Abstract
Identification of new markers associated with long-term efficacy in patients treated with CAR T cells is a current medical need, particularly in diseases such as multiple myeloma. In this study, we address the impact of CAR density on the functionality of BCMA CAR T cells. Functional and transcriptional studies demonstrate that CAR T cells with high expression of the CAR construct show an increased tonic signaling with up-regulation of exhaustion markers and increased in vitro cytotoxicity but a decrease in in vivo BM infiltration. Characterization of gene regulatory networks using scRNA-seq identified regulons associated to activation and exhaustion up-regulated in CARHigh T cells, providing mechanistic insights behind differential functionality of these cells. Last, we demonstrate that patients treated with CAR T cell products enriched in CARHigh T cells show a significantly worse clinical response in several hematological malignancies. In summary, our work demonstrates that CAR density plays an important role in CAR T activity with notable impact on clinical response.
High CAR molecule density affects CAR T cell activity and is associated with impaired clinical response.
INTRODUCTION
Chimeric antigen receptor (CAR) T cell therapies have emerged as a promising therapeutic tool against cancer, revolutionizing cancer immunotherapy (1). Second-generation CAR T cells have shown to induce impressive clinical responses in hematological malignancies, such as chemotherapy-resistant B cell leukemias and lymphomas (2–4) and multiple myeloma (MM) (5–7). Despite the high rates of remissions, not every patient achieves a complete response (CR) after CAR T cell therapy. In addition, a substantial number of patients experience a relapse of the disease. Particularly in patients with MM, despite their impressive responses, apparently, so far, there is no plateau in the survival curves after CAR T cell therapy (5–7), which contrasts with results obtained with CD19 CAR T cells in acute lymphoblastic leukemia and non-Hodgkin’s lymphomas.
It is well known that CAR T cells are heterogeneous products with multiple factors contributing to their efficacy. Several studies have demonstrated how extrinsic factors, such as antigen density or tumor burden, strongly influence efficacy of CAR T cells (8, 9). In addition, factors related to CAR structure, such as the costimulatory domain or the hinge length, can also affect the antitumoral potential of CAR T cells (10–12). Moreover, intrinsic cell factors, such as the differentiation state of T cells, the CD4/CD8 ratio, or the T cell polyfunctionality, have been correlated with the therapeutic efficacy and have led to the hypothesis that enrichment of CAR T cell products in T cells with a more immature phenotype may be associated with improvement in long-term responses (13–16). On the other hand, an increase in T cells with an effector or exhausted phenotype may result in a reduced persistence of CAR T cells with a decrease in clinical response (13). Previous studies identified that CAR signaling in the absence of antigen stimulation, denominated tonic signaling, is associated with early T cell exhaustion (17–20), suggesting its role in triggering premature T cell dysfunction. Additional factors related to the CAR construct may have an impact on functionality. For instance, recent studies have demonstrated that a more physiological expression of the CAR, by the integration of the transgene in the TRAC locus or the use of different promoters, can be associated with improved efficacy and reduced toxicity (21–23). These results also suggest that the density of CAR molecule in the membrane of CAR T cells might influence CAR signaling, affecting their antitumoral efficacy (23, 24). However, this hypothesis and the impact on clinical efficacy have not been formally explored.
Technological advances in genomics, such as single-cell sequencing, have allowed a notable progress toward understanding the genomic landscape of CAR T cells, providing some mechanistic insights into proper CAR T cell function (25–29). Moreover, functional commitment of CAR T cells is governed by complex gene regulatory networks (GRNs) that control CAR T cells at baseline as well as CAR T cell dynamics after antigen recognition being essential for CAR T functionality (30). Using single-cell RNA sequencing (scRNA-seq), recent studies have identified specific T cell signatures associated with efficacy and toxicity in patients with large B cell lymphomas (26), or molecular determinants of CAR T cell persistence such as IRF7-mediated regulation of chronic interferon signaling (29), establishing this technology as a useful tool to improve efficacy of CAR T.
In this study, we address the impact of CAR density on the functionality of B cell maturation antigen (BCMA) CAR T cells. Phenotypic, functional, transcriptomic, and epigenomic studies at bulk and single-cell level revealed different profiles between CAR T cells with high and low expression of the CAR molecule (CARHigh T and CARLow T cells). We show that CARHigh T cells are associated with tonic signaling and an exhausted phenotype, and identify the molecular mechanisms involved in the different functionalities of CARHigh T and CARLow T cells. We define a molecular signature associated with increased CAR density that, when applied to CAR T cell products, may predict clinical response. These results would provide a useful tool to understand the mechanisms behind proper CAR T cell function and identify biomarkers of response with potential clinical implications.
RESULTS
CAR T cells exhibit a wide range of CAR density on cell surface that influences CAR-mediated signaling
Current CAR T cell products are generated using retroviral/lentiviral (LV) vectors that render different levels of transduction and transgene expression within the cells, consequently observing a wide range of CAR molecule densities on the surface of transduced cells (fig. S1). We hypothesized that this heterogeneity in CAR density could affect CAR-mediated signaling and, hence, influence the efficacy of CAR T cell products. To address this question, we first used a triple parameter reporter (TPR) system in Jurkat cells (31) to measure, by flow cytometry, the CAR-mediated activation of the main signaling pathways [nuclear factor of activated T cells (NFAT), nuclear factor κB (NFκB), and activator protein 1 (AP1)] after tumor recognition using CAR T cells with different levels of CAR on the cell surface. Jurkat-TPR cells were infected with a second-generation CAR construct targeting BCMA, with 4-1BB as costimulatory domain, that was further modified to include a truncated epidermal growth factor receptor (EGFRt) reporter, facilitating the measurement of CAR level (fig. S2A). Then, subsets of Jurkat-TPR cells presenting different levels of CAR (termed CARHigh and CARLow cells) were selected according to the fluorescence intensity (FI) of EGFRt (top and bottom FI quartiles, respectively; see Materials and Methods) (fig. S2B) and analyzed after coculture with different BCMA-expressing MM cell lines. We observed significantly increased levels of activation in the three mentioned pathways within the CARHigh population (fig. S2C). Moreover, a significant increase of activation of CARHigh cells was also observed even in the absence of tumor cells, indicating an increase in tonic signaling at baseline (fig. S2D). We consistently observed this functional pattern with other CAR constructs targeting CD19, CD33, and HER2 (fig. S2, C and D), indicating that a higher density of CAR molecules in the cell surface increases both the tonic signaling and the signal transduction after tumor encountering.
CAR density influences antitumoral response of CAR T cells targeting BCMA
To further analyze the effect of CAR density on antitumoral efficacy, we characterized CAR T cells from 10 healthy donors that were generated using a BCMA-targeting CAR construct (derived from ARI-0002h) coexpressing blue fluorescent protein (BFP) as a reporter marker (fig. S3). CAR T cells were sorted into CARHigh and CARLow subpopulations based on the expression of BFP (Fig. 1A and fig. S4A). CARHigh T cells include those cells with a BFP FI > 1.2 × 104 (average FI, 26,861 ± 5795), while the CARLow T cell subpopulation was restricted to BFP FI < 4 × 103 (average FI, 2513 ± 388). These FI values corresponded to the top and bottom FI quartiles, respectively (fig. S4A). Then, BFP FI values were used to quantify the number of CAR molecules on the surface of these two CAR T cell subpopulations using an antibody-binding capacity bead assay. CARHigh T cells presented more than 5000 CAR molecules per cell, while the number of molecules per cell in CARLow T cells was below 1500 (fig. S4B). Because integration site could affect CAR expression, we performed an integration site analysis in CARHigh T and CARLow T cells. No differences were observed in the integration profile between cells with different CAR densities, being most of the integrations located at active sites (i.e., promoter and intronic regions) as expected for LV vectors (fig. S4, C and D) (32, 33). However, vector copy number (VCN) analysis revealed a significantly higher number of viral integrations within CARHigh T cells, with an average of 4.5 ± 1.3 integrations in CARHigh T cells versus 1.8 ± 0.3 in CARLow T cells (fig. S4E). This increased VCN resulted in a significant increase in CAR mRNA expression levels (fig. S4F), which could explain the increased CAR density observed in these cells rather than due to the specific integration sites. We found increased cytotoxic activity and greater levels of interferon-γ (IFN-γ), interleukin-2 (IL-2), tumor necrosis factor–α (TNFα), and granzyme B (GZMB) production in CARHigh T cells (Fig. 1, B and C, and fig. S4G). To determine the translational value of these findings, we examined CAR levels in CAR T cell products from an academic clinical trial (CARTBCMA-HCB-01; NCT04309981) (fig. S4H). We observed an increased in vitro lytic activity in those CAR T cell products enriched in CARHigh T cells (>30% of cells with >5000 CAR molecules per cell) (fig. S4I).
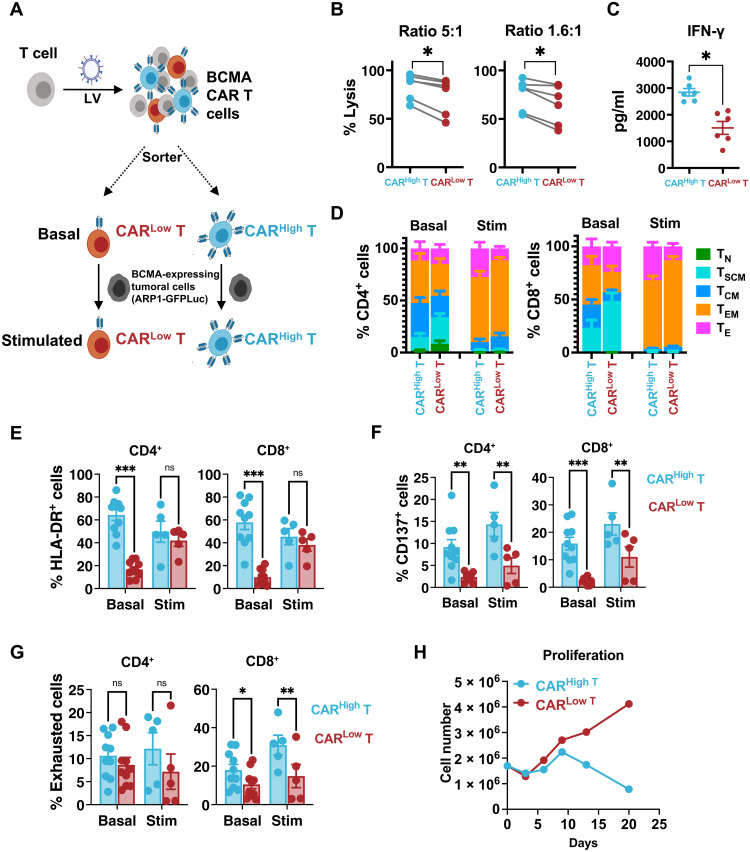
Fig. 1. CARHigh T cells present increased in vitro antitumoral efficacy and exhausted phenotype.
An in vitro functional and phenotypic characterization was performed on CAR T cells targeting BCMA presenting different densities of the CAR molecule. (A) Schematic representation of the procedure. CAR T cells were sorted into CARHigh T and CARLow T cell subpopulations via BFP expression. Analyses were performed at basal state or after stimulation with tumor cells expressing BCMA. (B) Quantification of the cytotoxic activity of CARHigh T and CARLow T cells against ARP1-GFPLuc at different effector to target (E:T) cell ratios. The percentage of specific lysis (average of three technical replicates) for each CAR T cell production (n = 6) is depicted. (C) Quantification of IFN-γ levels in supernatants from cytotoxic assays (ratio of 5:1) measured by enzyme-linked immunosorbent assay (ELISA). The cytokine concentration (pg/ml; average of three technical replicates) for each CAR T cell production (n = 6) is depicted. (D) Analysis of the phenotype of CARHigh T and CARLow T cell populations before (Basal; n = 10) and after stimulation (Stim; n = 5) with ARP1-GFPLuc tumor cells. Mean ± SEM of each T cell subpopulation within CARHigh T and CARLow T cells is depicted. TN, naïve; TSCM, stem central memory; TCM, central memory; TEM, effector memory; TE, effector. Analysis of the expression of HLA-DR (E), CD137 (F), and a combination of >2 exhaustion markers (LAG3, TIM3, and/or PD1) (G) in CARHigh T and CARLow T cells before (Basal; n = 10) and after stimulation (Stim; n = 5) with ARP1-GFPLuc tumor cells. ns, not significant. (H) Proliferation of CARHigh T and CARLow T cells after continuous repeated in vitro stimulation for 21 days with tumoral cells. Wilcoxon test for paired samples (B and C). Two-way analysis of variance (ANOVA) with Sidak’s multiple comparison (E to G). *P < 0.05, **P < 0.01, and ***P < 0.001.
Next, we analyzed the phenotype of CARHigh T and CARLow T cells before and after stimulation with tumor cells. No differences were observed in the CD4/CD8 ratio at any condition (fig. S5A). However, at baseline, we observed a statistically significant enrichment of central memory (TCM) and effector memory (TEM) phenotypes within CARHigh T cells, with concomitant reduction of naïve (TN) and stem central memory (TSCM) cells in both CD4+ and CD8+ subsets (Fig. 1D and fig. S5B). After antigen stimulation, both populations acquired a TEM-TE phenotype, although increased numbers of TE were observed in CARHigh T cells (Fig. 1D and fig. S5C). These results suggest a higher degree of differentiation in CARHigh T cells even in the absence of antigen stimulation. Moreover, we observed that CARHigh T cells presented an increase in basal activation, with significant higher levels of human leukocyte antigen (HLA)–DR+ and CD137+ cells, along with a higher percentage of CD8+ T cells expressing a combination of two or more markers of exhaustion (LAG3, TIM3, and/or PD1) (Fig. 1, E to G). These differences in cell exhaustion increased after antigen stimulation, with more than 30% of CD8+ CARHigh T cells expressing an exhausted phenotype (Fig. 1G). A continuous repeated in vitro stimulation for 21 days with tumoral cells revealed increased differences in the differentiation and exhausted phenotype, and an impaired proliferation potential of CARHigh T cells (Fig. 1H and fig. S5, D and E). Moreover, because Jurkat reporter system revealed signs of tonic signaling in CARHigh cells, we further analyze this phenomenon in CAR T cells using an NFAT–green fluorescent protein (GFP) reporter vector. In the absence of tumoral cells, we observed an increased number of cells expressing significantly higher levels (FI) of GFP within CARHigh T cells, corroborating the tonic signaling in this population (fig. S5F).
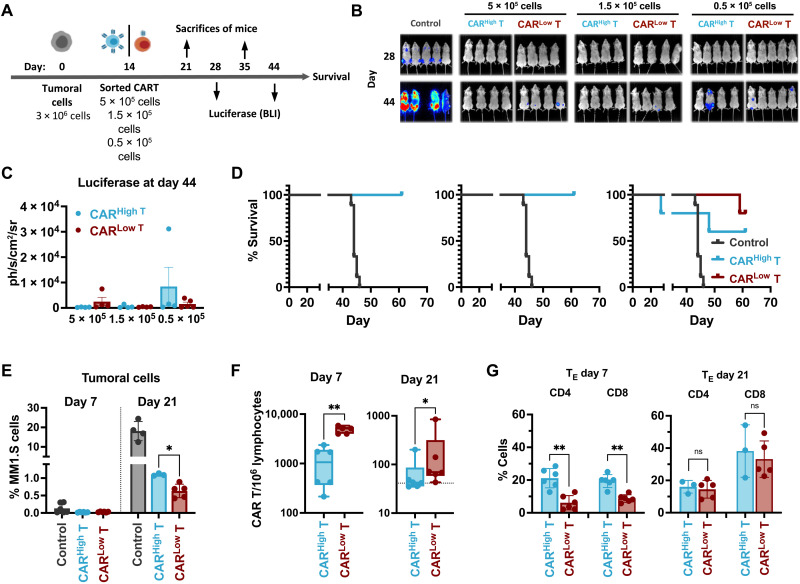
To determine whether different CAR densities would affect long-term antitumor potential, we further evaluated CAR T cell efficacy in vivo using a stress test in NOD-SCID-Il2rg−/− (NSG) mice (21, 34). Thus, MM1.S cells expressing luciferase (3 × 106 cells per animal) were transplanted in NSG mice, and after 14 days, different doses of fluorescence-activated cell sorting (FACS)–sorted CARHigh T or CARLow T cells (5 × 105, 1.5 × 105, and 0.5 × 105) were infused into the animals (Fig. 2A). All the animals treated with CAR T cells presented increased survival compared to controls with a dose-dependent response. At the lower dose, we observed an increased presence of tumoral cells in the animals treated with CARHigh T cells, as demonstrated by luciferase measurements (Fig. 2, B and C, and fig. S6A), which lead to a tendency of reducing the survival of the animals, although no statistical differences were observed (Fig. 2D). Increased tumor progression in animals treated with CARHigh T cells was confirmed by measuring the presence of tumoral cells in the bone marrow (BM) 21 days after treatment (Fig. 2E). We further characterized CAR T cell persistence, phenotype, and functionality at different time points after cell administration. First, we analyzed the infiltration of CAR T cells in the BM of the animals at 7 and 21 days after CAR T administration, and we observed a statistically significant reduced number of infiltrated CAR T cells in those animals treated with CARHigh T cells (Fig. 2F). Moreover, CARHigh T cells also presented a more differentiated phenotype at day 7 with increased percentage of terminally effector cells in both CD4+ and CD8+ subpopulations (Fig. 2G and fig. S6B). Last, functional ex vivo analysis of isolated CARHigh T and CARLow T cells 7 days after administration showed that CARHigh T cells were more cytotoxic with increased production of not only cytokines related to effector function (i.e., PRF1, GZMA, and GZMB) but also exhaustion markers (PD1, LAG3, and TIM3) (fig. S6, C and D).
Fig. 2. In vivo antitumoral efficacy of CAR T cells with different CAR densities.
(A) Schematic representation of the experimental procedure. NGS mice were injected intravenously on day 0 with 3 × 106 MM1.S-GFPLuc cells per animal. After 14 days, 5 × 105, 1.5 × 105, or 0.5 × 105 of CARHigh T or CARLow T cells were injected intravenously. Bioluminiscence analysis (BLI) was performed on days 28 and 44. Animal survival was monitored until the end of the experiment (day 67). On days 21 (7 days after CAR T administration) and 35 (21 days after CAR T administration), a subset of animals was sacrificed to analyze the presence of tumor and CAR T cells. (B) BLI images at the indicated days of control mice (n = 5) or treated with CARHigh T cells (n = 4 to 5) or CARLow T cells (n = 4 to 5). (C) Quantification of BLI (photons/s/cm2/sr) in the different group of animals as a measurement of tumor growth. (D) Survival of control mice (n = 5) or treated with CARHigh T cells (n = 4 to 5) or CARLow T cells (n = 4 to 5) at the indicated doses. (E) Quantification of the tumor cells present in the BM of control mice (n = 4) or treated with CARHigh T cells (n = 5) or CARLow T cells (n = 5) at days 7 and 21 after CAR T cell administration. (F) Quantification of the CAR T cells present in the BM of animals treated with CARHigh T cells (n = 6) or CARLow T cells (n = 6) at days 7 and 21 after CAR T cell administration. (G) Analysis of the phenotype of CARHigh T and CARLow T cell populations at days 7 and 21 after CAR T cells administration. Mean ± SEM of TE cell subpopulation within CD4+ and CD8+ subset of CARHigh T and CARLow T cells is depicted. Mantel-Cox (log-rank) test (D), Kruskal-Wallis test (E), Mann-Whitney test (F), and two-way ANOVA with Sidak’s multiple comparison (G). *P < 0.05 and **P < 0.01.
Overall, these results indicate that CARHigh T cells show increased activation and tonic signaling associated with increased in vitro cytotoxicity. Moreover, the more differentiated and exhausted phenotype observed in CARHigh T cells (both in vitro and in vivo) together with the decreased BM infiltration capacity resulted in lower in vivo antitumoral effect under stressed conditions, which could have an impact on the clinical response.
CARHigh T cells display different transcriptomic and chromatin landscape with increased tonic signaling and T cell activation
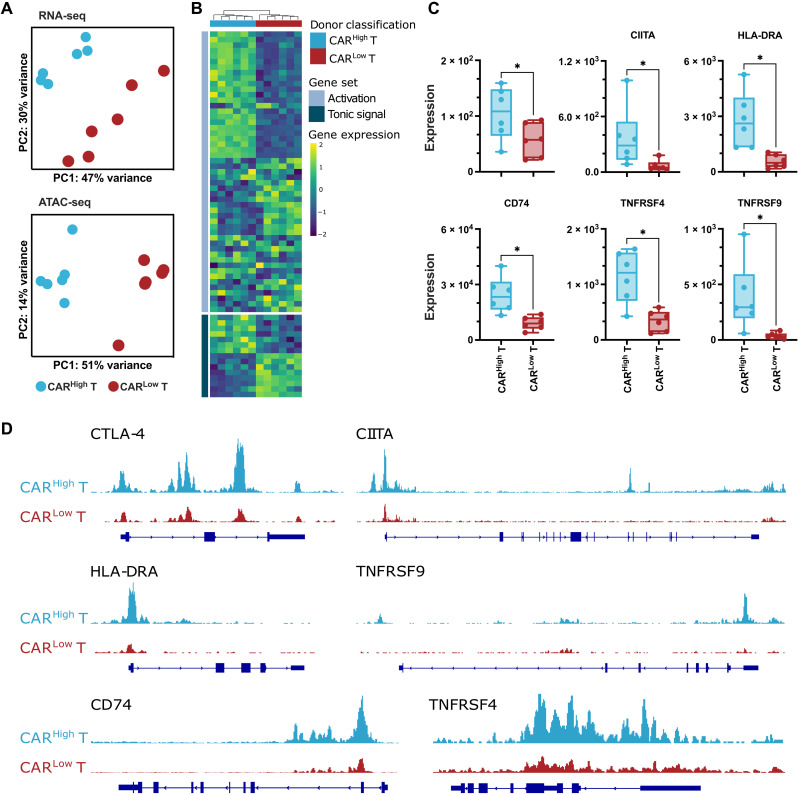
Given the differences observed in phenotype and persistence between CARHigh T and CARLow T cells, we delved into the transcriptomic and epigenetic landscape of the two subpopulations of CAR T cells. Sorted CD4+ and CD8+ CARHigh T and CARLow T cell populations from six different CAR T cell productions were profiled using high-throughput RNA-seq and assay for transposase-accessible chromatin with sequencing (ATAC-seq). Transcriptomic analysis revealed less than 100 differentially expressed genes [DEGs; false discovery rate (FDR) < 0.05, log2 fold change (Log2FC) > 2] between CARHigh T and CARLow T cells (table S1). Similarly, the analysis of the ATAC-seq data revealed (i) a similar peak distribution in both populations, with >60% of the peaks located within the promoter regions and the first intron, and (ii) a limited number of differential accessible regions identified between CARHigh T and CARLow T cells (fig. S7 and table S2). These small transcriptomic and epigenomic differences were enough to separate CARHigh T cells from CARLow T cells in a principal components analysis in both CD4+ and CD8+ T cell subsets (Fig. 3A and fig. S8A). DEGs between CARHigh T and CARLow T cells were associated with genes involved in tonic signaling and T cell activation (Fig. 3B and fig. S8B). In particular, CARHigh T cells showed increased expression of genes related to lymphocyte activation, such as HLA-DRA, CIITA, and CD74, as well as costimulatory molecules, such as CTLA-4, TNFRSF4 (OX40), and TNFRSF9 (4-1BB) (Fig. 3C and fig. S8C). These results corroborated our phenotypic observations (see previous sections). While reduced overlapping was observed between DEGs and differential peaks, those genes showing differential expression and chromatin accessibility were mainly related to T cell activation and costimulation (Fig. 3D and fig. S8, D and E). Together, these results suggest that small differences in gene expression and chromatin accessibility associated to increased CAR levels can substantially modify the overall phenotypic and functional profile of CAR T cells.
Fig. 3. Transcriptomic profile and chromatin landscape of CD8+ CARHigh T cells.
The transcriptomic and epigenetic landscape of sorted CD8+ and CD4+ (see fig. S8) CARHigh T and CARLow T cells (n = 6) was profiled using high-throughput RNA-seq and ATAC-seq. (A) RNA-seq and ATAC-seq principal components (PC) analysis, corrected by patient heterogeneity, of sorted CD8+ CAR T cell subsets. Percentage of variance explained by PC1 and PC2 is depicted. (B) Heatmap of DEGs between CD8+ CARHigh T and CARLow T cells associated to genes involved in tonic signaling and T cell activation. (C) Quantification of CTLA-4, CIITA, HLA-DRA, CD74, TNFRSF4 (OX40), and TNFRSF9 (4-1BB) gene expression in CD8+ CARHigh T and CARLow T cells. (D) UCSC genome browser tracks of CTLA-4, CIITA, HLA-DRA, TNFRSF9, CD74, and TNFRSF4 showing differential peaks from ATAC-seq analysis between CD8+ CARHigh T and CARLow T cells. Wilcoxon test for paired samples (C). *P < 0.05.
Single-cell sequencing reveals specific distribution of CARHigh T cells
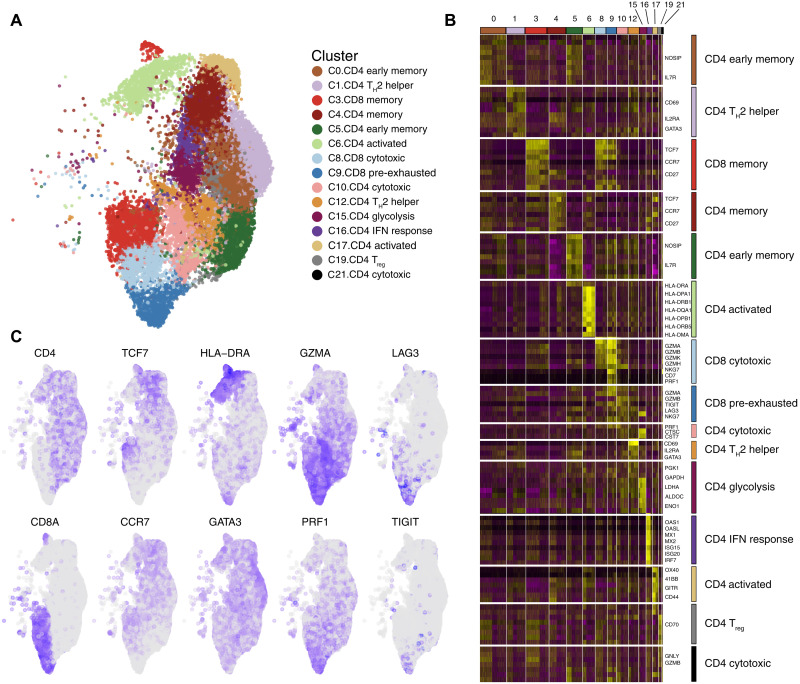
To better understand the heterogeneity of CAR T cells and the influence of CAR level on their transcriptomic profile, we performed single-cell transcriptomic analysis on 43,981 CAR T cells from three independent productions. After quality control and filtering, we performed an integrated analysis and identified 23 clusters of CAR T cell subpopulations (fig. S9A). Clusters identified based on cell cycle gene signatures, those containing high levels of mitochondrial genes, and clusters lacking expression of T cell markers were excluded from further analysis (fig. S9, B to F). Cell types and functional states of the remaining 15 clusters (containing 28,117 cells with range of 7416 to 105,093 cells per donor) were defined according to the expression of previously described canonical markers (Fig. 4, A and B) (26–28). Within CD4+ cells, we identified early memory (IL7R), memory (TCF7, CCR7, and CD27), activated (HLA-DRA and OX40), cytotoxic (GZMA and PRF1), and T helper 2 (TH2) (GATA3) CAR T cells, with a minority of other subtypes including cells expressing genes related to glycolysis and IFN response. Among CD8+ cells, we distinguished memory (TCF7 and CCR7) and cytotoxic (GZMA, PRF1, and NKG7) CAR T cells. Furthermore, in accordance with the phenotypical results, we identified a pre-exhausted cluster of CD8+ CAR T cells characterized by the expression of cytotoxic genes along with inhibitory receptors, such as LAG3 and TIGIT (Fig. 4, B and C) (35). Analysis of V(D)J rearrangement showed a polyclonal diversity of CAR T cells within all the clusters, indicating no specific enrichment of any particular clone (table S3).
Fig. 4. Characterization of CAR T cells at single-cell level.
CAR+ (BFP+) cells from three independent CAR T cell productions were assayed by scRNA-seq. (A) An overview of the 28,117 cells that passed quality control and filtering for subsequent analyses in this study. Uniform Manifold Approximation and Projection (UMAP) plot showing the 15 clusters that were analyzed. (B) Heatmap showing signature genes of each cluster and putative assignments to cell types according to canonical marker genes. (C) UMAP plot overlaid with mRNA expression of T cell markers (CD4 and CD8A), memory markers (TCF7 and CCR7), activation markers (HLA-DRA and GATA3), effector enzymes (GZMA and PRF1), and exhaustion markers (LAG3 and TIGIT).
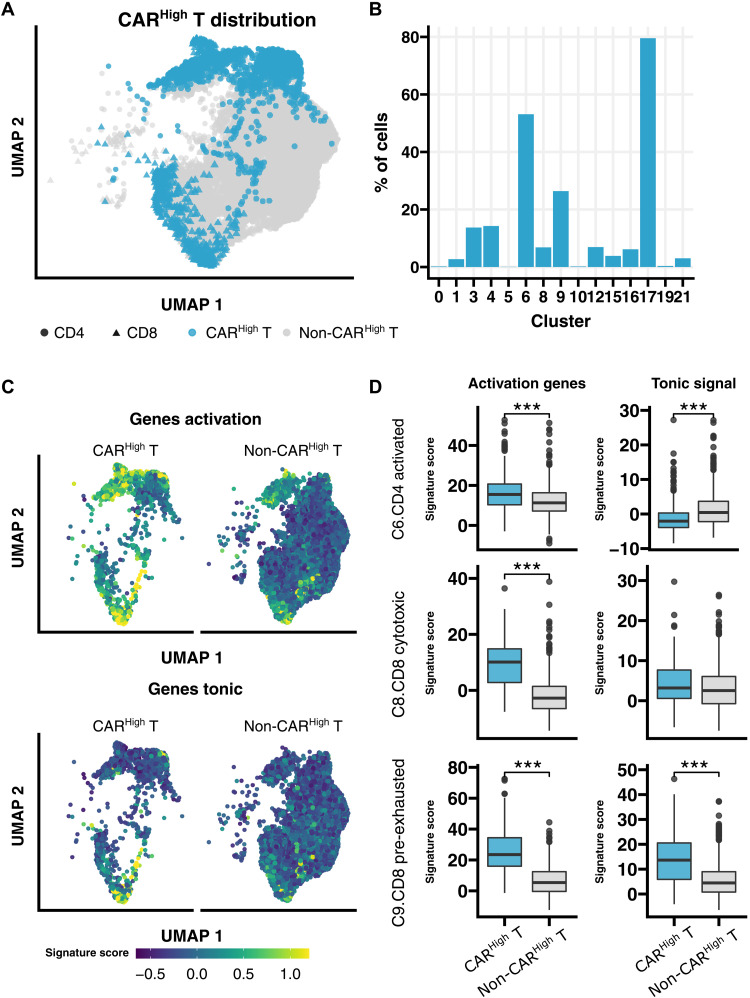
To robustly identify CAR T cells with high expression of the CAR construct in our single-cell data, we developed gene signatures associated to both CD4+ and CD8+ CARHigh T cells using all DEGs (Log2FC > 1, FDR < 0.05) from bulk RNA-seq analysis between CARHigh T and CARLow T cells (table S4 and fig. S10A), as these populations were sorted on the basis of CAR protein expression (see Materials and Methods). We found a better correlation between the gene signature and CAR protein level than the one observed between the expression of the CAR gene and its protein expression (fig. S10B). Moreover, the possibility to extrapolate these gene signatures to other datasets was assessed via leave-one-out cross-validation (fig. S10C). Then, we annotated CARHigh T cells in our single-cell data using the developed signatures (Fig. 5A), and we observed that CARHigh T cells mainly localized within activated CD4+ cells, representing more than 50% of the cells in cluster 6 and almost 80% of the cells in cluster 17 (Fig. 5B). Furthermore, in the CD8+ T cell compartment, CARHigh T cells were more represented in cluster 9, showing a pre-exhausted phenotype (Fig. 5B). Moreover, CARHigh T cells were significantly enriched in activation and tonic signaling signatures (Fig. 5, C and D), results that are in accordance with our previous phenotypic and transcriptomic analysis performed in FACS-sorted CARHigh T and CARLow T cell populations. In addition, we perform this analysis using an independent single-cell public dataset of CAR T products targeting CD19 from patients with diffuse large B cell lymphoma (DLBCL) (26), obtaining similar results (fig. S11). Together, our results would indicate a strong association among tonic signaling, CAR T cell activation, and cell exhaustion that is increased in CAR T cells with high CAR density defined by either the gene signature or the protein expression.
Fig. 5. Single-cell sequencing reveals specific distribution of CARHigh T cells.
Annotation and further analysis of CARHigh T in the single-cell data were performed by applying the gene signatures, developed in this work, associated to both CD4+ and CD8+ CARHigh T cells that showed higher correlation with the CAR protein level than that yielded by the CAR gene expression. (A) UMAP plot showing CARHigh T cell distribution across analyzed cells. (B) Quantification of the percentage of CARHigh T cell along the different clusters. In accordance with phenotypic results, CARHigh T cells are mainly localized within activated CD4+ cells (clusters 6 and 17) and pre-exhausted CD8+ T cells (cluster 9). (C) UMAP plots overlaid with the score of activation and tonic signaling signatures, showing their distribution across cells. CARHigh T cells were enriched in the score for both signatures. (D) Quantification of the signature score in CARHigh T cells from clusters 6 (CD4+ activated), 8 (CD8+ cytotoxic), and 9 (CD8+ pre-exhausted), in comparison with the rest of the cells, for both activation and tonic signaling signatures. CARHigh T cells presented a significant increase for both signatures in almost all three clusters analyzed. Wilcoxon test (D). ***P < 0.001.
CAR density is associated with differential activation of regulatory networks
To elucidate the molecular regulation of CARHigh T cells, we applied SimiC (36), a novel GRN inference algorithm for scRNA-seq data that imposes a similarity constraint when jointly inferring the GRNs for each specific cell state. On the basis of this analysis, we observed regulons [a transcription factor (TF) and its associated target genes] that were similarly activated between CARHigh T and the rest of the CAR T cells (fig. S12), such as regulons implicated in T cell differentiation (GATA3 and RUNX3) and signal transduction (STAT1, REL, RELA, and JUN/AP1) (37–39). On the other hand, we identified some regulons that were more active in CARHigh T cells, such as STAT3, a TF associated with development and maintenance of T cell memory (40), or ARID5A, a TF related to the control of the stability of STAT3 (41) (fig. S12), and other regulons that presented reduced activity in CARHigh T cells, such as BTG2, a TF related to the prevention of proliferation exacerbation and spontaneous activation (42) (fig. S12). These changes in regulon activity could explain the increased central memory phenotype and also provide a regulatory mechanism for the increased activation observed in CARHigh T cells.
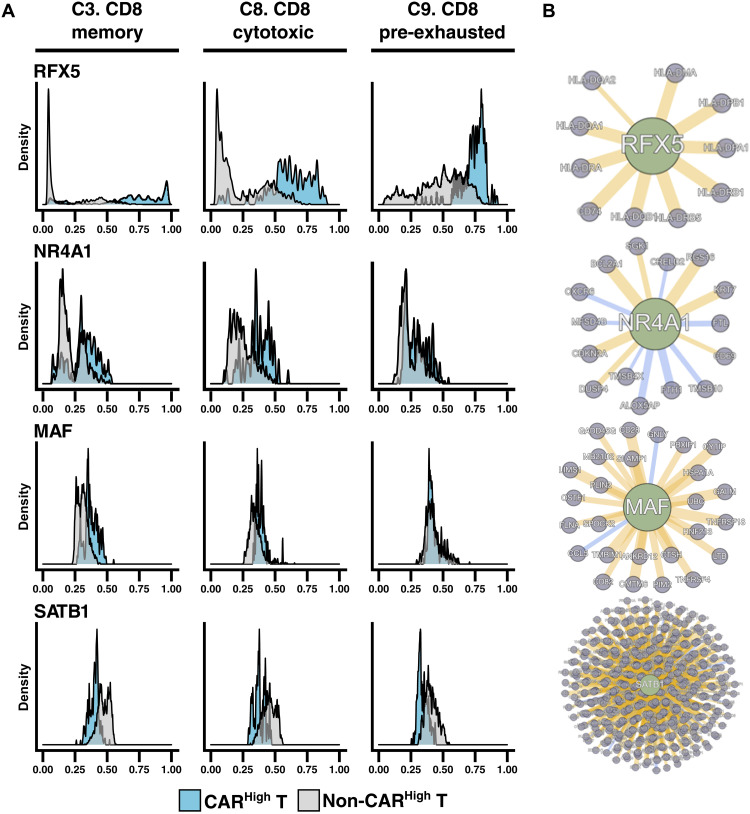
We also observed regulons presenting a multimodal activation profile. To determine whether this distribution might be related to different activity between clusters, we computed the distribution of the regulon activity in each cluster (provided it contains at least 5% of CARHigh T cells) (fig. S12). As an example, we observed that the activity of RFX5 regulon, a member of the RFX family that interacts with HLA class II genes and promotes their transcription (43, 44), was overexpressed in CD8+ CARHigh T cells independently of the T cell subtype. However, RFX5 regulon activity progressively increased through CD8+ differentiation when we analyzed CD8+ CAR T cells that were not CARHigh T cells from memory to cytotoxic and, finally, to pre-exhausted cells (Fig. 6). In addition, we searched for regulons that could explain the exhausted phenotype observed in CARHigh T cells. We observed that NR4A1 and MAF regulons, already described as drivers of T cell exhaustion (45–47), were more active in CARHigh T cells (Fig. 6). On the other hand, the SATB1 regulon, related to PD1 inhibition (48), presented lower activity in CARHigh T cells (Fig. 6). All these results may shed light on the molecular mechanism underpinning the exhausted phenotype observed in CARHigh T cells.
Fig. 6. CAR density is associated with differential activation of regulatory networks.
Dynamics in GRNs of CARHigh T cells were analyzed by SimiC, a novel GRN inference algorithm for scRNA-seq data that imposes a similarity constraint when jointly inferring the GRNs for each specific cell state. Histograms showing the activity score during CD8+ T cell differentiation in CARHigh T cells versus the rest of the cells (A) and the inferred gene network (B) of regulons RFX5, NR4A1, MAF, and SATB1. Regulon activity of RFX5, a member of the RFX family that interacts with HLA class II genes and promotes their transcription, and NR4A1 and MAF, which have been described as drivers of T cell exhaustion, was already high in CARHigh T cells independently of the phenotype; meanwhile, in the rest of the cells, their activity progressively increased from CD8+ memory (cluster 3) to CD8+ cytotoxic (cluster 8) and finally to CD8+ pre-exhausted (cluster 9) phenotypes. In contrast, the activity of SATB1 regulon, related to PD1 inhibition, was reduced in CARHigh T cells.
Moreover, we integrated the GRN analysis with our ATAC-seq data form sorted CARHigh T and CARLow T cells. We observed that the binding motifs of the TF from regulons presenting increased activity in CARHigh T cells, such as ARID5A, RFX5, or MAF, were enriched in regions with differential accessibility between CARHigh T and CARLow T cells (fig. S13 and table S8). These results would suggest a functional association between the epigenetic regulation and the regulons differentially active in CARHigh T cells.
Overall, our GRN analysis using SimiC provides mechanistic insights into the regulatory networks behind the phenotypic and functional differences observed in CARHigh T cells, identifying regulons that regulate T cell function previously described, supporting the usefulness of this methodology. The use of SimiC permits the generation of a hypothesis based on identified regulatory factors that could be modulated, ultimately to the design of optimized CAR T therapies.
CARHigh T gene signature is associated with clinical response
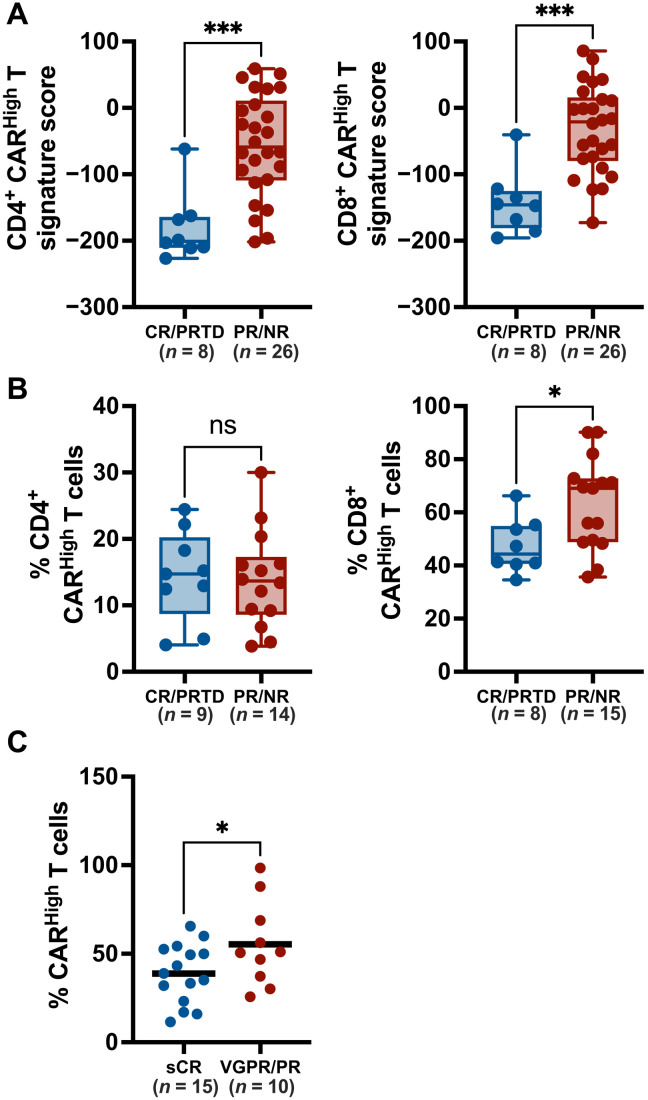
Given the differences in functionality between CARHigh T and CARLow T cells, we reasoned that CAR density might have an impact on the clinical response to CAR T cell therapies. To evaluate this hypothesis, we applied the gene signatures associated with CARHigh T cells to infusion products from several clinical trials with public transcriptomic data available (25, 26). We first applied our CD4+ and CD8+ signatures to bulk RNA-seq data of 34 infusion products from adult chronic lymphocytic leukemia (CLL) patients treated with CTL019 (25). We observed that products from patients with poor clinical response (partial responders and nonresponders) presented a significant higher score of both CD4+ and CD8+ CARHigh T signatures (Fig. 7A). We also assessed CARHigh T cell signature on an scRNA-seq dataset comprising anti-CD19 CAR T infusion products from 24 patients with DLBCL (26). We found that the products from nonresponder patients were significantly enriched in CD8+ CARHigh T cells (Fig. 7B), supporting the correlation between CAR density and clinical response. Last, we examined the correlation between the clinical response and the expression of CAR measured by FACS in the cell products of an academic clinical trial assessing ARI-0002h, a CAR T cell targeting BCMA (CARTBCMA-HCB-01; NCT04309981). Patients with partial response (PR) [less or equal than very good PR (VGPR)] showed an increase in the number of CARHigh T within the infusion product versus the patients presenting stringent CR (sCR) (Fig. 7C). Moreover, a shorter duration of response was observed in patients with increased percentage of CARHigh T cells (P = 0.04, as continuous variable in Cox regression model). No statistical correlation was observed with the development or grade of cytokine release syndrome (CRS), although the patients of this cohort presented only low-grade CRS (grades 1 and 2). Overall, our data suggest that CAR T therapeutic products enriched in T cells with high CAR density on the membrane would negatively affect the clinical response.
Fig. 7. Increased CAR levels negatively affect clinical response of CAR T therapies.
To evaluate the impact of increased CAR density on the clinical response, we quantify the presence of CARHigh T cells in the infusion products of several clinical trials. (A) The gene signatures developed in this work, associated to CD4+ and CD8+ CARHigh T cells, were applied to 34 infusion products from adult CLL patients treated with CTL019. Signature score for each CAR T cell product is represented for both CD4+ (left) and CD8+ (right) gene signatures, in patients divided according to clinical response into CR/PRTD (complete response/partial response with transformed disease) and PR/NP (partial response/nonresponse). Products from patients with poor clinical response (PR/NR) presented a significant higher score of both CARHigh T signatures. (B) Our gene signature was applied to available scRNA-seq dataset from 24 CAR T infusion products from adult DLBCL patients treated with axi-cel. Percentage of CD4+ (left) and CD8+ (right) CARHigh T cells present for each CAR T cell product is represented in patients divided according to clinical response as described in (A). Products from PR/NR patients were significantly enriched in CD8+ CARHigh T cells. (C) The number of CAR molecules per cell in 25 anti-BCMA CAR T infusion products from CARTBCMA-HCB-01 clinical trial was quantified by FACS. CARHigh T cells were defined as cells with >5000 molecules per cell (see Materials and Methods). Percentage of CARHigh T cells for each infusion product is represented in patients divided into sCR (stringent complete response) and VGPR (very good PR)/PR according to clinical response. Patients with sCR showed a significantly decreased number of CARHigh T cells. Unpaired t tests (A to C). *P < 0.05 and ***P < 0.001.
DISCUSSION
The functionality of CAR T cells relies on the interaction between the tumor and engineered T cells (1). However, as living drugs, CAR T cells are heterogeneous products in which intrinsic and extrinsic factors can influence their functionality having a significant impact on their clinical efficacy (8–16). Among others, the level of antigen expression has been associated with antitumor response (8, 24), with antigen loss representing one of the main mechanisms of resistance to CAR T therapies (49). Our study contributes to identify new determinants of CAR T cell function, demonstrating a clear role of the level of CAR expression on the functionality of CAR T cells. Our results indicate that high levels of CAR expression are associated with increased tonic signaling and a cell exhausted phenotype, characterized by the expression of multiple inhibitory receptors, such as PD1, CTLA4, LAG3, TIM3, and TIGIT, among others. This phenotype has been associated to reduced responses and worse long-term relapse-free survival (13, 25, 50), which is consistent with our findings demonstrating decreased responses in patients with increased levels of CARHigh T cells in different hematological malignancies.
Previous studies have demonstrated that constitutive signaling induced by multiple factors related to the different CAR moieties (51, 52) is associated to early T cell exhaustion (17–20). For instance, a recent study has shown fundamental differences in CAR signaling between CAR T cells with CD28 or CD8 transmembrane domains (TMDs) related to the heterodimerization potential of the different TMDs (53). Our results indicated that an increased density of the CAR molecule in the surface of the T cells (CARHigh T cells), produced by a higher number of viral integrations and subsequently increased expression, could be enough to trigger tonic signaling. Although the molecular mechanism should be further explored, the spontaneous clustering of the chimeric receptor molecules, leading to the CAR activation in the absence of antigen, could be a reasonable explanation. However, other mechanisms cannot be discarded including antigen shedding or inappropriate regulation of expression, as already reviewed (52). Tonic signaling was not restricted to CARHigh T cells targeting BCMA because it was also observed in CARHigh T cells with different specificities (CD19, CD33, and HER2), suggesting that this phenomenon was related to CAR density rather than CAR specificity.
Most CAR T constructs are generated using retro/LV vectors with strong promoters, such as human EF1a or the murine stem cell virus long terminal repeat, with no physiological regulation. These promoters induce high levels of CAR expression that can lead to tonic signaling and premature exhaustion. The possibility of reducing gene expression via weaker promoters as well as through more physiological promoters has been associated with decreased tonic signaling (22, 23, 54). The use of the MND promoter has been shown to reduce CAR surface density, while EF1a promoter increased its density, leading to a higher cytotoxic activity, cytokine production, and expression of exhaustion markers (23). Thus, the use of physiological promoters would be a strategy to prevent an increase in CARHigh T cells, thus limiting exhaustion of CAR T cells and favoring long-term persistence and antitumor efficacy (22).
The increased tonic signal and the concomitant basal activation of CARHigh T cells resulted in an increase in tumor cytotoxicity in vitro. The higher cytotoxicity and cytokine production observed in CARHigh T cells is also consistent with previous reports describing differences in antitumoral efficacy due to increased ligand-independent signaling (34, 55). Nevertheless, the pre-exhausted phenotype of CARHigh T cells could compromise their long-term persistence and function. Our results from the in vivo experiments based on “stress tests” (21, 34) support this hypothesis, because we observed reduced BM infiltration of CARHigh T cells. However, the use of CARLow T cells was not associated with improved survival of the animals because both CAR T populations were able to induce strong antitumor responses. Unfortunately, the use of immunodeficient mouse models is a substantial limitation for testing CAR T cell efficacy and toxicity. The use of humanized mice, or syngeneic mouse models together with mouse CAR T cells, should help to better characterize in vivo the impact of CAR expression of long-term efficacy of the different products. Nevertheless, the clinical correlation between response and decrease number of CARHigh T cells in the infusion product would also be consistent with the hypothesis that infusion of CARHigh T cells could compromise their long-term persistence and function.
The avidity of the single-chain variable fragment (scFv) against the antigen plays an important role on the functionality of the CAR T cells, and its control may result in more effective therapies (56, 57). Moreover, strategies combining the avidity of scFvs and the targeting of two molecules (or epitopes) may represent interesting approaches to increase the antitumoral efficacy and to reduce the toxicity (58, 59). This would be particularly relevant for MM where, despite impressive initial responses, a substantial number of patients experience a relapse after CAR T cell therapies (5, 6). However, the impact that CAR density could play on these approaches using CAR T cells with strong avidity and/or targeting several molecules would be an additional aspect that should be further analyzed.
A relevant question faced in this work was the development of specific methodology for proper identification of CARHigh T cells from transcriptomic data. We defined a molecular signature associated with increased CAR density, for both CD4+ and CD8+ CAR T cells, that showed a higher correlation with cell surface CAR levels than the RNA expression. The implications of these signatures were several fold: (i) This signature was key to annotate cells and identify clusters enriched in CARHigh T cells; (ii) in addition, these signatures allowed us to perform GRN analysis using single-cell data and identify key regulons associated to exhaustion of CAR T cells, such as NR4A1 and MAF (45–47), that were differentially activated in CARHigh T cells, providing mechanistic insights of the regulatory pathways driving the differences between CAR T cells with different CAR density; (iii) last, the definition of the signature associated with CARHigh T cells allowed us to apply the gene signature to transcriptomic data from CAR T cell products of patients undergoing CAR T treatment (25, 26), demonstrating a correlation between CAR expression and response in several cohorts of patients with different diseases (CLL and DLBCL). In the case of MM patients undergoing CAR T therapies, we also found a correlation between the CAR levels present on the therapeutic products and both the depth of response and its duration. These observations support our hypothesis that the presence of a high number of CARHigh T cells within the products limits the efficacy of CAR T therapies and suggests that the application of our gene signature may represent a new prognostic factor in patients treated with CAR T cells.
In summary, our data demonstrate that CAR density plays an important role in CAR T activity with an impact on clinical response. Moreover, the comprehension of regulatory mechanisms driven by CAR densities at the single-cell level offers an important tool for the identification of key regulatory factors that could be modulated for the development of improved therapies. Last, the application of a gene signature associated to increased CAR density, if validated in additional cohorts, may represent a valuable tool for predicting responses in patients undergoing CAR T cell therapy.
MATERIALS AND METHODS
Cell lines
Jurkat-TPR (provided by P. Steinberg, Medical University of Vienna), ARP-1-GFPLuc (provided by J. Epstein, University of Arkansas for Medical Sciences), MM1.S-GFPLuc, U266, MOLP8, K562, and K562-CD19 cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (FBS). MOLM13 and MV411 were cultured in RPMI 1640 supplemented with 20% FBS. Human embryonic kidney (HEK) 293T and BT474 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS. All media were supplemented with 1% penicillin/streptomycin and 1% l-glutamine. All cell lines were maintained at 37°C in 5% CO2.
LV vector construction and virus preparation
A third-generation self-inactivating LV vector (pCCL) was used to express under the EF1a promoter a second-generation CAR construct targeting BCMA (J22.9 clone from ARI-0002h), CD19 (FMC63 clone), CD33 (my96 clone), or HER2 (FRP5 clone). CAR structure comprised the scFv, a CD8 hinge and TMD, and the 4-1BB and CD3ζ endodomains fused to a truncated version of the EGFR or a BFP reporter gene. NFAT-GFP reporter LV vector was generated by subcloning a synthetic DNA fragment containing the NFAT regulatory element, from pGL4.30-Luc reporter plasmid (Promega), with the GFP sequence into the Eco RI sites of pHR_Gal4UAS_IRES_mC_pGK_tBFP vector (a gift from W. Lim; Addgene plasmid no. 79123). LV vectors were produced in HEK293T cells following standard procedures. Briefly, 6 × 106 cells were cotransfected with LV vector along with pMDLg/pRRE (Gag/Pol), pRSVRev, and pMD2.G (VSVG envelope) packaging plasmid using Lipofectamine 2000 (Invitrogen). Supernatants were collected 40 hours after transfection, filtered, concentrated using Lenti-X Concentrator (Takara) following the manufacturer’s specifications, and stored at −80°C until use.
Analysis of CAR signaling in Jurkat-TPR
Jurkat-TPR cells—transduced at a multiplicity of infection (MOI) of 1 with CARs targeting BCMA, CD33, CD19, or HER2—were cocultured in triplicate with ARP1-GFPLuc, MOLP8, and U266 cells for BCMA CARs, MOLM13 and MV4-11 cells for CD33, K562-CD19 cells for CD19 CARs, or BT474 cells for HER2 CARs at a 1:1 effector to tumor cell ratio. Nontransduced Jurkat-TPR cells were used as control. CARHigh and CARLow subpopulations were defined according to the FI using an EGFR-APC (allophycocyanin) antibody (clone AY13, BioLegend). Thresholds for CARHigh and CARLow were define as the top and bottom FI quartiles, respectively (CARHigh: FI > 1.5 × 105 and CARLow: FI < 2 × 104). Activation of the NFAT, NF-κB, and AP1 pathways was quantified before and 24 hours after coculture with tumor cells measuring enhanced GFP (eGFP), enhanced cyan fluorescent protein (eCFP), and mCherry emissions, respectively, using the CytoFLEX LX Flow Cytometer (Beckman Coulter) (fig. S1).
CAR T cell generation
CD4+ and CD8+ cells were isolated from peripheral blood mononuclear cells using CD4 and CD8 MicroBeads (Miltenyi Biotec) in the autoMACS Pro Separator (Miltenyi Biotec). Isolated T cells were activated with T cell TransAct (10 μl/ml; Miltenyi Biotec) for 48 hours and infected with the CAR LV vector at MOI of 2 with LentiBoost (10 μl/ml; Sirion Biotech). CAR T cells were expanded in RPMI 1640 culture medium supplemented with 3% human serum (Sigma-Aldrich), 1% penicillin/streptomycin, and human IL-7 (625 IU/ml) and human IL-15 (85 IU/ml; Miltenyi Biotec). CAR T cells were counted, and the concentration was adjusted to 1 × 106 cells/ml every 2 days.
Flow cytometry and CARHigh T and CARLow T cell isolation
Phenotypic characterization of T cells and CAR T cells was performed at days 0 and 13 of the production, respectively. All antibodies were purchased from BioLegend unless otherwise stated (table S5). Data were acquired on BD FACSCanto II (BD Biosciences) and analyzed using the FlowJo software version 10 (Treestar). CARHigh T and CARLow T cell subpopulations were sorted according to the BFP FI. Thresholds for CARHigh and CARLow were define as the top and bottom FI quartiles, respectively (CARHigh: FI > 1.2 × 104; CARLow: FI < 4 × 103) (fig. S3). All cells were sorted using a MoFlo Astrios EQ (Beckman Coulter). Tonic signaling in CARHigh T and CARLow T cells was analyzed using the NFAT-GFP reporter vector. Sorted CARHigh T and CARLow T cell populations were infected at MOI of 5 with LV vectors containing NFAT-GFP reporter, and the percentage of GFP cells and GFP intensity was measured in the absence of antigen stimulation.
Quantification of CAR density
The number of CAR molecules on the surface of CAR T cell products from healthy donors and MM patients was quantitated using Quantum Simply Cellular (Bangs Laboratories, USA, catalog no. 815), according to the manufacturer’s protocol. This methodology allows the conversion of FI value into absolute numbers of binding molecules using a calibration curve. Data acquisition was performed on BD FACSCanto II (BD Biosciences), and the results were analyzed using FlowJo software version 10 (Treestar). A threshold of 5000 and 1500 molecules was applied to define cells as CARHigh or CARLow, respectively, because these were a lower bound (5000 on CARHigh) and an upper bound (1500 on CARLow) on number of molecules present on the samples sorted for functional and transcriptomic experiments.
Viral copy number
Viral copy number (VCN) per cell was determined by quantitative polymerase chain reaction (qPCR). Genomic DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen). VCN per cell was quantified by duplex detection of the Psi sequence, normalized to albumin, using specific primers and detected with the TaqMan probes (table S6). qPCR was performed using the Absolute qPCR Mix Low ROX mix (Thermo Fisher Scientific) in a QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific). Results were analyzed in QuantStudio 3 Design and Analysis Software (Thermo Fisher Scientific).
Cytotoxicity assay and cytokine production
Cytotoxicity was determined using ARP1-GFPLuc as target tumor cells. Briefly, ARP1-GFPLuc cells were cultured with CARHigh T and CARLow T cells at different ratios in RPMI 1640 culture medium supplemented with 3% human serum (Sigma-Aldrich) and 1% penicillin/streptomycin in Nunc 96-well round bottom plates (Thermo Fisher Scientific). After 24 hours, luminescence was measured using the Bright-Glo Luciferase Assay System (Promega) according to the manufacturer’s instructions. IFN-γ, TNFα, and IL-2 cytokine production was quantified using BD Immunoassay enzyme-linked immunosorbent assay (ELISA) reagents (BD Biosciences) following the manufacturer’s protocol.
In vivo experiments
All experimental procedures were approved by the Ethics Committee of the University of Navarra and the Institute of Public Health of Navarra according to European Council Guidelines. NSG mice were purchased from The Jackson Laboratory (JAX) and bred and maintained in-house in a pathogen-free facility. Eight- to 12-week-old male or female mice were irradiated at 1.5 Gy at day −1, and 1 × 106 MM1.S-GFPLuc cells were intravenously injected the following day. Mice were randomized to ensure equal pretreatment tumor burden before CAR T cell treatment. At day 14, mice received intravenous injection of 5 × 105, 1.5 × 105, or 0.5 × 105 either CARHigh T or CARLow T cells. A subset of mice was sacrificed at days 7 and 21 after CAR T administration to analyze the presence of cells in BM and to perform phenotypic and functional characterizations as described above. Tumor progression was measured by bioluminescent imaging using the PhotonIMAGER (Biospace Lab). Signal was quantified using M3Vision Analysis Software (Biospace Lab). Mice were humanely euthanized when mice demonstrated signs of morbidity and/or hindlimb paralysis.
RNA-seq and bioinformatics analysis
RNA-seq was performed following massively parallel RNA single-cell sequencing (MARS)–seq protocol adapted for bulk RNA-seq (60, 61) with minor modifications. RNA–seq library quantification was done with Qubit 3.0 Fluorometer (Life Technologies), and size profiles were examined using Agilent’s 4200 TapeStation System. Libraries were sequenced in an Illumina NextSeq 500 at a sequence depth of 10 million reads per sample. Samples were aligned to the human genome (GRCh38) with STAR (v2.6.1). Gene expression was quantified with quant3p (github.com/ctlab/quant3p). Downstream analyses were performed in R (v3.6.2). Data transformation, normalization, and differential gene expression analysis were performed with DESeq2. T cell activation and tonic signaling gene signatures used in this work were obtained from previous publications (25–28) (table S7).
ATAC-seq and bioinformatics analysis
Accessible chromatin mapping was performed using FAST-ATAC-seq (62) with minor modifications. Libraries were quantified, and their size profiles were examined as described above. Sequencing was carried out in Illumina NextSeq 500 at a depth of 20 million reads per sample. ATAC-seq reads were aligned to the human genome (GRCh38) using Bowtie2 (v2.3.4). Peak calling from each individual replicate was performed with MACS2 (v.2.1.0). Differential accessible analysis was analyzed with csaw R package. The consensus peak set was derived from the union of all replicate peak sets for both conditions. For normalization, a nonlinear LOESS (locally estimated scatterplot smoothing)–based method was applied. Differential enrichment was analyzed using edgeR package. Last, ChIPseeker R package was used for annotation and visualization of genomics features. Transcription factor binding site enrichment was analyzed by HOMER.
Single-cell RNA-seq
scRNA-seq was performed in FACS-sorted CAR T cells (BFP+ cells) from three independent donors using the Chromium Single Cell 5′ Reagent Kit (10X Genomics) according to the manufacturer’s instructions. After quality control and quantification, single-cell libraries were sequenced at an average depth of at least 30,000 reads per cell. A total of 43,981 cells were analyzed, and 28,117 cells passed quality control with an average sequencing depth of 42,296 reads per cell, yielding an average of 2541 genes per cell. αβ T cell receptor (TCRα/β) sequencing was performed with 10X Genomics Single Cell V(D)J Immune Profiling solution (10X Genomics). After quality control and quantification, single-cell V(D)J-enriched libraries were pooled and sequenced at a minimum depth of 5000 reads per cell.
scRNA-seq data were demultiplexed and aligned to the human reference (GRCh38), and the feature-barcode matrix was quantified using Cell Ranger (v6.0.1) from 10X Genomics. Further computational analysis was performed using Seurat (v3.1.5). Cells were subjected to quality control filters based on the number of detected genes, number of unique molecular identifiers (UMIs), and proportion of UMIs mapped to mitochondrial and ribosomal genes per cell. Each dataset was subjected to normalization, identification of highly variable genes, and removal of unwanted sources of variation. Integration of all the dataset was based on Seurat’s canonical correlation analysis. Unsupervised clustering analysis with the resolution set to 0.8 yields a total of 23 cell clusters. Nonlinear dimensional reduction was performed using t-distributed stochastic neighbor embedding (t-SNE) and Uniform Manifold Approximation and Projection (UMAP). To describe the cell types and states defined by each cluster, we performed a manual review of the DEGs that were identified for each cell cluster by Seurat using canonical marker genes as reference. TCR reconstruction and paired TCR clonotype analysis were performed using Cell Ranger v6.0.2 for V(D)J sequence assembly.
Integration site analysis
Integration site analysis was performed in guide DNA isolated from FACS-sorted CARHigh T or CARLow T cells (three independent productions) using INSPIIRED pipeline with minor modifications (63, 64). Sequencing libraries prepared with specific primers (table S6) were quantified, and their size profiles were examined as described above. Sequencing was carried out in an Illumina MiSeq at a depth of 3 × 105 to 10 × 105 reads per sample. Samples were aligned to the human genome (GRCh38) using BLAT.
Generation of CARHigh signature
The CARHigh signature was generated using the DEGs (Log2FC > 1 and FDR < 0.05) between CARHigh T and CARLow T cell samples for each cell type (CD4+ and CD8+ cells; table S4). Specifically, the signature is created as
where Fi is the fold change of the gene i between the CARHigh T and CARLow T cell samples, 𝑧(.) is the function computing the z score, and Ei is the normalized gene expression for the ith gene. This signature takes the z score of the logarithm of the expression gene, allowing to compare the genes between them, and multiplying it by the logarithm of the fold change to imprint the directionality of the signature gene.
GRN analysis
Cells were labeled as CARHigh by applying the developed CARHigh signature. Then, using the most variable 300 TFs and 3000 genes, SimiC was run with the default parameters across CARHigh-labeled cells and the rest. GRNs were plotted using the GRN incidence matrices provided by SimiC. The histograms for the different regulons were computed from the “regulon activity score” provided by SimiC. This score was also used to compute the regulatory dissimilarity score for the selected cell clusters.
Patient samples and clinical data
Samples were obtained from patients with MM enrolled in the academic clinical trial CARTBCMA-HCB-01 (NCT04309981), led by C. Fernandez de Larrea (Hospital Clínic in Barcelona), assessing the BCMA-CAR T ARI-0002h, developed at IDIBAPS/Hospital Clínic in Barcelona. All subjects provided written informed consent. Clinical response was defined according to the MM Response Criteria (65) as sCR, VGPR, PR, and PD (progressive disease). The CARHigh signature was applied to gene expression data publicly available (25, 26), which allowed us to (i) quantify the number of CARHigh T cells within CD4+ and CD8+ population in CAR T cell products when scRNA-seq data were available and (ii) score the CAR T cell products according to CD4+ and CD8+ CARHigh signature when bulk RNA-seq data were available. The clinical data were judged the same way as in the original manuscript, grouping the different treatment responses into NR (nonresponse), PR, PRTD (PR with transformed disease), and CR.
Statistical analysis
Statistical analyses were performed using GraphPad Prism for Mac version 9.3.1. The different tests used in this work are indicated in the figure legend.
Acknowledgments
We thank the members of Hematology and Cell Therapy Department of the Clinica Universidad de Navarra for input throughout the course of the project and all the patients as well as families who made this study possible. We particularly acknowledge the patients for their participation in the Clinical trial CARTBCMA-HCB-01 (NCT04309981) and the Biobank of the University of Navarra for its collaboration.
Funding: This study was supported by the Instituto de Salud Carlos III co-financed by European Regional Development Fund-FEDER “A way to make Europe” (PI19/00922, PI19/00669, ICI19/00025, and ICI19/00069), Red de Terapia Celular TERCEL (RD16/0011/0005), Red de Terapias Avanzadas TERAV (RD21/0017/0009 and RD21/0017/0019), Centro de Investigación Biomédica en Red de Cáncer CIBERONC (CB16/12/00369 and CB16/12/00489), Ministerio de Ciencia e Innovación co-financed by European Regional Development Fund-FEDER “A way to make Europe” (RTC-2017-6578-1 and PID2019-108989RB-I00.), European Commission (H2020-JTI-IMI2-2019-18: Contract 945393; SC1-PM-08-2017: Contract 754658; and H2020-MSCA-IF-2019: Grant Agreement 898356), Gobierno de Navarra (AGATA: 0011-1411-2020-000011 and 0011-1411-2020-000010; DESCARTHeS: 0011-1411-2019-000079 and 0011-1411-2019-000072; alloCART-LMA: PC011-012), Fundacion La Caixa (CP042702), Asociacion Española Contra el Cáncer-AECC (LABAE21971FERN), and Paula and Rodger Riney Foundation. P.R.-M. was supported by FPU grant (FPU19/06160) from Ministerio de Universidades.
Author contributions: P.R.-M., M.E.C.-C., G.S., M.H., J.R.R.-M., and F.P. designed the experiments. P.R.-M., M.E.C.-C., G.S., A.M.-M., and C.Ca. conducted the experiments. P.R.-M., M.E.C.-C., G.S., A.O.-C., M.H., and J.R.R.-M. performed the data analysis. M.L.P.-B., P.R.-O., A.A.-P., and J.S.-M. provided clinical advice. A.O.-C., M.E.-R., C.Ce., M.C.V., M.R. M.P., M.J., B.M.-A., A.U.-I., C.F.d.L., and B.P. provided clinical samples and data. T.L., J.J.L., B.P., M.H., J.R.R.-M., and F.P. discussed the study design and the results. P.S.M.-U., A.V.-Z., S.R.-D., R.M.-T., and D.A. provided technical assistance. M.H., J.R.R.-M., and F.P. were responsible for research supervision, coordination, and strategy. P.R.-M. and J.R.R.-M. drafted the manuscript. J.J.L., J.S.-M., C.F.d.L., M.H., J.R.R.-M., and F.P. reviewed and edited the manuscript. All authors reviewed and approved the final version of the manuscript.
Competing interests: B.M.-A., A.U.-I., and M.J. are inventors on a patent application PCT/EP2020/071831 publication of WO2021023721A1 submitted by Fundacio Clinic Per A La Recerca Biomedica, Hospital Clinic De Barcelona, Institut d’Investigacions Biomèdiques August Pi I Sunyer (IDIBAPS) and Universitat de Barcelona that covers a chimeric antigen receptor structured by humanized scFv targeting BCMA in a general structure of different regions of CD8, 4-1BB, and CD3z. The authors declare no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The RNA-seq, ATAC-seq, and scRNA-seq data generated in this study have been deposited in the Gene Expression Omnibus database (GSE197851).
Supplementary Materials
This PDF file includes:
Extended Material and Methods
Figs. S1 to S13
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S8
REFERENCES AND NOTES
- 1.June C. H., Sadelain M., Chimeric antigen receptor therapy. N. Engl. J. Med. 379, 64–73 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schuster S. J., Bishop M. R., Tam C. S., Waller E. K., Borchmann P., McGuirk J. P., Jäger U., Jaglowski S., Andreadis C., Westin J. R., Fleury I., Bachanova V., Foley S. R., Ho P. J., Mielke S., Magenau J. M., Holte H., Pantano S., Pacaud L. B., Awasthi R., Chu J., Anak Ö., Salles G., Maziarz R. T.; JULIET Investigators , Tisagenlecleucel in adult relapsed or refractory diffuse large B-cell lymphoma. N. Engl. J. Med. 380, 45–56 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Maude S. L., Laetsch T. W., Buechner J., Rives S., Boyer M., Bittencourt H., Bader P., Verneris M. R., Stefanski H. E., Myers G. D., Qayed M., de Moerloose B., Hiramatsu H., Schlis K., Davis K. L., Martin P. L., Nemecek E. R., Yanik G. A., Peters C., Baruchel A., Boissel N., Mechinaud F., Balduzzi A., Krueger J., June C. H., Levine B. L., Wood P., Taran T., Leung M., Mueller K. T., Zhang Y., Sen K., Lebwohl D., Pulsipher M. A., Grupp S. A., Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 378, 439–448 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neelapu S. S., Locke F. L., Bartlett N. L., Lekakis L. J., Miklos D. B., Jacobson C. A., Braunschweig I., Oluwole O. O., Siddiqi T., Lin Y., Timmerman J. M., Stiff P. J., Friedberg J. W., Flinn I. W., Goy A., Hill B. T., Smith M. R., Deol A., Farooq U., McSweeney P., Munoz J., Avivi I., Castro J. E., Westin J. R., Chavez J. C., Ghobadi A., Komanduri K. V., Levy R., Jacobsen E. D., Witzig T. E., Reagan P., Bot A., Rossi J., Navale L., Jiang Y., Aycock J., Elias M., Chang D., Wiezorek J., Go W. Y., Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N. Engl. J. Med. 377, 2531–2544 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raje N., Berdeja J., Lin Y., Siegel D., Jagannath S., Madduri D., Liedtke M., Rosenblatt J., Maus M. V., Turka A., Lam L.-P., Morgan R. A., Friedman K., Massaro M., Wang J., Russotti G., Yang Z., Campbell T., Hege K., Petrocca F., Quigley M. T., Munshi N., Kochenderfer J. N., Anti-BCMA CAR T-cell therapy bb2121 in relapsed or refractory multiple myeloma. N. Engl. J. Med. 380, 1726–1737 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munshi N. C., Anderson L. D., Shah N., Madduri D., Berdeja J., Lonial S., Raje N., Lin Y., Siegel D., Oriol A., Moreau P., Yakoub-Agha I., Delforge M., Cavo M., Einsele H., Goldschmidt H., Weisel K., Rambaldi A., Reece D., Petrocca F., Massaro M., Connarn J. N., Kaiser S., Patel P., Huang L., Campbell T. B., Hege K., San-Miguel J., Idecabtagene vicleucel in relapsed and refractory multiple myeloma. N. Engl. J. Med. 384, 705–716 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Berdeja J. G., Madduri D., Usmani S. Z., Jakubowiak A., Agha M., Cohen A. D., Stewart A. K., Hari P., Htut M., Lesokhin A., Deol A., Munshi N. C., O’Donnell E., Avigan D., Singh I., Zudaire E., Yeh T. M., Allred A. J., Olyslager Y., Banerjee A., Jackson C. C., Goldberg J. D., Schecter J. M., Deraedt W., Zhuang S. H., Infante J., Geng D., Wu X., Carrasco-Alfonso M. J., Akram M., Hossain F., Rizvi S., Fan F., Lin Y., Martin T., Jagannath S., Ciltacabtagene autoleucel, a B-cell maturation antigen-directed chimeric antigen receptor T-cell therapy in patients with relapsed or refractory multiple myeloma (CARTITUDE-1): A phase 1b/2 open-label study. Lancet 398, 314–324 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Majzner R. G., Rietberg S. P., Sotillo E., Dong R., Vachharajani V. T., Labanieh L., Myklebust J. H., Kadapakkam M., Weber E. W., Tousley A. M., Richards R. M., Heitzeneder S., Nguyen S. M., Wiebking V., Theruvath J., Lynn R. C., Xu P., Dunn A. R., Vale R. D., Mackall C. L., Tuning the antigen density requirement for car T-cell activity. Cancer Discov. 10, 702–723 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Locke F. L., Rossi J. M., Neelapu S. S., Jacobson C. A., Miklos D. B., Ghobadi A., Oluwole O. O., Reagan P. M., Lekakis L. J., Lin Y., Sherman M., Better M., Go W. Y., Wiezorek J. S., Xue A., Bot A., Tumor burden, inflammation, and product attributes determine outcomes of axicabtagene ciloleucel in large B-cell lymphoma. Blood Adv. 4, 4898–4911 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawalekar O. U., O’Connor R. S., Fraietta J. A., Guo L., McGettigan S. E., Posey A. D., Patel P. R., Guedan S., Scholler J., Keith B., Snyder N., Blair I., Milone M. C., June C. H., Distinct signaling of coreceptors regulates specific metabolism pathways and impacts memory development in CAR T cells. Immunity 44, 380–390 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Guedan S., Posey A. D., Shaw C., Wing A., Da T., Patel P. R., McGettigan S. E., Casado-Medrano V., Kawalekar O. U., Uribe-Herranz M., Song D., Melenhorst J. J., Lacey S. F., Scholler J., Keith B., Young R. M., June C. H., Enhancing CAR T cell persistence through ICOS and 4-1BB costimulation. JCI Insight 3, e96976 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alabanza L., Pegues M., Geldres C., Shi V., Wiltzius J. J. W., Sievers S. A., Yang S., Kochenderfer J. N., Function of novel anti-CD19 chimeric antigen receptors with human variable regions is affected by hinge and transmembrane domains. Mol. Ther. 25, 2452–2465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garfall A. L., Dancy E. K., Cohen A. D., Hwang W. T., Fraietta J. A., Davis M. M., Levine B. L., Siegel D. L., Stadtmauer E. A., Vogl D. T., Waxman A., Rapoport A. P., Milone M. C., June C. H., Melenhorst J. J., T-cell phenotypes associated with effective CAR T-cell therapy in postinduction vs relapsed multiple myeloma. Blood Adv. 3, 2812–2815 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rossi J., Paczkowski P., Shen Y. W., Morse K., Flynn B., Kaiser A., Ng C., Gallatin K., Cain T., Fan R., Mackay S., Heath J. R., Rosenberg S. A., Kochenderfer J. N., Zhou J., Bot A., Preinfusion polyfunctional anti-CD19 chimeric antigen receptor T cells are associated with clinical outcomes in NHL. Blood 132, 804–814 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turtle C. J., Hanafi L. A., Berger C., Hudecek M., Pender B., Robinson E., Hawkins R., Chaney C., Cherian S., Chen X., Soma L., Wood B., Li D., Heimfeld S., Riddell S. R., Maloney D. G., Immunotherapy of non-Hodgkin’s lymphoma with a defined ratio of CD8+ and CD4+ CD19-specific chimeric antigen receptor-modified T cells. Sci. Transl. Med. 8, 355ra116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D. Sommermeyer, M. Hudecek, P. L. Kosasih, T. Gogishvili, D. G. Maloney, C. J. Turtle, S. R. Riddell, Chimeric antigen receptor-modified T cells derived from defined CD8+ and CD4+ subsets confer superior antitumor reactivity in vivo. Leukemia 30, 492–500 (2016). [DOI] [PMC free article] [PubMed]
- 17.Long A. H., Haso W. M., Shern J. F., Wanhainen K. M., Murgai M., Ingaramo M., Smith J. P., Walker A. J., Kohler M. E., Venkateshwara V. R., Kaplan R. N., Patterson G. H., Fry T. J., Orentas R. J., Mackall C. L., 4-1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat. Med. 21, 581–590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frigault M. J., Lee J., Basil M. C., Carpenito C., Motohashi S., Scholler J., Kawalekar O. U., Guedan S., McGettigan S. E., Posey A. D., Ang S., Cooper L. J. N., Platt J. M., Johnson F. B., Paulos C. M., Zhao Y., Kalos M., Milone M. C., June C. H., Identification of chimeric antigen receptors that mediate constitutive or inducible proliferation of T cells. Cancer Immunol. Res. 3, 356–367 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sukumaran S., Watanabe N., Bajgain P., Raja K., Mohammed S., Fisher W. E., Brenner M. K., Leen A. M., Vera J. F., Enhancing the potency and specificity of engineered T cells for cancer treatment. Cancer Discov. 8, 972–987 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomes-Silva D., Mukherjee M., Srinivasan M., Krenciute G., Dakhova O., Zheng Y., Cabral J. M. S., Rooney C. M., Orange J. S., Brenner M. K., Mamonkin M., Tonic 4-1BB costimulation in chimeric antigen receptors impedes T cell survival and is vector-dependent. Cell Rep. 21, 17–26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eyquem J., Mansilla-Soto J., Giavridis T., van der Stegen S. J. C., Hamieh M., Cunanan K. M., Odak A., Gönen M., Sadelain M., Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 543, 113–117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.M. Tristán-Manzano, N. Maldonado-Pérez, P. Justicia-Lirio, P. Muñoz, M. Cortijo-Gutiérrez, K. Pavlovic, R. Jiménez-Moreno, S. Nogueras, Md. Carmona, S. Sánchez-Hernández, A. Aguilar-González, M. Castella, M. Juan, C. Marañón, K. Benabdellah, C. Herrera, F. Martin, Physiological (TCR-like) regulated lentiviral vectors for the generation of improved CAR-T cells. medRxiv 2021.03.17.21253300 (2021); 10.1101/2021.03.17.21253300. [DOI]
- 23.Ho J. Y., Wang L., Liu Y., Ba M., Yang J., Zhang X., Chen D., Lu P., Li J., Promoter usage regulating the surface density of CAR molecules may modulate the kinetics of CAR-T cells in vivo. Mol. Ther. Methods Clin. Dev. 21, 237–246 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker A. J., Majzner R. G., Zhang L., Wanhainen K., Long A. H., Nguyen S. M., Lopomo P., Vigny M., Fry T. J., Orentas R. J., Mackall C. L., Tumor antigen and receptor densities regulate efficacy of a chimeric antigen receptor targeting anaplastic lymphoma kinase. Mol. Ther. 25, 2189–2201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fraietta J. A., Lacey S. F., Orlando E. J., Pruteanu-Malinici I., Gohil M., Lundh S., Boesteanu A. C., Wang Y., O’Connor R. S., Hwang W. T., Pequignot E., Ambrose D. E., Zhang C., Wilcox N., Bedoya F., Dorfmeier C., Chen F., Tian L., Parakandi H., Gupta M., Young R. M., Johnson F. B., Kulikovskaya I., Liu L., Xu J., Kassim S. H., Davis M. M., Levine B. L., Frey N. V., Siegel D. L., Huang A. C., Wherry E. J., Bitter H., Brogdon J. L., Porter D. L., June C. H., Melenhorst J. J., Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nat. Med. 24, 563–571 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng Q., Han G., Puebla-Osorio N., Ma M. C. J., Strati P., Chasen B., Dai E., Dang M., Jain N., Yang H., Wang Y., Zhang S., Wang R., Chen R., Showell J., Ghosh S., Patchva S., Zhang Q., Sun R., Hagemeister F., Fayad L., Samaniego F., Lee H. C., Nastoupil L. J., Fowler N., Davis R. E., Westin J., Neelapu S. S., Wang L., Green M. R., Characteristics of anti-CD19 CAR T cell infusion products associated with efficacy and toxicity in patients with large B cell lymphomas. Nat. Med. 26, 1878–1887 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheih A., Voillet V., Hanafi L. A., DeBerg H. A., Yajima M., Hawkins R., Gersuk V., Riddell S. R., Maloney D. G., Wohlfahrt M. E., Pande D., Enstrom M. R., Kiem H. P., Adair J. E., Gottardo R., Linsley P. S., Turtle C. J., Clonal kinetics and single-cell transcriptional profiling of CAR-T cells in patients undergoing CD19 CAR-T immunotherapy. Nat. Commun. 11, 219 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boroughs A. C., Larson R. C., Marjanovic N. D., Gosik K., Castano A. P., Porter C. B. M., Lorrey S. J., Ashenberg O., Jerby L., Hofree M., Smith-Rosario G., Morris R., Gould J., Riley L. S., Berger T. R., Riesenfeld S. J., Rozenblatt-Rosen O., Choi B. D., Regev A., Maus M. V., A distinct transcriptional program in human CAR T cells bearing the 4-1BB signaling domain revealed by scRNA-seq. Mol. Ther. 28, 2577–2592 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen G. M., Chen C., Das R. K., Gao P., Chen C. H., Bandyopadhyay S., Ding Y. Y., Uzun Y., Yu W., Zhu Q., Myers R. M., Grupp S. A., Barrett D. M., Tan K., Integrative bulk and single-cell profiling of premanufacture T-cell populations reveals factors mediating long-term persistence of CAR T-cell therapy. Cancer Discov. 11, 2186–2199 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Q., Hu H., Chen S. Y., Liu C. J., Hu F. F., Yu J., Wu Y., Guo A. Y., Transcriptome and regulatory network analyses of CD19-CAR-T immunotherapy for B-ALL. Genomics Proteomics Bioinformatics 17, 190, –200 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosskopf S., Leitner J., Paster W., Morton L. T., Hagedoorn R. S., Steinberger P., Heemskerk M. H. M., A Jurkat 76 based triple parameter reporter system to evaluate TCR functions and adoptive T cell strategies. Oncotarget 9, 17608–17619 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monjezi R., Miskey C., Gogishvili T., Schleef M., Schmeer M., Einsele H., Ivics Z., Hudecek M., Enhanced CAR T-cell engineering using non-viral Sleeping Beauty transposition from minicircle vectors. Leukemia 31, 186–194 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Holstein M., Mesa-Nuñez C., Miskey C., Almarza E., Poletti V., Schmeer M., Grueso E., Ordóñez Flores J. C., Kobelt D., Walther W., Aneja M. K., Geiger J., Bonig H. B., Izsvák Z., Schleef M., Rudolph C., Mavilio F., Bueren J. A., Guenechea G., Ivics Z., Efficient non-viral gene delivery into human hematopoietic stem cells by minicircle sleeping beauty transposon vectors. Mol. Ther. 26, 1137–1153 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feucht J., Sun J., Eyquem J., Ho Y. J., Zhao Z., Leibold J., Dobrin A., Cabriolu A., Hamieh M., Sadelain M., Calibration of CAR activation potential directs alternative T cell fates and therapeutic potency. Nat. Med. 25, 82–88 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blank C. U., Haining W. N., Held W., Hogan P. G., Kallies A., Lugli E., Lynn R. C., Philip M., Rao A., Restifo N. P., Schietinger A., Schumacher T. N., Schwartzberg P. L., Sharpe A. H., Speiser D. E., Wherry E. J., Youngblood B. A., Zehn D., Defining ‘T cell exhaustion’. Nat. Rev. Immunol. 19, 665–674 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.J. Peng, G. Serrano, I. M. Traniello, M. E. Calleja-Cervantes, U. V. Chembazhi, S. Bangru, T. Ezponda, J. Roberto Rodriguez-Madoz, A. Kalsotra, F. Prosper, I. Ochoa, M. Hernaez, A single-cell gene regulatory network inference method for identifying complex regulatory dynamics across cell phenotypes. bioRxiv 2020.04.03.023002 [Preprint]. 6 April 2021. 10.1101/2020.04.03.023002. [DOI]
- 37.Ho I.-C., Tai T.-S., Pai S.-Y., GATA3 and the T-cell lineage: Essential functions before and after T-helper-2-cell differentiation. Nat. Rev. Immunol. 9, 125–135 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang D., Diao H., Getzler A. J., Rogal W., Frederick M. A., Milner J., Yu B., Crotty S., Goldrath A. W., Pipkin M. E., The transcription factor Runx3 establishes chromatin accessibility of cis-regulatory landscapes that drive memory cytotoxic T lymphocyte formation. Immunity 48, 659–674.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaud G., Lesourne R., Love P. E., Regulatory mechanisms in T cell receptor signalling. Nat. Rev. Immunol. 18, 485–497 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Siegel A. M., Heimall J., Freeman A. F., Hsu A. P., Brittain E., Brenchley J. M., Douek D. C., Fahle G. H., Cohen J. I., Holland S. M., Milner J. D., A critical role for STAT3 transcription factor signaling in the development and maintenance of human T cell memory. Immunity 35, 806–818 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masuda K., Ripley B., Nyati K. K., Dubey P. K., Zaman M. M. U., Hanieh H., Higa M., Yamashita K., Standley D. M., Mashima T., Katahira M., Okamoto T., Matsuura Y., Takeuchi O., Kishimoto T., Arid5a regulates naive CD4+ T cell fate through selective stabilization of Stat3 mRNA. J. Exp. Med. 213, 605–619 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hwang S. S., Lim J., Yu Z., Kong P., Sefik E., Xu H., Harman C. C. D., Kim L. K., Lee G. R., Li H.-B., Flavell R. A., mRNA destabilization by BTG1 and BTG2 maintains T cell quiescence. Science 367, 1255–1260 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Villard J., Peretti M., Masternak K., Barras E., Caretti G., Mantovani R., Reith W., A functionally essential domain of RFX5 mediates activation of major histocompatibility complex class II promoters by promoting cooperative binding between RFX and NF-Y. Mol. Cell. Biol. 20, 3364–3376 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rousseau P., Masternak K., Krawczyk M., Reith W., Dausset J., Carosella E. D., Moreau P., In vivo, RFX5 binds differently to the human leucocyte antigen-E, -F, and -G gene promoters and participates in HLA class I protein expression in a cell type-dependent manner. Immunology 111, 53–65 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J., López-Moyado I. F., Seo H., Lio C. W. J., Hempleman L. J., Sekiya T., Yoshimura A., Scott-Browne J. P., Rao A., NR4A transcription factors limit CAR T cell function in solid tumours. Nature 567, 530–534 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu X., Wang Y., Lu H., Li J., Yan X., Xiao M., Hao J., Alekseev A., Khong H., Chen T., Huang R., Wu J., Zhao Q., Wu Q., Xu S., Wang X., Jin W., Yu S., Wang Y., Wei L., Wang A., Zhong B., Ni L., Liu X., Nurieva R., Ye L., Tian Q., Bian X. W., Dong C., Genome-wide analysis identifies NR4A1 as a key mediator of T cell dysfunction. Nature 567, 525–529 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giordano M., Henin C., Maurizio J., Imbratta C., Bourdely P., Buferne M., Baitsch L., Vanhille L., Sieweke M. H., Speiser D. E., Auphan-Anezin N., Schmitt-Verhulst A., Verdeil G., Molecular profiling of CD 8 T cells in autochthonous melanoma identifies Maf as driver of exhaustion. EMBO J. 34, 2042–2058 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stephen T. L., Payne K. K., Chaurio R. A., Allegrezza M. J., Zhu H., Perez-Sanz J., Perales-Puchalt A., Nguyen J. M., Vara-Ailor A. E., Eruslanov E. B., Borowsky M. E., Zhang R., Laufer T. M., Conejo-Garcia J. R., SATB1 expression governs epigenetic repression of PD-1 in tumor-reactive T cells. Immunity 46, 51–64 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.da Vià M. C., Dietrich O., Truger M., Arampatzi P., Duell J., Heidemeier A., Zhou X., Danhof S., Kraus S., Chatterjee M., Meggendorfer M., Twardziok S., Goebeler M. E., Topp M. S., Hudecek M., Prommersberger S., Hege K., Kaiser S., Fuhr V., Weinhold N., Rosenwald A., Erhard F., Haferlach C., Einsele H., Kortüm K. M., Saliba A. E., Rasche L., Homozygous BCMA gene deletion in response to anti-BCMA CAR T cells in a patient with multiple myeloma. Nat. Med. 27, 616–619 (2021). [DOI] [PubMed] [Google Scholar]
- 50.N. W. C. J. van de Donk, M. Themeli, S. Z. Usmani, Determinants of response and mechanisms of resistance of CAR T-cell therapy in multiple myeloma. Blood Cancer Discov. 2, 302–318 (2021). [DOI] [PMC free article] [PubMed]
- 51.Jayaraman J., Mellody M. P., Hou A. J., Desai R. P., Fung A. W., Pham A. H. T., Chen Y. Y., Zhao W., CAR-T design: Elements and their synergistic function. EBioMedicine 58, 102931 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ajina A., Maher J., Strategies to address chimeric antigen receptor tonic signaling. Mol. Cancer Ther. 17, 1795–1815 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muller Y. D., Nguyen D. P., Ferreira L. M. R., Ho P., Raffin C., Valencia R. V. B., Congrave-Wilson Z., Roth T. L., Eyquem J., van Gool F., Marson A., Perez L., Wells J. A., Bluestone J. A., Tang Q., The CD28-transmembrane domain mediates chimeric antigen receptor heterodimerization with CD28. Front. Immunol. 12, 639818 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rad S. M. A. H., Poudel A., Tan G. M. Y., McLellan A. D., Promoter choice: Who should drive the CAR in T cells? PLOS ONE 15, e0232915 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guedan S., Madar A., Casado-Medrano V., Shaw C., Wing A., Liu F., Young R. M., June C. H., Posey A. D., Single residue in CD28-costimulated CAR-T cells limits long-term persistence and antitumor durability. J. Clin. Investig. 130, 3087–3097 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greenman R., Pizem Y., Haus-Cohen M., Goor A., Horev G., Denkberg G., Sinik K., Elbaz Y., Bronner V., Levin A. G., Horn G., Shen-Orr S., Reiter Y., Shaping functional avidity of CAR T Cells: Affinity, avidity, and antigen density that regulate response. Mol. Cancer Ther. 20, 872–884 (2021). [DOI] [PubMed] [Google Scholar]
- 57.Salzer B., Schueller C. M., Zajc C. U., Peters T., Schoeber M. A., Kovacic B., Buri M. C., Lobner E., Dushek O., Huppa J. B., Obinger C., Putz E. M., Holter W., Traxlmayr M. W., Lehner M., Engineering AvidCARs for combinatorial antigen recognition and reversible control of CAR function. Nat. Commun. 11, 4166 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernández de Larrea C., Staehr M., Lopez A. V., Ng K. Y., Chen Y., Godfrey W. D., Purdon T. J., Ponomarev V., Wendel H.-G., Brentjens R. J., Smith E. L., Defining an optimal dual-targeted CAR T-cell therapy approach simultaneously targeting BCMA and GPRC5D to prevent BCMA escape–driven relapse in multiple myeloma. Blood Cancer Discov. 1, 146–154 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berdeja J. G., Madduri D., Usmani S. Z., Singh I., Zudaire E., Yeh T.-M., Allred A. J., Olyslager Y., Banerjee A., Goldberg J. D., Schecter J., Geng D., Wu X., Carrasco-Alfonso M., Rizvi S., Fan F. (X.), Jakubowiak A. J., Jagannath S., Update of CARTITUDE-1: A phase Ib/II study of JNJ-4528, a B-cell maturation antigen (BCMA)-directed CAR-T-cell therapy, in relapsed/refractory multiple myeloma. J. Clin. Oncol. 38, 8505 (2020). [Google Scholar]
- 60.Jaitin D. A., Kenigsberg E., Keren-Shaul H., Elefant N., Paul F., Zaretsky I., Mildner A., Cohen N., Jung S., Tanay A., Amit I., Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lavin Y., Kobayashi S., Leader A., Amir E.-A. D., Elefant N., Bigenwald C., Remark R., Sweeney R., Becker C. D., Levine J. H., Meinhof K., Chow A., Kim-Shulze S., Wolf A., Medaglia C., Li H., Rytlewski J. A., Emerson R. O., Solovyov A., Greenbaum B. D., Sanders C., Vignali M., Beasley M. B., Flores R., Gnjatic S., Pe’er D., Rahman A., Amit I., Merad M., Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell 169, 750–765.e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Corces M. R., Buenrostro J. D., Wu B., Greenside P. G., Chan S. M., Koenig J. L., Snyder M. P., Pritchard J. K., Kundaje A., Greenleaf W. J., Majeti R., Chang H. Y., Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet. 48, 1193–1203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berry C. C., Nobles C., Six E., Wu Y., Malani N., Sherman E., Dryga A., Everett J. K., Male F., Bailey A., Bittinger K., Drake M. J., Caccavelli L., Bates P., Hacein-Bey-Abina S., Cavazzana M., Bushman F. D., INSPIIRED: Quantification and visualization tools for analyzing integration site distributions. Mol. Ther. Methods Clin. Dev. 4, 17–26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sherman E., Nobles C., Berry C. C., Six E., Wu Y., Dryga A., Malani N., Male F., Reddy S., Bailey A., Bittinger K., Everett J. K., Caccavelli L., Drake M. J., Bates P., Hacein-Bey-Abina S., Cavazzana M., Bushman F. D., INSPIIRED: A pipeline for quantitative analysis of sites of new DNA integration in cellular genomes. Mol. Ther. Methods Clin. Dev. 4, 39–49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Durie B. G. M., Harousseau J. L., Miguel J. S., Bladé J., Barlogie B., Anderson K., Gertz M., Dimopoulos M., Westin J., Sonneveld P., Ludwig H., Gahrton G., Beksac M., Crowley J., Belch A., Boccadaro M., Cavo M., Turesson I., Joshua D., Vesole D., Kyle R., Tricot G., Alexanian R., Attal M., Merlini G., Powles R., Richardson P., Shimizu K., Tosi P., Rajkumar S. V., Morgan G.; International Myeloma Working Group , International uniform response criteria for multiple myeloma. Leukemia 20, 1467–1473 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Extended Material and Methods
Figs. S1 to S13
Tables S1 to S8