Abstract
Background
Androgen receptor (AR) is a potential therapeutic target in triple-negative breast cancer (TNBC). We aimed to elucidate the association of AR expression with glucose metabolic features in TNBC.
Methods
Two independent datasets were analyzed: FDG PET data of our institution and a public dataset of GSE135565. In PET analysis, patients with TNBC who underwent pretreatment PET between Jan 2013 and Dec 2017 were retrospectively enrolled. Clinicopathologic features and maximum standardized uptake value (SUVmax) of tumors were compared with AR expression. In GSE135565 dataset, glycolysis score was calculated by the pattern of glycolysis-related genes, and of which association with SUVmax and AR gene expression were analyzed.
Results
A total of 608 female patients were included in the PET data of our institution. SUVmax was lower in AR-positive tumors (P < 0.001) and correlated with lower AR expression (rho = –0.26, P < 0.001). In multivariate analysis, AR was a deterministic factor for low SUVmax (P = 0.012), along with other key clinicopathologic features. In the GSE135565 dataset, AR expression also exhibited a negative correlation with SUVmax (r = –0.34, P = 0.001) and the glycolysis score (r = –0.27, P = 0.013).
Conclusions
Low glucose metabolism is a signature of AR expression in TNBC. It is suggested that evaluation of AR expression status needs to be considered in clinical practice particularly in TNBC with low glucose metabolism.
Introduction
Triple-negative breast cancer (TNBC) is defined as breast cancer without detectable estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2)/neu gene overexpression [1]. Although TNBC comprises different disease entities, it is associated with poor outcomes because of aggressive features of cancer cells and absence of effective targeted therapy [2–4]. Androgen receptor (AR) is deemed to be associated with tumorigenesis in the breast. However, AR-mediated androgenic stimulation has diverse effects in growth of breast cancer cells [5] and the role of AR in breast cancer progression is unclear. Novel treatment targets have been investigated in TNBC, and AR was recently suggested as a potential therapeutic target and a marker for subgrouping of the disease. Recent studies demonstrated that AR-negative TNBC is related to poor disease-free survival and overall survival [6–9] which demonstrates that AR can be a prognostic marker in TNBC. Additionally, AR can be a target of specific treatment and there are several ongoing clinical trials of anti-androgenic agents for breast cancers expressing [10–12].
Glucose metabolism of cancer can be easily evaluated by 18F-fluorodeoxyglucose (FDG) positron emission tomography (PET)/computed tomography (CT) in clinical practice. Glucose metabolism is one of the key characteristics and a significant prognostic marker in many cancers. Thus, the association of glucose metabolism with a specific gene expression or mutation has been investigated regarding crucial genes, such as EGFR or ALK [12, 13]. Although little is known about the exact mechanism, AR is presumed to be related to glucose metabolism in cancer cells, as well as normal tissues [14, 15]. In case of prostate cancer, most cancer cells show enhanced AR expression [16], and AR-driven gene expression enhances β-oxidation of fatty acid (FA) to supply energy source and ATP, and finally, reduces glucose consumption of tumor cells [17, 18]. In clinical setting, the association between AR expression and glucose metabolism in TNBC has not been investigated. Because luminal AR (LAR) subtype of TNBC was reported to show increased lipid metabolism such as FA oxidation [6], we hypothesized that AR expression would affect glucose metabolism in TNBC.
The present study aims to evaluate the association of AR with glucose metabolic features in TNBC by analyzing image phenotypes from patient data of our institution and gene expressions in a public dataset.
Materials and methods
Patients for FDG PET
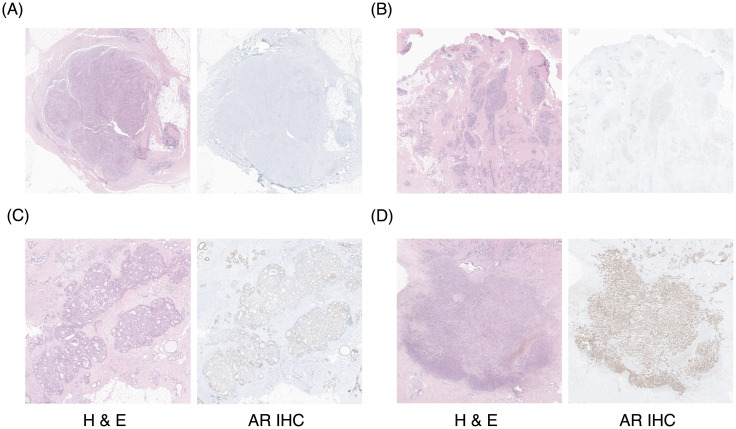
Clinicopathologic data for patients who received surgery for TNBC between January 2013 and December 2017 were retrospectively retrieved from the patient archive of our institution. The inclusion criteria were; baseline FDG PET/CT before surgical resection and any neoadjuvant treatment, and available clinicopathologic data including TNBC and AR status of surgical specimens. Evaluation of ER, PR, HER2, and AR expression for pathologic specimens were routinely performed. Cutoff values for estrogen receptor and progesterone receptor positivity were determined as ≥ 1% on immunohistochemistry, according to the current guidelines [19]. HER2 status was evaluated according to the ASCO/CAP guideline [20], using 4B5 Ventana assay and PathVysion HER-2 DNA probe Kit (Vysis). Immunohistochemistry for AR was carried out from each paraffin block, applying the specific antibody (clone SP107, Rabbit Monoclonal, Roche Diagnostics; ready to use) on automated system (Ventana Benchmark Ultra), according to the manufacturer’s instructions. Representative immunohistochemistry images of various AR expressions are shown in Fig 1. The cutoff value for AR positivity was determined as ≥ 1%. Information on clinicopathologic features such as age, tumor size, lymph node status, pathologic stage, and histologic grade was obtained from medical records review.
Fig 1. Representative immunohistochemistry images of AR expression.
(A) AR expression < 1%, (B) 3%, (C) 65%, (D) 90%.
The protocol of this retrospective study was approved and informed consent from each patient was waived by the Institutional Review Board of Seoul National University Hospital (IRB No.: 1905-136-1035). All methods were performed in accordance with the relevant guidelines and regulations.
PET image acquisition and analysis
All patients fasted for at least 6 hours, and blood glucose levels were confirmed to be < 140 mg/dL. 5.18 MBq/kg (0.14 mCi/kg) of 18F-FDG was intravenously injected, and PET/CT was performed 60 minutes after injection using dedicated PET/CT scanners (Biograph mCT40 or mCT64, Siemens Healthcare). A low-dose CT scan (120 kVp, 50 mAs) was performed first for attenuation correction and anatomical localization, and PET images were obtained from the skull base to the proximal thigh for 1 minute per bed position (6–7 bed positions for a patient). PET images were reconstructed by an iterative algorithm (ordered subset expectation maximization, iteration 2, subset 21). Images were reviewed by two specialists (J.C.P. with 20-year experience and R.L. with 5-year experience), who were unaware of patient and clinical information. On PET/CT fusion images, a volume of interest was drawn carefully to encircle the primary breast tumor and SUVmax was measured using an analysis software package (Syngo.via, Siemens Healthcare) as the index for glucose metabolism.
GEO GSE135565 gene expression dataset analysis
The correlations between SUV, gene expressions for glucose metabolism and AR were additionally tested using a public dataset, the Gene Expression Omnibus (GEO) Series, GSE135565 (https://www.ncbi.nlm.nih.gov/geo/download/?acc=GSE135565). This dataset includes clinical, microarray, and SUVmax data of 84 TNBC patients [21]. Glycolysis signatures were analyzed by using the single sample gene set enrichment analysis (ssGSEA) [22, 23] and metabolic pathway gene information obtained from the Reactome database [24]. ssGSEA for glycolysis score was performed by the Gene Set Variation Analysis (GSVA) package of R/Bioconductor [25]. The glycolysis score (enrichment score) was calculated from the REACTOME_GLYCOLYSIS [26], in which the glycolysis score indicates how close gene expression pattern of a sample is, to expect expression pattern of the glycolysis-related gene set (S1 Table). The enrichment score of FA β-oxidation was calculated from the REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION [27].
Statistical analysis
Values were expressed as mean ± standard deviation (SD) for parametric test or median [interquartile range] for non-parametric test. Chi-square test was used for comparison of AR positivity according to the clinical characteristics. Mann-Whitney U test were used for comparison of SUVmax according to the various clinicopathological features. Tumors were classified into high and low SUVmax groups using the median SUVmax as a cutoff value, and univariate and multivariate logistic regression analyses were performed to select significant clinicopathologic factors for determining tumors of high SUVmax. Pearson’s correlation test was performed to evaluate correlations among AR, SUVmax, and glycolysis-associated genes expressions. SUVmax and AR expression of our institution dataset were analyzed using Spearman correlation test, because of non-normality. Data were analyzed using the R program (ver. 3.4.5). P-values less than 0.05 were deemed to be statistically significant.
Results
Patients for FDG PET analysis
A total of 608 female patients (age 54.2 ± 11.7 y, range 26–93 y) were included in the analysis. Most of the cases were invasive ductal carcinoma (534 patients, 87.8%), and 473 cases (77.8%) were histological grade III. Lymph node metastasis was present in 314 patients (51.6%). Tumor size was larger than 2.0 cm in 402 patients (66.1%). Patient characteristics are summarized in Table 1. Among 608 cases, 216 (35.5%) were AR-positive, and the other 392 (64.5%) were AR-negative. The rate of AR positivity was higher in old age (> 50 years, P = 0.001) and low histologic grade (grade I/II, P < 0.001) (Table 2). There was no significant difference in AR positivity according to tumor size, lymph node metastasis, and overall cancer stage.
Table 1. Patient characteristics.
| Characteristics | N (%) |
|---|---|
| Age | |
| ≤ 50 | 238 (39.1%) |
| > 50 | 370 (60.9%) |
| Tumor stage | |
| I | 152 (25.0%) |
| II | 274 (45.1%) |
| III | 163 (26.8%) |
| IV | 19 (3.1%) |
| T-stage | |
| T1 | 194 (31.9%) |
| T2 | 334 (54.9%) |
| T3 | 40 (6.6%) |
| T4 | 40 (6.6%) |
| N-stage | |
| N0 | 294 (48.4%) |
| N1 | 154 (25.3%) |
| N2 | 101 (16.6%) |
| N3 | 59 (9.7%) |
| Pathologic subtypes | |
| Ductal | 534 (87.8%) |
| Lobular | 5 (0.8%) |
| Metaplastic | 44 (7.2%) |
| Others | 25 (4.2%) |
| Histologic grade | |
| I | 4 (0.7%) |
| II | 127 (21.0%) |
| III | 473 (77.8%) |
Table 2. Androgen receptor expression according to clinicopathologic characteristics.
| Characteristic | AR positivity, N (%) | P |
|---|---|---|
| Overall | 216/608 (35.5%) | |
| Age | 0.001 | |
| ≤ 50 | 66/238 (27.7%) | |
| > 50 | 150/370 (40.5%) | |
| Tumor size | 0.064 | |
| ≤ 2 cm | 84/206 (40.8%) | |
| > 2 cm | 132/402 (32.8%) | |
| Lymph node metastasis | 0.304 | |
| Negative | 111/294 (37.8%) | |
| Positive | 105/314 (33.4%) | |
| Stage | 1.000 | |
| I/II | 151/426 (35.4%) | |
| III/IV | 65/182 (35.7%) | |
| Histologic grade | < 0.001 | |
| I/II | 72/131 (55.0%) | |
| III | 142/473 (30.0%) |
AR: androgen receptor
Association of AR expression with FDG PET findings
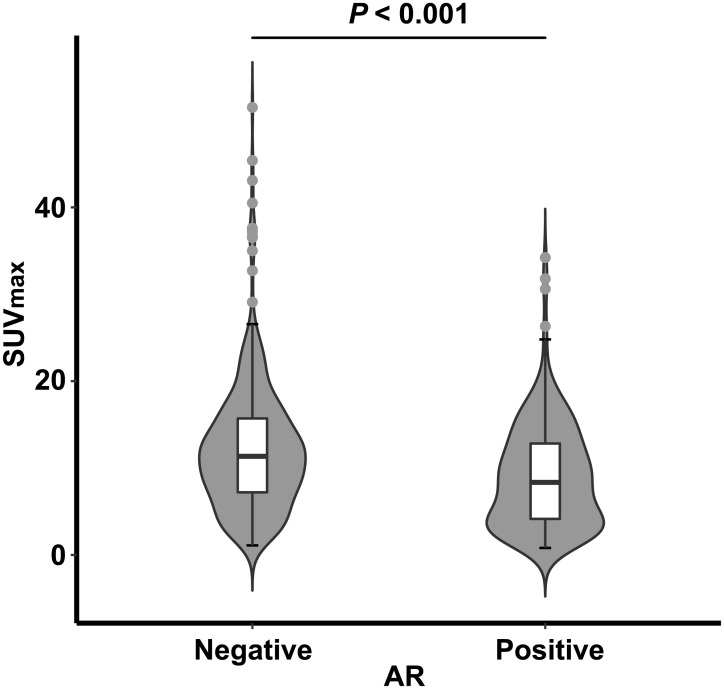
Almost all breast cancer lesions showed high glucose metabolism and average SUVmax was 11.0 ± 7.1 (range 0.8–51.5). SUVmax was significantly higher in tumors of large size, positive lymph node metastasis, high histologic grade, and high Ki-67 index (P < 0.001 for all in Mann-Whitney U test, Table 3). SUVmax was also significantly higher in AR-negative tumors than in AR-positive tumors (P < 0.001, Fig 2 and Table 3). There was a weak but significant negative correlation between SUVmax and the degree of AR positivity (rho = –0.26, P < 0.001 in Spearman test). SUVmax was significantly different between AR-positive and AR-negative tumors, regardless of cutoff values for AR positivity (S2 Table). In a multivariate analysis to select independent factors for determining tumors of low SUVmax (cutoff 10.3, median value of SUVmax), AR expression was selected as a significant factor for determining SUVmax (P = 0.012), along with tumor size (P < 0.001), presence of lymph node metastasis (P < 0.001), and histologic grade (P < 0.001) (Table 4). In additional analysis using a cutoff of 10% for AR positivity, AR was still a significant determinant of SUVmax (S3 Table).
Table 3. FDG uptake according to various clinicopathologic factors.
| Factors | N | Median SUVmax [interquartile range] | P |
|---|---|---|---|
| Age | 0.002 | ||
| ≤ 50 | 238 | 11.0 [7.2–16.1] | |
| > 50 | 370 | 9.7 [5.2–13.8] | |
| Tumor size (cm) | < 0.001 | ||
| ≤ 2.0 | 206 | 5.8 [3.3–10.0] | |
| > 2.0 | 402 | 12.3 [8.5–16.1] | |
| Lymph node metastasis | < 0.001 | ||
| Positive | 314 | 12.0 [8.3–16.1] | |
| Negative | 294 | 8.2 [4.0–12.4] | |
| Stage | < 0.001 | ||
| I/II | 426 | 8.9 [4.8–13.4] | |
| III/IV | 182 | 12.8 [9.3–17.0] | |
| Histologic grade | < 0.001 | ||
| I/II | 131 | 6.4 [3.3–11.5] | |
| III | 473 | 11.1 [7.2–15.7] | |
| Ki-67 | < 0.001 | ||
| High | 264 | 11.4 [7.3–16.1] | |
| Low | 333 | 9.4 [4.7–13.5] | |
| AR expression | < 0.001 | ||
| Positive | 216 | 8.4 [4.1–12.9] | |
| Negative | 392 | 11.4 [7.2–15.7] |
AR, androgen receptor; SUVmax, maximum standardized uptake value
Fig 2. Difference of SUVmax according to AR status in FDG PET analysis.
SUVmax is higher in AR-negative than in AR-positive TNBC. Violin plots indicate the distribution of SUVmax according to the AR positivity. Box plot inside the violin plot denotes median value as a central line within a box, 25 percentile and 75 percentile value as an outline of a box and minimum and maximal number are marked as error bar with outliers marked separately. AR, androgen receptor expression on immunohistochemistry.
Table 4. Univariate and multivariate analyses for determining SUVmax.
| Variables | Univariate analysis | Multivariate analysis (backward deletion) | ||
|---|---|---|---|---|
| Odds ratio (95% CI) | P | Odds ratio (95% CI) | P | |
| Age > 50 y vs. ≤ 50 y) | 0.67 (0.49–0.94) | 0.0184 | eliminated | NA |
| Tumor size (> 2 cm vs. ≤ 2 cm) | 5.27 (3.62–7.68) | < 0.001 | 4.0 (2.65–6.03) | < 0.001 |
| Lymph node metastasis (positive vs. negative) | 2.94 (2.11–4.09) | < 0.001 | 2.07 (1.42–3.02) | < 0.001 |
| Histologic grade (III vs. I/II) | 2.62 (1.74–3.95) | < 0.001 | 2.38 (1.50–3.80) | < 0.001 |
| Ki-67 (> 15 vs. ≤ 15) | 1.72 (1.24–2.38) | 0.001 | 1.36 (0.93–1.97) | 0.112 |
| AR expression (≥ 1% vs. < 1%) | 0.49 (0.35–0.68) | < 0.001 | 0.61 (0.41–0.90) | 0.012 |
CI, confidence interval; AR, androgen receptor; SUVmax, maximum standardized uptake value; Bold p value: statistically significant (p < 0.05) on logistic regression analyses
Association between expressions of AR, glycolysis, and FA metabolism-related genes
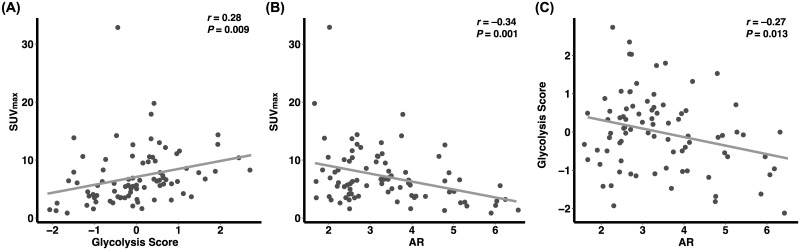
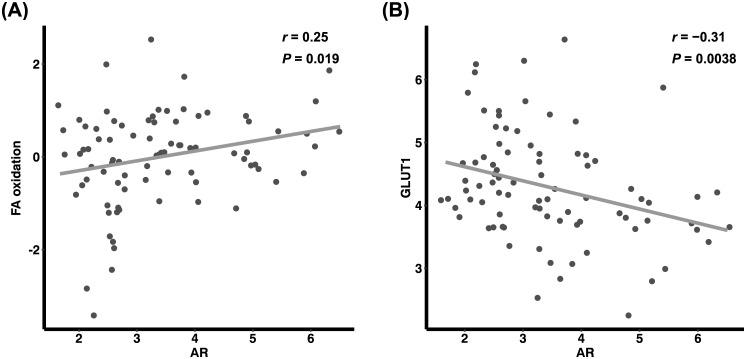
In the analysis of the GSE135565 dataset, glycolysis score exhibited a significant correlation with SUVmax of tumors (r = 0.28, P = 0.009, Fig 3A). In accordance with the results from our patient data analysis, there was a significant negative correlation between SUVmax and AR expression (r = –0.34, P = 0.001, Fig 3B). Additionally, AR expression also exhibited a significant negative correlation with glycolysis score (r = –0.27, P = 0.013, Fig 3C). AR expression showed a significant positive correlation with enrichment score of FA β-oxidation (r = 0.25, P = 0.019, Fig 4A), and a negative correlation with GLUT1 (SLC2A1) expression (r = –0.31, P = 0.004, Fig 4B).
Fig 3. Correlations among AR, SUVmax, and glycolysis-associated genes expressions analyzed using GSE135565 database.
SUVmax was well correlated with glycolysis score (A). A negative correlation between AR expression and SUVmax was observed (B). The AR expression is inversely correlated with glycolysis score (C). AR, expression level of androgen receptor gene; Glycolysis score indicates how close gene expression pattern of a sample is, to expected expression pattern of the glycolysis-related gene set.
Fig 4. Correlations of AR with expression of FA β-oxidation-associated genes and GLUT1.
AR expression show positive correlation with FA oxidation (A) and negative correlation with GLUT1 expression (B). AR, expression level of androgen receptor gene; FA, fatty acid; FA oxidation, gene set enrichment score of FA β-oxidation-associated genes; GLUT1, expression level of GLUT1 (SLC2A1) gene.
Discussion
In this study, it was demonstrated that AR expression is consistently related to low glucose metabolic features. In our patient data, SUVmax was significantly higher in AR-negative tumors than in AR-positive tumors. Additionally, in both datasets, negative correlations were observed between SUVmax and AR expression. In GSE135565 dataset, AR expression also exhibited a negative correlation with the glycolysis score, which is determined by the expression pattern of glycolysis-related genes.
TNBC exhibit aggressive features and poor outcomes [28]. The poor outcome is caused by innate biological features of TNBC, as well as lack of response to common targeted therapies, including anti-hormonal and anti-HER2 agents. FDG PET is a well-known imaging biomarker for glucose metabolic activity, which is related to aggressive features of cancers and poor prognosis. In most cancers, high glucose metabolic activity and high SUVmax on FDG PET is a significant marker for poor prognosis. Breast cancers with high glucose metabolism also have poor prognosis, and TNBC tends to exhibit higher SUVmax than other types of breast cancer [29].
AR is a member of the steroid hormone receptor family, and there are emerging pieces of evidence on the associations between androgen effect and tumorigenesis in breast cancer [30]. Recently, the subclassification of TNBC by AR expression has been suggested [7–9, 31–33]. In the current study, AR was positive in 35.4% of TNBC, and related to young age and low histologic grade of tumors, which is in accordance with previous studies [7, 31–33].
Currently, there is not much information on the relation between AR status and glucose metabolism. Humbert et al. reported that there is a trend that TNBC with positive AR exhibits lower FDG uptake than those with negative AR, in a small study, including 50 TNBC patients [34]. However, statistical significance was not reached in the study, probably due to the small case number. In our analysis of 608 patients, FDG uptake was significantly lower in AR-positive TNBC. AR was an independent factor for determining low SUVmax in multivariate analysis, including other key pathologic factors. The association between FDG uptake and AR status was not affected by varying cutoff value for AR positivity. The results suggest that AR negativity is independently related to aggressive biological features and poor prognosis in TNBC. GSE135565 dataset was additionally analyzed in the present study as a crosscheck for the association between AR expression and glucose metabolism at the gene level. In the analysis, there was a negative correlation between AR expression and SUVmax, which is in agreement with the results from our patient data. Furthermore, a negative correlation was observed between AR and glycolysis score, which suggests that AR expression decreases the glucose metabolism at the gene and cellular level in TNBC. In this dataset, glycolysis score was also well correlated with the image phenotype of SUVmax.
Many studies have reported that positive AR expression is a good prognostic marker and is related to favorable clinical outcomes in breast cancer [7–9, 32, 35]. The findings of the present study also suggest that AR expression is a good prognostic marker by presenting lower glucose metabolism, and presumably, less aggressive biological features. However, there have been some conflicting results on the prognostic effect of AR [32, 36, 37], which needs further investigations. At present, AR testing and AR-targeted therapy are not yet standard of care in TNBC patients. The recognition of low glucose metabolism in a known TNBC might allow additional immunohistochemical evaluation for AR expression and AR-targeted therapy as part of a clinical trial. Hence, another implication of our study is that AR status needs to be considered in TNBC, particularly in cases with low SUVmax.
Specific mechanism or signaling pathway for the correlation between AR and glucose metabolism is unclear. In this study, there was a significant positive correlation between AR expression and FA β-oxidation, which is consistent with the increased lipid metabolism in LAR subtype of TNBC [38]. The shift of metabolic substrate from glycolysis to FA oxidation may be one of the causes of reduced GLUT1 expression, glucose consumption, and decrease in FDG uptake [39]. In case of prostate cancer that expresses abundant androgen receptors, it is well-known that the tumor shows low FDG avidity and its grade is not well correlated with FDG accumulation [17]. Further studies are required on the specific mechanism for the AR effect on glucose metabolism.
There are a few limitations in the present study. First, the patient image analysis was performed in a retrospectively collected cohort, and there might have been an unexpected selection bias. However, because FDG PET/CT and AR immunohistochemistry were performed without specific selection, the possible bias would have been fairly controlled. Second, the prognostic effect of AR expression and FDG uptake was not evaluated because of the insufficient follow-up time. Based on the current study, further prospective studies for the prognostic role of AR expression and FDG uptake are warranted.
Conclusion
AR-positive TNBC tumors show low FDG uptake on PET, which is associated with low expression of glycolysis-related genes. In our multivariate analyses, AR expression was an independent determinant for low glucose metabolism. The results suggest that low glucose metabolism is a signature of AR expression in TNBC, and that AR expression status needs to be considered in clinical practice particularly in TNBC with low glucose metabolism.
Supporting information
(DOCX)
AR: androgen receptor.
(DOCX)
AR: androgen receptor.
(DOCX)
Data Availability
The public datasets we used are available from the NCBI GEO public database (accession number GSE135565). Data of our institution cannot be shared publicly because of the confidentiality restrictions imposed by the approved ethics of study. Data are available from the Institutional Review Board of Seoul National University Hospital (contact via cris@bri.snuh.org) for researchers who meet the criteria for access to confidential data.
Funding Statement
This research was supported the Korea Health Technology R&D Project through Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare, Republic of Korea in the form of a grant (HI14C1277) awarded to JCP. This study was also funded by Chung-Ang University in the form of 2022 research grants awarded to RL. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010; 363: 1938–1948. doi: 10.1056/NEJMra1001389 [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012; 490: 61–70. doi: 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007; 13: 4429–4434. doi: 10.1158/1078-0432.CCR-06-3045 [DOI] [PubMed] [Google Scholar]
- 4.Malorni L, Shetty PB, De Angelis C, Hilsenbeck S, Rimawi MF, Elledge R et al. Clinical and biologic features of triple-negative breast cancers in a large cohort of patients with long-term follow-up. Breast Cancer Res Treat. 2012; 136: 795–804. doi: 10.1007/s10549-012-2315-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birrell SN, Bentel JM, Hickey TE, Ricciardelli C, Weger MA, Horsfall DJ et al. Androgens induce divergent proliferative responses in human breast cancer cell lines. J Steroid Biochem Mol Biol. 1995; 52: 459–467. doi: 10.1016/0960-0760(95)00005-k [DOI] [PubMed] [Google Scholar]
- 6.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rakha EA, El-Sayed ME, Green AR, Lee AH, Robertson JF, Ellis IO. Prognostic markers in triple-negative breast cancer. Cancer. 2007; 109: 25–32. doi: 10.1002/cncr.22381 [DOI] [PubMed] [Google Scholar]
- 8.Asano Y, Kashiwagi S, Goto W, Tanaka S, Morisaki T, Takashima T et al. Expression and clinical significance of androgen receptor in triple-negative breast cancer. Cancers. 2017; 9: 4. doi: 10.3390/cancers9010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sutton LM, Cao D, Sarode V, Molberg KH, Torgbe K, Haley B et al. Decreased Androgen Receptor Expression Is Associated With Distant Metastases in Patients With Androgen Receptor–Expressing Triple-Negative Breast Carcinoma. Am J Clin Pathol. 2012; 138: 511–516. doi: 10.1309/AJCP8AVF8FDPTZLH [DOI] [PubMed] [Google Scholar]
- 10.Traina TA, Miller K, Yardley DA, Eakle J, Schwartzberg LS, O’Shaughnessy J et al. Enzalutamide for the Treatment of Androgen Receptor-Expressing Triple-Negative Breast Cancer. J Clin Oncol. 2018; 36: 884–890. doi: 10.1200/JCO.2016.71.3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bardia A, Gucalp A, DaCosta N, Gabrail N, Danso M, Ali H et al. Phase 1 study of seviteronel, a selective CYP17 lyase and androgen receptor inhibitor, in women with estrogen receptor-positive or triple-negative breast cancer. Breast Cancer Res Treat. 2018; 171: 111–120. doi: 10.1007/s10549-018-4813-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi H, Paeng JC, Kim DW, Lee JK, Park CM, Kang KW et al. Metabolic and metastatic characteristics of ALK-rearranged lung adenocarcinoma on FDG PET/CT. Lung Cancer. 2013; 79: 242–247. doi: 10.1016/j.lungcan.2012.11.021 [DOI] [PubMed] [Google Scholar]
- 13.Park S, Ha S, Lee SH, Paeng JC, Keam B, Kim TM et al. Intratumoral heterogeneity characterized by pretreatment PET in non-small cell lung cancer patients predicts progression-free survival on EGFR tyrosine kinase inhibitor. PLoS One. 2018; 13: e0189766. doi: 10.1371/journal.pone.0189766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Navarro G, Xu W, Jacobson DA, Wicksteed B, Allard C, Zhang G et al. Extranuclear Actions of the Androgen Receptor Enhance Glucose-Stimulated Insulin Secretion in the Male. Cell Metab. 2016; 23: 837–851. doi: 10.1016/j.cmet.2016.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Massie CE, Lynch A, Ramos-Montoya A, Boren J, Stark R, Fazli L et al. The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J. 2011; 30: 2719–2733. doi: 10.1038/emboj.2011.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bluemn EG, Coleman IM, Lucas JM, Coleman RT, Hernandez-Lopez S, Tharakan R et al. Androgen Receptor Pathway-Independent Prostate Cancer Is Sustained through FGF Signaling. Cancer Cell. 2017; 32: 474–489.e6. doi: 10.1016/j.ccell.2017.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y. Fatty acid oxidation is a dominant bioenergetic pathway in prostate cancer. Prostate Cancer and Prostatic Dis. 2006; 9: 230–234. doi: 10.1038/sj.pcan.4500879 [DOI] [PubMed] [Google Scholar]
- 18.Zha S, Ferdinandusse S, Hicks JL, Denis S, Dunn TA, Wanders RJ et al. Peroxisomal branched chain fatty acid Beta-oxidation pathway is upregulated in prostate cancer. The Prostate. 2005; 63: 316–323. doi: 10.1002/pros.20177 [DOI] [PubMed] [Google Scholar]
- 19.Allison KH, Hammond MEH, Dowsett M, McKernin SE, Carey LA, Fitzgibbons PL et al. Estrogen and Progesterone Receptor Testing in Breast Cancer: ASCO/CAP Guideline Update. J Clin Oncol. 2020; 38: 1346–1366. doi: 10.1200/JCO.19.02309 [DOI] [PubMed] [Google Scholar]
- 20.Wolff AC, Hammond MEH, Schwartz JN, Hagerty KL, Allred DC, Cote RJ et al. American Society of Clinical Oncology/College of American Pathologists Guideline Recommendations for Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer. J Clin Oncol. 2007; 25: 118–145. doi: 10.1200/JCO.2006.09.2775 [DOI] [PubMed] [Google Scholar]
- 21.Kim SK, Ahn SG, Mun JY, Jeong MS, Bae SJ, Lee JS et al. Genomic Signature of the Standardized Uptake Value in 18F-Fluorodeoxyglucose Positron Emission Tomography in Breast Cancer. Cancers (Basel). 2020;12:497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009; 462: 108–112. doi: 10.1038/nature08460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005; 102: 15545–15550. doi: 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014; 42: D472–7. doi: 10.1093/nar/gkt1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanzelmann S CR, Guinney J. GSVA: The Gene Set Variation Analysis package for microarray and RNA-seq data. 2019. http://bioconductor.org/packages/release/bioc/vignettes/GSVA/inst/doc/GSVA.pdf. [DOI] [PMC free article] [PubMed]
- 26.Gene Set: REACTOME_GLYCOLYSIS. https://www.gsea-msigdb.org/gsea/msigdb/cards/REACTOME_GLYCOLYSIS.
- 27.Gene Set: REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION. https://www.gsea-msigdb.org/gsea/msigdb/cards/REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION.
- 28.Cheang MC, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK et al. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008; 14: 1368–1376. doi: 10.1158/1078-0432.CCR-07-1658 [DOI] [PubMed] [Google Scholar]
- 29.Basu S, Chen W, Tchou J, Mavi A, Cermik T, Czerniecki B et al. Comparison of triple-negative and estrogen receptor-positive/progesterone receptor-positive/HER2-negative breast carcinoma using quantitative fluorine-18 fluorodeoxyglucose/positron emission tomography imaging parameters: a potentially useful method for disease characterization. Cancer. 2008; 112: 995–1000. doi: 10.1002/cncr.23226 [DOI] [PubMed] [Google Scholar]
- 30.Gucalp A, Traina TA. Triple-negative breast cancer: role of the androgen receptor. Cancer J. 2010; 16: 62–65. doi: 10.1097/PPO.0b013e3181ce4ae1 [DOI] [PubMed] [Google Scholar]
- 31.Park S, Koo J, Park HS, Kim J-H, Choi S-Y, Lee JH et al. Expression of androgen receptors in primary breast cancer. Ann Oncol. 2009; 21: 488–492. doi: 10.1093/annonc/mdp510 [DOI] [PubMed] [Google Scholar]
- 32.McGhan LJ, McCullough AE, Protheroe CA, Dueck AC, Lee JJ, Nunez-Nateras R et al. Androgen receptor-positive triple negative breast cancer: a unique breast cancer subtype. Ann Surg Oncol. 2014;21:361–367. doi: 10.1245/s10434-013-3260-7 [DOI] [PubMed] [Google Scholar]
- 33.Vidula N, Yau C, Wolf D, Rugo HS. Androgen receptor gene expression in primary breast cancer. NPJ Breast Cancer. 2019;5:47. doi: 10.1038/s41523-019-0142-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Humbert O, Riedinger JM, Charon-Barra C, Berriolo-Riedinger A, Desmoulins I, Lorgis V et al. Identification of Biomarkers Including 18FDG-PET/CT for Early Prediction of Response to Neoadjuvant Chemotherapy in Triple-Negative Breast Cancer. Clin Cancer Res. 2015; 21: 5460–5468. doi: 10.1158/1078-0432.CCR-15-0384 [DOI] [PubMed] [Google Scholar]
- 35.Lehmann BD, Jovanović B, Chen X, Estrada MV, Johnson KN, Shyr Y et al. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS One. 2016; 11: e0157368. doi: 10.1371/journal.pone.0157368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dieci MV, Tsvetkova V, Griguolo G, Miglietta F, Mantiero M, Tasca G et al. Androgen Receptor Expression and Association With Distant Disease-Free Survival in Triple Negative Breast Cancer: Analysis of 263 Patients Treated With Standard Therapy for Stage I-III Disease. Front Oncol. 2019; 9: 452. doi: 10.3389/fonc.2019.00452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu M, Yuan Y, Yan P, Jiang J, Ma P, Niu X et al. Prognostic Significance of Androgen Receptor Expression in Triple Negative Breast Cancer: A Systematic Review and Meta-Analysis. Clin Breast Cancer. 2020; 20: e385–e396. doi: 10.1016/j.clbc.2020.01.002 [DOI] [PubMed] [Google Scholar]
- 38.Gong Y, Ji P, Yang Y-S, Xie S, Yu T-J, Xiao Y et al. Metabolic-Pathway-Based Subtyping of Triple-Negative Breast Cancer Reveals Potential Therapeutic Targets. Cell Metab. 2021; 33: 51–64.e9. doi: 10.1016/j.cmet.2020.10.012 [DOI] [PubMed] [Google Scholar]
- 39.Smith TA. Facilitative glucose transporter expression in human cancer tissue. Br J Biomed Sci. 1999; 56: 285–292. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
AR: androgen receptor.
(DOCX)
AR: androgen receptor.
(DOCX)
Data Availability Statement
The public datasets we used are available from the NCBI GEO public database (accession number GSE135565). Data of our institution cannot be shared publicly because of the confidentiality restrictions imposed by the approved ethics of study. Data are available from the Institutional Review Board of Seoul National University Hospital (contact via cris@bri.snuh.org) for researchers who meet the criteria for access to confidential data.