Abstract
Mutualism between Synechococcus and heterotrophic bacteria has been found to support their prolonged survival in nutrient-depleted conditions. However, environmental interference on the fate of their mutualism is not understood. Here, we show that exogenous nutrients disrupt their established mutualism. Once the exogenous nutrients were exhausted, Synechococcus and heterotrophic bacteria gradually reestablished their metabolic mutualism during 450 days of culture, which revived unhealthy Synechococcus cells. Using metagenomics, metatranscriptomics, and the 15N tracer method, we reveal that the associated bacterial nitrogen fixation triggered the reestablishment of the mutualism and revival of Synechococcus health. During this process, bacterial community structure and functions underwent tremendous adjustments to achieve the driving effect, and a cogeneration of nitrogen, phosphorus, iron, and vitamin by the heterotrophic bacteria sustained Synechococcus’s prolonged healthy growth. Our findings suggest that Synechococcus and heterotrophic bacteria may have an inherent tendency toward mutualism despite environmental interference. This may exhibit their coevolutionary adaptations in nutrient-deficient environments.
Synechococcus and bacteria tend to be mutually beneficial in nature when nutrients are limited.
INTRODUCTION
Marine phytoplankton and heterotrophic bacteria are actively involved in both inter- and intraspecies interactions, and the fate of these interactions can directly or indirectly influence the biota and biogeochemical cycles in their immediate environment (1). Heterotrophic bacteria that feast upon phytoplankton-derived organic matter govern all the major biogeochemical cycles of Earth (1). They are known to influence the growth of phytoplankton by regulating the nutrient cycle (1) and by processes such as algicide secretion, auxin secretion, and parasitism (1). Picocyanobacteria of the Synechococcus and Prochlorococcus genera are the most omnipresent phytoplanktonic group in the ocean, contributing up to 25% of the oceanic primary production (2). Their sheer overall mean abundance (i.e., 7.0 ± 0.3 × 1026 and 2.9 ± 0.1 × 1027 cells ml−1, respectively) demonstrates their dominance in the global ocean, and with increasing global warming, their role is expected to be more important in the near future (2).
Compared with Prochlorococcus, Synechococcus are biogeographically more diverse, distributed from cold polar waters to warm tropical waters (3, 4). Various factors have been attributed to the successful proliferation of Synechococcus in nutrient-deficient or oligotrophic waters (2, 4). Nevertheless, the survivability of Synechococcus in oligotrophic regions largely remains a mystery. Synechococcus are one of the most ancient photosynthetic microbes to exist and may have developed complex mechanisms and adaptations to cope with extreme environmental stress (5), such that Synechococcus can survive prolonged nutrient deprivation by undergoing chlorosis (6).
Recent studies have demonstrated that heterotrophic bacteria can support prolonged Synechococcus growth by establishing metabolic mutualism for nutrient exchange (3, 7). For example, bacterial mineralization of Synechococcus-derived nitrogen-rich organic matter sustained Synechococcus sp. WH7803 growth for approximately 200 days (8). Similarly, in our previous study, a laboratory-established coculture system of Synechococcus sp. PCC7002 and its associated bacteria were found to thrive for more than 2 years without any external nutrient support (3). These hints that microbial interactions may sustain long-term Synechococcus growth in natural oligotrophic ecosystems, wherein persistent stratification limits the nutrient import from the deep ocean (9) and nutrient circulation in the microbial loop predominates the flow of energy (8).
However, the ocean is not a static system and experiences spatiotemporal changes in environmental factors (9) that can greatly influence the surrounding biota, reshaping the interactions between Synechococcus and heterotrophic bacteria. Mutualistic interactions can be disturbed and even disrupted if the essential benefits can be procured cheaply or freely from other sources. Environmental factors such as external nutrient availability would strongly affect their nutrient cycle–based symbiotic interactions, which is relevant even to open ocean oligotrophic regions, that experience intermittent or irregular nutrient pulses both vertically and horizontally (10, 11).
To assess this, we exogenously supplied sterile inorganic nutrients to a long-term established stable mutualistic coculture system consisting of Synechococcus and heterotrophic bacteria from our previous study (3). In our previous study, we cultured an axenic Synechococcus sp. PCC7002 with a natural heterotrophic bacterial community (3). To our surprise, the heterotrophic bacterial community underwent community succession that changed its antagonistic relationship with the host Synechococcus to commensalism and later to mutualism over a period of ~1 year. The initial bacterial community, which consisted of diverse genera, was replaced by the genera Hyphobacterium and Erythrobacter, which together accounted for 83% of the total bacterial population (3). This mutualistic coculture system thrived for ~2 years without any external nutrient support, by establishing internal microbial nutrient cycling (3). In this present study, we show that the availability of exogenous nutrients strongly affects the established Synechococcus–heterotrophic bacteria mutualism as well as the community structure and function of Synechococcus-associated bacteria. Nevertheless, with long-term coexistence, Synechococcus and heterotrophic bacteria could reestablish the metabolic mutualism, reviving Synechococcus health mediated by heterotrophic nitrogen fixation and the provision of other essential nutrients.
RESULTS
Distinct phases of Synechococcus sp. PCC7002 growth during long-term coculture with heterotrophic bacteria
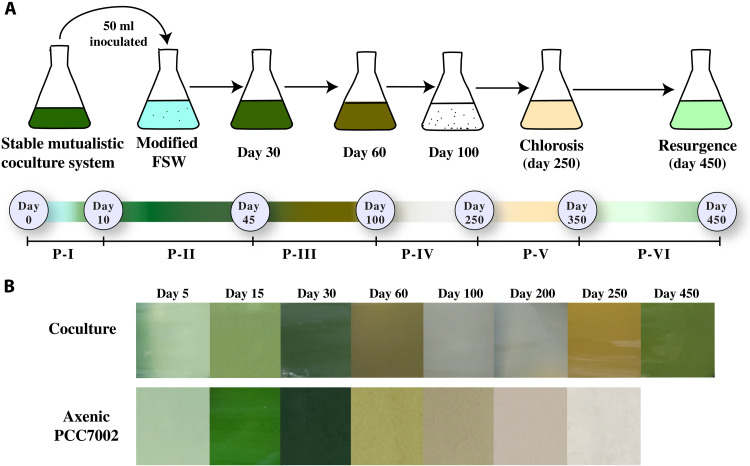
The Synechococcus–heterotrophic bacterial symbiotic system was supplied with exogenous nutrients [modified filtered sea water (FSW), table S1]. The growth status of Synechococcus sp. PCC7002 in the coculture system and in the axenic culture was assessed by monitoring the cell abundance, chlorophyll intensity, and quantum efficiency of photosystem II [Fv (variable fluorescence)/Fm (maximum yield of fluorescence under ambient light)] throughout the experimental period. On the basis of these parameters and the observed visual color change of the coculture systems, the coculture systems demonstrated six distinct phases of growth (P-I to P-VI; Figs. 1, A and B, and 2A).
Fig. 1. Experimental design and visual color change within the Synechococcus and heterotrophic bacteria coculture system.
(A) Illustration showing the experimental setup. Exogenous nutrients were added to an existing Synechococcus–heterotrophic bacterial mutualistic coculture system, and Synechococcus growth was monitored for 450 days. The time scale below the culture flasks illustrates the start and end of each phase with their corresponding observed (visually) color change. (B) Real-time visual color change within the coculture system and in the axenic Synechococcus sp. PCC7002 culture.
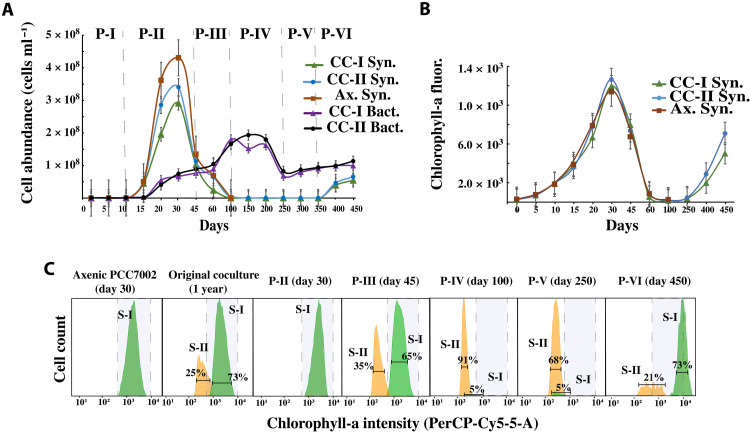
Fig. 2. Growth of Synechococcus and heterotrophic bacteria within the coculture system.
(A) Flow cytometric measurement showing Synechococcus and heterotrophic bacterial cell abundance over the 450 days of coculture period. Cocultures I and II are replicate culture systems. Each data point is an average of two replicates. The vertical dotted line approximately demarcates the start of each of the observed phases (P-I to P-VI). (B) Chlorophyll-a fluorescence, as an indicator of Synechococcus growth, was measured at 440/680 nm. Each data point is an average of three replicates. (C) Histogram showing the intensity of chlorophyll-a of the two subpopulations (S-I and S-II) related to Synechococcus sp. PCC7002. The error bars in each plot indicate the standard error of the mean. CC-I Syn., Synechococcus sp. PCC7002 abundance in coculture-I; CC-II Syn., Synechococcus sp. PCC7002 abundance in coculture II; Ax. Syn., Synechococcus sp. PCC7002 abundance in axenic culture; CC-I Bact., heterotrophic bacterial abundance in coculture I; CC-II Bact., heterotrophic bacterial abundance in coculture II.
The first two phases (P-I and P-II) of the coculture systems resembled those of the axenic Synechococcus (Fig. 2, A and B). During this period, the cell abundance of Synechococcus in all the culture systems increased by three orders of magnitude, reaching maximal abundance in 30 days with visible dark green color and chlorophyll-a intensity the same as that of the healthy Synechococcus sp. PCC7002 (Fig. 2, A and B). Moreover, from days 45 to 100 (P-III), the cocultured Synechococcus abundance along with the axenic culture rapidly declined by three orders of magnitude, with the culture system briefly becoming murky and eventually settling to a white debris deposition (Figs. 1B and 2A). The chlorophyll-a intensity was also reduced to its lowest point (Fig. 2B).
Phase P-IV lasted approximately from day 100 to day 250 without any further drop in cell abundance, and in contrast, the abundance of Synechococcus increased by 25.4% on day 200 compared to that on day 100 in the coculture systems but not in the axenic system. The coculture system then entered the phase P-V around day 250, with the system turning into a murky brownish yellow liquid (Fig. 1B), a peculiar characteristic of chlorosis and with a slight drop in the cell abundance of Synechococcus (13% as compared to that on day 100). Compared with the chlorophyll-a intensity at P-IV, here, chlorophyll-a intensity showed a slight improvement but was still lower than that of the healthy coculture system (Fig. 2B). From day 250 to day 350, the Synechococcus cell abundance was almost stable. However, in phase VI (the resurgence phase), i.e., from day 350 to day 450, the Synechococcus cell abundance suddenly increased by a magnitude of two (compared to P-IV and V) and continued to exhibit positive growth in cell abundance till the end of the experiment on day 450. The chlorophyll-a intensity was also substantially improved, with the coculture system displaying the same chlorophyll-a intensity as exponentially growing Synechococcus cells (Fig. 2B). Visually, both cocultures (I and II) showed signs of regaining healthy green coloration, which intensified from day 400 to day 450 (Fig. 1B). In contrast, the axenic Synechococcus system did not exhibit any notable change in cell abundance or chlorophyll-a intensity after P-III.
Furthermore, we also analyzed the change in Synechococcus pigment intensities [chlorophyll-a measured via PerCP (Peridinin chlorophyll)-Cy5-5A and allophycocyanin measured via allophycocyamin (APC)] by flow cytometer fluorophores. In the later growth period (after 45 days of culture) of all the long-term cocultured systems (cocultures I and II) and the original stable mutualistic coculture system (>1 year old), two subpopulations (S-I and S-II) were identified (having the same cell size but different PerCP-Cy5-5-A and APC intensities). S-I exhibited high APC and chlorophyll-a intensity, depicted as light green color, whereas S-II showed low APC and chlorophyll-a intensity, depicted as light orange color (Fig. 2C). However, S-II did not appear during the initial 30 days of culture, either in the axenic Synechococcus sp. PCC7002 culture or in the coculture systems (Fig. 2C), indicating that S-II might be unhealthy or stressed Synechococcus cells. Further, the abundance of S-II increased highly at P-III (Fig. 2C), whereas that of S-I decreased by 40 to 45% and 80 to 95% at P-III and P-IV, respectively (Fig. 2C). This reduction in S-I abundance remained unchanged till the end of P-V, during which both the coculture systems entered the chlorosis phase (P-V). Although the S-II population was present at P-III and P-IV, we did not see any visible symptoms of chlorosis, indicating that the emergence of S-II is not a direct reflection of chlorosis. Moreover, at P-VI, the abundance of S-I rose to 70 to 73% (while that of S-II declined to 20 to 25%) in both the coculture systems.
The quantum yield of photosystem II (Fv/Fm) followed a similar path as that of the Synechococcus abundance (fig. S1), maintaining a higher Fv/Fm (0.23 to 0.35) during the first two phases, followed by a decline in the Fv/Fm with the lowest value recorded at P-III and P-IV (>0.05). During the chlorosis phase (P-V), there was a slight increase in the Fv/Fm (0.13 c.a.), but the Synechococcus cells were still under stress. Furthermore, at P-VI (i.e., from days 350 to 450), the Fv/Fm was substantially increased. On day 450, the Synechococcus cells exhibited an Fv/Fm similar to that of the healthy initial phase (fig. S1).
Meanwhile, the heterotrophic bacterial abundance remained lower in the first two phases (107 cells ml−1), increasing gradually at P-III (108 cells ml−1; Fig. 2A). This increase in the bacterial abundance was maintained until day 200 and declined to 107 cells ml−1 from days 200 to 250. Furthermore, at P-VI, the bacterial abundance showed a gradual increase in abundance (108 cells ml−1; Fig. 2A).
Substantial increase in inorganic nutrients during the resurgence phase of Synechococcus
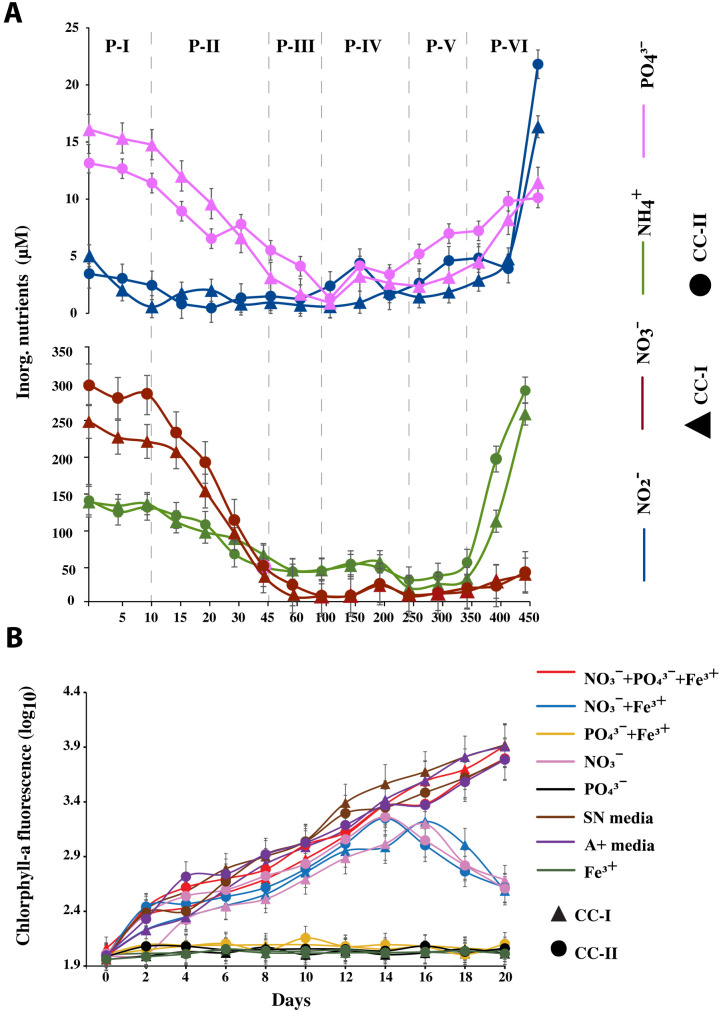
The concentration of inorganic nitrogen (NO3−, NO2−, and NH4+) species and PO43− were greatly reduced by the end of P-III (day 100). Of the inorganic nitrogen sources, NO3− was reduced by 99%, NH4+ was reduced by 68%, while PO43− was reduced by 90% of the initial concentration (Fig. 3A). NO2− was reduced by 93.5 and 33.4% in cocultures I and II, respectively. Moreover, from day 100 to day 200, both the cocultures experienced a slight increase in inorganic N species and PO43− concentration (by 5 to 13%; Fig. 3A), decreasing again on day 250. Astonishingly, at phase P-VI, the system showed a significantly higher concentration of inorganic N (inorganic N = NO3− + NO2− + NH4+, P = 0.0052) and PO43− (P = 0.0466) without any exogenous input. Here, the NH4+ and NO2− concentrations were even higher by ~2- and 3.5-fold, respectively, than their initial (P-I) concentration (Fig. 3A).
Fig. 3. Nutrient concentration and key nutrients that limited Synechococcus growth within the coculture system.
(A) Changes in the concentration of inorganic nutrients (NO2−, NO3−, NH4+, and PO43−) during different phases of Synechococcus growth. Each data point is an average of two replicates. (B) Effect of exogenous inorganic nutrients on the chlorotic Synechococcus cells. Synechococcus growth was determined as a measure of chlorophyll-a fluorescence. Each data point is an average of three replicates. Error bars in each plot depict the standard error of the mean. CC-I, coculture I; CC-II, coculture II.
N and P colimited the growth of Synechococcus sp. PCC7002 within the coculture system
On inoculation with different nutrient treatments (table S2), the unhealthy, chlorotic Synechococcus cells from the P-V phase showed an increase in chlorophyll-a fluorescence with a healthy green pigmentation (visually) in all the treatments containing NO3− within the first 4 days (Fig. 3B and fig. S2). Synechococcus cells inoculated in nutrient treatments without any nitrogen source remained unchanged, suggesting nitrogen as the critical driver of the growth of Synechococcus. Furthermore, although the growth was resumed in all the treatments with NO3−, the treatments lacking PO43− as a conutrient collapsed after 16 to 18 days (Fig. 3B). Only the chlorotic cells inoculated with NO3− + PO43− + Fe3+ sustained healthy growth as that of the chlorotic cells inoculated with nutrient-rich media (SN and A+ media). However, Fe3+ either in combination with NO3− or alone, did not show any substantial improvement in chlorophyll-a or green coloration in comparison to treatments with NO3− alone.
DOC content in the coculture systems
Our data showed that dissolved organic carbon (DOC) content was inversely proportional to inorganic nutrients in both the coculture systems (fig. S3). The DOC was highest (46 to 51 mg ml−1) during the P-III to P-IV phase, followed by a 67 to 70% reduction at P-V, implying that there was high bacterial consumption of DOC. Moreover, the reduction in DOC concentration was slower after the P-V phase and saw a further reduction of 37 to 50% by the end of the experiment (at day 450).
Changes in Synechococcus-associated bacterial community composition
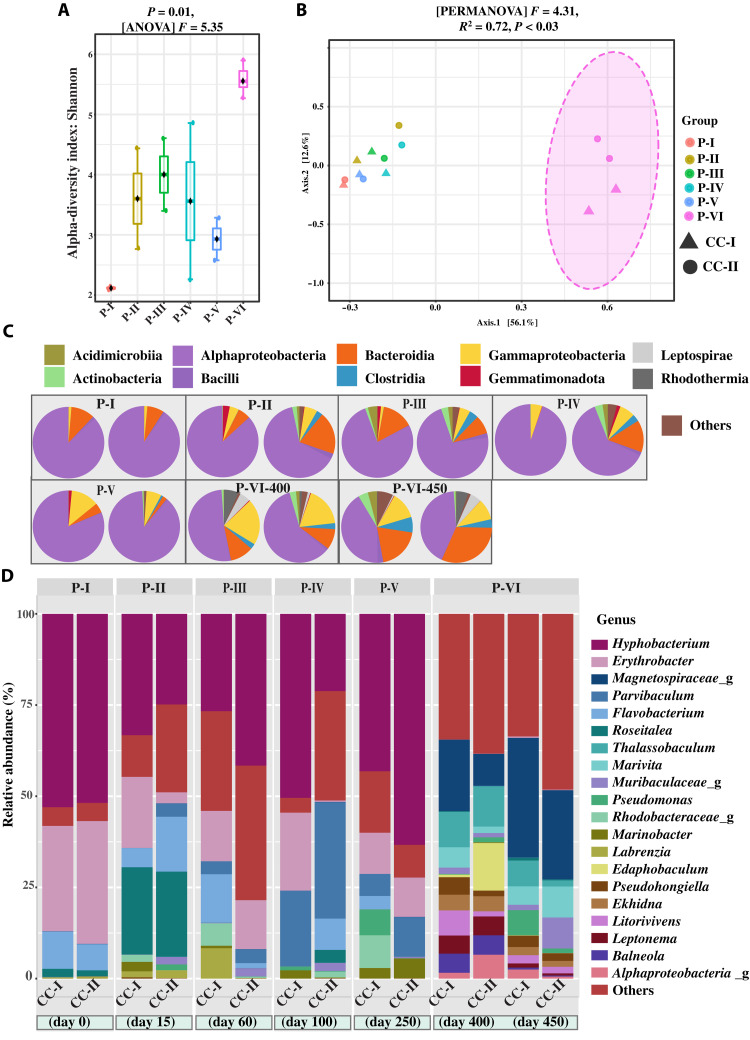
We examined the bacterial community composition in all six phases (PI to P-VI) by 16S ribosomal RNA (rRNA) gene sequencing. After filtering and rarefaction, a total of 1271 unique amplicon sequence variants (ASVs) were examined for differences among the six phases. In general, the P-VI showed a higher ASV richness (Shannon index, P = 0.01; F = 5.35) than P-I to P-V (Fig. 4A). From principal coordinate analysis (PCoA) with a Permutational multivariate analysis of variance is the expanded form of PERMANOVA (999 permutations) test and the Bray-Curtis distance metrics, it was clear that the bacterial composition of P-I to P-V was more similar than the bacterial composition of P-VI (Fig. 4B). Moreover, a Bray-Curtis dissimilarity test showed that both coculture systems (cocultures I and II) had similar bacterial compositions during healthy Synechococcus growth and followed the same bacterial successional pattern (fig. S4). (3).
Fig. 4. Changes in the composition of the heterotrophic bacterial community as revealed by 16S rRNA gene amplicon sequencing.
(A) Box plots illustrating the alpha diversity (Shannon diversity index) for samples at different phases (P = 0.01, F = 5.35). (B) PCoA of heterotrophic bacterial communities in different phases: PERMANOVA: F = 4.3125; R2 = 0.72939; P < 0.003. Ordination method: PCoA, distance method: Bray-Curtis, taxonomic level: ASV, statistical method: PERMANOVA. (C) The relative abundance of 16S rRNA gene amplicon sequences assigned to the top 10 most abundant bacterial classes. (D) The relative abundance of 16S rRNA gene amplicon sequences assigned to the top 20 most abundant bacterial genera. CC-I, coculture I; CC-II, coculture II.
The initial mutualistic system (3) that served as the starting point (P-I) of cocultures I and II was composed of the heterotrophic bacterial community dominated by Alphaproteobacteria (~90%, P-I; Fig. 4C) and the genera Hyphobacterium and Erythrobacter (~83% relative abundance, P-I; Fig. 4D). In our previous study, it was revealed that this bacterial community did not show any notable changes in their community composition over a period of 2 months and was considered a fairly stable community (3). Furthermore, upon exposure to fresh medium (modified FSW), the composition of this otherwise fairly stable heterotrophic bacterial community began to fluctuate with an increase in the abundance of the genera Roseitalea, Flavobacterium, Marinobacter, and Parvibaculum (P-II to P-V; Fig. 4D). Nonetheless, the relative abundance of Hyphobacterium did not change considerably from P-I to P-V, while the relative abundance of Erythrobacter was reduced to half of what it was at P-I. Moreover, at P-VI, the abundance of Alphaproteobacteria was reduced to 50%, while the abundance of other classes, such as Bacteriodia, Clostridia, Gammaproteobacteria, Leptospirae, Actinobacteria, and Rhodothermia increased highly (Fig. 4C). The dominant (top 20) heterotrophic bacterial genera at P-I to P-V were entirely replaced by other genera (whose abundance was negligible at P-I to P-V) at P-VI (Fig. 4D). The most abundant genera at P-VI were Thalassobaculum, Litorivivens, Marivita, Balneola, and genera related to the families Muribaculaceae and Magnetospiraceae (Fig. 4D).
In addition, a redundancy analysis between the dominant taxa, inorganic nutrients, and Synechococcus abundance indicated that NH4+ (P = 0.005), NO2− (P = 0.010), and Synechococcus cell abundance (P = 0.043) were the significant contributors that drove the compositional change of the bacterial community (fig. S5). Members of the class clostridia were highly correlated to the NH4+ and NO2− variance, while Actinobacteria, Rhodothermia, Leptospirae, and Acidimicrobia were related to Synechococcus cell abundance.
Nitrogen and phosphorus generation capability of the heterotrophic bacterial community
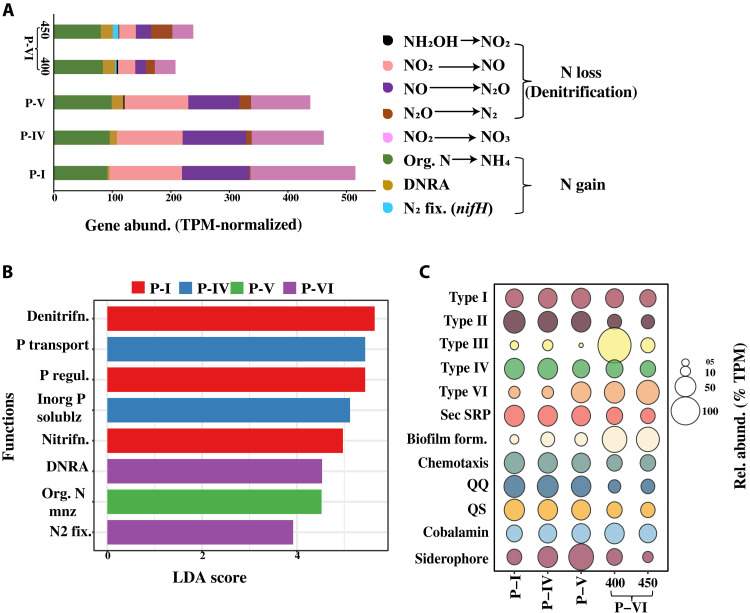
Shotgun metagenomic analysis was used to explore the microbial nutrient cycling capability of the coculture systems and the potential contributors at P-I, P-IV, P-V and P-VI (see table S3 for sequence statistics). Because of sample volume limitations, we could not sequence other phases. Here, the bacterial metagenome had all major genes involved in the nitrogen (N) and phosphorus (P) cycles (Fig. 5A, fig. S6, and tables S4 and S5). Regarding the N-cycle genes, there was a substantial increase in the gene abundance of N-gain processes, which are dissimilatory nitrate reduction to ammonium (DNRA) (nirBD/nrfAH) and N2 fixation (nifH) at the P-VI phase (Fig. 5A). Gene abundance is reported as transcripts (DNA in this case) per million (TPM). Notably, the nifH (biomarker gene for N2 fixation), which was not detected in the P-I, P-IV, and P-V metagenomes, covered up to 4% (gene abundance = 22.9 TPM) of all the N-cycle genes at P-VI (day 450; Fig. 5A). The P-VI metagenome also showed the presence of organic N mineralization (gudB, GLUD1_2, gdh, and gdhD), with a gene abundance similar to that of P-V (Fig. 5A). In contrast, there was no noticeable difference at P-cycle potential among all the sequenced phases and showed the presence of genes related to organic P mineralization, solubilization of labile inorganic P, P regulation, and transportation (fig. S6). In addition, a linear discriminant analysis (LDA) among the N and P genes clearly showed the significance of potential N2 fixation and DNRA at P-VI (LDA scores of 3.92 and 4.53 and P values of 0.049 and 0.040, respectively; Fig. 5B). Moreover, there was no discernible difference in the abundance of cobalamin (vitamin B12)–related genes across all sequenced phases (cobU), while the abundance of iron (Fe) siderophore synthesis genes decreased at P-VI (Fig. 5C and tables S6 and S7).
Fig. 5. Comparative analysis of the functional capability of the heterotrophic bacteria at different phases.
(A) Gene abundance (TPM-normalized) of all detected genes from the nitrogen cycle across the analyzed metagenomic samples. (B) LDA plot showing logarithmic LDA score (effect size) of the most significant (false discovery rate adjusted P value of 0.01, log LDR score of 2) genes involved in the nitrogen and phosphorus cycles across the analyzed metagenomic samples. Group P-VI includes samples from days 400 and 450. (C) Bubble plot showing the relative abundance (TPM-normalized) of other functional genes [bacterial secretory systems, biofilm formation, chemotaxis, QS-QQ, cobalamin (vitamin B12), and Fe siderophore synthesis] across the analyzed metagenomic samples.
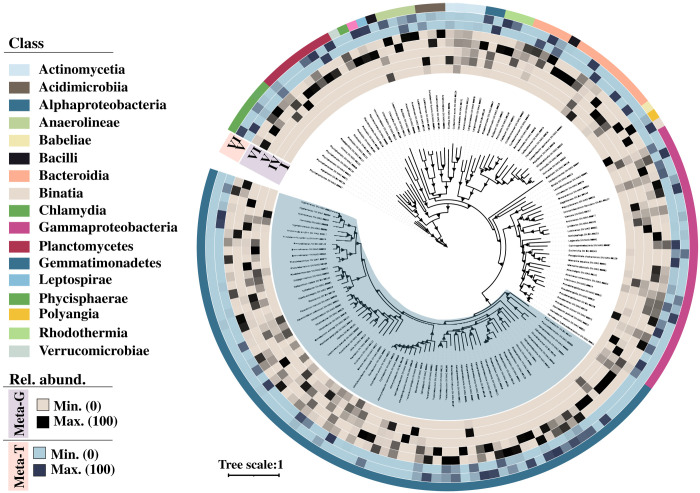
Meanwhile, 134 quality bins were recovered from these metagenomic samples (Fig. 6 and table S8). Comparatively, the coverage (relative abundance across all samples) of most of these bins was higher at P-VI than at the other phases, suggesting a potential relationship between phase P-VI and these bins (Fig. 6). To investigate this potential relationship, we explored the metabolic capability of these bins. As expected, most of these bins contained genes related to essential nutrient cycling (N and P), vitamin B12, and siderophore synthesis (fig. S7A and tables S8 and S9). The relative abundance (across metagenomic samples) of bins having N-cycling potential related to classes Alphaproteobacteria, Actinomycetia, Phycisphaerae, Planctomycetes, Rhodothermia, Gammaproteobacteria, and Bacteroidia increased highly at P-VI (table S10). Among phosphorus (P) cycle processes, all assembled bins had complete or partial sets of genes for organic P mineralization, solubilization of labile inorganic P, P regulation, and transportation (tables S8 and S9).
Fig. 6. Unrooted maximum-likelihood phylogenetic tree of the assembled bacterial bins.
The tree was generated using a concatenated alignment of 120 conserved bacterial markers. The first five concentric rings moving outward from the tree show the relative abundance of the bins across the metagenomic samples [P-I, P-IV, P-V, P-VI (day 400), and P-VI (day 450)]. The following two rings illustrate the relative abundance of these bins across the metatranscriptomic samples [P-V and P-VI (day 450)]. The outermost ring assigns color codes to the bins according to their respective taxonomic classes.
In addition, 57% of bins associated with Alphaproteobacteria, 46% of bins associated with Gammaproteobacteria, and 50% of bins associated with Actinomycetia and Anearolineae had vitamin B12 synthesis genes (fig. S7A). On the other hand, 2% of Alphaproteobacteria, 14% of Gammaproteobacteria, and 25% of the Actinomycetia bins had genes for siderophore synthesis (fig. S7A). Overall, all the bins carrying genes for essential nutrient cycles had higher coverage at P-VI than at other analyzed phases (tables S9 and S10).
Distinct carbohydrate-degrading enzyme profiles of the heterotrophic bacterial community
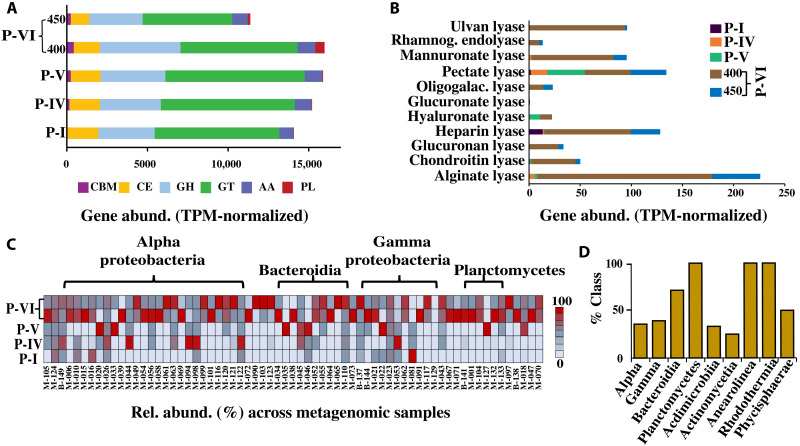
It is well established that phytoplankton exudate can shape the composition of the associated microbiome (12). We compared the Carbohydrate-Active enZYmes profiles of the metagenomic samples and the extracted bins to better understand the correlation between the change in bacterial community composition and carbohydrate metabolism at different phases. As expected, the gene abundance of the polysaccharide degrading family [polysaccharide lyase (PL)] increased significantly (P < 0.0001, one-way ANOVA test) at P-VI (Fig. 7A and table S11) than at other phases. Among these PL genes, the abundance of genes encoding for alginate lyase (PL6, PL14, PL15, and PL17), chondroitin lyase (PL35, PL29, and PL37), poly(β-d-mannuronate) lyase (PL7), heparin lyase (PL12), and ulvan lyase (PL24, PL25, PL28, and PL40) were higher at P-VI by 29.5, 7.4, 13.8, 14.6, and 16.1%, respectively (Fig. 7B) than at the other phases. Furthermore, it was observed that the coverage (relative abundance across all samples) of all the assembled bins containing these PL family genes was noticeably high at P-VI (Fig. 7C and table S9), with a higher percentage of bins from Planctomycetes, Bacteroidia, Anearolineae, and Rhodothermia carrying these genes (Fig. 7D). Classes Alphaproteobacteria, Gammaproteobacteria, Actinomycetia, Acedomicrobia, and Phycisphaerae also showed the presence of these genes. In addition, the abundance of genes related to the glycoside hydrolase family (which also participates in the breakdown of complex carbohydrates) and the carbohydrate-binding module family also followed a slight increase in their abundance at P-VI (Fig. 7A).
Fig. 7. CAZy profiles of the metagenomic community.
(A) Gene abundance (TPM-normalized) of the detected CAZy family genes across the analyzed metagenomic samples. The CAZy genes are categorized into the auxiliary activity (AA) family, carbohydrate-binding module (CBM) family, carbohydrate esterase (CE) family, glycoside hydrolase (GH) family, and polysaccharide lyase (PL) family. (B) The abundance of the genes (TPM-normalized) encoding potential PL family enzymes across the metagenomic samples. (C) Relative abundance of the bins containing PL family genes across the analyzed metagenomic samples. The number label corresponds to MAG/BIN number, M = MAG and B = Bin. (D) Percentage of taxonomic classes (from the assembled bins) having PL family genes.
Bacterial genes involved in various phycospheric interactions within the coculture system
We speculated that the interaction between heterotrophic bacteria and Synechococcus might be complex and need a multidimensional investigation. For this purpose, we created Kyoto Encyclopedia of Genes and Genomes (KEGG) profiles of all the bacterial genes that may be prevalent in phycospheric interactions after a rigorous literature review (tables S12 to S14). These genes encode vital functions such as the bacterial secretory systems, quorum sensing (QS), quorum quenching (QQ), chemotaxis, and biofilm formation.
It was seen that the abundance of genes necessary for the bacterial secretory systems, QS, QQ, and chemotaxis was higher during P-I, P-IV, and P-V and then decreased at P-VI, except for the type III and type VI bacterial secretory systems (Fig. 5C and tables S12 to S14). Compared to the earlier phases, the abundance of genes involved in biofilm formation increased significantly during P-VI (P < 0.0001, one-way ANOVA test; Fig. 5C and table S14).
Similarly, most of the bins carried genes that were potentially necessary for these phycospheric interactions, with a considerable increase in the relative abundance of bins involved in chemotaxis and biofilm formation at the P-VI phase than at other phases (fig. S7A and tables S8 and S9). The percentage of bins carrying genes for chemotaxis and biofilm formation was much higher among the Actinomycetia, Bacteroidia, Gammaproteobacteria, and Planctomycetes classes (fig. S7B). Moreover, almost all the isolated bins had a partial or complete set of genes encoding QS and QQ as well as the bacterial secretory systems (tables S8 and S9).
Nitrogen fixers within the coculture systems
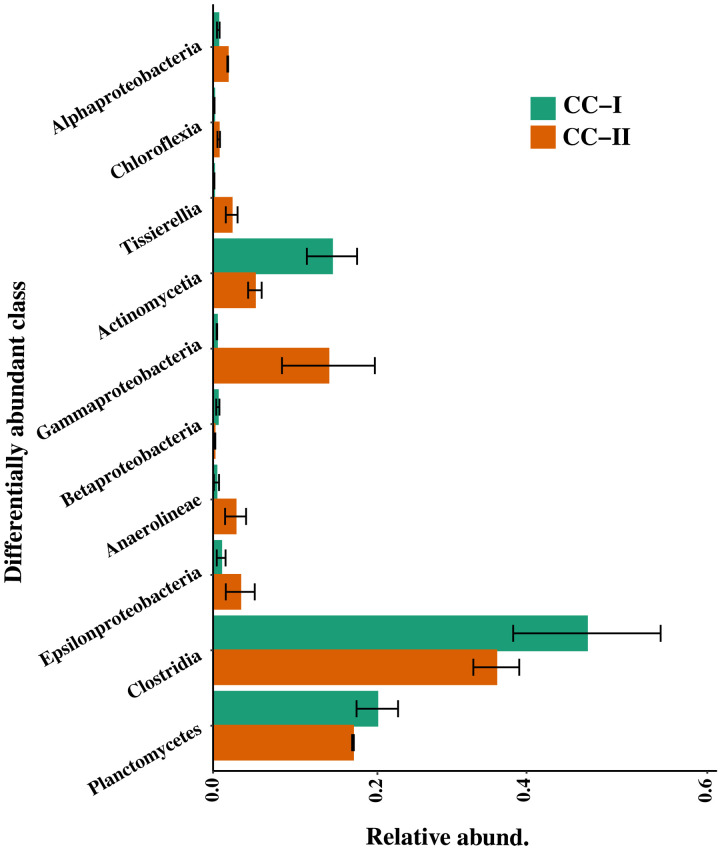
In addition, we extracted RNA from the P-VI phase (i.e., from day 450 culture) of cocultures I and II and validated the expression of the nifH gene via reverse transcription-quantitative polymerase chain reaction (RT-qPCR) (table S15). Once validated, the nifH gene fragments were amplified and sequenced. To determine the classes that were differentially abundant between cocultures I and II, a total of 212 ASVs were statistically tested using metastat (P = 0.05; Fig. 8). No unique classes were observed showing nifH expression in either coculture I or II. Classes Clostridia, Planctomycetes, Gammaproteobacteria, and Actinomycetia were highly abundant in both the coculture systems (Fig. 8). Unfortunately, because of the lack of a standard and robust nifH gene/transcript database, we were unable to classify these amplified sequences at lower taxonomic levels.
Fig. 8. Relative abundance of differentially abundant classes showing nifH expression at P-VI.
Differentially abundant classes were identified by multivariate statistical analysis of Metastat (P = 0.05). Error bars in each plot depict the standard error of the mean. CC-I, coculture I; CC-II, coculture II.
Significant changes in the gene expression of the coculture system from chlorosis (P-V) to resurgence (P-VI)
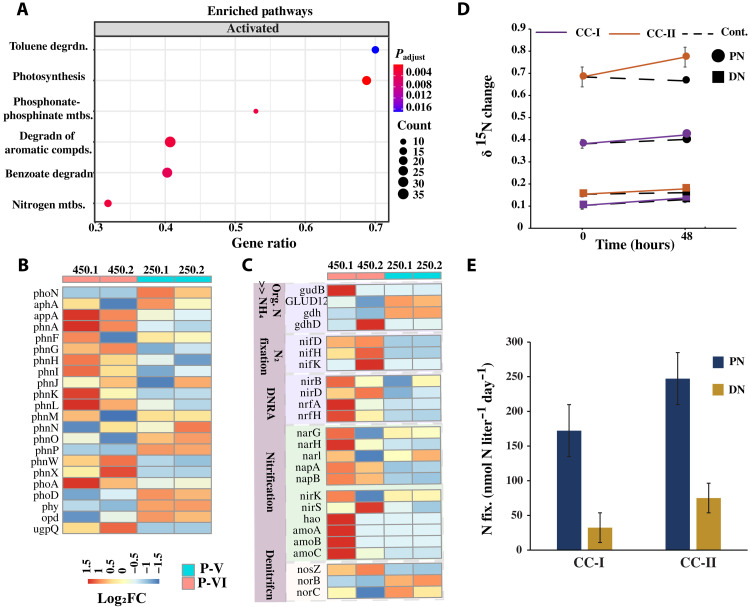
We also conducted a metatranscriptomics analysis (see table S3 for sequence statistics) to examine the differential gene expression, followed by a gene enrichment analysis between the P-V (chlorosis) and P-VI (resurgence) phases of the bacterial community using a ±1.5-log2 fold change threshold (corresponding to an adjusted P value of 0.05). At P-VI, we observed a significant enrichment in the expression of transcripts associated with genes related to organic phosphorus mineralization (phosphonate and phosphinate metabolism), nitrogen cycle, and photosynthesis (Fig. 9A). When zoomed in, several transcripts associated with the organic P-mineralization genes (appA, phnAGHIKLWX, phoAD, and ugpQ) were strongly expressed with a log2 fold change value of 1 to 1.5 (Fig. 9B), whereas most of the transcripts participating in the N-gain processes such as nitrogen fixation (nifHDK) and DNRA (nirBD and nrfAH), ammonia oxidation (amoABC), and nitrification (hao and narH) were all highly up-regulated at P-VI (log2 fold change of 1 to 1.5; Fig. 9C). These findings strongly supported the role of the heterotrophic bacteria to the increase in inorganic nutrients at P-VI. Transcripts corresponding to denitrification (N-loss) genes were also differentially expressed at P-VI (Fig. 9C), which suggested an active nitrogen cycle activity.
Fig. 9. Differential gene expression and nitrogen fixation rate at P-VI of the Synechococcus and heterotrophic bacteria coculture system.
(A) GSEA of the significantly up-regulated genes at P-VI [P = 0.05, number of permutation (nPerm) = 10,000]. Heatmap representation of expression levels for differentially expressed transcripts (log2 fold change, adjusted P value of 0.05) related to (B) inorganic phosphorus mineralization and (C) the nitrogen cycle processes. (D) Changes in δ15N incorporation in PN and DN in the coculture system over 48 hours. CC-I and CC-II represent cocultures I and II with the addition of the 15N tracer. The accompanying dashed line represents the control set (without the addition of the 15N tracer) for CC-I and CC-II, respectively. (E) Nitrogen fixation rates (after subtracting their respective controls) within the PN and DN pools of CC-I and CC-II upon incubation for 48 hours. Each data point is an average of two replicates, with error bars showing the standard error of the mean. CC-I, coculture I; CC-II, coculture II.
Furthermore, at P-VI, there was also a substantial increase in the expression of transcripts involved in vitamin B12 synthesis and biofilm formation processes when compared with P-V (fig. S8, A and B). Several of the transcripts necessary for carbohydrate (polysaccharide) degradation and related processes were also found to be highly expressed at P-VI (fig. S8C). These transcripts encode enzymes such as alginate lyase (PL5, PL-7, PL-34, and PL-39) and pectate lyase (PL-9). In addition, the transcripts involved in the activity of enzymes heparin lyase (PL-12), hyaluronate lyase (PL-33), and poly(β-d-mannuronate) lyase (PL-7) were expressed in both phases (fig. S8C). Transcripts related to the PL-14 and PL-1 CAZy family that encode for enzymes alginate lyase and pectate lyase, respectively, were also up-regulated in both phases (fig. S8C). In contrast, transcripts related to the PL-22, PL-25, PL-29, PL-35, PL-38, and PL-11 family, corresponding to enzyme oligogalacturonate lyase, ulvan lyase, chondroitin-sulfate ABC endolyase, chondroitin lyase, endo-β-1,4-glucuronan lyase, and rhamnogalacturonan endolyase, respectively, were up-regulated at P-V compared to P-VI (fig. S8C). Among the GH family, most of the transcripts required for cell wall degradation (lysozyme, glucosidase, and peptidoglycan lytic transglycosylase) were up-regulated at P-VI (fig. S8D).
15N incorporation and N2 fixation rate within the coculture system
We further quantified the N2 fixation rate by determining the incorporation of 15N within the particulate nitrogen (PN) and dissolved nitrogen (DN) pools at P-VI (day 450). It was found that on enrichment with 15N, the incorporation of 15N (δ15N) within the particulate and DN pools of cocultures I and II was substantially increased when compared with the control set (without the addition of tracer 15N, Fig. 9D). The δ15N value within the PN pool (δ15N PN) of cocultures I and II was increased by 10.5 ± 2% and 13.1 ± 1%, respectively, within 48 hours, which is two times higher than that in the control set (Fig. 9D). Simultaneously, the DN pools of cocultures I and II also showed an increased incorporation of 15N (δ15N DN) by 33.0 ± 3% (i.e., 1.2 times higher than the control) and by 15.6 ± 2% (i.e., 3.5 times higher than the control) within 48 hours (Fig. 9D). Furthermore, cocultures I and II showed a total nitrogen fixation rate of 172.2 and 247.3 nmol N liter−1 day−1, respectively, for the PN pool and 33.4 and 84.1 nmol N liter−1 day−1, respectively, for the DN pool (Fig. 9E). As the 15N tracer method can underestimate the N2 fixation rate (13), the data reported here should be considered as the minimal rate.
DISCUSSION
Microscale interactions between heterotrophic bacteria and phytoplankton underpin ocean biogeochemistry. However, these interactions between the phytoplankton and heterotrophic bacteria are not stable and can be disturbed or even disrupted by external nutrient supply (14). In this study, we show that despite disturbances from environmental factors (inorganic nutrients), Synechococcus and heterotrophic bacteria can form recurrent mutualistic interactions that could drive the survival of Synechococcus in nutrient-depleted conditions.
Complex organic matter and microbial interspecies and intraspecies interactions drive the heterotrophic bacterial community changes
We observed that although the abundance of Synechococcus and bacteria changed considerably in the first five phases, the bacterial community composition did not change much during this time (Fig. 4, B to D). However, at P-VI, when the growth of Synechococcus was revived, the heterotrophic bacterial members showed obvious changes in community composition with a sudden increase in the members of Gammaproteobacteria, Bacteriodia, Clostridia, Rhodothermia, and others. Increased organic matter concentration provides a favorable environment for the rapid boom in the specialist bacterial population and such a niche frequently (but not always) favors the Bacteroidia, Gammaproteobacteria, and some Alphaproteobacterial taxa (15). These bacterial taxa, especially Bacteroidia and Gammaproteobacteria, are known to exhibit rapid chemotactic behavior toward the phytoplankton-derived dissolved organic matter, prefer particle association, and are frequently found in high abundance during phytoplankton blooms and in phytoplankton phycosphere (16, 17).
In addition, phytoplankton are known to actively or passively exude organic matter and specific metabolites that can attract selective bacterial groups and form symbiotic relationships (2, 12, 16). While polysaccharides make up a substantial portion of this leaked matter, the PL enzyme group plays a major role in cleaving these complex polysaccharides into simple oligo or monomers (12). The increase in the abundance of genes encoding PL enzymes at P-VI mirrors this (Fig. 7C). Similarly, from the gene abundance and expression analysis, it was seen that most of these PL genes target specific high-molecular weight (HMW) sulfated polysaccharides (Fig. 7B and fig. S8C), such as alginate, ulvan, etc., which a variety of cyanobacterial species are known to produce (18, 19), and such polysaccharides, especially alginate, are known to favor copiotrophic bacteria such as the Gammaproteobacteria (20).
Moreover, the Bacteroidetes (mainly composed of the Bacteroidia) are highly specialized in consuming such HMW polysaccharides due to the presence of polysaccharide utilization loci, specific HMW substrate-binding proteins, and an efficient TonB-dependent transporter system (21). Likewise, the Gammaproteobacteria, Planctomycetes, Phycispaerae, Rhodothermia, and Anearoineae are also known to consume HMW polysaccharides (21–24), and these, along with the Bacteroidetes, are usually abundant in algal organic matter-rich environments such as algal blooms (22–24). Thus, the high abundance and expression of alginate, ulvan, heparin, and other similar sulfated HMW organic substrate degrading PL genes seen at the P-VI phase (Fig. 7B) hints toward the role of specific Synechococcus-related organic matter in filtering its associated bacterial members. However, it is not conclusive whether the specific polysaccharide degradation activity was driven by Synechococcus’s exudation of selective polysaccharides or a reduction in the labile DOC pool (fig. S3) within the coculture systems.
Nonetheless, several PL family transcripts were also found to be expressed at P-V (fig. S8C). This is not unexpected considering the presence of the abovementioned taxa (i.e., Bacteroidia, Gammaproteobacteria, and Rhodothermia) at P-V (Fig. 4C). Thus, we speculated that the change in the heterotrophic bacterial community composition at P-VI may be due to not only the organic matter degradation but also the bacteria-bacteria interactions (3). In the host-associated environment, a complex interplay of factors shapes the microbiome. It was seen that all the essential genes involved in inter- and intraspecies interactions (including bacterial secretory systems, QS and QQ, and chemotaxis) were highly represented at P-I, P-IV, and P-V (Fig. 5C). In bacteria, the protein secretory systems play an essential role in interacting with their host or with other bacteria, aiding in adhesion, aggregation, motility, and the secretion of various enzymes (25). While the role of the bacterial secretory systems in interactions with phytoplankton is yet to be studied, sporadic research suggests that they might play an important role in inter- and intraspecies communications (26). The gene abundance of type III and type VI bacterial secretory systems was higher at P-VI (Fig. 5C), suggesting their potential role in the reestablishment of mutualism. These two secretory systems are known to be involved in mutualistic interactions (27).
The genes corresponding to the bacterial protein secretory systems (except types III and VI) and QS and QQ followed a trend similar to those of chemotaxis, indicating a strong correlation between these three processes (Fig. 5C). QS signaling molecule has been shown to alter the bacterial composition (28), and QS-QQ are known to regulate bacterial chemotaxis, biofilm formation, and adhesion to phytoplankton environment (29). With this potential for such a profound effect, the contribution of these genes to the shift in bacterial community composition and the reestablishment of mutualism within the coculture system cannot be denied.
Moreover, the highly abundant classes at P-VI carried a high percentage of bins with genes necessary for both chemotaxis and biofilm formation or chemotaxis alone (fig. S7, A and B, and tables S8 and S9). Thus, we conclude that the change in the heterotrophic bacterial community composition might be attributed to the reduction in niche dimensions (i.e., reduction in the labile organic carbon) and/or the specificity of niche dimension (related to the specific polysaccharide substrates) as well as the various bacteria-bacteria and bacteria-Synechococcus interactions.
Nitrogen fixation by heterotrophic bacteria revives Synechococcus cells and reestablishes metabolic mutualism
Most of the phytoplankton productivity in the oligotrophic regions is limited by nitrogen and nitrogen deficiency has a more prominent effect on phytoplankton growth and the rate of primary production than phosphorus (9, 30). As Synechococcus sp. PCC7002 cannot fix bioavailable nitrogen (3), nitrogen limitation may lead to Synechococcus chlorosis (6). Here, Synechococcus sp. PCC7002 chlorosis observed at P-V was strongly related to the lack of inorganic nitrogen within the coculture system (Fig. 3A) and external provision of nitrogen (NO3−) induced revival of Synechococcus growth (Fig. 3B). Chlorosis may be a vital adaptive strategy of Synechococcus to survive in nutrient-depleted conditions (6, 31), and such nitrogen limitation-induced chlorosis has been widely observed and studied among Synechococcus species (6, 31).
In our study, the axenic counterpart did not exhibit chlorosis (Figs. 1B and 2A) and did not revive on the addition of exogenous nutrients (fig. S9). It was recently reported that Prochlorococcus strain MIT9313 relies on interactions with heterotrophic bacteria for long-term nutrient starvation by undergoing a chlorosis-like phenomenon (32). Such bacterial interactions with chlorotic autotrophs have also been widely studied in soil plants (33). Despite such profound evidence, the role of autotroph-associated bacteria in phytoplankton chlorosis is still not well explored.
Furthermore, we found that after 350 days in coculture with heterotrophic bacteria (P-VI), the Synechococcus cells recovered from chlorosis (Figs. 1, A and B, and 2, A to D) and the coculture system exhibited an increased concentration of inorganic nitrogen compounds (NO2−, NO3−, NH4+;Fig. 3A). This, along with the substantial increase in gene abundance (Fig. 5, A and B) and high up-regulation of transcripts involved in N-gain processes (Fig. 9, A and C), particularly nitrogen fixation genes (nifHDK), at P-VI, clearly demonstrates that heterotrophic bacterial nitrogen fixation greatly contributed to the revival of Synechococcus cells from chlorosis. This was further verified by the 15N-labeled nitrogen fixation experiment, which showed measurable nitrogen fixation activity within both the coculture systems at P-VI (Fig. 9E).
However, the nitrogenase enzyme complex that carries out nitrogen fixation is irreversibly deactivated in oxic conditions (34), so how do the heterotrophic bacteria fix nitrogen here? We found that the higher abundance of genes and up-regulation of transcripts related to nitrogen fixation (i.e., nifHDK) at P-VI astonishingly coincided with an increase in the abundance of genes and up-regulation of transcripts involved in biofilm formation (i.e., pgaABC, mcbAR, and bssS; Fig. 5C and fig. S8B). This peculiar trend was not observed in the other phases. Biofilm formation has previously been found to support heterotrophic bacterial nitrogen fixation by creating oxygen-deficient microzones, especially in particle-attached environments (34). Such anoxic conditions have been observed in phytoplankton and cyanobacterial aggregates supporting nitrogen fixation (35). While nitrogen fixation is an energy-intensive process with the hydrolysis of 16 mol of adenosine triphosphate (ATP) for the reduction of 1 mol of N2 (34), nitrogen fixation in this manner (by creating oxygen-deficient microzones) will increase the energy cost even further. Thus, a labile organic matter–rich environment is imperative for such a process (34, 36). The high abundance of genes corresponding to the PL enzyme family, the significant expression of the related transcripts, and the higher relative abundance of bins carrying these genes during the early P-VI phase (Fig. 7, A to D, and fig. S8C) strongly indicate that the high-energy requirement for nitrogen fixation could be fulfilled by the utilization of rich organic matter exudates from Synechococcus. Increasing polysaccharide concentration has been found to promote nitrogen fixation (36) and the addition of polysaccharides such as alginate boosted nitrogen fixation by 10-fold (36).
The differentially abundant classes showing expression of the nifH transcript at P-VI (i.e., Clostridia, Gammaproteobacteria, Actinomycetia, Planctomycetes, and Anaerolinea; Fig. 8) were also the ones highly enriched at P-VI (Fig. 4C). Among these, classes Clostridia and Actinomycetia were statistically related to the change in NH4+, NO2−, and Synechococcus abundance, whereas the Gammaproteobacteria, Bacteroidia, and Rhodothermia were also at least partially related (fig. S5), strongly supporting their role in nitrogen fixation within the coculture systems. Members of these taxa have been widely found to be associated with nitrogen fixation in diverse marine environments (37, 38). Furthermore, these same classes were also potentially involved in chemotaxis and biofilm formation (fig. S7, A and B), as well as exhibiting potential PL activity (Fig. 7, C and D) within the cocultures. This implies that the members of these classes were actively involved in reestablishing the mutualism and resurrecting Synechococcus cells from chlorosis.
In addition to N2 fixation, genomic carriers for other N-gain processes (N mineralization and DNRA) were also present at P-VI (Figs. 5, A and B, and 9C). Genes encoding organic nitrogen mineralization (gudB, gdh, gdhD, and GLUD1_2) are also known to carry out the same reaction in reverse order, i.e., generating amino acid glutamate from NH4+ (39), therefore confounding its influence on bioavailable N generation. At the same time, the significant up-regulation of DNRA in the P-VI phase also cannot be ignored. As nitrogen fixation is an energy-consuming activity (34), lone and long-term dependence on nitrogen fixation may not be feasible for mutualistic cohabitants. Therefore, coactivation of N-gain processes or an eventual shift toward other less energy-intensive N-gain processes (DNRA and N mineralization) will ease the pressure off the nitrogen-fixing community. Last, the high up-regulation of most of the N-cycle transcripts at P-VI suggested a balanced and robust N-cycle activity at this phase, which is very crucial in maintaining N-gain and N-loss balance, ultimately determining the nutrient stoichiometry, as well as conversion/removal of any harmful intermediate substrate (40).
A coavailability of nitrogen, phosphorus, iron, and vitamin B12 is essential for the long-term healthy growth of Synechococcus
Although nitrogen is vital for phytoplankton growth, the effect of other essential nutrients cannot be neglected (9). A colimitation of nitrogen and phosphorus has been shown to have a far more drastic effect on phytoplankton productivity than that of nitrogen alone (41). In oligotrophic waters, phytoplankton productivity is often limited by nitrogen or a colimitation of nitrogen and phosphorus or nitrogen, phosphorus, and iron (9, 41). In this study, the addition of NO3− alone revived Synechococcus from chlorosis but could not support healthy Synechococcus growth for an extended period (Fig. 3B). As we did not observe any change in chlorotic Synechococcus physiology on the addition of PO43− or Fe3+ alone (Fig. 3B), we believe that the generation of inorganic nitrogen is the key factor in reviving Synechococcus cells from chlorosis. Although a cosupplementation of NO3−, PO43−, and Fe3+ sustained healthy Synechococcus growth, the cosupplementation lacking PO43− (i.e., treatment with NO3− and Fe3+) did not (Fig. 3B). In addition, the transcripts related to genes involved in phosphorus mineralization were also found to be significantly up-regulated at the P-VI phase (Fig. 9B). This suggested that phosphorus was also an influential factor in reviving Synechococcus cells. Moreover, Fe is an essential micronutrient for Synechococcus growth and Fe limitation has been shown to affect growth rate, metabolic regulation of cofactor expression, and activation of antioxidative stress responses in Synechococcus sp. PCC 7002 (42). Although we did not observe the expression of Fe siderophore synthesis transcripts at P-VI, the heterotrophic bacterial metagenome had Fe siderophore synthesis capability (Fig. 5C), hinting toward the role of iron in cosustaining the coculture system.
Moreover, many algae, including Synechococcus sp. PCC7002, require an exogenous supply of vitamin B12 for healthy growth (43) and was evidenced by the up-regulation of transcripts involved in vitamin B12 synthesis (fig. S8A) at P-VI. The expression of vitamin B12 transcripts during the revival of Synechococcus from chlorosis (P-VI) is not a surprise, as many bacteria, including Synechococcus, also use vitamin B12 as a cofactor for the vitamin B12–dependent methionine synthase (MetH) enzyme (43). Thus, further investigations are required to discern its significant effect on Synechococcus growth limitation and revival from chlorosis.
In summary, we assessed the effect of exogenous nutrients on existing, nutrient exchange–based Synechococcus–heterotrophic bacteria mutualism. Our findings demonstrated that the addition of exogenous nutrients disrupts the mutualistic relationship between Synechococcus and heterotrophic bacteria, but they can reestablish the disrupted mutualism in long-term coexistence. The heterotrophic bacterial population found enriched on reestablishment of the mutualism exhibited various unique behavioral tactics, such as chemotaxis and biofilm formation, and had the potential to consume specific polysaccharides, which might have aided in the shift in the bacterial community composition. Once mutualism was reestablished between the Synechococcus and heterotrophic bacterial community, the provision of bioavailable nitrogen (specifically by nitrogen fixation) by the heterotrophic bacteria revived the Synechococcus cells from chlorosis. However, the final long-term healthy sustainability of Synechococcus cells was due to not only the provision of nitrogen but also a cogeneration of all the essential nutrients, including nitrogen, phosphorus, iron, and vitamin B12. In exchange, the heterotrophic bacteria could use the organic nutrient–rich Synechococcus exudates. These findings suggest that nutrient stoichiometry has a consequential impact on Synechococcus–heterotrophic bacteria mutualism. However, because of their coexistence for billions of years, Synechococcus and heterotrophic bacteria might have developed evolutionary adaptations toward beneficial mutual interactions to thrive in unfavorable conditions, which was manifested through their recurrent establishment of reciprocal metabolic interaction on long-term coexistence in our study.
MATERIALS AND METHODS
Cocultivation of Synechococcus and heterotrophic bacteria
In our previous study, we established a long-term, mutualistic coculture system between Synechococcus sp. PCC7002 and a natural heterotrophic bacterial community (3). In brief, an exponentially growing axenic Synechococcus sp. PCC7002 culture was inoculated with a natural heterotrophic bacterial community and serially subcultured for 20 generations. On subsequent long-term static cocultivation (kept in the same flask), the coculture system was found to thrive for ~2 years without any external nutrient input (3). Furthermore, in this present study, we were interested in investigating how this existing Synechococcus–heterotrophic bacteria mutualistic system will react to environmental interference such as the provision of exogenous nutrients. To assess this, we transferred 50 ml of the above Synechococcus–heterotrophic bacteria mutualistic system into each of two culture flasks (labeled as cocultures I and II) containing 3.5 liters of 0.2-μm pore-size sterile modified FSW (at a final concentration of Synechococcus cells = 105 cells ml−1). The modified FSW was amended with inorganic nutrients and was composed of 132.6 μM NH4+, 263.5 μM NO3, and 14.3 μM PO43−, as well as other essential nutrients (for details, see table S1). In addition, an axenic set was also established by inoculating 50 ml of exponentially growing axenic Synechococcus sp. PCC7002 into 3.5 liters of modified FSW (at a final concentration of Synechococcus cells = 105 cells ml−1) in duplicate. All the culture bottles were incubated under natural daylight conditions at 25°C for 450 days with regular gentle shaking.
Synechococcus sp. PCC7002 sterility was checked throughout the experimental period as described previously (3). Periodic samples were collected at different phases of Synechococcus growth (i.e., from P-I to P-VI; Fig. 1A) and Synechococcus and heterotrophic bacterial cell abundance and Synechococcus pigment intensity were monitored by flow cytometry (BD FACSAria II). Chlorophyll-a fluorescence, as an indicator of Synechococcus growth, was measured by a Biotek Synergy HT microplate reader (excitation and emission wavelengths of 440 and 680 nm, respectively) and the Fv/Fm (quantum yield of photosystem II) was quantified by using an AquaPen-C 100 [photon systems instruments (PSI), Czech Republic] following standard protocols (3). All experiments were performed with independent biological duplicates unless mentioned otherwise in the text. Algal contamination was checked before all experiments, as described in the Supplementary Text.
Measurement of inorganic nutrients and determination of the key nutrients limiting Synechococcus sp. PCC7002 growth in the coculture system
As mentioned above, measurements of samples for inorganic nutrients (NO2−, NO3−, NH4+, and PO43−) were collected periodically in triplicate. A volume of 20 ml was filtered through a 0.7-μm pore-size GF/F filter (precombusted at 450°C for 5 hours) and then stored in polyethylene plastic bottles at −20°C before use. During analysis, the nutrient concentrations were determined by spectrophotometry using an AutoAnalyzer (Bran and Luebbe AA3, Germany). Furthermore, to elucidate the key nutrients that limited Synechococcus sp. PCC7002 growth within the coculture systems, subsamples (30 ml each) from both the coculture systems (at chlorosis phase, P-VI) were inoculated with 30 μl of different inorganic nutrient treatments in duplicate. These nutrient treatments consisted of NO3−, PO43−, and Fe3+ nutrients either as individual constituents or in combinations (for details, see table S2). The culture bottles were incubated under natural daylight conditions at 25°C for 20 days. Chlorophyll-a fluorescence was measured every 2 days with the Biotek Synergy HT microplate reader at excitation/emission wavelengths of 440 and 680 nm.
Determination of DOC in the coculture systems
To quantify the DOC in each coculture system, 20-ml samples of each were filtered through a precombusted (450°C for 5 hours) 0.7-μm pore-size glass-fiber GF/F filter and then collected into precombusted glass vials in duplicate. The vials were stored at −20°C until further analysis. Before analysis, the frozen samples were thawed and acidified with 40 μl of 4 N hydrochloric acid (HCl). DOC was analyzed through high-temperature catalytic oxidation using a Shimadzu TOC-L analyzer (Shimadzu, Japan) attached with an ASI-V autosampler following standard procedures.
16S rRNA gene-based microbial community profiling of the Synechococcus–heterotrophic bacteria coculture
High-throughput 16S rRNA amplicon sequencing was used to profile the bacterial community at different phases of Synechococcus growth (P-I to P-VI; Fig. 1A), separately from cocultures I and II. Briefly, 50 ml of the samples from both the coculture systems (I and II) from P-I to P-VI was collected separately, gently vortexed, and then prefiltered through a 2-μm pore-size polycarbonate filter (Merck Millipore, USA) to reduce the host Synechococcus cells. The 2-μm filtered samples were then collected on a 0.2-μm pore-size polycarbonate filter (Merck Millipore, USA). DNA was extracted separately from cocultures I and II, representing the biological replicates of the experiment using a FastDNA SPIN kit (MP Biomedicals). Technical replicates (of the extracted DNA samples) from cocultures I and II were used to amplify the V3-V4 region of the 16S rRNA gene by primers 343F (5′-TACGGRAGGCAGCAG-3′) and 798R (5′-AGGGTATCTAATCCT-3′). The quality of the amplicon was checked using gel electrophoresis and later purified using AMPure XP beads (Beckman Coulter, CA, USA). The amplicon was amplified again with the addition of Illumina i5/i7 dual-index adapters and purified as described above. The final amplicon was quantified using a Qubit double-stranded DNA assay kit (Thermo Fisher Scientific, MA, USA) and sequenced on the Illumina MiSeq platform at OE Biotech Co. Ltd. (Shanghai, China). The sequenced reads were then preprocessed using Sunbeam v.2.1.0 (44) to discard ambiguous bases (N), adapters, primers, and low-quality sequences (average quality score below 25). The resulting cleaned 16S rRNA paired-end reads were analyzed using the DADA2 package v.1.16 (45). Briefly, the reads were quality-filtered, and sequences below a Phred score of 25 (if any) were trimmed off. Further filters were applied via the DADA2 package [maxN = 0, maxEE = c (5,4), truncQ = 2, rm.phix = TRUE, minLen = 50]. The filtered R1 and R2 reads were merged, grouped into ASVs and passed through the default DADA2 chimera removal script (see table S16 for the number of reads passed at each step). The final nonchimeric ASVs were classified by the Silva small subunit rRNA nr database v.138.1 of the DADA2 package. The raw counts of technical replicate samples were averaged. The ASV table was then fed into the MicrobiomeAnalyst web-based package (https://microbiomeanalyst.ca) for statistics and visualization using default filter settings. The default filter settings of the MicrobiomeAnalyst package that were used are as follows: The raw ASV count matrix for assigned bacterial taxa was filtered for features containing all zeros or appearing in only one sample, low abundant features (20% prevalence for a minimum count of four), and features that exhibit low variance based on interquantile range. The filtered ASVs were then normalized by rarefying to the minimum library size and transformed via the total sum scaling method.
Metagenomic and metatranscriptomic analysis
Because of sample volume limitations, we did not collect samples from the P-II and P-III phases for metagenomic and metatranscriptomics analysis. In addition, for the same reason and to achieve sufficient sequencing depth, we pooled the samples from cocultures I and II before DNA/RNA extraction. Briefly, 50 ml of samples from the original (initial) culture and 100 ml each from day 100, day 250, day 400, and day 450 cultures were collected and processed as described above. For metatranscriptomics, the 0.2-μm pore-size polycarbonate filter containing the microbial biomass was immediately transferred to a 2-ml RNAlater solution (Invitrogen), incubated for 24 hours at 4°C, and later transferred to −80°C until further analysis. Before RNA extraction, the surface of the working bench and pipettes were thoroughly cleaned, first with a laboratory-prepared ribonuclease (RNase) decontaminating solution and then with 70% ethanol. All the materials used were RNase and deoxyribonuclease (DNase) free. RNA extraction and DNase treatment were conducted using the RNAqueous Micro Kit (Invitrogen) following the manufacturer’s protocol. The quality and integrity of RNA were determined by the NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, MA, USA) and by gel electrophoresis, respectively.
DNA and RNA concentrations were measured using a Qubit v.2.0 fluorometer (Invitrogen, Carlsbad, CA). For metagenomics, libraries were prepared using a TruSeq Nano DNA LT library prep kit (Illumina, CL, USA) according to the manufacturer’s instructions. In brief, 1 μg of the extracted DNA was sheared through the Covaris S220 (Covaris, MA, USA) ultrasonicator and purified using AMPure XP beads (Beckman Coulter, CA, USA). Overhangs were repaired into blunt ends using the end-repair mix 2. The 3′ ends of the repaired DNA fragments were adenylated with A-bases and ligated with adapters. Libraries were then prepared with an insertion size of 550 or 350 base pairs (bp).
For metatranscriptomics, Illumina-compatible libraries were prepared using the NEBNext Ultra nondirectional RNA library prep kit (New England Biolabs, MA, USA) according to the manufacturer’s instructions. Briefly, the bacterial 16S and 23S rRNA transcripts from the total extracted RNA were reduced using an ALFA-SEQ rRNA depletion kit. One microgram of the RNA was fragmented using NEBNext first-strand synthesis reaction buffer. The first strand of complementary DNA (cDNA) was synthesized using NEBNext random primers and Moloney Murine Leukemia Virus (M-MuLV) reverse transcriptase (RNase H), while the second strand of cDNA was synthesized with DNA polymerase I and RNase H. After adenylation of the 3′ ends, the DNA fragments were ligated using NEBNext adapters. Further, 150- to 200-bp fragments were selected with SpeedBead magnetic carboxylate modified particles (Global Life Sciences Solutions, MA, USA), and PCR was performed with Phusion High-Fidelity DNA polymerase, universal PCR primers, and NEBNext index PCR primers. The resulting PCR products were purified with AMPure XP beads, and library insert size was assessed on the Qsep400 high-throughput nucleic acid protein analysis system (Houze Biological Technology Co., Hangzhou, China).
Metagenomic and metatranscriptomic samples were sequenced on Illumina NovaSeq 6000 at OE Biotech Co. Ltd. (Shanghai, China). However, because of RNA quality and quantity issues, we were unable to sequence samples from the P-I and P-IV phases. The resulting raw reads were passed through Sunbeam v.2.1.0 (44) for initial adapter removal, quality trimming, and host Synechococcus reads removal [by aligning against the published Synechococcus sp. PCC7002 genome, National Center for Biotechnology Information (NCBI) GeneBank accession number GCA_000019485.1]. In addition, rRNA sequences from the metatranscriptomic reads were removed through SortMeRNA v.4.2.0 (46). All the cleaned R1 and R2 reads of metagenomic and metatranscriptomic samples were concatenated into two separate files (i.e., R1 and R2 files of metagenomes were concatenated separately, and R1 and R2 files of metatranscriptomes were concatenated separately). Later, the concatenated reads were coassembled via SPAdes (for metagenomic reads) and rnaSPAdes (for metatranscriptomic reads) v.3.11.1 (47) with default settings and using k-mers 21, 33, 55, 77, 9, and 127. Assembly statistics were generated using MetaQUAST v.5.0.2 (48).
For binning, the coassembled scaffolds from the metagenomic assembly were used to extract bins using metaBAT2, CONCOCT, and MaxBin2 of the MetaWRAP pipeline v.1.3.2 (49) from the metagenomic samples. CheckM v.1.1.3 (50) was used to compute the bin statistics, and quality bins with >50% completion and <10% redundancy were filtered for further study (see table S8). These filtered bins were then passed through Anvi’o v.7 (51) for further refining and calculating bin statistics (see table S8). All bins with >70% completeness and <10% redundancy were defined as Metagenome-assembled genomes (MAGs). The Genome Taxonomy Database Toolkit (GTDB-Tk) v.1.5.0 (52) package with GTDB database v.R06-RS202 was then used to assign taxonomy to each MAG, and phylogenetic analysis of the GTDB-Tk aligned protein sequences [Multiple Sequence Alignment (MSA)] file was carried out using IQ-TREE v.2.0 (http://iqtree.org) with default settings and ITOL v.6.1.2 (https://itol.embl.de).
The coassembled scaffolds (from metagenomics), transcripts (from metatranscriptomics), and quality bins were used separately for protein-coding region prediction using Prodigal v.2.6.3 (53), and the translated proteins were then functionally annotated through a GHOSTX search of the GhostKOALA server against a nonredundant set of KEGG genes (54). For further confirmation, random annotations obtained from the GhostKOALA were cross-verified using eggNOG-mapper v.2.1.0 (55). Secondary metabolite biosynthesis gene clusters (for siderophore synthesis annotation) were identified with antiSMASH v.5.0 (56). In addition, genes involved in carbohydrate metabolism were annotated using the dbCAN2 package v.2.0.11 via the CAZy database (57). Protein-coding gene reads were quantified using FeatureCounts v.2.0.1 (58) and were later normalized for gene length and sequencing depth using the TPM (DNA, in this case) normalization method. For metatranscriptomics, a differential gene expression analysis was used using the nonnormalized (raw) transcript counts (quantified by FeatureCounts) via DESeq2 v.1.28.1 (59). The output of DESeq2 was then used for gene set enrichment analysis (GSEA) via clusterProfiler v.4.0.5 (60) with a P value cutoff of 0.05 and 10,000 permutations.
nifH transcript RT-qPCR validation and sequencing
For reverse transcription, RNA extraction and DNase treatment were performed as described above for the samples collected from P-VI on day 450 of cocultures I and II. One hundred nanograms of extracted RNA was used for cDNA conversion using the POLF/POLR primers targeting the nifH gene with the HiScript III First-Strand cDNA Synthesis Kit (Vazyme Biotech., Nanjing, China). Negative control was prepared without the reverse transcriptase enzyme to check for gDNA contamination. qPCR was performed on three replicates each of cocultures I and II to quantify the expression of the nifH gene. The qPCR reaction mixture was set to 20 μl and consisted of 1 μl of cDNA, 1 μl each of POLF/POLR primers, 10 μl of ChamQ Universal SYBR qPCR Master Mix (Vazyme Biotech., Nanjing, China), and 7 μl of RNase-free distilled water. The reaction was performed on a Roche LightCycler 480 instrument (Roche Diagnostics, SZ) with the following profile: 1 cycle of predenaturation at 95°C for 1 min.; 34 cycles of 95°C for 10 s (denaturation), 55°C for 30 s (annealing), and 60°C for 30 s (extension), followed by a melting curve (95°C for 15 s, 60°C for 1 min, and 95°C for 15 s). The quality of the amplified nifH gene-specific cDNA was determined by gel electrophoresis and purified using an Axygen AxyPrep DNA gel extraction kit (Corning, NY, USA). Libraries were constructed using a NEXTFLEX Rapid DNA-Seq library prep kit (PerkinElmer, MA, USA) following the manufacturer’s instructions. Sequencing was carried out by Majorbio Bio-pharm (Shanghai, China) on an Illumina MiSeq PE300 sequencer. The sequenced reads were processed as previously described (see table S16 for the number of reads passed at each step). Here, the DADA2 filters were kept the same as mentioned above except for the maxEE and minFoldParentOverAbundance parameters, which were increased to (5,5) and 2, respectively. The final nonchimeric ASVs were checked for sequence length, and only sequences within a 240- to 380-bp range were taxonomically classified using the nifH database v.1.1.0 (61) with a minimum bootstrap value of 50.
Determination of δ15N and N2 fixation within particulate and dissolved N pool
δ15N and the nitrogen fixation rate were determined following published protocols (13). In brief, duplicate samples of 125 ml from the P-VI (day 450) phase of cocultures I and II were injected with 1 ml of atmospheric pressure equilibrated tracer 15N2 gas (99.8%; Cambridge Isotope Laboratories) in airtight serum bottles. For efficient dissolution of the tracer 15N2 gas into the seawater, the bottles were gently mixed by inversion for 5 min following the 15N2 bubble method (13). An initial sample (T0) was also collected and filtered immediately without any incubation on a precombusted (450°C for 5 hours) 0.3-μm pore-size glass-fiber filter paper (25 mm in diameter, Advantec GF75, Advantec, CA) under <400 mbar. Simultaneously, a control group was prepared without the addition of tracer 15N2 gas. The treatment (with 15N2 tracer gas) and the control groups were incubated for 48 hours under natural daylight conditions and later filtered as described above for the determination of particulate incorporation of 15N (PN). Simultaneously, 40 ml of the filtrate (i.e., passed through 0.3 μm) from each sample was collected and stored at −20°C until further analysis to measure the incorporation of 15N in the DN pool.
For PN incorporation of 15N, all the filters containing the coculture biomass were vacuum-dried for 24 hours, decarbonated by adding a few drops of 6 N HCl (Merck), and again dried in an oven at 50°C for 24 hours. The dried filters were introduced into an elemental analyzer (FLASH 2000 CHNS/O analyzer, Thermo Fisher Scientific) to determine total PN. The resulting gas from the elemental analyzer was then injected into the isotope ratio mass spectrometer (IsoPrime100, Elementar, Germany) for determining the nitrogen stable isotope ratios. For the DN incorporation of 15N, samples were oxidized to nitrate using a persulfate oxidation reagent (purified three to four times by recrystallization), and the concentration was determined using the chemiluminescent method (62). The 15N DN-derived nitrate was analyzed using the denitrifier method (63). Last, the nitrogen fixation rate was calculated (13, 64) as
| (1) |
where NFR is the N2 fixation rate (nM day−1), and APNc and APNf represent the isotopic ratio of 15N of the nitrogen pool of the control and the 15N tracer added bottles after the incubation, reported in units of atom % (i.e., 15N/[15N + 14N] × 100). PNc and PNf represent the concentrations of PN/DN of the nitrogen pool of the control and the treatment bottles after the incubation, respectively. AN2 is the isotopic ratio of 15N of the source gas in units of atom%, and Δt is the incubation period in days.
Statistical analysis
For the statistical significance of inorganic nutrients within the coculture system, a Student’s t test was used, with P < 0.05 indicating significance. In addition, a PCoA with a PERMANOVA as a statistical method and Bray-Curtis distance matrix was applied to 16S rRNA amplicon ASVs to investigate the relationship between the different phases of Synechococcus growth and the bacterial taxa using the Microbiomanalyst web-based package (https://microbiomeanalyst.ca). For species richness within samples (α diversity), the Shannon diversity index was measured using the ANOVA approach of the Microbiomanalyst web-based package. The correlation between inorganic nutrients and Synechococcus abundance with the bacterial taxa identified by 16S amplicon sequencing was performed by distance-based redundancy analysis, and the significance of the correlating factors was determined by the Mantel test using the Microeco package v.0.6.0 (65). The statistical significance among the analyzed metagenomes for the genes related to biofilm formation and PL function was established via a one-way ANOVA method. In addition, to investigate the effect size of significantly differentially abundant genes among the nitrogen and phosphorus cycles, an LDA analysis with a nonparametric factorial Kruskal-Wallis test was carried out using the Microbiomanalyst web-based package. Last, differentially abundant classes from nifH amplicon ASVs were identified using Metastat and a Student’s t test with a P value cutoff of 0.05 (65).
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China 42188102 and U1906216 (China); National Key Research and Development Program 2020YFA0608304 (China); and Senior User Project RV KEXUE KEXUE2019GZ03 supported by the Center for Ocean Mega-Science, Chinese Academy of Sciences (China).
Author contributions: Conceptualization: Y.Z. Methodology: Z.Z., S.N., H.Z., H.L., and H.S. Investigation: Z.Z. and S.N. Visualization: S.N. Supervision: Y.Z., Z.Z., S.-J.K., and N.J. Writing—original draft: S.N. Writing—review and editing: Y.Z., Z.Z., and S.N.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The sequences reported in this paper have been deposited in the NCBI Sequence Read Archive (www.ncbi.nlm.nih.gov) under accession no. PRJNA752793. The bioinformatics analysis workflow and code that supports the findings of this paper are available at https://doi.org/10.5281/zenodo.6872635.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S9
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S16
REFERENCES AND NOTES
- 1.Seymour J. R., Amin S. A., Raina J. B., Stocker R., Zooming in on the phycosphere: The ecological interface for phytoplankton-bacteria relationships. Nat. Microbiol. 2, 17065 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Flombaum P., Gallegos J. L., Gordillo R. A., Rincón J., Zabala L. L., Jiao N., Karl D. M., Li W. K. W., Lomas M. W., Veneziano D., Vera C. S., Vrugt J. A., Martiny A. C., Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. U.S.A. 110, 9824–9829 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Z., Nair S., Tang L., Zhao H., Hu Z., Chen M., Zhang Y., Kao S.-J., Jiao N., Zhang Y., Long-term survival of Synechococcus and heterotrophic bacteria without external nutrient supply after changes in their relationship from antagonism to mutualism. MBio 12, e0161421 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dvořák P., Casamatta D. A., Poulíčková A., Hašler P., Ondřej V., Sanges R., Synechococcus: 3 billion years of global dominance. Mol. Ecol. 23, 5538–5551 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Rachedi R., Foglino M., Latifi A., Stress signaling in cyanobacteria: A mechanistic overview. Life 10, 312 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sauer J., Schreiber U., Schmid R., Völker U., Forchhammer K., Nitrogen starvation-induced chlorosis in Synechococcus PCC 7942. Low-level photosynthesis as a mechanism of long-term survival. Plant Physiol. 126, 233–243 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christie-Oleza J. A., Sousoni D., Lloyd M., Armengaud J., Scanlan D. J., Nutrient recycling facilitates long-term stability of marine microbial phototroph-heterotroph interactions. Nat. Microbiol. 2, 17100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pomeroy L. R., le Williams P. J. B., Azam F., Hobbie J. E., The microbial loop. Oceanography 20, 28–33 (2007). [Google Scholar]
- 9.Bristow L. A., Mohr W., Ahmerkamp S., Kuypers M. M. M., Nutrients that limit growth in the ocean. Curr. Biol. 27, R474–R478 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Barcelos Ramos E. J., Schulz K. G., Voss M., Narciso Á., Müller M. N., Reis F. V., Cachão M., Azevedo E. B., Nutrient-specific responses of a phytoplankton community: A case study of the North Atlantic Gyre, Azores. J. Plankton Res. 39, 744–761 (2017). [Google Scholar]
- 11.Kurian S., Chndrasekhararao A. V., Vidya P. J., Shenoy D. M., Gauns M., Uskaikar H., Aparna S. G., Role of oceanic fronts in enhancing phytoplankton biomass in the eastern Arabian Sea during an oligotrophic period. Mar. Environ. Res. 160, 105023 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Mühlenbruch M., Grossart H. P., Eigemann F., Voss M., Mini-review: Phytoplankton-derived polysaccharides in the marine environment and their interactions with heterotrophic bacteria. Environ. Microbiol. 20, 2671–2685 (2018). [DOI] [PubMed] [Google Scholar]
- 13.White A. E., Granger J., Selden C., Gradoville M. R., Potts L., Bourbonnais A., Fulweiler R. W., Knapp A. N., Mohr W., Moisander P. H., Tobias C. R., Caffin M., Wilson S. T., Benavides M., Bonnet S., Mulholland M. R., Chang B. X., A critical review of the 15N2 tracer method to measure diazotrophic production in pelagic ecosystems. Limnol. Oceanogr. Methods 18, 129–147 (2020). [Google Scholar]
- 14.Cao X., Li H., Zhou Y., Song C., The shift of mutualistic relationships among algae, free-living and attached bacteria through different nutrient addition mode: A mesocosm study. J. Freshwater Ecol. 35, 535–548 (2020). [Google Scholar]
- 15.Buchan A., LeCleir G. R., Gulvik C. A., González J. M., Master recyclers: Features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 12, 686–698 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Seymour J. R., Ahmed T., Durham W. M., Stocker R., Chemotactic response of marine bacteria to the extracellular products of Synechococcus and Prochlorococcus. Aquat. Microb. Ecol. 59, 161–168 (2010). [Google Scholar]
- 17.Kearney S. M., Thomas E., Coe A., Chisholm S. W., Microbial diversity of co-occurring heterotrophs in cultures of marine picocyanobacteria. Environ. Microbiome 16, 1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.S. Colliec-Jouault, C. Bavington, C. Delbarre-Ladrat, Heparin-Like Entities from Marine Organisms in Handbook of Experimental Pharmacology (Springer, 2012), vol 207, pp. 423–449. [DOI] [PubMed] [Google Scholar]
- 19.Maeda K., Okuda Y., Enomoto G., Watanabe S., Ikeuchi M., Biosynthesis of a sulfated exopolysaccharide, synechan and bloom formation in the model cyanobacterium Synechocystis sp. strain PCC6803. Elife 10, e66538 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas F., Le Duff N., Di Wu T., Cébron A., Uroz S., Riera P., Leroux C., Tanguy G., Legeay E., Guerquin-Kern J. L., Isotopic tracing reveals single-cell assimilation of a macroalgal polysaccharide by a few marine Flavobacteria and Gammaproteobacteria. ISME J. 15, 3062–3075 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ben Francis T., Bartosik D., Sura T., Sichert A., Hehemann J. H., Markert S., Schweder T., Fuchs B. M., Teeling H., Amann R. I., Becher D., Changing expression patterns of TonB-dependent transporters suggest shifts in polysaccharide consumption over the course of a spring phytoplankton bloom. ISME J. 15, 2336–2350 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reintjes G., Arnosti C., Fuchs B., Amann R., Selfish, sharing and scavenging bacteria in the Atlantic Ocean: A biogeographical study of bacterial substrate utilisation. ISME J. 13, 1119–1132 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francis B., Urich T., Mikolasch A., Teeling H., Amann R., North Sea spring bloom-associated Gammaproteobacteria fill diverse heterotrophic niches. Environ. Microbiome 16, 15 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou J., Richlen M. L., Sehein T. R., Kulis D. M., Anderson D. M., Cai Z., Microbial community structure and associations during a marine dinoflagellate bloom. Front. Microbiol. 9, 1201 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green E. R., Mecsas J., Bacterial secretion systems: An overview. Microbiol. Spectr. 4, 10.1128/microbiolspec.VMBF-0012-2015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abby S. S., Touchon M., Jode A. E., Grimsley N., Piganeau G., Bacteria in Ostreococcus tauri cultures—Friends, foes or hitchhikers? Front. Microbiol. 5, 505 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tseng T.-T., Tyler B. M., Setubal J. C., Protein secretion systems in bacterial-host associations and their description in the Gene Ontology. BMC Microbiol. 9, S2 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whalen K. E., Becker J. W., Schrecengost A. M., Gao Y., Giannetti N., Harvey E. L., Bacterial alkylquinolone signaling contributes to structuring microbial communities in the ocean. Microbiome 7, 93 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fei C., Ochsenkühn M., Shibl A., Isaac A., Wang C., Amin S., Quorum sensing regulates “swim-or-stick” lifestyle in the phycosphere. Environ. Microbiol. 11, 4761–4778 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang W., Cerdán-García E., Berthelot H., Polyviou D., Wang S., Baylay A., Whitby H., Planquette H., Mowlem M., Robidart J., Cassar N., New insights into the distributions of nitrogen fixation and diazotrophs revealed by high-resolution sensing and sampling methods. ISME J. 14, 2514–2526 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forchhammer K., Schwarz R., Nitrogen chlorosis in unicellular cyanobacteria—A developmental program for surviving nitrogen deprivation. Environ. Microbiol. 21, 1173–1184 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Roth-Rosenberg D., Aharonovich D., Luzzatto-Knaan T., Vogts A., Zoccarato L., Eigemann F., Nago N., Grossart H. P., Voss M., Sher D., Prochlorococcus cells rely on microbial interactions rather than on chlorotic resting stages to survive long-term nutrient starvation. MBio 11, e01846-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kong W. L., Wu X. Q., Zhao Y. J., Effects of Rahnella aquatilis JZ-GX1 on treat chlorosis induced by iron deficiency in Cinnamomum camphora. J. Plant Growth Regul. 39, 877–887 (2020). [Google Scholar]
- 34.Wang D., Xu A., Elmerich C., Ma L. Z., Biofilm formation enables free-living nitrogen-fixing rhizobacteria to fix nitrogen under aerobic conditions. ISME J. 11, 1602–1613 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stief P., Kamp A., Thamdrup B., Glud R. N., Anaerobic nitrogen turnover by sinking diatom aggregates at varying ambient oxygen levels. Front. Microbiol. 7, 98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohen R. A., Walker K., Carpenter E. J., Polysaccharide addition effects on rhizosphere nitrogen fixation rates of the California cordgrass, Spartina foliosa. Wetlands 29, 1063–1069 (2009). [Google Scholar]
- 37.Shiozaki T., Ijichi M., Kodama T., Takeda S., Furuya K., Heterotrophic bacteria as major nitrogen fixers in the euphotic zone of the Indian Ocean. Global Biogeochem. Cycles 28, 1096–1110 (2014). [Google Scholar]
- 38.Delmont T. O., Quince C., Shaiber A., Esen Ö. C., Lee S. T., Rappé M. S., MacLellan S. L., Lücker S., Eren A. M., Nitrogen-fixing populations of Planctomycetes and Proteobacteria are abundant in surface ocean metagenomes. Nat. Microbiol. 3, 804–813 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chuckran P. F., Fofanov V., Hungate B. A., Morrissey E. M., Schwartz E., Walkup J., Dijkstra P., Rapid response of nitrogen cycling gene transcription to labile carbon amendments in a soil microbial community. mSystems 6, e00161-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wyman M., Bird C., Lack of control of nitrite assimilation by ammonium in an oceanic Picocyanobacterium, Synechococcus sp. strain WH 8103. Appl. Environ. Microbiol. 73, 3028–3033 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Browning T. J., Achterberg E. P., Rapp I., Engel A., Bertrand E. M., Tagliabue A., Moore C. M., Nutrient co-limitation at the boundary of an oceanic gyre. Nature 551, 242–246 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Blanco-Ameijeiras S., Cosio C., Hassler C. S., Long-term acclimation to iron imitation reveals new insights in metabolism regulation of Synechococcus sp. PCC7002. Front. Mar. Sci. 4, 247 (2017). [Google Scholar]
- 43.Pérez A. A., Liu Z., Rodionov D. A., Li Z., Bryant D. A., Complementation of cobalamin auxotrophy in Synechococcus sp. strain PCC 7002 and validation of a putative cobalamin riboswitch in vivo. J. Bacteriol. 198, 2743–2752 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clarke E. L., Taylor L. J., Zhao C., Connell A., Lee J. J., Fett B., Bushman F. D., Bittinger K., Sunbeam: An extensible pipeline for analyzing metagenomic sequencing experiments. Microbiome 7, 46 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P., DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kopylova E., Noé L., Touzet H., SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28, 3211–3217 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Bushmanova E., Antipov D., Lapidus A., Prjibelski A. D., RnaSPAdes: A de novo transcriptome assembler and its application to RNA-seq data. Gigascience 8, giz100 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mikheenko A., Saveliev V., Gurevich A., MetaQUAST: Evaluation of metagenome assemblies. Bioinformatics 32, 1088–1090 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Uritskiy G. V., DiRuggiero J., Taylor J., MetaWRAP-a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 6, 158 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P., Tyson G. W., CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells and metagenomes. Genome Res. 25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eren A. M., Esen Ö. C., Quince C., Vineis J. H., Morrison H. G., Sogin M. L., Delmont T. O., Anvi’o: An advanced analysis and visualization platform for omics data. Peer J. 3, e1319 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chaumeil P.-A., Mussig A. J., Hugenholtz P., Parks D. H., GTDB-Tk: A toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36, 1925–1927 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hyatt D., Chen G. L., LoCascio P. F., Land M. L., Larimer F. W., Hauser L. J., Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kanehisa M., Goto S., KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huerta-Cepas J., Szklarczyk D., Heller D., Hernández-Plaza A., Forslund S. K., Cook H., Mende D. R., Letunic I., Rattei T., Jensen L. J., von Mering C., Bork P., EggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kai B., Shaw S., Steinke K., Villebro R., Ziemert N., Lee S. Y., Medema M. H., Weber T., antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 47, W81–W87 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang H., Yohe T., Huang L., Entwistle S., Wu P., Yang Z., Busk P. K., Xu Y., Yin Y., DbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 46, W95–W101 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liao Y., Smyth G. K., Shi W., FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu G., Wang L.-G., Han Y., He Q.-Y., clusterProfiler: An R package for comparing biological themes among gene clusters. Omi. A J. Integr. Biol. 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moynihan M. A., Goodkin N. F., Morgan K. M., Phyllis Kho Y. Y., Lopes dos Santos A., Federico M. L., Baker D. M., Martin P., Coral-associated nitrogen fixation rates and diazotrophic diversity on a nutrient-replete equatorial reef. ISME J. 16, 233–246 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu Y., Wen Z., Shi D., Chen M., Zhang Y., Bonnet S., Li Y., Tian J., Kao S. J., Effect of light on N2 fixation and net nitrogen release of Trichodesmium in a field study. Biogeosciences 15, 1–12 (2018). [Google Scholar]
- 63.Sigman D. M., Casciotti K. L., Andreani M., Barford C., Galanter M., Böhlke J. K., A bacterial method for the nitrogen isotopic analysis of nitrate in seawater and freshwater. Anal. Chem. 73, 4145–4153 (2001). [DOI] [PubMed] [Google Scholar]
- 64.Xu M. N., Zhang W., Zhu Y., Liu L., Zheng Z., Wan X. S., Qian W., Dai M., Gan J., Hutchins D. A., Kao S.-J., Enhanced ammonia oxidation caused by lateral Kuroshio intrusion in the boundary zone of the Northern South China Sea. Geophys. Res. Lett. 45, 6585–6593 (2018). [Google Scholar]
- 65.Liu C., Cui Y., Li X., Yao M., microeco: An R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 97, fiaa255 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S9
Tables S1 to S16