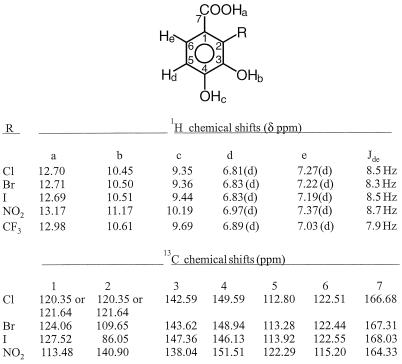
Products of most of the color-forming transformations, after purification by chromatography on Sephadex LH-20, were identified by 13C and 1H NMR spectroscopy (Fig. 2) and GC-MS (Table 2) as 2-substituted protocatechuic acids (3,4-dihydroxybenzoic acids [Fig. 1, compound IX]). As with previously identified 2-substituted protocatechuates produced by strain 12B, the absence of a carboxyl group at the 2 position prevents them from serving as substrates for the next enzyme of the pathway, 3,4-dihydroxyphthalate 2-decarboxylase (Fig. 1, enzyme C2). Color production is likely to be due to the formation of iron chelates by these ortho-dihydroxylated products. Colored chelates were not observed in previous biotransformations of phthalate analogs because those transformations were carried out in phosphate buffer in the absence of iron (23, 25, 26).
FIG. 2.
NMR spectral data for selected 2-substituted protocatechuic acids produced from 2-substituted benzoates by phthalate-grown A. keyseri 12B.
TABLE 2.
GC-MS data for TMS derivatives of products formed by phthalate-induced A. keyseri 12B
| Substrate/TMS-derivatized product (retention time) | m/z of major ion peaks (% intensity, proposed composition)a |
|---|---|
| 2-Chlorobenzoate/2-chloro-3,4-dihydroxybenzoate (18.24 min) | 406 (6, M+); 404 (11, M+); 391 (5, [M − CH3]+); 389 (11, [M − CH3]+); 317 (2, [M − OTMS]+); 315 (6, [M − OTMS]+); 301 (2); 257 (2); 237 (10, [M − TMS − CO2 −Cl − CH3]+); 227 (100, [M − OTMS − TMS − CH3]+); 193 (3); 171 (6); 147 (3); 73 (86, [TMS]+) |
| 2-Bromobenzoate/2-bromo-3,4-dihydroxybenzoate (19.25 min) | 450 (16 M+); 448 (14, M+); 435 (14, [M − CH3]+); 433 (12, [M − CH3]+); 361 (7, [M − OTMS]+); 359 (6, [M − OTMS]+); 347 (4, [M − TMS − CH3 − CH3]+); 345 (3, [M − TMS − CH3 − CH3]+); 303 (4, [M − TMS − TMS − H]+); 301 (3, [M − TMS − TMS − H]+); 273 (100, [M − OTMS − TMS − TMS − CH3]+); 271 (94, [M − OTMS − TMS − CH3]+); 237 (13, [M − TMS − CO2 − Br − CH3]+); 217 (4); 215 (4); 209 (2); 207 (2); 193 (4); 179 (2); 73 (89, [TMS]+) |
| 2-Iodobenzoate/2-iodo-3,4-dihydroxybenzoate (20.50 min) | 496 (17, M+); 481 (10, [M − CH3]+); 407 (4, [M − OTMS]+); 393 (2, [M − TMS − CH3 − CH3]+); 354 (3, [M − I − CH3]+); 339 (6, [M − I − CH3 − CH3]+); 319 (100, [M − OTMS − TMS − CH3]+); 266 (6, [M − I − TMS − CH3 − CH3]+); 237 (3, [M − I − TMS − CO2 −CH3]+); 207 (3); 193 (3); 164 (7); 147 (3); 133 (3); 73 (50, [TMS]+) |
| 2-Nitrobenzoate/2-nitro-3,4-dihydroxybenzoate (26.06 min) | 415 (4, M+); 400 (38, [M − CH3]+); 385 (1, [M − CH3 − CH3]+); 312 (4, [M − TMS − CH3 − CH3]+); 238 (30, [M − OTMS − TMS − CH3]+); 164 (9, [M − OTMS − OTMS − TMS]+); 147 (9, [M − OTMS − TMS − CO2 − NO2]+); 133 (6); 73 (100, [TMS]+) |
| 2-Trifluoromethylbenzoate/2-trifluoromethyl-3,4-dihydroxybenzoate (16.47 min) | 438 (9, M+); 423 (3, [M − CH3]+); 349 (1, [M − OTMS]+); 331 (10); 309 (1, [M − TMS − CO2]+; 261 (11, [M − OTMS − TMS − CH3]+); 239 (100, [M − TMS − TMS − CH3 − F − F]+); 217 (3); 203 (2); 155 (6); 77); 73 (65, [TMS]+) |
| 2-Acetylbenzoate/2-acetyl-3,4-dihydroxybenzoate (25.33 min) | 412 (7, M+); 397 (51, [M − CH3]+); 353 (3, [M − CH3 − CH3CO − H]+); 323 (5, [M − OTMS]+); 307 (23, [M − OTMS − CH3CH3 − H]+); 279 (10, [M − OTMS − CH3CO − H]+); 235 (10, [M − OTMS − TMS − CH3]+); 147 (14); 133 (5); 73 (100, [TMS]+) |
| o-Anisate/2-methoxy-3,4-dihydroxybenzoate (21.76 min) | 400 (11, M+); 385 (19, [M − CH3]+); 355 (6, [M − CH3 − CH3 − CH3]+); 311 (6, [M − OTMS]+); 295 (18, [M − TMS − CH3OH]+); 281 (8, [M − OTMS − CH3 − CH3]+); 223 (100, [M − OTMS − TMS − CH3]+); 208 (8); 73 (94, [TMS]+) |
| 2-Chloro-6-fluorobenzoate/2-chloro-(3 or 4)- hydroxy-6-fluorobenzoate (14.135 min) [major product, > 90%] | 336 (3, M+); 334 (9, M+); 321 (23, [M − CH3]+); 319 (52, [M − CH3]+); 247 (2, [M − OTMS]+); 245 (3, [M − OTMS]+); 241 (4); 211 (7, [M − TMS − CH3 − Cl]+); 209 (6); 183 (5); 179 (6); 167 (6, M − TMS − CO2 − Cl − CH3]+); 153 (6, [M − TMS − TMS − Cl]+); 137 (7, [M −OTMS − TMS − Cl]+); 105 (6); 93 (14, [M − OTMS − TMS − CO2 − Cl]+); 77 (91); 73 (100, [TMS]+) |
| 2-Chloro-6-fluorobenzoate/2-chloro-3,4-dihydroxy-6-fluorobenzoate (17.50 min) | 424 (6, M+); 422 (14, M+); 409 (7, [M − CH3]+); 407 (16, [M − CH3]+); 333 (1, [M − OTMS]+); 321 (2, [M − TMS − CH3 − CH3]+); 319 (6, [M − TMS − CH3 −CH3]+); 255 (2, [M − TMS − CO2 − Cl − CH3]+); 247 (27, [M − OTMS − TMS − CH3]+); 245 (74, [M − OTMS − TMS − CH3]+); 189 (2); 147 (2); 93 (4); 77, (6); 73 (100, [TMS]+) |
| 2,6-Dichlorobenzoate/2,6-dichloro-(3 or 4)-hydroxybenzoate (20.97 min) [major product], LH-20 peak 1 | 352 (8, M+); 350 (10, M+); 337 (33, [M − CH3]+); 335 (46, [M − CH3]+); 263 (17, [M − OTMS]+); 261 (23, [M − OTMS]+); 227 (11, [M − TMS − CH3 − Cl]+); 225 (10, [M − TMS − CH3 − Cl]+); 185 (5); 183 (12); 95 (14); 93 (38); 73 (100, [TMS]+) |
| 2,6-Dichlorobenzoate/2-chloro-3,4-dihydroxybenzoate (23.37 min), LH-20 peak 1 | 406 (10, M+); 404 (19, M+); 391 (5, [M − CH3]+); 389 (10, [M − CH3]+); 317 (4, [M − OTMS]+); 315 (9, [M − OTMS]+); 229 (19, [M − OTMS − TMS − CH3]+); 227 (52, M − OTMS − TMS − CH3]+); 199 (4); 179 (4); 147 (4); 93 (3); 73 (100, [TMS]+) |
| 2,6-Dichlorobenzoate/2,6-dichloro-3,4-dihydroxy-3,4-dihydrobenzoate (23.53 min), LH-20 peak 1 | 442 (2, M+); 440 (3, M+); 407 (10, [− Cl]+); 405 (20, [M − Cl]+); 337 (4, [M − TMS − CH3 − OH]+); 335 (5, [M − TMS − CH3 − OH]+); 325 (2, [M − TMS − CO2]+); 323 (3, [M − TMS − CO2]+); 265 (3); 263 (7, [M − OTMS − TMS − OH]+); 261 (8, [M − OTMS − TMS − OH]+); 243 (3); 227 (3); 187 (3); 161 (5); 147 (21); 93 (11); 75 (16); 73 (100, [TMS]+) |
| 2,6-Dichlorobenzoate/2,6-dichloro-(3 or 4)-hydroxybenzoate (20.87 min), LH-20 peak 2 | 352 (8, M+); 350 (11, M+); 337 (36, [M − CH3]+); 335 (48, [M − CH3]+); 263 (17, [M − OTMS]+); 261 (24, [M − OTMS]+); 227 (12, [M − TMS − CH3 − Cl]+); 225 (12, [M − TMS − CH3 − Cl]+); 183 (13); 160 (6); 153 (6); 123 (3); 95 (13); 93 (34); 73 (100, [TMS]+) |
| 2,6-Dichlorobenzoate/2,6-dichloro-3,4-dihydroxybenzoate (25.26 min), LH-20 peak 2 | 440 (7, M+); 438 (9, M+); 425 (3, [M − CH3]+); 423 (5, [M − CH3]+; 351 (4, [M − OTMS]+); 349 (5, [M − OTMS]+); 273 (2, [M − TMS − CO2 − CH3 − Cl]+); 271 (5, [M − TMS − CO2 − CH3 − Cl]+); 265 (6); 263 (32, [M − OTMS − TMS − CH3]+); 261 (42, [M − OTMS − TMS − CH3]+); 147 (4); 93 (5); 73 (100, [TMS]+) |
Paired ions differing by two mass units result from major chlorine isotopes of mass 35 and 37 and bromine isotopes of mass 79 and 81.
In transformations of substrates having a substituent at the 6 position (2,6-dichlorobenzoate and 2-chloro-6-fluorobenzoate), product mixtures were more complex and the major products identified were monohydroxylated derivatives of the starting compounds. These were probably formed by dehydration of dihydrodiol products during acidification and extraction. Lesser quantities of 2,6-disubstituted protocatechuates were formed and identified as their TMS derivatives. Also formed from 2,6-dichlorobenzoate and identified as its TMS derivative was the 3,4-dihydrodiol which is presumed to have given rise to the major phenolic product. Accumulation of these dihydrodiols suggests that substitutions at the 6 position may reduce activity of the dihydrodiol dehydrogenase (Fig. 1, enzyme B2) toward these dihydrodiol substrates.
Although a carboxyl group is required at C-1 for a compound to serve as substrate for phthalate 3,4-dioxygenase and cis-3,4-dihydroxy-3,4-dihydrophthalate dehydrogenase in A. keyseri 12B (23, 24), a variety of electron-withdrawing substituents can replace the carboxyl group at C-2 of phthalate. Substitutions in other positions can have a negative effect; thus, a substituent at C-5, as present in 2,5-dichlorobenzoate and dicamba, prevents activity of phthalate 3,4-dioxygenase, while chlorine or fluorine at C-6 reduces the ability of the dihydrodiol dehydrogenase to act. Some of the substrate analogs acted on by enzymes of the phthalate pathway may occur as intermediates or products of other catabolic pathways. 2-Chlorobenzoate and 2,6-dichlorobenzoate are formed by the biotransformation of certain polychlorinated biphenyls (28), while 2-nitrobenzoate may be formed during metabolism of nitrotoluene explosives (7). Genes encoding phthalate pathway enzymes (described below) are therefore potentially useful for the construction of strains having extended or altered pathways for the metabolism of these compounds.
Isolation of cured strain 12B-C14.
Strain 12B carries three plasmids. All of the phthalate-positive strains derived in curing experiments contained the largest, pRE1. One of these strains which contained only that plasmid was designated 12B-C14. While pRE1 is present in strains 12B and 12B-C14, it is not present in a previously isolated cured derivative strain 12B-C1 (24) or other phthalate-negative cured strains. By determining and adding together the sizes of fragments generated by digesting each of the plasmids with various restriction enzymes (data not shown), plasmid sizes were estimated to be 130 kbp (pRE1), 80 kbp (pRE2), and 70 kbp (pRE3).
Cloning phthalate catabolism genes.
The demonstration that enzymes of the phthalate catabolic pathway could convert substrates to products forming colored chelates immediately suggested a method for identifying recombinant bacteria carrying genes encoding those enzymes. 2-Trifluoromethylbenzoate was chosen as the substrate for cloning phthalate catabolism genes, although other 2-substituted benzoates could have served equally well. Of 192 microtiter wells in which recombinant E. coli JM109 strains had been inoculated into 2-trifluoromethylbenzoate-supplemented minimal medium, two became a light red color. Plasmids isolated from the responsible bacteria were composed of identical 8.14-kbp PstI fragments inserted in pLV59. One of these plasmids was designated pRE824. The 8.14-kbp PstI fragment (Fig. 3, map coordinates 17.2 to 25.4) carries genes encoding not only the conversion of phthalate to 3,4-dihydroxyphthalate, as indicated by the conversion of 2-trifluoromethylbenzoate to 2-trifluoromethylprotocatechuate, but also the decarboxylation of 3,4-dihydroxyphthalate to protocatechuate (see below; Fig. 1, enzymes A2, B2, and C2). It should be noted that the transformation of 2-trifluoromethylbenzoate is not a generic screening method since only a few enzyme systems may act in a similar manner toward this or related substrates.
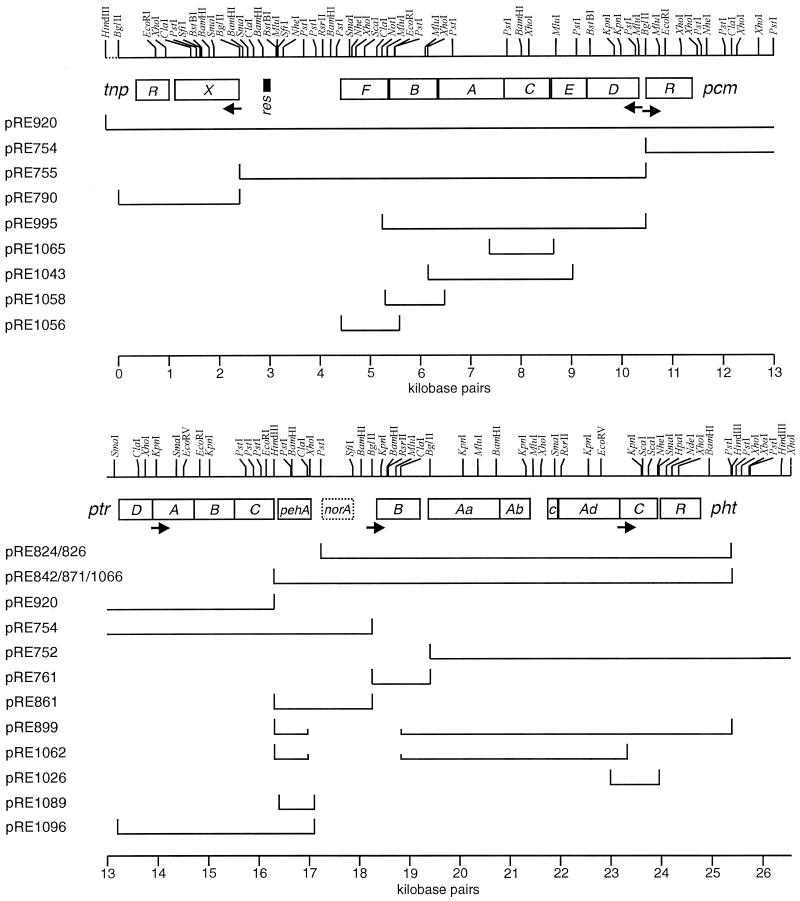
FIG. 3.
DNA of pRE1 that encodes phthalate and protocatechuate catabolism. This is a restriction map of a 26,274-bp segment divided into two parts. Below the restriction map are boxes indicating the locations of genes (described in Table 3). Below these are DNA fragments that have been cloned or subcloned (Table 1). Names of the plasmids carrying these fragments are indicated at the left. Arrows indicate possible promoters, while the small black box indicates a resolvase binding (res) site.
Thirteen different BglII fragments representing most or all of pRE1 (isolated from strain 12B-C14) were cloned in pLV59. However, the relative locations of the cloned BglII fragments in pRE1 and the functions that they encode were not identified until after the isolation of pRE824. At that time, all of the plasmids generated using BglII were examined for homology to the phthalate catabolic pathway-encoding 8.14-kbp PstI fragment insert of pRE824 by Southern hybridization (81) using the 32P-labeled PstI fragment as a probe (not shown). Cloned BglII fragments in pRE754 (7.79 kbp), pRE761 (1.15 kbp), and pRE 752 (14.1 kbp) all hybridized to the PstI fragment. Restriction endonuclease cleavage maps of pRE824 and these BglII-generated recombinant plasmids were constructed (Fig. 3). Using these maps, DNA probes were chosen for identifying additional recombinant bacteria carrying cloned overlapping fragments by colony hybridization (30). This led to the cloning and restriction mapping of a region of pRE1 of 33.5 kbp. From these clones, a collection of subclones was generated; some of these are described here (Table 1; Fig. 3).
Nucleotide sequence.
Using many of these clones and subclones, the sequences of both strands of a 26,274-bp segment were determined (Fig. 3). Genes and deduced gene product sequences were then compared to those in GenBank (Table 3). Sequencing and sequence comparisons revealed that the DNA segment carries several different genetic units: the pht operon encoding the conversion of phthalate to protocatechuate; the pcm operon encoding enzymes that carry out the further metabolism of protocatechuate to pyruvate and oxaloacetate; a putative ptr operon encoding a possible phthalate, protocatechuate, or phthalate ester transporter; a possible phthalate ester hydrolase gene, pehA; and a transposon resolvase gene, tnpR.
TABLE 3.
Genes and gene products
| Gene | Location (bp) | Gene product | Deduced mol wt (amino acid residues) | Related proteins, accession no., % identity/amino acid residues |
|---|---|---|---|---|
| tnpR | 999–349 | Transposon resolvase | 24,495 (216) | Resolvases for a transposon from Enterobacter cloacaeY09025, 75%; Tn1721 and Tn4653, P06692, 75%, Tn501, K01725, 73%; Tn21, M10791, 73% |
| tnpX | 2379–1114 | 44,552 (421) | ||
| pcmF | 5384–4410 | Dehydrogenase | 34,077 (324) | Putative aldo-keto reductases from: Streptomyces clavuligerus, AAD30468, 43%; Deinococcus radiodurans, AE002058, 42% |
| pcmB | 6334–5381 | 2-Hydroxy-4-carboxymuconic semialdehyde dehydrogenase | 34,665 (317) | 2-Hydroxy-4-carboxymuconate semialdehyde dehydrogenase (LigC) from Sphingomonas paucimobilis SYK-6, BAA97119, 71% |
| pcmA | 7661–6360 | Protocatechuate 4,5-dioxygenase | 48,131 (433) | Putative protocatechuate-4,5-dioxygenase from Sphingomonas sp. strain LB126: α subunit (FldV), CAB87562, 61% in 90 residues; β subunit (FldU), CAB 87561, 63% in 276 residues; protocatechuate 4,5-dioxygenase from S. paucimobilis SYK-6: α subunit (LigA), P22635, 62% in 117 residues; β subunit (LigB), P22636, 55% in 283 residues |
| pcmC | 8601–7663 | 2-Pyrone-4,6-dicarboxylate hydrolase | 35,362 (312) | Putative 2-pyrone-4,6-dicarboxylate hydrolase (FldB) from Sphingomonas sp. strain LB126, CAB87568, 57% in 284 residues; 2-pyrone-4,6-dicarboxylate hydrolase (LigI) from S. paucimobilis SYK-6, BAA33799, 52% in 286 residues |
| pcmE | 9293–8601 | 4-Oxalocitramalate aldolase | 24,442 (230) | Putative acyl transferase (FldZ) from Sphingomonas sp. strain LB126, CAB875 66, 59%; putative transferase from Streptomyces coelicolor A3(2), CAA16197, 32% in 132 residues; putative d-arabino-3-hexulose-6-phosphate formaldehyde lyase from Archaeoglobus fulgidus, AAB90381, 36% in 113 residues |
| pcmD | 10333–9305 | 4-Oxalomesaconate hydratase | 38,096 (342) | 4-Oxalmesaconate hydratase (LigJ) from S. paucimobilis SYK-6, BAA97116, 63%; putative hydratase (FldW) from Sphingomonas sp. strain LB126, CAB87563, 62% |
| pcmR | 10479–11384 | pcm operon regulator | 33,365 (301) | Putative LysR family regulator from S. coelicolor A3(2), CAB76357, 38% in 281 residues; salicylate degradation regulator (SalR) from Acinetobacter sp. strain ADP1, AAF04311, 27% in 285 residues |
| ptrD | 13233–13895 | Transporter, substrate-binding protein | 23,574 (220) | Sulfate ester binding protein (AtsR) from Pseudomonas putida S-313, AAD31785, 21% in 220 residues; putative sulfate ester binding protein (AtsR) from P. aeruginosa PAO, CAA88422, 21% in 173 residues |
| ptrA | 13892–14719 | Transporter, ATPase | 30,375 (275) | Transporter ATPases: putative from Methanococcus jannaschii Q57855, 49% in 250 residues; putative from Synechocystis sp. strain PC6803, P73265, 46% in 222 residues; chromate resistance (ChrD) from Ralstonia eutropha CH3 4, AAD21772, 44% in 229 residues; taurine transport (TauB) from Escherichia coli, Q47538, 44% in 208 residues; sulfate ester transporter (AtsC) from P. putida S-313, AAD31787, 42% in 251 residues |
| ptrB | 14716–15516 | Transporter, permease 1 | 29,049 (266) | Transporter permeases: PtrC (below), 29% in 240 residues; putative from D. radiodurans, AAF10090, 27% in 238 residues; putative from Thermatoga maritima, AAD35570, 25% in 236 residues; sulfate ester transporter (AtsB) from P. putida, AAD31786, 23% in 248 residues |
| ptrC | 15513–16307 | Transporter, permease 2 | 27,800 (264) | Transporter permeases: PtrB (above), 29% in 240 residues; sulfonate transporter (SrpM) from Synechococcus sp. strain PCC942, AAD53164, 30% in 233 residues; sulfonate transporter (SsuC) from P. putida S-313, AAC31906, 29% in 239 residues; sulfate ester transporter (AtsB) from P. putida, AAD31786, 27% in 248 residues |
| pehA | 16382–17038 | Putative phthalate ester hydrolase | 23,898 (218) | N-Carbamoylsarcosine amidohydrolase from Arthrobacter sp., P32400, 34% in 213 residues |
| norA | 17260–17880 | Antibiotic resistance transporter, fragment | Hypothetical membrane transporter from S. coelicolor A3(2), CAB46807, 27% in 219 residues; quinolone resistance efflux transporter (NorA) from Staphylococcus aureus, AB019536, 22% in 134 residues | |
| phtB | 18347–19210 | 3,4-Dihydroxy-3,4-dihydrophthalate dehydrogenase | 31,839 (287) | Putative oxidoreductase from S. coelicolor A3(2), CAA22355, 44% in 264 residues; morphine 6-dehydrogenase from P. putida M10, Q02198, 42% in 276 residues; 2,5-diketo-d-gluconate reductase from Zymomonas mobilis ZM4, AAD42404, 42% in 276 residues |
| phtAa | 19376–20797 | Phthalate dioxygenase, large subunit | 53,416 (473) | Dioxygenases, large subunits: aromatic dioxygenase (NidA) from Rhodococcus sp. strain 124, AF121905, 45% in 427 residues; naphthalene dioxygenase (NarAa) from Rhodococcus sp. strain NCIMB12038, AF082663, 43% in 457 residues |
| phtAb | 20801–21400 | Phthalate dioxygenase, small subunit | 22,611 (199) | Dioxygenases, small subunits: naphthalene dioxygenase (RnoA4) from Rhodococcus sp. strain CIR2, AB024936, 44% in 155 residues; aromatic dioxygenase (NidB) from Rhodococcus sp. strain I24, AF121905, 44% in 155 residues |
| phtAc | 21752–21946 | Phthalate dioxygenase, ferredoxin | 6,768 (64) | 3Fe-4S ferredoxins: SubB, from Streptomyces griseolus, P18325, 42%; from Thermococcus litoralis, P29604, 42%; phenanthrene dioxygenase subunit (PhdC) from Nocardioides sp. strain KP7, BAA94713, 34% |
| phtAd | 21968–23179 | Phthalate dioxygenase, ferredoxin reductase | 42,920 (403) | Hypothetical ferredoxin reductase from Mycobacterium tuberculosis H37Rv, CAB06451, 36% in 394 residues; rhodocoxin reductase from Rhodococcus erythropolis, P43494, 36% in 387 residues; phenanthrene dioxygenase ferredoxin reductase subunit from Nocardioides sp. strain KP7, BAA84715, 34% in 391 residues |
| phtC | 23179–23925 | 3,4-Dihydroxyphthalate decarboxylase | 26,011 (248) | Fuculose-1-phosphate aldolases: from Methanobacterium thermoautotrophicum, AAB85883, 30% in 187 residues; S. coelicolor A3(2), AL132644, 30% in 208 residues |
| phtR | 23982–24773 | pht operon regulator | 28,691 (263) | Hypothetical regulator from E. coli K-12 MG1655, P77300, 30% in 241 residues; repressor of the aceBAK operon (IclR) from E. coli K-12, AAA50561, 28% in 249 residues |
The pht operon.
The pht operon, phtBAaAbAcAdCR (Fig. 3), encodes the conversion of phthalate to protocatechuate (Fig. 4). Plasmid pRE1066 carries the complete pht operon on a 9.1-kbp HindIII fragment which encompasses the original 8.14-kbp PstI fragment of pRE824.
FIG. 4.
Phthalate catabolic pathway in A. keyseri 12B. Enzymes: PehA, phthalate ester hydrolase (esterase); PhtA, phthalate 3,4-dioxygenase; PhtB, cis-3,4-dihydroxy-3,4-dihydrophthalate dehydrogenase; PhtC, 3,4-dihydroxyphthalate 2-decarboxylase; PcmA, protocatechuate 4,5-dioxygenase; PcmB, 2-hydroxy-4-carboxymuconic semialdehyde dehydrogenase; PcmC, 2-pyrone-4,6-dicarboxylate hydrolase; PcmD, 4-oxalomesaconate hydratase; PcmE, 4-oxalocitramalate aldolase. Chemicals: X, phthalate ester; I, o-phthalate; V, cis-3,4-dihydroxy-3,4-dihydrophthalate; VI, 3,4-dihydroxyphthalate; IV, protocatechuate; XI, 2-hydroxy-4-carboxymuconic semialdehyde; XII, 2-hydroxy-4-carboxymuconic semialdehyde-hemiacetal; XIII, 2-pyrone-4,6-dicarboxylate; XIV, 4-oxalomesaconate; XV, 4-oxalocitramalate; XVI, oxaloacetate; and XVII, pyruvate.
Biotransformation of 2-bromobenzoate by enzymes of the pht operon.
Incubation of E. coli JM109(pRE1066) with the phthalate analog 2-bromobenzoate yielded a single biotransformation product, identified as 2-bromoprotocatechuic acid. Its tri-TMS derivative, analyzed by GC-MS, eluted at 19.29 min and had major ions at m/z (percentage intensity, proposed composition) 450 (36, M+), 448 (30, M+), 435 (18, [M − CH3]+), 433 (15, [M − CH3)+), 361 (6, [M − OTMS]+), 359 (6, [M − OTMS]+), 347 (5, [M − TMS − CH3 − CH3]+), 345 (5, [M − TMS − CH3 − CH3]+), 273 (92, [M − CH3 − TMS − OTMS]+), 271 (84, [M − CH3 − TMS − OTMS]+), 237 (13, [M − CH3 − Br − TMS − CO2]+), 73 (100, TMS+). The proton NMR spectrum in d6-DMSO contained two aromatic doublets, at δ ppm 6.82 (H5) and 7.20 (H6), JH5,H6 = 9.4 Hz. This product is identical to that isolated and described from the transformation of 2-bromobenzoate by phthalate-grown cells of A. keyseri 12B (see above) and is formed by the sequential activities of phthalate 3,4-dioxygenase and 3,4-dihydroxy-3,4-dihydrophthalate dehydrogenase (Fig. 1, enzymes A2 and B2). It accumulates because, unlike the 2-carboxyl group of 3,4-dihydroxyphthalate, the 2-bromine is not removed by 3,4-dihydroxyphthalate decarboxylase (Fig. 1, enzyme C2), the product of phtC.
Plasmid pRE1062 carries phtAaAbAcAd but lacks phtB, encoding 3,4-dihydroxy-3,4-dihydrophthalate dehydrogenase. A strain carrying pRE1062 would be expected to accumulate cis-dihydrodiol intermediates from phthalate and phthalate analogs. Following incubation of E. coli JM109(pRE1062) with 2-bromobenzoate (1 g in 1 liter), cells were removed by centrifugation and the culture supernatant was acidified and extracted with ethyl acetate. Chromatography on Sephadex LH-20 separated three compounds; peak fractions were pooled, and the solvent was removed. Peak A (fractions 67 to 77) contained cis-3,4-dihydroxy-3,4-dihydro-2-bromobenzoic acid (440 mg); peak B (fractions 104 to 122) contained the starting compound, 2-bromobenzoic acid (185 mg); peak C (fractions 138 to 151) contained a monohydroxy-2-bromobenzoic acid (164 mg) (this is probably 2-bromo-3-hydroxybenzoic acid produced by dehydration of the dihydrodiol [see below]).
The proton NMR spectrum of peak A, in d6-DMSO, contained four doublets at δ ppm 4.08 (H3), 4.33 (H4), JH3,H4 = 6 Hz; 6.04 (H5), 5.95 (H6), JH5,H6 = 11 Hz, JH4,H5 = 1 to 2 Hz, as well as two broad hydroxyl proton peaks at δ ppm 5.28 and 5.5. The coupling constant JH3,H4 = 6 Hz is as expected for cis protons, while trans protons would have a larger coupling constant (J = 10 to 16) (39). Different preparations contained various amounts of a contaminant. Its proton NMR spectrum in d6-DMSO, which had a doublet at δ ppm 7.08 (H4 and H6) and a triplet at δ ppm 7.26 (H5) (JH4,H5 = JH6,H5 = 8.6 Hz), indicates that it is 2-bromo-3-hydroxybenzoic acid, the product of dihydrodiol dehydration.
Trimethylsilylation of the dihydrodiol in peak A yielded the di-TMS derivative of the dehydration product, 2-bromo-3-hydroxybenzoic acid, which eluted from the GC at 15.64 min and had major ions at m/z (percentage intensity, proposed composition) 362 (20, M+), 360 (18, M+), 347 (100, [M − CH3]+), 345 (92, [M − CH3]+), 273 (26, [M − OTMS]+), 271 (26, [M − OTMS]+), 266 (26, [M − CH3 − Br]+), 191 (16), 166 (15), 165 (14), 149 (28, [M − CH3 − Br − TMS − CO2]+), 139 (13), 137 (15), 119 (9), 73 (87, TMS+).
Like other dihydrodiols having electron-withdrawing substituents, 3,4-dihydroxy-3,4-dihydro-2-bromobenzoic acid is relatively stable in acid and can survive acidification and extraction of culture supernatants. However, it will eventually dehydrate to form more stable phenolic products as shown here.
Biotransformation of phthalate by enzymes of the pht operon.
Phthalate was not transformed by any E. coli clones at neutral pH. This may be due to the inability of the dicarboxylate anion to enter these cells. The pKas of phthalic acid and some of its analogs which are transformed by E. coli clones at neutral pH are as follows: phthalic acid, 2.89 and 5.51; 2-bromobenzoic acid, 3.86; 2-chlorobenzoic acid, 1.92; 2-iodobenzoic acid, 2.85; and 2-nitrobenzoic acid, 2.16 (87). At neutral pH, the analogs exist as monocarboxylate anions, while phthalate is a dianion. However, at a pH below 5.51, a significant fraction of phthalate should have one protonated carboxyl group.
Transformation of phthalate by E. coli JM109(pRE1066) at pH 4.7 gave two products which could be separated on Sephadex LH-20. These were identified as 3,4-dihydroxyphthalic acid (112 mg) and protocatechuic acid (22 mg). 3,4-Dihydroxyphthalic acid was identified by GC-MS analysis of its tetra-TMS derivative, which eluted at 20.07 min and gave major ions at m/z (percentage intensity, proposed composition) 486 (1.3, M+), 471 (27, [M − CH3]+), 383 (3), 353 (2), 309, (31, [M − CH3 − OTMS − TMS]+), 147 (33), 133 (4), and 73 (100, TMS). The product, like authentic 3,4-dihydroxyphthalate (see below), could be converted to protocatechuate by 3,4-dihydroxyphthalate decarboxylase-containing extracts of E. coli BL21(DE3)(pLysS)(pRE1026). The protocatechuic acid from phthalate was initially identified by thin-layer chromatography on silica gel plates using ethyl acetate as the solvent (product, Rf = 0.41; authentic protocatechuate, Rf = 0.39; mixture, Rf = 0.42). GC-MS analysis of its tri-TMS derivative which eluted at 15.91 min gave major ions at m/z (percentage intensity, proposed composition) 370 (32, M+), 355 (15, [M − CH3]+), 311 (11), 281 (7), 223 (7), 193 (100, [M − CH3 − OTMS − TMS]+), 165 (7), 137 (6), 133 (3.5), 73 (72, TMS). This product, like authentic protocatechuate (see below), could be converted to 2-hydroxy-4-carboxymuconic semialdehyde by protocatechuate 4,5-dioxygenase-containing extracts of E. coli BL21(DE3)(pLysS)(pRE1043). When A. keyseri 12B grows with phthalate (at pH 6.8), it does not accumulate intermediates (24); therefore, it was somewhat surprising that the intermediate 3,4-dihydroxyphthalate accumulated here. While the low pH of the incubation may be responsible, another possible reason for this occurrence is discussed below in the section on regulation.
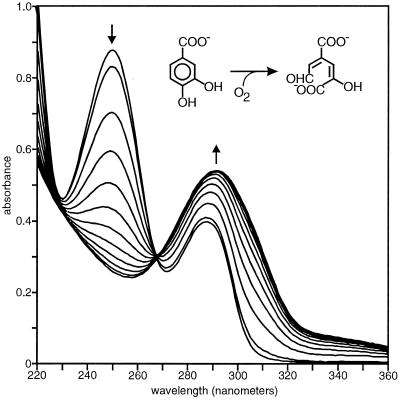
Plasmid pRE1026 carries phtC, encoding 3,4-dihydroxyphthalate 2-decarboxylase. Cell extracts of E. coli BL21(DE3)(pLysS)(pRE1026) transformed (authentic) 3,4-dihydroxyphthalate to protocatechuate (Fig. 5).
FIG. 5.
Conversion of 3,4-dihydroxyphthalate (λmax = 309 nm) to protocatechuate (λmax = 250 and 290 nm) by cell extracts of E. coli strain BL21 (DE3)(pLysS)(pRE1026) at 30°C. The sample and reference cuvettes contained 50 mM potassium-sodium-phosphate buffer (pH 6.8) in 1-ml volumes. The sample cuvette also contained 100 nmol of 3,4-dihydroxyphthalate. Spectra were recorded before the addition of 5 μl of extract (14 μg of protein) and after 0.17, 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, and 26 min.
Enzymes encoded by the pht operon. (i) Phthalate 3,4-dioxygenase.
The first step in the catabolism of phthalate is catalyzed by a three-component (class II) reductive dioxygenase. In similar dioxygenases, electrons are transferred from NADH through a flavoprotein reductase to a ferredoxin and then to a two-subunit terminal dioxygenase. The reduced terminal dioxygenase then interacts with its aromatic substrate and molecular oxygen to introduce two hydroxyl groups into the aromatic ring (37). The terminal dioxygenase and reductase components of phthalate 3,4-dioxygenase are most closely related (Table 3) to those of a branch of the class II reductive dioxygenase phylogenetic tree that includes naphthalene and indene dioxygenases from Rhodococcus sp. (51, 85) and a phenanthrene dioxygenase from Nocardioides sp. strain KP7 (78). The ferredoxin is related to several 3Fe-4S ferredoxins, most closely to a component of the sulfonylurea monooxygenase of Streptomyces griseolus (69), and retains three cysteines at positions 8, 14, and 52 present in those 3Fe-4S ferredoxins. The related 3Fe-4S ferredoxin (PhdC) recently demonstrated to be a component of phenanthrene dioxygenase in Nocardioides sp. strain KP7 (78) is possibly the first example of this type of ferredoxin in a reductive dioxygenase. Phthalate 3,4-dioxygenase differs significantly from the two-component, class I phthalate 4,5-dioxygenases (61, 63).
(ii) cis-3,4-Dihydroxy-3,4-dihydrophthalate dehydrogenase.
cis-3,4-Dihydroxy-3,4-dihydrophthalate dehydrogenase is a member of a superfamily of oxidoreductases that includes morphine dehydrogenase (10). It has 19 of 22 amino acid residues that are invariant in this superfamily with the substitutions: Met for Leu at residue 114, Gln for Glu at 174, and Trp for Arg at 246. This group was proposed primarily on the basis of sequence comparisons of 19 superfamily members and crystallographic studies of one member, human aldose reductase; its members lack homology to the zinc-requiring and short-chain alcohol dehydrogenases (10, 70, 71) as well as to 4,5-dihydroxy-4,5-dihydrophthalate dehydrogenase (12). As the biotranformations of phthalate analogs have revealed, this enzyme can act on a variety of dihydrodiols derived from 2-substituted benzoates but has reduced activity toward those dihydrodiols having substitutions in the 6 position.
(iii) 3,4-Dihydroxyphthalate decarboxylase.
3,4-Dihydroxyphthalate decarboxylase is unrelated to the three 4,5-dihydroxyphthalate decarboxylases for which sequence information is available (GenBank Q59727, AAD03553, and Q05185). The deduced amino acid sequence of the decarboxylase most closely resembles those of aldolases which catalyze the cleavage of fuculose 1-phosphate to yield dihydroxyacetone phosphate and l-lactaldehyde. This sequence similarity includes a conserved glutamate (residue 90) and three conserved histidines (residues 109, 111, and 177) shown in fuculose 1-phosphate aldolase (20, 38) to act as acid and base in catalysis and in the coordination of a catalytic Zn2+, respectively. The reaction mechanism of the decarboxylase, although not an aldol cleavage, may thus resemble one to some degree. Tautomerization of the 3-hydroxyl group of 3,4-dihydroxyphthalate to form an intermediate Zn2+-stabilized enolizable β-keto acid (1,2-dicarboxy-3-keto-4-hydroxycyclohexa-4,6-diene) would lead to the ready elimination of the β-carboxy substituent as carbon dioxide.
(iv) PhtR.
The proposed regulatory protein encoded by phtR is related to a family of regulatory proteins that include IcIR, a repressor controlling genes encoding enzymes of the glyoxalate cycle in E. coli (65, 83). It contains a putative DNA-binding region having the helix-turn-helix motif (amino acid residues 59-LTDASNYLGVASSTAHRLMG-78) (9) corresponding to that found in other members of the IcIR family. Its activity in regulating expression of the pht operon has not yet been demonstrated (see below).
The pcm operon.
Protocatechuate is converted to pyruvate and oxaloacetate by a five-step enzyme-catalyzed pathway (Fig. 4) encoded in A. keyseri 12B by the pcm operon, pcmDECABF (Fig. 5). Current understanding of the protocatechuate meta-cleavage pathway is a result of the work of several laboratories (15, 16, 18, 24, 32, 41, 53–60, 66, 74, 84), beginning with the demonstration that enzyme-catalyzed insertion of molecular oxygen between carbons 4 and 5 of protocatechuate, with the resulting opening of the aromatic ring (meta cleavage), occurs (15, 74). One of the more significant contributions has been by Maruyama and colleagues, who carried out a detailed study of the protocatechuate catabolic pathway in phthalate-grown Pseudomonas ochraceae (53–58), including the purification and characterization of the enzymes catalyzing the final four steps of the pathway and the demonstration of the central importance of 2-pyrone-4,6-dicarboxylate (Fig. 4, compound XIII).
Biotransformation of protocatechuate by enzymes of the pcm operon.
Plasmid pRE1043 carries pcmA, encoding protocatechuate 4,5-dioxygenase. Cell extracts of E. coli BL21(DE3)(pLysS)(pRE1043) incubated with protocatechuate (Fig. 6) converted it to 2-hydroxy-4-carboxymuconic semialdehyde. pcmB, encoding the next enzyme of the pathway, 2-hydroxy-4-carboxymuconic semialdehyde dehydrogenase, is located on pRE1058. Following addition of NAD+ and extract of E. coli JM109(pRE1058) to the spectrophotometer cuvettes at the end of the reaction shown in Fig. 6, the spectrum of 2-hydroxy-4-carboxymuconic semialdehyde disappeared, with the formation of a product having a spectrum with maximum at 310 nm (data not shown). This spectrum is characteristic of 2-pyrone-4,6-dicarboxylate (Fig. 4, compound XIII) formed from 2-hydroxy-4-carboxymuconic semialdehyde, probably by dehydrogenation of an intermediate hemiacetal (Fig. 4, compound XII) (40).
FIG. 6.
Conversion of protocatechuate (λmax = 250 and 290 nm) to 2-hydroxy-4-carboxymuconic semialdehyde (λmax = 293 nm) by cell extracts of E. coli strain BL21 (DE3)(pLysS)(pRE1043) at 30°C. The sample and reference cuvettes contained 50 mM potassium-sodium-phosphate buffer (pH 6.8) in 1-ml volumes. The sample cuvette also contained 100 nmol of protocatechuate. Spectra were recorded before the addition of 5 μl of extract (11 μg of protein) and after 0.17, 4, 8, 12, 16, 20, 24, 28, 32, 36, and 40 min.
The hydrolysis of 2-pyrone-4,6-dicarboxylate to 4-oxalomesaconate (Fig. 4, compound XIV) is a reversible enzyme-catalyzed reaction. At pH 7,2-pyrone-4,6-dicarboxylate is 87% of the equilibrium mixture, while at pH 8.5, it is only 21% (41, 54). Assays of the hydrolase in the forward direction (for which the substrate was available) was therefore more appropriately performed at the higher pH. The gene pcmC, encoding the 2-pyrone-4,6-dicarboxylate hydrolase, is carried on pRE1065. The hydrolase in extracts of IPTG-induced E. coli BL21(DE3)(pLysS)(pRE1065) was assayed spectrophotometrically in pH 8.5 Tris-Cl buffer. The sample cuvette contained 0.15 mM 2-pyrone-4,6-dicarboxylate and various volumes of extract. A rate of hydrolysis of 3.7 (±0.1) nmol min−1 mg of protein−1 was determined from the decrease in absorbance at 312 nm over time, using ɛ312 = 6,600 (60). Extracts of E. coli BL21(DE3)(pLysS) lacking pRE1065 failed to act on 2-pyrone-4,6-dicarboxylate under similar conditions.
Most of the pcm operon (pcmDECAB) except for pcmF is carried on a 5.4-kbp ClaI-BglII fragment in pRE995. Extract (2 ml) of E. coli BL21(DE3)(pLysS)(pRE995) incubated with 0.5 mmol of protocatechuate in 50 ml of 30 mM Tris-Cl buffer (pH 8.5) containing 20 μM MgCl2 and 120 μM NAD+ converted protocatechuate to pyruvate and oxaloacetate, which were extracted and identified in two ways. Analysis of DNPHs of transformation products by thin-layer chromatography showed DNPHs with Rf values of 59, 32, 19, and 5, which compared well with those of pyruvate-DNPH (Rf = 30 and 17), oxaloacetate-DNPH (Rf = 31, 17, and 3 to 7), and DNPH (Rf = 65). Incubation of products with NADH and lactate dehydrogenase or malic dehydrogenase caused a decrease in absorbance at 340 nm due to enzyme-catalyzed oxidation of NADH to NAD+ coupled to the reduction of pyruvate to lactate or oxaloacetate to malate. The responses indicated a ratio of pyruvate to oxaloacetate of 3:1 in the product mixture. This nonequivalence is not surprising since the β-keto acid, oxaloacetate, is readily decarboxylated, either enzymatically or spontaneously. This lability was noted in the product of DNPH derivatization of authentic oxaloacetate, which also contains a significant proportion of the pyruvate-DNPHs.
E. coli cells and extracts of cells lacking relevant recombinant plasmids did not act on any of the substrates tested here. This suggests that all of the enzymes of the phthalate catabolic pathway except 4-oxalocitramalate aldolase, for which there was not an available substrate, are absent from E. coli host strains.
Enzymes encoded by the pcm operon.
The enzymes of the protocatechuate meta-cleavage pathway in A. keyseri 12B are closely related to enzymes of the same pathway in Sphingomonas sp. strain LB126 and Sphingomonas paucimobilis SYK-6 (32, 59, 60, 66), having between 50 and 70% amino acid sequence identity (Table 3).
Protocatechuate 4,5-dioxygenase catalyzes the insertion of a molecule of oxygen between carbons 4 and 5 of protocatechuate, opening the aromatic ring. Its deduced amino acid sequence is similar to that of other protocatechuate 4,5-dioxygenases. However, those related dioxygenases are synthesized from contiguous genes as small and large subunits (139 and 302 amino acids, respectively, in S. paucimobilis SYK-6 [66]), while in A. keyseri 12B, the contiguous DNA segments corresponding to the large and small subunit genes are joined to form a single gene, pcmA, encoding a 433-amino-acid protocatechuate 4,5-dioxygenase peptide. The pcmA gene from A. keyseri 12B and surrounding DNA have been resequenced and examined for evidence of errors that might have resulted in an overlooked stop codon, but the sequence appears to be correct.
PcmE, 4-oxalocitramalate aldolase, most resembles an enzyme in S. paucimobilis SYK-6 proposed to be an acyltransferase but more likely having the same function as PcmE. PcmE also resembles a group of aldolases involved in the metabolism of C1 compounds, 3-hexulose-6-phosphate synthases (d-arabino-3-hexulose-6-phosphate formaldehyde lyase) (90). These enzymes catalyze aldol condensation of formaldehyde and d-ribulose-5-phosphate. The molecular weight of PcmE, 24,442, is similar to the subunit molecular weight of 26,000 (enzyme molecular weight, 160,000) determined for the P. ochraceae enzyme (57).
PcmF is an oxidoreductase without a known function. It is not required for the conversion of protocatechuate to pyruvate and oxaloacetate (above), and extracts of E. coli strains carrying the pcmF gene (on pRE1056, e.g.) did not have activity toward such substrates as 2-hydroxy-4-carboxymuconic semialdehyde and 2-pyrone-4,6-dicarboxylate. PcmF is a member of the aldo-keto reductase superfamily (35).
PcmR.
PcmR is a member of the LysR family of regulatory proteins (79). Its role in regulation of pcm operon expression has not been demonstrated.
ABC transporter.
The putative ptr operon, located between pcm and pht operons, encodes polypeptides similar to those of an ABC (ATP-binding cassette) transport system (33) (Table 3) consisting of an ATPase (PtrA) and two permeases. Together, they are most similar to sulfate ester transporters (42). Upstream of these genes is a fourth gene having a product, PtrD, that is similar to putative sulfate ester-binding proteins (86). The PtrB and PtrC permeases are most closely related to each other but share only 29% identity in 240 amino acid residues. The function of the Ptr system has not been established. It has not been possible to show transport activity toward phthalate, protocatechuate, or any of the diesters (dimethylphthalate, diethylphthalate, or dibutylphthalate) in E. coli clones carrying ABC transporter genes on such plasmids as pRE754 or pRE1096 (data not shown).
Phthalate probably requires a transport system to enter A. keyseri 12B cells. The Ptr transporter, because of the location of the ptr genes between pht and pcm operons, seems a likely if unproven candidate. Another phthalate-degrading strain, Burkholderia cepacia ATCC 17616, has two phthalate-inducible phthalate transporters; one of these, OphD, encoded by a recombinant plasmid in E. coli JM109, allowed that strain to take up phthalate which, as also shown here, it is otherwise unable to do at neutral pH (13).
Putative phthalate ester hydrolase.
The product of pehA is related to a hydrolase (CSHase) from Arthrobacter sp. which catalyzes the hydrolysis of N-carbamoylsarcosine to sarcosine, CO2, and NH3 (77). The cysteine at residue 159 proposed to be the catalytic nucleophile in CSHase (as residue 177) is conserved. It has not been possible to demonstrate hydrolase activity in cells or cell extracts of recombinant E. coli strains carrying pehA (on, e.g., pRE754, pRE842, pRE861, pRE1089, or pRE1096) toward dimethyl-, diethyl-, or dibutylphthalate, all substrates for a constitutive plasmid-encoded esterase previously demonstrated in A. keyseri 12B (24).
Between pehA and phtB is a gene remnant encoding a fragment of a protein similar to NorA (conferring resistance to the quinolone norfloxacin) (67, 92) and other antibiotic efflux transporters.
Transposon functions.
Near the beginning of the DNA sequence is a gene encoding a transposon resolvase that is closely related to resolvases of the Tn21 family. This is preceded by a large, unidentified open reading frame (tnpX, 421 bp), a possible promoter, and a sequence (bp 2888 to 3011) corresponding to the resolvase binding (res) sites of Tn21 family transposons (76, 93). The res sequence has 58% homology to the Tn1721 res site in 122 bp and includes a 30-bp segment (bp 2964 to 2993) of perfect dyad symmetry. The Tn3-like transposons including the Tn21 family, transpose in two steps, through the formation of a cointegrate and its resolution into two molecules. The sequence of the DNA that has been characterized here does not extend to include a cointegrate-forming transposase gene or terminal inverted repeats. However, the presence of resolvase gene and res site suggests that the phthalate catabolism region of pRE1 is associated with a transposable element, a common attribute of many catabolic operons (89).
Regulation.
Upstream of each operon and regulatory gene lie sequences having recognizable similarity to the E. coli ς70 promoter consensus (Fig. 3). However, there was no evidence of expression from these putative promoters present in recombinant plasmids in E. coli. Expression of an A. keyseri 12B gene in E. coli was detectable only when the gene was located downstream from a vector-specified promoter. This has made it difficult to study regulation of A. keyseri 12B genes in E. coli.
Expression of phthalate catabolism genes is inducible by phthalate in A. keyseri 12B. Upstream of the pht operon and overlapping the putative pht promoter is a 32-bp segment having 75% dyad symmetry which may be the pht operator recognized by PhtR. An additional putative promoter was identified within the pht operon, upstream of phtCR. In a previous study of A. keyseri 12B (24), production of 3,4-dihydroxyphthalate decarboxylase was shown to be constitutive (300 nmol min−1 mg of protein−1) but also further inducible threefold by growth with phthalate. Constitutive expression of phtC from the putative phtCR promoter with additional phthalate-induced expression from a pht operon promoter would explain the previously observed variations in decarboxylase activities in extracts of A. keyseri 12B (24). This could also explain why 3,4-dihydroxyphthalate accumulated in incubations of E. coli JM109(pRE1066) with phthalate (above); since phtC expression was from only the vector promoter, the levels of the decarboxylase relative to the preceding enzymes in the pathway were reduced.
The putative pcm operon regulatory system has features typical of regulators of the LysR family (79). The putative promoters for pcmR and the pcm operon are divergent and overlap. Between the pcm operator and pcmR (and upstream of the putative pcm operon promoter) is a region of dyad symmetry containing the T-N11-A motif suggested (79) to be essential for regulatory protein binding in other LysR-type systems. This possible pcm operator is located such that PcmR binding to it could repress expression of pcmR while activating expression of the pcm operon.
Conclusion.
By assaying enzymes in strains 12B and 12B-C1, the phthalate ester catabolic pathway was previously shown (24) to be divided into at least four different plasmid-specified units: (i) a constitutive phthalate ester hydrolase (esterase); (ii) phthalate 3,4-dioxygenase and 3,4-dihydroxy-3,4-dihydrophthalate dehydrogenase, inducible (19-fold) by phthalate; (iii) 3,4-dihydroxyphthalate decarboxylase, constitutive but also slightly (3-fold) inducible by phthalate; and (iv) protocatechuate meta-cleavage pathway, inducible by protocatechuate. Analysis of the phthalate catabolism region of pRE1 supports these observations. The pht and pcm operons (Fig. 3), specifying the conversion of phthalate to protocatechuate and of protocatechuate to pyruvate and oxaloacetate (Fig. 4), have been identified and characterized. Within the pht operon, phtC, encoding 3,4-dihydroxyphthalate decarboxylase, may be expressed not only from the phthalate-inducible promoter but also from a second constitutive promoter located near the end of the upstream phtAd gene. The roles of the neighboring ABC transporter and hydrolase genes have not been established. Although their location between the pht and pcm operons and the requirement for esterase and transport activities suggest that they could be involved in phthalate ester catabolism, no activities of their gene products toward phthalate esters, phthalate, or protocatechuate have been demonstrated.
The reactions catalyzed by enzymes of the phthalate catabolic pathway in A. keyseri 12B provide novel and convenient routes to a family of 2-substituted protocatechuates (Fig. 1, compound IX). By using recombinant E. coli strains carrying genes encoding phthalate 3,4-dioxygenase but not the dihydrodiol dehydrogenase (as on pRE1066), it is also possible to produce a corresponding family of novel cis-dihydrodiols. These cis-dihydrodiols have two asymmetric (chiral) carbons, which can make them attractive starting compounds in organic syntheses of natural products (8, 35). In this context, the dihydrodiol produced from 2-iodobenzoate may be particularly interesting since the ready removal of iodine by catalytic hydrogenolysis would yield cis-3,4-dihydroxy-3,4-dihydrobenzoate, an enantiomerically pure 1,2-dihydrodiol unusual because it contains a substituent at C-4.
ACKNOWLEDGMENTS
I thank Jerome Gurst, Chemistry Department, University of West Florida, Pensacola, for Nuclear Magnetic Resonance Spectroscopy; Wallace Gilliam, U.S. EPA, Gulf Breeze, Fla., for GC-MS; Heron Yu, ACGT, Inc., and Ernesto Almira and Savita Shanker, ICBR DNA Sequencing Core Lab University of Florida, Gainesville, for DNA sequence analysis; Diane Yates for analysis of fatty acid methyl esters; Richard Devereux, U.S. EPA, Gulf Breeze, Fla., and A. Schramm, University of Bayreuth, Bayreuth, Germany, for advice on primers for 16S rDNA amplification and sequencing; and Peter Chapman, U.S. EPA, Gulf Breeze, Fla., for critical reading of the manuscript.
Partial support for the purchase of the NMR spectrometer at University of West Florida was provided by grant USE-9050802 from the National Science Foundation.
Footnotes
Contribution 1130 from the Gulf Ecology Division, National Health and Environmental Effects Laboratory, U.S. Environmental Protection Agency, Gulf Breeze, Fla.
REFERENCES
- 1.Alting-Mees M A, Short J M. pBluescript II: gene mapping vectors. Nucleic Acids Res. 1989;17:9494. doi: 10.1093/nar/17.22.9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnsley E A. Phthalate pathway of phenanthrene metabolism: formation of2-carboxybenzalpyruvate. J Bacteriol. 1983;154:113–117. doi: 10.1128/jb.154.1.113-117.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batie C J, LaHaie E, Ballou D P. Purification and characterization of phthalate oxygenase and phthalate oxygenase reductase from Pseudomonas cepacia. J Biol Chem. 1987;262:1510–1518. [PubMed] [Google Scholar]
- 5.Bauer M J, Hermann R. Estimation of environmental contamination by phthalic acid esters leaching from household wastes. Sci Total Environ. 1997;208:49–57. doi: 10.1016/s0048-9697(97)00272-6. [DOI] [PubMed] [Google Scholar]
- 6.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boopathy R, Manning J F. Characterization of partial anaerobic metabolic pathway for 2,4,6-trinitrotoluene degradation by a sulfate-reducing bacterial consortium. Can J Microbiol. 1996;42:1203–1208. doi: 10.1139/m96-155. [DOI] [PubMed] [Google Scholar]
- 8.Boyd D R, Sheldrake G N. The dioxygenase-catalyzed formation of vicinal cis-diols. Nat. Prod. Rep. 1998. pp. 309–324. [Google Scholar]
- 9.Brennan R G, Matthews B W. The helix-turn-helix DNA binding motif. J Biol Chem. 1989;264:1903–1906. [PubMed] [Google Scholar]
- 10.Bruce N C, Willey D L, Coulson A F W, Jeffrey J. Bacterial morphine dehydrogenase further defines a distinct superfamily of oxidoreductases with diverse functional activities. Biochem J. 1994;299:805–811. doi: 10.1042/bj2990805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cadogan D F, Papez M, Poppe A C, Pugh D M, Scheubel J. An assessment of the release, occurrence and possible effects of plasticizers in the environment. Prog Rubber Plastics Technol. 1993;10:1–19. [Google Scholar]
- 12.Chang H-K, Zylstra G J. Novel organization of the genes for phthalate degradation from Burkholderia cepaciaDB01. J Bacteriol. 1998;180:6529–6537. doi: 10.1128/jb.180.24.6529-6537.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang H-K, Zylstra G J. Characterization of the phthalate permease OphD from Burkholderia cepaciaATCC 17616. J Bacteriol. 1999;181:6197–6199. doi: 10.1128/jb.181.19.6197-6199.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clewell D B, Helinski D R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an open circular DNA form. Proc Natl Acad Sci USA. 1969;62:1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dagley S, Evans W C, Ribbons D W. New pathways in the oxidative metabolism of aromatic compounds by micro-organisms. Nature. 1960;188:560–566. doi: 10.1038/188560a0. [DOI] [PubMed] [Google Scholar]
- 16.Dagley S, Geary P J, Wood J M. The metabolism of protocatechuate by Pseudomonas testosteroni. Biochem J. 1968;109:559–568. doi: 10.1042/bj1090559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 18.Dennis D A, Chapman P J, Dagley S. Degradation of protocatechuate in Pseudomonas testosteroni by a pathway involving oxidation of the product of meta-fission. J Bacteriol. 1973;113:521–523. doi: 10.1128/jb.113.1.521-523.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dretzen G, Bellard M, Sassone-Corsi P, Chambon P. A reliable method for the recovery of DNA fragments from agarose and acrylamide gels. Anal Biochem. 1981;112:295–298. doi: 10.1016/0003-2697(81)90296-7. [DOI] [PubMed] [Google Scholar]
- 20.Dreyer M K, Schulz G E. Catalytic mechanism of the metal-dependent fuculose aldolase from Escherichia colias derived from the structure. J Mol Biol. 1996;259:458–466. doi: 10.1006/jmbi.1996.0332. [DOI] [PubMed] [Google Scholar]
- 21.Eaton R W. p-Cumate catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA carrying the cmtoperon. J Bacteriol. 1996;178:1351–1362. doi: 10.1128/jb.178.5.1351-1362.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eaton R W, Nitterauer J D. Biotransformation of benzothiophene by isopropylbenzene-degrading bacteria. J Bacteriol. 1994;176:3992–4002. doi: 10.1128/jb.176.13.3992-4002.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eaton R W, Ribbons D W. The transformation of phthalaldehydate by phthalate-grown Micrococcusstrain 12B. Arch Biochem Biophys. 1982;215:289–295. doi: 10.1016/0003-9861(82)90213-2. [DOI] [PubMed] [Google Scholar]
- 24.Eaton R W, Ribbons D W. Metabolism of dibutylphthalate and phthalate by Micrococcussp. strain 12B. J Bacteriol. 1982;151:48–57. doi: 10.1128/jb.151.1.48-57.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eaton R W, Ribbons D W. Metabolism of dimethylphthalate by Micrococcussp. strain 12B. J Bacteriol. 1982;151:465–467. doi: 10.1128/jb.151.1.465-467.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eaton R W, Ribbons D W. Biotransformation of 3-methylphthalate by Micrococcussp. strain 12B. J Gen Microbiol. 1987;133:2473–2476. doi: 10.1099/00221287-133-9-2473. [DOI] [PubMed] [Google Scholar]
- 27.Eaton R W, Timmis K N. Characterization of a plasmid-specified pathway for catabolism of isopropylbenzene in Pseudomonas putidaRE204. J Bacteriol. 1986;168:123–131. doi: 10.1128/jb.168.1.123-131.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furukawa K. Microbial degradation of polychlorinated biphenyls (PCBs) In: Chakrabarty A M, editor. Biodegradation and detoxification of environmental pollutants. Boca Raton, Fla: CRC Press; 1982. pp. 33–57. [Google Scholar]
- 29.Grifoll M, Selifonov S A, Chapman P J. Evidence for a novel pathway in the degradation of fluorene by Pseudomonassp. strain F274. Appl Environ Microbiol. 1994;60:2438–2449. doi: 10.1128/aem.60.7.2438-2449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grunstein M, Hogness D. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci USA. 1975;72:3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen J H, Olsen R H. Isolation of large bacterial plasmids and characterization of the P2 incompatibility group plasmids, pMG1 and pMG5. J Bacteriol. 1978;135:227–238. doi: 10.1128/jb.135.1.227-238.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hara H, Masai E, Katayama Y, Fukuda M. The 4-oxalomesaconate hydratase gene, involved in the protocatechuate 4,5-cleavage pathway, is essential to vanillate and syringate degradation in Sphingomonas paucimobilisSYK-6. J Bacteriol. 2000;182:6950–6957. doi: 10.1128/jb.182.24.6950-6957.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higgins C F. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 34.Holmes D S, Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981;114:193–199. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- 35.Hudlicky T, Gonzalez D, Gibson D T. Enzymatic dihydroxylation of aromatics in enantioselective synthesis: expanding asymmetric methodology. Aldrichim Acta. 1999;32:35–62. [Google Scholar]
- 36.Jez J M, Bennett M J, Schlegel B P, Lewis M, Penning T M. Comparative anatomy of the aldo-keto reductase family. Biochem J. 1997;326:625–636. doi: 10.1042/bj3260625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang H, Parales R E, Lynch N A, Gibson D T. Site-directed mutagenesis of conserved amino acids in the alpha subunit of toluene dioxygenase: potential mononuclear non-heme iron coordination sites. J Bacteriol. 1996;178:3133–3139. doi: 10.1128/jb.178.11.3133-3139.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joerger A C, Gosse C, Fessner W-D, Schulz G E. Catalytic action of fuculose 1-phosphate aldolase (class II) as derived from structure-directed mutagenesis. Biochemistry. 2000;39:6033–6041. doi: 10.1021/bi9927686. [DOI] [PubMed] [Google Scholar]
- 39.Karplus M. Vicinal proton coupling in nuclear magnetic resonance. J Am Chem Soc. 1963;85:2870–2871. [Google Scholar]
- 40.Keith L M. Environmental endocrine disruptors. New York, N.Y: John Wiley & Sons, Inc.; 1997. [Google Scholar]
- 41.Kersten P J, Dagley S, Whittaker J W, Arciero D M, Lipscomb J D. 2-Pyrone-4,6-dicarboxylic acid, a catabolite of gallic acids in Pseudomonasspecies. J Bacteriol. 1982;152:1154–1162. doi: 10.1128/jb.152.3.1154-1162.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kertesz M A. Riding the sulfur cycle—metabolism of sulfonates and sulfate esters in Gram-negative bacteria. FEMS Microbiol Rev. 1999;24:135–175. doi: 10.1016/S0168-6445(99)00033-9. [DOI] [PubMed] [Google Scholar]
- 43.Keyser P. Aerobic metabolism of the phthalates by selected pseudomonads. M.S. dissertation. Miami, Fla: University of Miami; 1974. [Google Scholar]
- 44.Keyser P. Phthalic acid-4,5-dioxygenase from Pseudomonas fluorescens PHK. Ph.D. dissertation. Miami, Fla: University of Miami; 1976. [Google Scholar]
- 45.Keyser P, Pujar B G, Eaton R W, Ribbons D W. Biodegradation of phthalates and their esters by bacteria. Environ Health Perspect. 1976;18:159–166. doi: 10.1289/ehp.7618159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kiyohara H, Nagao K. The catabolism of phenanthrene and anthracene by bacteria. J Gen Microbiol. 1978;105:69–75. [Google Scholar]
- 47.Koch C, Schumann P, Stackebrandt E. Reclassification of Micrococcus agilis (Ali-Cohen 1889) to the genus Arthrobacter as Arthrobacter agilis comb. nov. and emendation of the genus Arthrobacter. Int J Syst Bacteriol. 1995;45:837–839. doi: 10.1099/00207713-45-4-837. [DOI] [PubMed] [Google Scholar]
- 48.Kovach M E, Elzer P H, Hill D S, Robertson G T, Farris M A, Roop II R M, Peterson K M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 49.Kulik I, Stevens J, Toledano M B, Storz G. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for DNA binding and multimerization. J Bacteriol. 1995;177:1285–1291. doi: 10.1128/jb.177.5.1285-1291.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lane D J. 16S/23S rDNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons; 1991. pp. 115–175. [Google Scholar]
- 51.Larkin M J, Allen C C R, Kulakov L A, Lipscomb D A. Purification and characterization of a novel naphthalene dioxygenase from Rhodococcussp. strain NCIMB12038. J Bacteriol. 1999;181:6200–6204. doi: 10.1128/jb.181.19.6200-6204.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lawrence W H. Phthalate esters: the question of safety. Clin Toxicol. 1978;13:89–139. doi: 10.3109/15563657808988230. [DOI] [PubMed] [Google Scholar]
- 53.Maruyama K. Isolation and identification of the reaction product of α-hydroxy-γ-carboxymuconic ɛ-semialdehyde dehydrogenase. J Biochem. 1979;86:1671–1677. doi: 10.1093/oxfordjournals.jbchem.a132687. [DOI] [PubMed] [Google Scholar]
- 54.Maruyama K. Purification and properties of 2-pyrone-4,6-dicarboxylate hydrolase. J Biochem. 1983;93:557–565. doi: 10.1093/oxfordjournals.jbchem.a134210. [DOI] [PubMed] [Google Scholar]
- 55.Maruyama K. Enzymes responsible for degradation of 4-oxalmesaconic acid in Pseudomonas ochraceae. J Biochem. 1983;93:567–574. doi: 10.1093/oxfordjournals.jbchem.a134211. [DOI] [PubMed] [Google Scholar]
- 56.Maruyama K. Purification and properties of γ-oxalmesaconate hydratase from Pseudomonas ochraceaegrown with phthalate. Biochem Biophys Res Commun. 1985;128:271–277. doi: 10.1016/0006-291x(85)91674-2. [DOI] [PubMed] [Google Scholar]
- 57.Maruyama K. Purification and properties of 4-hydroxy-4-methyl-2-oxoglutarate aldolase from Pseudomonas ochraceaegrown on phthalate. J Biochem. 1990;108:327–333. doi: 10.1093/oxfordjournals.jbchem.a123201. [DOI] [PubMed] [Google Scholar]
- 58.Maruyama K, Ariga N, Tsuda M, Deguchi K. Purification and properties of α-hydroxy-γ-carboxymuconic ɛ-semialdehyde dehydrogenase. J Biochem. 1978;83:1125–1134. doi: 10.1093/oxfordjournals.jbchem.a132002. [DOI] [PubMed] [Google Scholar]
- 59.Masai E, Momose K, Hara H, Nishikawa S, Katayama Y, Fukuda M. Genetic and biochemical characterization of 4-carboxy-2-hydroxymuconate-6-semialdehyde dehydrogenase and its role in the protocatechuate 4,5-cleavage pathway in Sphingomonas paucimobilisSYK-6. J Bacteriol. 2000;182:6651–6658. doi: 10.1128/jb.182.23.6651-6658.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Masai E, Shinohara S, Hara H, Nishikawa S, Katayama Y, Fukuda M. Genetic and biochemical characterization of a 2-pyrone-4,6-dicarboxylic acid hydrolase involved in the protocatechuate 4,5-cleavage pathway of Sphingomonas paucimobilisSYK-6. J Bacteriol. 1999;181:55–62. doi: 10.1128/jb.181.1.55-62.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mason J R, Cammack R. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu Rev Microbiol. 1992;46:277–305. doi: 10.1146/annurev.mi.46.100192.001425. [DOI] [PubMed] [Google Scholar]
- 62.Moore N P. The oestrogenic potential of the phthalate esters. Reprod Toxicol. 2000;14:183–192. doi: 10.1016/s0890-6238(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 63.Nakatsu C H, Straus N A, Wyndham R C. The nucleotide sequence of the Tn5271 3-chlorobenzoate 3,4-dioxygenase genes (cbaAB) unites the class IA oxygenases in a single lineage. Microbiology. 1995;141:485–495. doi: 10.1099/13500872-141-2-485. [DOI] [PubMed] [Google Scholar]
- 64.Nakazawa T, Hayashi E. Phthalate metabolism in Pseudomonas testosteroni: accumulation of 4,5-dihydroxyphthalate by a mutant strain. J Bacteriol. 1977;131:42–48. doi: 10.1128/jb.131.1.42-48.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nègre D, Cortay J-C, Old I G, Galinier A, Richaud C, Saint Girons I, Cozzone A J. Overproduction and characterization of the iclR gene product of Escherichia coli K-12 and comparison with that of Salmonella typhimuriumLT2. Gene. 1991;97:29–37. doi: 10.1016/0378-1119(91)90006-w. [DOI] [PubMed] [Google Scholar]
- 66.Noda Y, Nishikawa S, Shiozuka K-I, Kadokura H, Nakajima H, Yoda K, Katayama Y, Morohoshi N, Haraguchi T, Yamasaki M. Molecular cloning of the protocatechuate 4,5-dioxygenase genes of Pseudomonas paucimobilis. J Bacteriol. 1990;172:2704–2709. doi: 10.1128/jb.172.5.2704-2709.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Noguchi N, Hase M, Kitta M, Sasatsu M, Deguchi K, Kono M. Antiseptic susceptibility and distribution of antiseptic-resistance genes in methicillin-resistant Staphylococcus aureus. FEMS Microbiol Lett. 1999;172:247–253. doi: 10.1111/j.1574-6968.1999.tb13475.x. [DOI] [PubMed] [Google Scholar]
- 68.O'Connor C D, Humphries G O. Expression of the EcoRI restriction-modification system and the construction of positive-selection cloning vectors. Gene. 1982;20:219–229. doi: 10.1016/0378-1119(82)90041-5. [DOI] [PubMed] [Google Scholar]
- 69.O'Keefe D P, Gibson K J, Emptage M H, Lenstra R, Romesser J A, Little P J, Omer C A. Ferredoxins from two sulfonylurea herbicide monooxygenase systems in Streptomyces griseolus. Biochemistry. 1991;30:447–455. doi: 10.1021/bi00216a021. [DOI] [PubMed] [Google Scholar]
- 70.Persson B, Krook M, Jörnvall H. Characteristics of short-chain alcohol dehydrogenases and related enzymes. Eur J Biochem. 1991;200:537–543. doi: 10.1111/j.1432-1033.1991.tb16215.x. [DOI] [PubMed] [Google Scholar]
- 71.Reid M F, Fewson C A. Molecular characterization of microbial alcohol dehydrogenases. Crit Rev Microbiol. 1994;20:13–56. doi: 10.3109/10408419409113545. [DOI] [PubMed] [Google Scholar]
- 72.Ribbons D W, Eaton R W. Chemical transformations of aromatic hydrocarbons that support the growth of microorganisms. In: Chakrabarty A M, editor. Biodegradation and detoxification of environmental pollutants. Boca Raton, Fla: CRC Press; 1982. pp. 59–84. [Google Scholar]
- 73.Ribbons D W, Evans W C. Oxidative metabolism of phthalic acid by soil pseudomonads. Biochem J. 1960;76:310–318. doi: 10.1042/bj0760310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ribbons D W, Evans W C. Oxidative metabolism of protocatechuic acid by certain soil pseudomonads: a new ring-fission mechanism. Biochem J. 1962;83:482–492. doi: 10.1042/bj0830482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ribbons D W, Keyser P, Kunz D A, Taylor B F, Eaton R W, Anderson B N. Microbial degradation of phthalates. In: Gibson D T, editor. Microbial degradation of organic compounds. New York, N.Y: Marcel Dekker, Inc; 1984. pp. 371–397. [Google Scholar]
- 76.Rogowsky P, Halford S E, Schmitt R. Definition of three resolvase binding sites at the res loci of Tn21 and Tn1721. EMBO J. 1985;8:2135–2141. doi: 10.1002/j.1460-2075.1985.tb03904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Romão M J, Turk D, Gomis-Rüth F-X, Huber R. Crystal structure analysis, refinement, and enzymatic reaction mechanism of N-carbamoylsarcosine amidohydrolase from Arthrobacter sp. at 2.0 Åresolution. J Mol Biol. 1992;226:1111–1130. doi: 10.1016/0022-2836(92)91056-u. [DOI] [PubMed] [Google Scholar]
- 78.Saito A, Iwabuchi T, Harayama S. A novel phenanthrene dioxygenase from Nocardioides sp. strain KP7: expression in Escherichia coli. J Bacteriol. 2000;182:2134–2141. doi: 10.1128/jb.182.8.2134-2141.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 80.S̆epic̆ E, Bricelj M, Leskovs̆ek H. Degradation of fluoranthene by Pasteurella sp. IFA and Mycobacteriumsp. PYR-1: isolation and identification of metabolites. J Appl Microbiol. 1998;85:746–754. doi: 10.1111/j.1365-2672.1998.00587.x. [DOI] [PubMed] [Google Scholar]
- 81.Smith G E, Summers M D. The bidirectional transfer of DNA and RNA to nitrocellulose or diazobenzyloxymethyl paper. Anal Biochem. 1980;109:123–129. doi: 10.1016/0003-2697(80)90019-6. [DOI] [PubMed] [Google Scholar]
- 82.Studier F W, Rosenberg A H, Dunn J J. Use of T7 polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 83.Sunnarborg A, Klumpp D, Chung T, LaPorte D C. Regulation of the glyoxalate bypass operon: cloning and characterization of iclRJ. Bacteriol. 1990;172:2642–2649. doi: 10.1128/jb.172.5.2642-2649.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tack B F, Chapman P J, Dagley S. Purification and properties of 4-hydroxy-4-methyl-2-oxoglutarate aldolase. J Biol Chem. 1972;247:6444–6449. [PubMed] [Google Scholar]
- 85.Treadway S L, Yanagimachi K S, Lankenau E, Lessard P A, Stephanopoulos G, Sinskey A J. Isolation and characterization of indene bioconversion genes from Rhodococcusstrain I24. Appl Microbiol Biotechnol. 1999;51:786–793. doi: 10.1007/s002530051463. [DOI] [PubMed] [Google Scholar]
- 86.Vermeij P, Weitek C, Kahnert A, Wüest T, Kertesz M A. Genetic organization of sulphur-controlled aryl desulphonation in Pseudomonas putidaS-313. Mol Microbiol. 1999;32:913–926. doi: 10.1046/j.1365-2958.1999.01398.x. [DOI] [PubMed] [Google Scholar]
- 87.Weast R C, editor. Handbook of chemistry and physics. 45th ed. Cleveland, Ohio: The Chemical Rubber Co.; 1964. [Google Scholar]
- 88.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S Ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wyndham R C, Cashore A E, Nakatsu C H, Peel M. Catabolic transposons. Biodegradation. 1994;5:323–342. doi: 10.1007/BF00696468. [DOI] [PubMed] [Google Scholar]
- 90.Yanase H, Ikeyama K, Mitsui R, Ra S, Kita K, Sakai Y, Kato N. Cloning and sequence analysis of the gene encoding 3-hexulose-6-phosphate synthase from the methylotrophic bacterium, Methylomonas aminofaciens 77a, and its expression in Escherichia coli. FEMS Microbiol Lett. 1996;135:201–205. doi: 10.1111/j.1574-6968.1996.tb07990.x. [DOI] [PubMed] [Google Scholar]
- 91.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 92.Yoshida H, Bogaki M, Nakamura S, Ubukata K, Konno M. Nucleotide sequence and characterization of the Staphylococcus aureus norAgene, which confers resistance to quinolones. J Bacteriol. 1990;172:6942–6949. doi: 10.1128/jb.172.12.6942-6949.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yurieva O, Kholodii G, Minakhin L, Gorlenko Z, Kalyaeva E, Mindlin S, Nikiforov V. Intercontinental spread of promiscuous mercury-resistance transposons in environmental bacteria. Mol Microbiol. 1997;24:321–329. doi: 10.1046/j.1365-2958.1997.3261688.x. [DOI] [PubMed] [Google Scholar]