Abstract
The rpoS gene encodes the sigma factor which was identified in several gram-negative bacteria as a central regulator during stationary phase. rpoS gene regulation is known to respond to cell density, showing higher expression in stationary phase. For Pseudomonas aeruginosa, it has been demonstrated that the cell-density-dependent regulation response known as quorum sensing interacts with this regulatory response. Using the rpoS promoter of P. putida, we identified a genomic Tn5 insertion mutant of P. putida which showed a 90% decrease in rpoS promoter activity, resulting in less RpoS being present in a cell at stationary phase. Molecular analysis revealed that this mutant carried a Tn5 insertion in a gene, designated psrA (Pseudomonas sigma regulator), which codes for a protein (PsrA) of 26.3 kDa. PsrA contains a helix-turn-helix motif typical of DNA binding proteins and belongs to the TetR family of bacterial regulators. The homolog of the psrA gene was identified in P. aeruginosa; the protein showed 90% identity to PsrA of P. putida. A psrA::Tn5 insertion mutant of P. aeruginosa was constructed. In both Pseudomonas species, psrA was genetically linked to the SOS lexA repressor gene. Similar to what was observed for P. putida, a psrA null mutant of P. aeruginosa also showed a 90% reduction in rpoS promoter activity; both mutants could be complemented for rpoS promoter activity when the psrA gene was provided in trans. psrA mutants of both Pseudomonas species lost the ability to induce rpoS expression at stationary phase, but they retained the ability to produce quorum-sensing autoinducer molecules. PsrA was demonstrated to negatively regulate psrA gene expression in Pseudomonas and in Escherichia coli as well as to be capable of activating the rpoS promoter in E. coli. Our data suggest that PsrA is an important regulatory protein of Pseudomonas spp. involved in the regulatory cascade controlling rpoS gene regulation in response to cell density.
The rpoS gene codes for sigma factor RpoS (also called ςs and ς38) (12, 18), which was identified as a central regulator during stationary phase in Escherichia coli; this factor is involved in the survival of famine conditions and in cross-protection against osmotic, acidic, oxidative, and heat stresses (19, 22). Since then, it has been identified in various gram-negative bacteria, including several species belonging to the fluorescent pseudomonads; these findings demonstrate that in these bacteria as well this factor has an important regulatory role, including adaptation to nutrient-limiting conditions, survival in the presence of several environmental stresses, and the production of virulence factors (16, 17, 34, 37, 42, 43).
RpoS is an alternative sigma factor, resulting in the alteration of RNA polymerase core specificity and thereby switching gene expression at stationary phase. The levels of RpoS within a bacterial cell are carefully controlled, since perturbations in the relative amounts can have severe consequences. Thus, the regulation of RpoS levels is of crucial importance (8). In E. coli, the levels of RpoS are extremely low in exponential growth phase but increase markedly upon entry into stationary phase (15). The regulation of RpoS in E. coli remains a subject of extensive investigation, since regulation occurs at the transcriptional, posttranscriptional, and protein levels (6, 13, 20). At the transcriptional level, it has been observed that in rich media there is a considerable increase in transcription at the transition to stationary phase and that the cyclic AMP-cyclic AMP receptor protein complex is involved either directly or indirectly in this regulation. However, RpoS levels in E. coli appear to be regulated mainly at the posttranscriptional level through susceptibility to proteolysis. RpoS is rapidly degraded by the ClpXP protease (29, 38); this degradation absolutely requires the phosphorylated form of a two-component response regulator called SprE or RssB (33), which in turn is modulated by a LysR regulatory family protein called LrhA (13). It is still unclear which signals and effector molecules trigger the regulators responsible for the regulation of this RpoS proteolysis.
The regulation of rpoS in Pseudomonas has also been addressed recently. The cell-cell communication device, called quorum sensing (10), used by gram-negative bacteria to regulate several physiological processes has been demonstrated to be involved in the control of rpoS transcription in Pseudomonas aeruginosa (21). There are at least two chemically and genetically independent quorum-sensing systems in P. aeruginosa, designated the LasR-LasI and the RhlR-RhlI systems, each having a cognate N-acylhomoserine lactone (AHL) (21); these systems are involved in the regulation of a large number of exoproducts in response to cell density. It has been reported that the LasR-LasI elements regulate the activation at the transcriptional level of the RhlR-RhlI system, which then directs the regulation of rpoS transcription, resulting in RpoS accumulation in response to cell density (21). A recent study, however, reports that RpoS regulates rhlI transcription (45); thus, RpoS and the quorum-sensing system in P. aeruginosa are part of the same regulatory network. Future work is required to precisely define the molecular events leading to rpoS regulation. In P. fluorescens Pf-5, the GacS-GacA two-component regulatory system positively influences rpoS expression (44). This two-component system is well conserved in Pseudomonas spp. and regulates the production of several secondary metabolites.
The rpoS gene of P. putida WCS358 has been identified (17). In this study, we used rpoS-reporter gene transcriptional fusions to identify a Tn5 mutant of strain WCS358 that had considerably reduced rpoS expression. This mutant had a Tn5 insertion in a regulatory gene (designated psrA) coding for a protein (designated PsrA) belonging to the TetR regulatory family. A homolog of this gene in P. aeruginosa encodes a protein having 90% identity with PsrA of P. putida. This gene was inactivated by transposon mutagenesis and homologous recombination; the resulting mutant also showed a 90% reduction in rpoS promoter activity. Our data suggest that in Pseudomonas spp., psrA plays an important role in rpoS expression.
MATERIALS AND METHODS
Strains, plasmids, media, and chemicals.
The strains used in this study included E. coli HB101 (35), DH5α (14), and XL1-Blue (4); P. putida WCS358, a plant-growth-promoting strain isolated from the rhizosphere of potato roots (11); P. aeruginosa PAO1 (Holloway collection); and Chromobacterium violaceum CVO26, a double mini-Tn5 mutant derived from ATCC 31532. This mutant is nonpigmented, and production of the purple pigment can be induced by providing exogenous AHL inducer molecules (27). E. coli and P. aeruginosa PAO1 were grown in LB medium (28) at 37°C, whereas P. putida WCS358 was cultured in LB medium or in M9 minimal medium (35) at 30°C. The following antibiotic concentrations were used: tetracycline, 10 μg/ml (E. coli), 40 μg/ml (strain WCS358), and 300 μg/ml (PAO1); kanamycin, 100 and 300 μg/ml (PAO1); ampicillin, 100 μg/ml; nalidixic acid, 25 μg/ml; chloramphenicol, 25 μg/ml (E. coli), 250 μg/ml (strain WCS358), and 500 μg/ml (PAO1); and gentamicin, 10 μg/ml (E. coli), 40 μg/ml (strain WCS358), and 60 μg/ml (PAO1). The plasmids used in this study are listed in Table 1.
TABLE 1.
Plasmids used
| Plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| pUC18 | Apr; ColE1 replicon | 47 |
| pBluescript KS | Apr; ColE1 replicon | Stratagene |
| pBluescript SK | Apr; ColE1 replicon | Stratagene |
| pQE30 | Apr; ColE1 replicon; His6 expression vector | Qiagen |
| pQE31 | Apr; ColE1 replicon; His6 expression vector | Qiagen |
| pREP-4 | lacI; Kmr; p15A replicon | Qiagen |
| pRK2013 | Kmr Tra+ Mob+; ColE1 replicon | 9 |
| pPH1JI | IncP1; Gmr | 1 |
| pMP220 | Promoter probe vector; IncP1; Tcr | 39 |
| pMP77 | Promoter probe vector; IncQ; Cmr | 39 |
| pLAFR3 | Broad-host-range cloning vector; IncP1; Tcr | 41 |
| pH3.5 | rpoS gene of strain WCS358 in pBluescript | 17 |
| pSB1075 | Apr; ColE1 replicon; AHL biosensor | 46 |
| pMK962 | rpoS promoter cloned in pCU18 | This study |
| pRPO77 | rpoS promoter cloned in pMP77 | This study |
| pRPO220A | rpoS incomplete promoter cloned in pMP220 | This study |
| pRPO220B | rpoS promoter cloned in pMP220 | This study |
| pPPSR18 | psrA promoter cloned in pUC18 | This study |
| pPPSR220 | psrA promoter cloned in pMP220 | This study |
| pCOS17 | pLAFR3 containing WCS358 DNA | This study |
| pCOS18 | pLAFR3 containing WCS358 DNA | This study |
| pAPC17 | pUC18 containing 680 bp of P. putida MT17 DNA bordering Tn5 | This study |
| pBSH10 | pBluescript containing 10-kb HindIII fragment from pCOS17 | This study |
| pBS25 | pBluescript containing 2.5-kb PstI fragment from pCOS17 | This study |
| pLM17E | pLAFR3 containing 5-kb EcoRI fragment from pCOS17 | This study |
| pLM17P | pLAFR3 containing 2.5-kb PstI fragment from pCOS17 | This study |
| pMKP25 | pMP77 containing 2.5-kb PstI fragment from pCOS17 | This study |
| pPSRPAO1 | 2.2-kb PCR EcoRI fragment from PAO1 genome harboring psrA in pBluescript | This study |
| pPSRPAO1::Tn5 | pPSRPAO1 with Tn5 insertion in psrA | This study |
| pSRE8 | pLAFR3 harboring 8-kb EcoRI fragment from pPSRPAO1::Tn5 | This study |
| pBS25::Tn5R | pBS25 with Tn5 insertion in psrA | This study |
| pCOS17::Tn5R | pCOS17 with Tn5 insertion in psrA | This study |
| pQEPSRA | psrA cloned in pQE30 | This study |
| pQERPOS | rpoS cloned in pQE31 | This study |
Apr, Kmr, Smr, Tcr, Gmr, and Cmr, resistant to ampicillin, kanamycin, streptomycin, tetracycline, gentamicin, and chloramphenicol, respectively.
The rpoS promoter transcriptional fusions were constructed as follows. First, a 135-bp fragment consisting of the −9 to −144 DNA region, where position 0 is the ATG codon of the rpoS gene of P. putida WCS358 (17), was cloned into promoter probe vector pMP220 by making use of two synthetic oligonucleotides (5′-CCTTTGCTGCAGTTCGAACTCAGA-3′ and 5-′CCCGTGGATCCACTCCAGTTCCTG-3′). One oligonucleotide had a PstI restriction site inserted and the other had a BamHI site, and they were cloned into the BglII and PstI sites in pMP220 to yield pRPO220A. Second, a HindIII-AatII DNA fragment of 962 bp obtained from plasmid pH3.5 and containing 920 bp of DNA upstream of the ATG of the rpoS gene of strain WCS358 (17) was end filled and cloned into the SmaI site of pUC18 to yield pMK962. pMK962 was then digested with BamHI and KpnI, and the 974-bp fragment was cloned into pMP220 and pMP77, both digested with BglII and KpnI, to yield pRPO220B and pRPO77, respectively.
Plasmid pPSRPAO1 harbors a 2.2-kb fragment of PAO1 genomic DNA. It was constructed by amplifying by PCR, using the PAO1 genome as a template and two oligonucleotides (5′-ACCTTGCTGAATTCGCGCTTGAAGCG-3′ and 5′-CGCCACATGGGAATTCGGCCTCGGCC-3′), a 2.2-kb fragment located at positions 3368363 to 3370563 in the PAO1 genome (www.pseudomonas.com). This fragment was cloned as an EcoRI (the two oligonucleotides used harbored EcoRI restriction sites) fragment of pBluescript to yield pPSRPAO1.
The psrA promoter was cloned into promoter probe vector pMP220 as follows. An XmnI-SacII fragment of 435 bp (Fig. 1) containing the psrA promoter was blunted by end filling and cloned into the SmaI site of pUC18 to generate pPPSR18. The promoter was then removed as a BamHI-KpnI fragment and cloned into BglII-KpnI-digested pMP220 to yield pPPSR220. The psrA gene was cloned into expression vector pQE30 as follows. The gene was amplified by PCR using two oligonucleotides (psrA-BamHI-start, 5′-GGAATAATCGGATCCCAATCGGAAACCG-3′, and psrA-HindIII-end, 5′-GCGCAAGCTTAGCCGAAGCGCCCTGCCCC-3′) and cloned as a BamHI-HindIII fragment into the corresponding sites of pQE30, yielding pQEPSRA and resulting in psrA being in frame with the six histidines. The P. putida WCS358 rpoS gene was cloned into expression vector pQE31 as follows. The gene was amplified by PCR using two oligonucleotides (rpoS-BamHI-start, 5′-CTATAACAATGGATCCCAATAAAGAAGCGCC-3′ and rpoS-HindIII-end, 5′-GTCTTAAGCTTGCGAACAGCGTATTACTGG-3′) and cloned as a BamHI-HindIII fragment into the corresponding sites of pQE31, yielding pQERPOS.
FIG. 1.
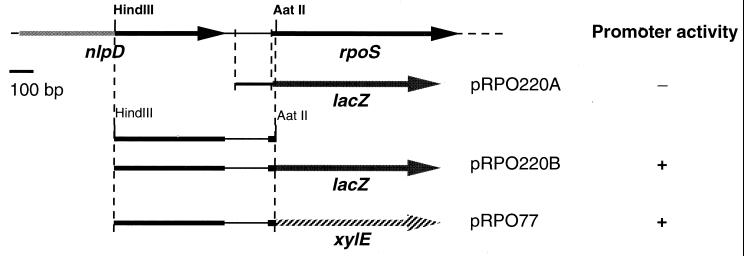
Strategy for construction of rpoS-lacZ and rpoS-xylE transcriptional fusions. The genetic map shows the location of rpoS in the P. putida WCS358 genome, as described by Kojic et al. (17). Plasmid constructs pRPO220A and pRPO220B are derivatives of promoter probe vector pMP220. pRPO77 is derived from pMP77 (see the text for details). Also shown is whether the transcriptional fusion had promoter activity, as detected using the reporter gene. Plasmid construct pRPO220A contains the rpoS-nlpD intergenic region cloned upstream of a promoterless lacZ gene. Plasmid constructs pRPO220B and pRPO77 contain the indicated 962-bp fragment cloned upstream of promoterless lacZ and xylE genes, respectively (see the text for details).
Recombinant DNA techniques.
Digestion with restriction enzymes, agarose gel electrophoresis, purification of DNA fragments, ligation with T4 DNA ligase, end filling with the Klenow fragment of DNA polymerase, Southern hybridization, transformation of E. coli, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis were performed as described by Sambrook et al. (35). Analytical amounts of plasmids were isolated as described by Birnboim (3), whereas preparative amounts were purified with Qiagen columns. Total DNA from Pseudomonas was isolated by Sarkosyl-pronase lysis as described by Better et al. (2). Triparental matings from E. coli to Pseudomonas were performed with an E. coli(pRK2013) helper strain (9). The DNA sequence flanking transposon mutant P. putida MT17 was determined using arbitrary PCR. In this procedure, the DNA flanking the Tn5 insertion site was enriched in two rounds of amplification using primers specific to the ends of the Tn5 element and primers of random sequence which annealed to chromosomal sequences flanking the transposon as described by O' Toole and Kolter (31).
Protein expression, analysis, and purification and antibodies against PsrA and RpoS.
Expression and purification of His6-PsrA (pQEPSRA) and His6-RpoS (pQERPOS) were carried out with E. coli M15(pREP-4) according to the instructions of the supplier (Qiagen, Hilden, Germany). Proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (Immobilon-P; Millipore Corp.) using a tank system according to the manufacturer's instructions. The membrane was subjected to Western blot analysis using polyclonal antibodies against either RpoS or PsrA. After incubation with the secondary horseradish peroxidase-labeled antibody, the proteins were detected with 3-3′-diaminobenzidine tetrahydrochloride tablets (Sigma, St. Louis, Mo.). Antibodies against PsrA and RpoS were produced in rabbits by injecting the purified protein. No significant cross-reaction was observed in this study.
Reporter gene fusion assays; purification, detection, and visualization of autoinducer molecules; pyocyanin quantification; and stress response assays.
β-Galactosidase activity was determined essentially as described by Miller (28) with the modifications of Stachel et al. (40). The purification, detection, and visualization of AHL inducer molecules from culture supernatants were performed essentially as described by McClean et al. (27) and Kojic et al. (17). C. violaceum CVO26 and E. coli(pSB1075) were used as the indicator strains for C4-homoserine lactone (HSL) and 3-oxo-C12-HSL, respectively, on thin-layer chromatography plates to detect the presence of AHL molecules (27, 46). As controls, chemically synthesized C4-HSL and 3-oxo-C12-HSL (kindly donated by P. Williams) were used. Pyocyanin production was measured as described by Essar et al. (7) The measurement of cell viability to survive heat stress and osmotic stress and sensitivity to hydrogen peroxide were determined as described by Kojic et al. (17).
Identification of rpoS regulatory mutants and their complementation with a WCS358 gene bank.
About 200 cells per plate of a Tn5 mutant bank of strain WCS358 (26), harboring promoter fusion pRPO77, were spread on LB plates containing kanamycin (100 μg/ml) and chloramphenicol (250 μg/ml). These plates were incubated for 2 days at 30°C before being sprayed with a 0.1 M catechol solution. Colonies with a decrease in yellow color were purified and further studied. Tn5 mutants which had reduced promoter activity were complemented for promoter activity as follows. About 4 × 109 cells each of E. coli HB101 harboring the P. putida WCS358 gene bank in pLAFR3 and E. coli(pRK2013) and 2 × 108 cells of mutant MT17(pRPO77) were mixed, and the suspension was applied to a 0.45-μm-pore-size membrane filter (Millipore) on an LB plate. The plate was incubated overnight at 30°C before the cells were resuspended and plated on LB plates containing nalidixic acid (25 μg/ml), ampicillin (100 μg/ml), kanamycin (100 μg/ml), chloramphenicol (250 μg/ml), and tetracycline (40 μg/ml). These plates were incubated for 2 days at 30°C, and the cells were resuspended and pooled. About 100 cells were spread on LB plates containing chloramphenicol (250 μg/ml), kanamycin (100 μg/ml), and tetracycline (40 μg/ml). These plates were incubated at 30°C for 2 days before being sprayed with a 0.1 M catechol solution. Transconjugants that turned yellow were purified and further assayed.
Construction of a psrA mutant of P. putida WCS358.
Transposon Tn5 insertions within recombinant plasmid pBS25 were obtained as described by Magazin et al. (25) with E. coli HB101::Tn5 as the source of the transposon. E. coli HB101 cells containing Tn5 insertions within plasmid pBS25 were identified by purifying plasmid DNA from HB101::Tn5(pBS25), using it to transform E. coli DH5α, and selecting for plasmids having ampicillin and kanamycin resistance. Transposon insertions in the psrA gene of P. putida were then mapped by restriction and Southern analyses and DNA sequencing using a Tn5-based primer (5′-GAACGTTACCATGTTAGGAGGTC-3′). One Tn5 insertion in the psrA gene in pBS25 was identified (the plasmid was designated pBS25::Tn5R) and was located 428 bp downstream of the putative ATG codon of the P. putida psrA gene (see Fig. 3A). This Tn5 insertion in pBS25 was transferred to the psrA gene harbored in pCOS17 by homologous recombination in the following way. pBS25::Tn5 was used to transform HB101(pCOS17), and the resulting construct, HB101(pCOS17)(pBS25::Tn5), was grown overnight. The culture was used for a triparental conjugation into P. putida WCS358 with E. coli(pRK2013) as a helper. After appropriate selection, pCOS17::Tn5R was selected. It was verified that Tn5 had been transferred by double-crossover homologous recombination from pBS25::Tn5 to pCOS17. Plasmid pCOS17::Tn5 was then used in a marker exchange technique (5) in order to introduce insertion mutations site specifically with the psrA gene of P. putida; in this experiment, pPH1JI was used as the incoming incompatible plasmid as previously described (1). The fidelity of the marker exchange event in the P. putida psrA::Tn5 mutant was confirmed by Southern analysis (data not shown). This mutant was designated P. putida M17R.
FIG. 3.
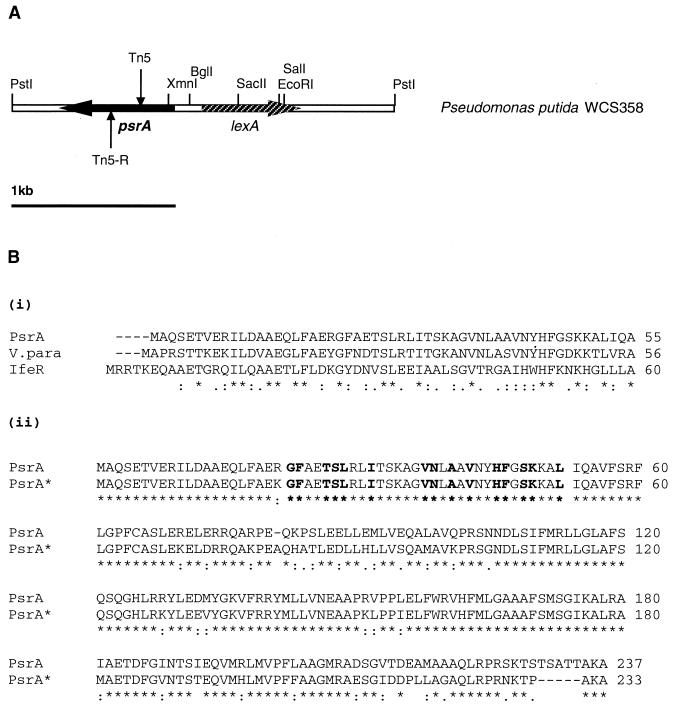
Gene map and protein sequence alignments. (A) Genetic organization of the region (PstI fragment; see text for details) containing the psrA regulatory gene in P. putida WCS358. Shown is the Tn5 insertion position located in mutant MT17. Also shown is the Tn5 insertion (shown as Tn5-R) located in the regenerated psrA mutant, designated MT17R. The DNA sequence of this 2.5-kb fragment can be found in GenBank accession number AJ293485. (B) (i) Protein sequence alignment, using the one-letter code, of the first 60 amino acids of PsrA, a TetR regulator of V. parahaemolyticus (V.para) (GenBank accession number Q56726), and IfeR of A. tumefaciens (GenBank accession number O68442). (ii) Protein sequence alignment, using the one-letter code, of the PsrA proteins from P. putida (PsrA) and P. aeruginosa (PsrA*) (this study). In bold are shown the amino acids which constitute the TetR family signature pattern (Prosite accession number PS01081). A star indicates conservation of identical amino acids, a colon indicates a conserved substitution, and a period indicates a semiconserved substitution.
Construction of a psrA::Tn5 mutant of P. aeruginosa PAO1.
Transposon Tn5 insertions within recombinant plasmid pPSRPAO1 were obtained as described by Magazin et al. (25) with E. coli HB101::Tn5 as the source of the transposon as described above. One Tn5 insertion in the psrA gene in pPSRPAO1 was identified (the plasmid was designated pPSRPAO::Tn5) and was located 478 bp downstream of the putative ATG codon of the PAO1 psrA gene. Plasmid pPSRPAO1::Tn5 was digested with EcoRI, and the 8-kb fragment harboring the 2.2-kb EcoRI fragment of PAO1 DNA with a Tn5 insertion was cloned into the corresponding site of pLAFR3 to yield pPSRE8. This plasmid was conjugated into P. aeruginosa PAO1 and used in a marker exchange technique (5) in order to introduce insertion mutations site specifically with the psrA gene of P. aeruginosa PAO1 (1). The fidelity of the marker exchange event in the PAO1 psrA::Tn5 mutant was confirmed by Southern analysis (data not shown).
DNA sequence determination and analysis.
DNA segments of various sizes were created from plasmid pBS25 using a nested deletion kit (Amersham-Pharmacia). These constructs were either encapsidated as single-stranded DNA upon infection with helper phage VCSM13 (Stratagene Co.) or used directly for DNA sequencing. Nucleotide sequences were determined by the dideoxy chain termination method (36) using [α-35S]dATP for labeling and 7-deaza-dGTP (Amersham-Pharmacia) instead of dGTP. The DNA fragments were separated with a Bio-Rad electrophoresis kit. The nucleotide sequences presented here were determined in both orientations and across all restriction sites.
Nucleotide sequence accession number.
The GenBank/EMBL/DDBJ accession number for the sequence reported in this paper is AJ293485.
RESULTS
Isolation and characterization of P. putida WCS358 regulatory mutants affected in rpoS expression.
Two transcriptional fusions were constructed using the P. putida WCS358 rpoS promoter and a promoterless lacZ gene as described in Materials and Methods (Fig. 1). The first fusion (pRPO220A) contained a 135-bp fragment of the rpoS promoter starting from 9 bp upstream of the ATG and reaching 17 bp away from the stop codon of the nlpD gene, which is located upstream and transcribed in the same orientation as the rpoS gene (Fig. 1). The second fusion (pRPO220B) contained 920 bp upstream of the ATG codon, as depicted in Fig. 1. The first fusion did not show any β-galactosidase activity; thus, there was no promoter element in the nlpD-rpoS intergenic region. However, the second, larger construct displayed significant promoter activity, demonstrating that the rpoS promoter was contained in this fragment within the nlpD gene (Fig. 1). This same fragment was cloned in promoter probe vector pMP77, yielding pRPO77 (Fig. 1), which contained a promoterless xylE gene. This procedure provided a convenient plate assay for detecting promoter activity compared to the more sensitive detection of β-galactosidase activity.
A transposon Tn5 insertion mutant bank of P. putida strain WCS358 harboring promoter fusion pRPO77 was assayed for promoter activity. With this construct, promoter activity can be conveniently detected on plates by assaying for the xylE gene product, catechol 2,3-dioxygenase (XylE), which converts catechol to a yellow product. Bacterial colonies exhibiting promoter activity become yellow when sprayed with a catechol solution. Ten thousand mutants were screened, and 1 mutant, designated MT17, showed very little yellow coloration after being sprayed with a catechol solution. In order to confirm the reduced rpoS promoter activity, plasmid pRPO77 was cured from mutant MT17 by growth for several generations in the absence of antibiotics and then selection for a chloramphenicol-sensitive colony. pRPO77 was then conjugated into the mutants, and it was reconfirmed that MT17(pRPO77) displayed less yellow color than wild-type WCS358(pRPO77) when sprayed with a catechol solution.
The activity of the rpoS promoter was quantified in mutant MT17 after the introduction of plasmid pRPO220B. This plasmid contains the same promoter as pRPO77 but cloned upstream of a promoterless lacZ gene, providing a more convenient way to quantify promoter activity through assaying for β-galactosidase activity. The rpoS promoter showed strong activity in parent strain WCS358, whereas in mutant strain MT17 it displayed only 10% the activity shown in parent strain WCS358 (Fig. 2). It was concluded that mutant MT17 had a Tn5 insertion in a locus affecting rpoS gene expression.
FIG. 2.
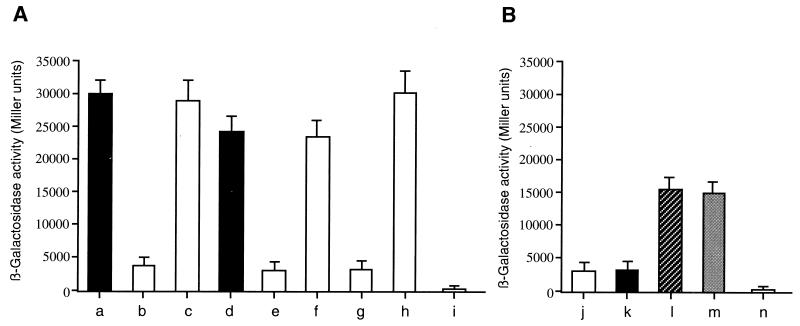
β-Galactosidase activities. (A) β-Galactosidase activities driven by the rpoS-lacZ fusion pRPO220B. All measurements were done in triplicate; the mean and standard error are shown. Cells harboring plasmids were grown for 16 h in LB medium in the presence of appropriate antibiotics. P. putida MT17 is a psrA null mutant of parent strain WCS358, P. putida MT17R is also a psrA null mutant, and P. aeruginosa MKV8 is a psrA null mutant of parent strain PAO1. Plasmid pRPO220B (IncP; Tcr) is an rpoS-pacZ transcriptional fusion, whereas pMKP25 (IncQ; Cmr) harbors a 2.5-kb PstI insert of P. putida WCS358 DNA carrying the complete psrA gene. (B) β-Galactosidase activities driven by the psrA-lacZ fusion pPPSR220. Cells harboring plasmids were grown for 16 h in LB medium in the presence of appropriate antibiotics. See the text for details. Columns: a, P. putida WCS358(pRPO220B); b, P. putida MT17(pRPO220B); c, P. putida MT17(pRPO220B)(pMKP25); d, P. aeruginosa PAO1(pRPO220B); e, P. aeruginosa MKV8(pRPO220B); f, P. aeruginosa MKV8(pRPO220B)(pMKP25); g, P. putida MT17R(pRPO220B); h, P. putida MT17R(pRPO220B)(pMKP25); i, P. putida WCS358(pMP220); j, P. putida WCS358(pPPSR220); k, P. putida rpoS::Tn5 WCS358(pPPSR220); l, P. putida MT17(pPPSR220); m, P. putida MT17R(pPPSR220); n, P. putida WCS358(pMP220).
Complementation of P. putida MT17 Tn5 regulatory mutants.
A gene bank containing partially digested HindIII chromosomal DNA fragments of strain WCS358 cloned in the corresponding site in cosmid vector pLAFR3 was introduced into the regulatory mutants. The transconjugants were complemented for the restoration of the promoter activity of plasmid pRPO77 as described in Materials and Methods. Two cosmids, pCOS17 and pCOS18, were isolated that complemented mutant MT17 for promoter activity. Restriction analysis revealed that pCOS17 and pCOS18 shared a 10-kb HindIII fragment.
A portion of the DNA sequence flanking Tn5 was cloned using an arbitrary PCR method as described in Materials and Methods. This allowed the cloning into pUC18 of a 680-bp fragment (pAPC17) flanking Tn5 in mutant MT17. This fragment was sequenced and used as a probe in Southern analysis, which allowed the localization of the complementing region within the 10-kb HindIII fragment of pCOS17 and pCOS18. This 10-kb HindIII fragment was cloned into the corresponding site in pBluescript to yield pBSH10. Further mapping using the 680-bp PCR fragment from pAPC17 as a probe revealed that within the 10-kb HindIII fragment, a 5-kb EcoRI fragment and a 2.5-kb PstI fragment contained the DNA flanking the Tn5 transposon in mutant MT17. Both of these fragments were cloned into the corresponding site in vector pLAFR3, yielding pLM17E and pLM17P, respectively, and were conjugated into P. putida MT17(pRPO77). Both of these subclones restored rpoS promoter activity (data not shown). The 2.5-kb PstI fragment was also cloned into the IncQ plasmid pMP77 to yield pMKP25, which was transferred to P. putida MT17(pRPO220B); this procedure restored wild-type levels of rpoS promoter activity, as detected by β-galactosidase activity (Fig. 2). It was concluded that the 2.5-kb PstI fragment could complement the mutant and contained all the necessary information to restore rpoS promoter activity in mutant MT17.
Characterization of the complementing DNA.
The 2.5-kb PstI fragment from pCOS17 and pCOS18 (i.e., construct pBS25) was sequenced in both directions (GenBank accession number AJ293485). The genetic map of the region is presented in Fig. 3A. Tn5 in mutant MT17 was inserted within an open reading frame (ORF) of 714 bp and coding for a putative protein of 237 amino acids and having a predicted molecular mass of 26,359 Da (Fig. 3B). The precise position of the Tn5 insertion in P. putida MT17 was 178 bp downstream of the putative ATG of this ORF. The gene representing this ORF was designated psrA (Pseudomonas sigma regulator). Western analysis using anti-PsrA antibodies revealed that mutant MT17 no longer produced PsrA (Fig. 4). This ORF was preceded by a potential Shine-Dalgarno sequence and was located 216 bp upstream of and in the orientation opposite that of the first codon of the lexA gene (GenBank accession number AJ293485). The LexA repressor protein identified here displayed a high level of identity to the characterized LexA proteins of other gram-negative bacteria (data not shown).
FIG. 4.
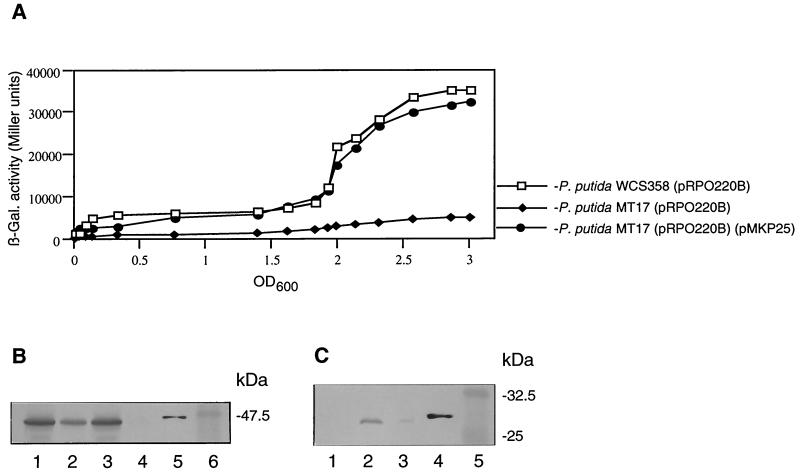
β-Galactosidase activities and Western analyses. (A) Growth and β-galactosidase (β-Gal.) activities of P. putida WCS358(pRPO220B), psrA null mutant MT17(pRPO220B), and MT17(pRPO220B)(pMKP25). The values were determined with LB medium, and the means of triplicate experiments are shown. OD600, OD at 600 nm. (B) Cellular RpoS levels of overnight cultures (OD, 2.3) of P. putida WCS358 (lane 1), P. putida MT17 (lane 2), P. putida MT17(pMKP25) (lane 3), rpoS:: Tn5 P. putida (lane 4), purified His6-RpoS (lane 5), and prestained molecular mass markers (lane 6). Immunoblot analysis was performed with 30 μg of total cellular protein per lane, 20 ng of purified His6-RpoS (lane 5), and anti-RpoS antibodies. (C) Cellular PsrA levels of overnight cultures (OD, 2.4) of P. putida MT17 (lane 1), P. putida MT17(pMKP25) (lane 2), P. putida WCS358 (lane 3), and purified His6-PsrA (lane 4). Immunoblot analysis was performed with 30 μg of total cellular protein per lane, 20 ng of purified His6-PsrA (lane 4), prestained molecular mass markers (lane 5), and anti-PsrA antibodies.
Regeneration of the psrA::Tn5 mutant of P. putida WCS358.
In order to unequivocally confirm that the psrA gene alone was responsible for the phenotype in mutant MT17, this gene was insertionally inactivated in P. putida WCS358 as described in Materials and Methods. In the regenerated psrA null mutant, rpoS promoter activity was reduced by 90%, just as in P. putida MT17, as identified by genetic selection. It was concluded that the ORF coding for the PsrA protein was responsible for the phenotype of reduced rpoS gene expression.
PsrA belongs to the TetR family of bacterial regulators.
A computer-assisted homology search between PsrA and proteins deposited in data banks revealed significant homology to bacterial regulators belonging to the TetR family (Prosite accession number PS01081). Many of the proteins of this family appear to be repressors, and their targets are often genes encoding proteins involved in cell envelope permeability. They have similar molecular masses (from 21 to 27 kDa). As a signature pattern, there is a conserved region that starts four residues before the helix-turn-helix motif and ends six residues after it and that is located near the N terminus. For PsrA, the signature is between amino acids 10 and 56, as depicted in Fig. 3B. The two proteins which displayed the highest level of identity to PsrA were a 175-amino-acid TetR regulator of Vibrio parahaemolyticus (30), showing 40% identity, and a regulator called IfeR (207 amino acids) of Agrobacterium tumefaciens (32), showing 34% identity. Both of these regulators are genetically linked to operons encoding efflux pumps. An alignment of the first 60 amino acids of PsrA with these two proteins is depicted in Fig. 3B. Apart from these proteins, PsrA displayed significant identity in the first 60 amino acids (approximately 30%) to 18 putative TetR family regulators (data not shown). We have found a similar gene upstream of the lexA gene in P. aeruginosa (www.pseudomonas.com); the genetic organization is like that in P. putida. This putative gene codes for a protein which is 90% identical to PsrA (Fig. 3B); it has not been characterized but has been identified through sequencing of the P. aeruginosa genome (see below).
Growth phase dependence of rpoS expression in P. putida
The growth phase dependence of rpoS expression was assessed using the pRPO220B rpoS-lacZ fusion (Fig. 4). Expression was relatively constant during early and exponential growth and then increased approximately fivefold in late exponential and early stationary phases. In psrA::Tn5 mutant MT17, rpoS expression was low throughout growth. The induction observed in the parent strain was not detected; rpoS expression was approximately 10% that seen in strain WCS358 (Fig. 4). P. putida MT17 harboring psrA in plasmid pMKP25 regained promoter activity and inducibility in response to cell density.
The psrA gene of P. aeruginosa is also involved in rpoS gene expression.
To investigate the role of the putative psrA gene of P. aeruginosa PAO1, we insertionally inactivated this locus in P. aeruginosa. A psrA null mutant was constructed and designated P. aeruginosa MKV8. Similar to what was observed for P. putida WCC358, a 90% reduction in rpoS promoter activity was observed in the mutant strain relative to the wild-type strain (Fig. 2). This reduction in promoter activity was restored when the P. putida psrA gene harbored in pMKP25 was introduced in trans into P. aeruginosa MKV8(pRPO220B) (Fig. 2). It was concluded that this gene also plays a role in rpoS gene expression in P. aeruginosa.
Phenotype of the Pseudomonas psrA::Tn5 mutants.
RpoS is known to confer cross-protection against heat and osmotic shock as well as hydrogen peroxide. It was reported that P. putida WCS358 containing rpoS::Tn5 was more sensitive to these three environmental stresses (17). When stationary-phase cultures of mutant MT17 were exposed to a sudden shift in temperature from 30 to 50°C, it was found to be twofold more sensitive at 50°C than the parent strain (data not shown). Similarly, when mutant MT17 was subjected to an increase in osmotic pressure caused by the addition of a high concentration of salt, the mutant was almost twofold more sensitive than strain WCS358 (data not shown). We also tested protection against hydrogen peroxide. In exponentially growing cells, there was no difference in sensitivity between the psrA mutant and the parent strain; however, in stationary-phase cells, the zone of inhibition was approximately 10% larger (data not shown), most probably due to a decrease in catalase activity (42).
It was reported that in P. aeruginosa, RpoS negatively regulates the production and secretion of pyocyanin pigment (42), the rpoS mutants showing a clear dark-blue coloration compared to the wild type. The psrA mutants of P. aeruginosa PAO1 also displayed a dark-blue coloration compared to the wild type. Therefore, we determined pyocyanin production in P. aeruginosa MKV8 and in PAO1 and found that the psrA mutant produces 2.5-fold more pyocyanin than the wild type (data not shown).
Finally, we examined levels of RpoS proteins in 24-h-old stationary-phase cultures of P. putida WCS358 and the psrA knockout P. putida MT17 using anti-RpoS antibodies. As depicted in Fig. 4, RpoS levels in P. putida MT17 were significantly lower than those in the wild type and the complemented mutant. Using a densitometer (Pharmacia LKB Ultroscan), we determined that RpoS levels in overnight cultures of the psrA mutant were approximately 50% those in parent strain WCS358 or the complemented mutant (Fig. 4).
Pseudomonas psrA mutants produce autoinducer molecules.
In P. aeruginosa, quorum sensing was implicated in rpoS gene expression and/or vice versa (21, 45). We tested the production of autoinducer (AHL) molecules in the psrA null mutant of P. aeruginosa. P. aeruginosa PAO1 synthesized the two identified and characterized AHLs, namely, N-3-oxododecanoyl-l-homoserine lactone, encoded by the lasI gene, and N-butanoyl-l-homoserine lactone, encoded by the rhlI gene, as detected by violacein production with the biosensor C. violaceum CVO26 and by light production with E. coli(pSB1075), respectively. AHL molecules were purified in a volume corresponding to 5 × 108 CFU at optical densities (ODs) of 0.1, 0.2, 0.4, 0.8, 1.2, 1.6, 2, and 2.5 from culture supernatants of P. aeruginosa PAO1 and the psrA mutant P. aeruginosa MKV8 grown on LB plates. The purified extracts were placed on thin-layer chromatography plates and overlaid with the bacterial sensor. No significant differences were observed for the psrA mutant P. aeruginosa MKV8 (data not shown). It was previously reported that P. putida WCS358 produces at least three different autoinducer molecules, as detected using the biosensor CVO26 (17). Purifying autoinducer molecules from spent supernatants of P. putida WCS358 and P. putida MT17R revealed that in the psrA null mutant, there was no difference in the production of these molecules. It was concluded that PsrA of P. aeruginosa and P. putida does not play a major role in the regulation of autoinducer biosynthesis.
psrA is autoregulated.
In order to establish whether PsrA regulated its own synthesis, the promoter of psrA fused to a promoterless β-galactosidase gene in construct pPPSR220 was introduced into wild-type strain WCS358, psrA mutant MT17, and WCS358 containing rpoS::Tn5 (rpoS mutant) (17). As depicted in Fig. 2, in mutant MT17 psrA gene expression was considerably increased (approximately fourfold), indicating that PsrA had a negative effect on its own synthesis. No difference was observed between the rpoS mutant and the wild type.
Effect of PsrA on rpoS and psrA promoters in E. coli.
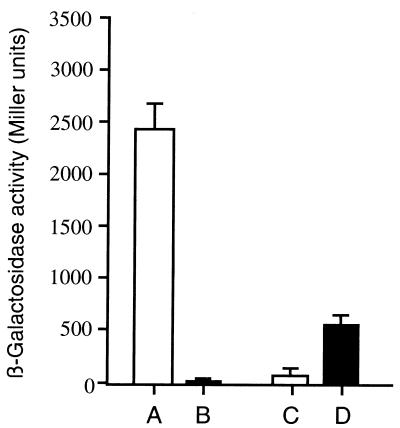
We determined whether PsrA had an effect on psrA and rpoS expression in a heterologous background (E. coli). The psrA gene was cloned into expression vector pQE30 (construct designated pQEPSRA), resulting in His6-PsrA production in E. coli, as observed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis (data not shown). For E. coli(pQEPSRA), the psrA-lacZ fusion (pPPSR220) and the rpoS-lacZ fusion (pRPO220B) were introduced in independent experiments, and β-galactosidase activity was determined. As shown in Fig. 5, the psrA promoter in E. coli was active, giving approximately 2,445 Miller units; this activity was repressed in the presence of pQEPSRA. On the other hand, the rpoS promoter in E. coli produced only 60 Miller units; this activity was increased by approximately 10-fold to 550 Miller units in the presence of pQEPSRA. As expected from the results obtained with Pseudomonas, PsrA appeared to act as a repressor of psrA expression and as an activator of rpoS expression.
FIG. 5.
β-Galactosidase activities in E. coli DH5α driven by the rpoS-lacZ (pRPO220B) and psrA-lacZ (pPPSRA220) fusions. Plasmid pQEPSRA contains the psrA gene and expresses the PsrA protein, whereas plasmid pQE30 is the vector (see text for details). All measurements were done in triplicate; the mean and standard error are shown. Cells harboring plasmids were grown for 16 h in LB medium in the presence of appropriate antibiotics. Columns: A, E. coli(pQE30)(pPPSR220); B, E. coli(pQEPSRA)(pPPSR220); C, E. coli(pQE30)(pRPO220B); D, E. coli(pQEPSRA)(pRPO220B).
DISCUSSION
The regulation of rpoS expression in P. putida WCS358 was investigated. Using an rpoS transcriptional fusion, we identified a Tn5 genomic mutant, MT17, which showed a 90% decrease in rpoS promoter activity. This mutant had a Tn5 insertion in an ORF of 714 bp and coding for a protein (PsrA) of 26.3 kDa. The PsrA protein contains, near its N terminus, a helix-turn-helix motif and belongs to the TetR family of bacterial regulators (Fig. 3B). Many of the regulators belonging to the TetR regulatory family act as repressors of transcription, their targets very often being genes which encode proteins involved in membrane permeability (23, 24). PsrA likely acts as a positive regulator of rpoS gene expression, since a null mutant resulted in a considerable decrease in rpoS gene expression and in less RpoS protein being present in the cell (Fig. 4). It is not known whether this regulatory action is direct or indirect, through another regulatory component(s). PsrA acts as a repressor of its own synthesis, since psrA was expressed at a much higher level in P. putida MT17 than in wild-type WCS358, indicating that PsrA regulates its own synthesis in a negative way. The observation that PsrA represses psrA and activates rpoS in E. coli is an indication that it might do so directly. The repression was strong and clear, whereas the activation of rpoS was rather weak compared to promoter activities in Pseudomonas. Thus, it is likely that some other factors are missing in E. coli.
To our knowledge, this study represents the first genetic screen of Pseudomonas spp. using an rpoS transcriptional fusion and selecting for down-regulated mutants. Our selection led to the identification of only one mutant having Tn5 in the psrA gene. The screen made use of the xylE reporter gene, which is less sensitive than other reporter systems (e.g., lacZ). However, mutants not showing a strong decrease in promoter activity cannot be detected in plate assays. The fact that the P. putida rpoS promoter exhibited strong promoter activity made the selection even more difficult. It is therefore possible that other mutants which showed lower levels of rpoS expression were not detected. The fact that a mutant exhibiting no rpoS promoter activity was not isolated could indicate that the regulation of RpoS occurs at different levels, as in E. coli, or that more transcriptional activators are involved. Our data showed that in Pseudomonas, there is an important regulator controlling rpoS transcription. However, additional control could take place in response to various stresses or to the stringent response. At present we do not know to which effector molecule(s) or environmental stimuli PsrA responds.
Previous studies have indicated that two other regulatory systems are involved in the regulation of rpoS expression in P. aeruginosa, namely, the LasR-LasI and RhlI-RhlR quorum-sensing systems and the GacA-GacS two-component regulatory system (see above) (21, 45). Surprisingly, no mutations in either of these systems were isolated in the genetic selection. We have demonstrated that the rpoS regulatory gene psrA is also present in P. aeruginosa PAO1 and that, as in P. putida, it plays a role in rpoS expression, since a psrA null mutant of PAO1 displayed a 90% reduction in rpoS promoter activity. By analysis of the production of quorum-sensing autoinducer molecules, it was verified that psrA mutants of both P. putida and P. aeruginosa still produced autoinducer molecules. Thus, it does not appear that PsrA influences rpoS gene expression through the biosynthesis of these molecules. Similarly, a reduction of rpoS expression in the psrA mutants did not result in any alteration in the production of autoinducer molecules. Following the contradictory reports of Latifi et al. (21) and Whiteley et al. (45), in which RpoS was linked to autoinducer production, we did not conclude in this study that PsrA plays a major role. However, it must be stressed that small quantities of RpoS were still made in psrA mutants.
Experiments thus far have indicated that in Pseudomonas, unlike in E. coli, rpoS appears to be extensively regulated at the transcriptional level through the use of various regulators. More work is needed to show whether the regulation of rpoS expression by PsrA occurs directly or indirectly or through quorum sensing and/or other regulatory components and to determine which effector molecules act on which regulators.
ACKNOWLEDGMENTS
M.K. is a scientist on leave from the Institute of Molecular Genetics and Genetic Engineering, Belgrade, Yugoslavia, and is benefiting from an ICGEB fellowship.
We are grateful to G. Degrassi and C. Aguilar for interest and support. We thank P. Williams and his coworkers for providing bacterial strains, plasmid constructs, and purified autoinducers.
REFERENCES
- 1.Beringer J E, Beynon J L, Buchanan-Wollaston A V, Johnston A W B. Transfer of the drug-resistance transposon Tn5 to Rhizobium. Nature. 1978;276:633–634. [Google Scholar]
- 2.Better M, Lewis B, Corbin D, Ditta G, Helinsky D R. Structural relationships among Rhizobium meliloti promoters. Cell. 1983;35:479–485. doi: 10.1016/0092-8674(83)90181-2. [DOI] [PubMed] [Google Scholar]
- 3.Birnboim H C. A rapid alkaline method for the isolation of plasmid DNA. Methods Enzymol. 1983;100:243–255. doi: 10.1016/0076-6879(83)00059-2. [DOI] [PubMed] [Google Scholar]
- 4.Bullock W O, Fernandez J M, Short J M. XL1-Blue: a high efficency plasmid transforming recA Escherichia coli strain with β-galactosidase selection. BioTechniques. 1987;5:376–382. [Google Scholar]
- 5.Corbin D, Ditta G, Helinski D R. Clustering of nitrogen fixation (nif) genes in Rhizobium meliloti. J Bacteriol. 1982;149:4759–4764. doi: 10.1128/jb.149.1.221-228.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunning C, Brown L, Elliott T. Promoter substitution and deletion analysis of upstream region required for rpoS translational regulation. J Bacteriol. 1998;180:4564–4570. doi: 10.1128/jb.180.17.4564-4570.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Essar D W, Eberly L, Hadero A, Crawford I P. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol. 1990;172:884–900. doi: 10.1128/jb.172.2.884-900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farewell A, Kvint K, Nystrom T. Negative regulation by RpoS: a case of sigma factor competition. Mol Microbiol. 1998;29:1039–1051. doi: 10.1046/j.1365-2958.1998.00990.x. [DOI] [PubMed] [Google Scholar]
- 9.Figurski D H, Helinski D R. Replication of an origin containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuqua W C, Winans S C, Greenberg E P. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geels F P, Schippers B. Reduction in yield depression in high frequency potato cropping soil after seed tuber treatments with antagonistic fluorescent Pseudomonas spp. Phytopathol Z. 1983;108:207–221. [Google Scholar]
- 12.Gerard F, Dri A M, Moreau P L. Role of Escherichia coli RpoS, LexA and H-NS global regulators in metabolism and under aerobic, phosphate-starvation conditions. Microbiology. 1999;145:1547–1562. doi: 10.1099/13500872-145-7-1547. [DOI] [PubMed] [Google Scholar]
- 13.Gibson K E, Silhavy T J. The LysR homolog LrhA promotes RpoS degradation by modulating activity of the response regulator sprE. J Bacteriol. 1999;181:563–571. doi: 10.1128/jb.181.2.563-571.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanahan D. Studies of transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 15.Jishage M, Iwata A, Ueda S, Ishihama A. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J Bacteriol. 1996;178:5447–5451. doi: 10.1128/jb.178.18.5447-5451.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jorgensen F, Bally M, Chapon-Herve V, Michel G, Lazdunski A, Williams P, Stewart G S. RpoS-dependent stress tolerance in Pseudomonas aeruginosa. Microbiology. 1999;145:835–844. doi: 10.1099/13500872-145-4-835. [DOI] [PubMed] [Google Scholar]
- 17.Kojic M, Degrassi G, Venturi V. Cloning and characterisation of the rpoS gene from plant growth-promoting Pseudomonas putida WCS358: RpoS is not involved in siderophore and homoserine lactone production. Biochim Biophys Acta. 1999;1489:413–420. doi: 10.1016/s0167-4781(99)00210-9. [DOI] [PubMed] [Google Scholar]
- 18.Lange R, Fischer D, Hengge-Aronis R. Identification of transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the sigma S subunit of RNA polymerase in Escherichia coli. J Bacteriol. 1995;177:4676–4680. doi: 10.1128/jb.177.16.4676-4680.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 20.Lange R, Hengge-Aronis R. The cellular concentration of the sigma S subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 1994;8:1600–1612. doi: 10.1101/gad.8.13.1600. [DOI] [PubMed] [Google Scholar]
- 21.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 22.Loewen P C, Hengge-Aronis R. The role of the sigma factor ςs (KatF) in bacterial global regulation. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 23.Lucas C E, Balthazar J T, Hagman K E, Shafer W M. The MtrR repressor binds the DNA sequence between the mtrR and mtrC genes of Neisseria gonorrhoeae. J Bacteriol. 1997;179:4123–4128. doi: 10.1128/jb.179.13.4123-4128.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma D, Alberti M, Lynch C, Nikaido H, Hearst J E. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol Microbiol. 1996;19:101–112. doi: 10.1046/j.1365-2958.1996.357881.x. [DOI] [PubMed] [Google Scholar]
- 25.Magazin M, Moores J C, Leong J. Cloning of the gene coding for the outer membrane receptor protein for ferric pseudobactin, a siderophore from plant growth-promoting Pseudomonas strain. J Biol Chem. 1986;261:795–799. [PubMed] [Google Scholar]
- 26.Marugg J D, van Spanje M, Hoekstra W P, Schippers B, Weisbeek P J. Isolation and analysis of genes involved in siderophore biosynthesis in plant-growth-stimulating Pseudomonas putida. J Bacteriol. 1985;164:563–570. doi: 10.1128/jb.164.2.563-570.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McClean K H, Winson M K, Fish L, Taylor A, Chhabra S R, Camara M, Daykin M, Lamb J H, Swift S, Bycroft B W, Stewart G S, Williams P. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143:3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 28.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 29.Muffler A, Fischer D, Altuvia S, Storz G, Hengge-Aronis R. The response regulator RssB controls stability of the sigma(S) subunit of RNA polymerase in Escherichia coli. EMBO J. 1996;15:1333–1339. [PMC free article] [PubMed] [Google Scholar]
- 30.Nozaki K, Inaba K, Kuroda T, Tsuda M, Tsuchiya T. Cloning and sequencing of the gene for Na+/H+ antiporter of Vibrio parahaemolyticus. Biochem Biophys Res Commun. 1996;222:774–779. doi: 10.1006/bbrc.1996.0820. [DOI] [PubMed] [Google Scholar]
- 31.O'Toole G A, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 32.Palumbo J D, Kado C I, Phillips D A. An isoflavonoid-inducible efflux pump in Agrobacterium tumefaciens is involved in competitive colonization of roots. J Bacteriol. 1998;180:3107–3113. doi: 10.1128/jb.180.12.3107-3113.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pratt L A, Silhavy T J. The response regulator SprE controls the stability of RpoS. Proc Natl Acad Sci USA. 1996;93:2488–2492. doi: 10.1073/pnas.93.6.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramos-Gonzalez M I, Molin S. Cloning, sequencing, and phenotypic characterization of the rpoS gene from Pseudomonas putida KT2440. J Bacteriol. 1998;180:3421–3431. doi: 10.1128/jb.180.13.3421-3431.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarniguet A, Kraus J, Henkels M D, Muehlchen A M, Loper J E. The sigma factor sigma s affects antibiotic production and biological control activity of Pseudomonas fluorescens Pf-5. Proc Natl Acad Sci USA. 1995;92:12255–12259. doi: 10.1073/pnas.92.26.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schweder T, Lee K H, Lomovskaya O, Matin A. Regulation of Escherichia coli starvation sigma factor (sigma s) by ClpXP protease. J Bacteriol. 1996;178:470–476. doi: 10.1128/jb.178.2.470-476.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spaink H P, Okker R J H, Wijffelmann C A, Pees E, Lugtenberg B J J. Promoter in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol Biol. 1987;9:27–39. doi: 10.1007/BF00017984. [DOI] [PubMed] [Google Scholar]
- 40.Stachel S E, An G, Flores C, Nester E W. A Tn3 lacZ transposon for the random generation of β-galactosidase gene fusions: application to the analysis of gene expression of Agrobacterium tumefaciens. EMBO J. 1985;4:891–898. doi: 10.1002/j.1460-2075.1985.tb03715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suh S J, Silo-Suh L, Woods D E, Hassett D J, West S E, Ohman D E. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J Bacteriol. 1999;181:3890–3897. doi: 10.1128/jb.181.13.3890-3897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka K, Takahashi H. Cloning, analysis and expression of an rpoS homologue gene from Pseudomonas aeruginosa PAO1. Gene. 1994;150:81–85. doi: 10.1016/0378-1119(94)90862-1. [DOI] [PubMed] [Google Scholar]
- 44.Whistler C A, Corbell N A, Sarniguet A, Ream W, Loper J E. The two-component regulators GacS and GacA influence accumulation of the stationary-phase sigma factor sigmaS and the stress response in Pseudomonas fluorescens Pf-5. J Bacteriol. 1998;180:6635–6641. doi: 10.1128/jb.180.24.6635-6641.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whiteley M, Parsek M R, Greenberg E P. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J Bacteriol. 2000;182:4356–4360. doi: 10.1128/jb.182.15.4356-4360.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winson M K, Swift S, Fish L, Throup J P, Jørgensen F, Chhabra S R, Bycroft B W, Williams P, Stewart G S. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol Lett. 1998;163:185–192. doi: 10.1111/j.1574-6968.1998.tb13044.x. [DOI] [PubMed] [Google Scholar]
- 47.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]