Background:
Chloride intracellular channel 1 (CLIC1) plays an important role in the process of cell epithelial transport, and is also involved in tumor invasion and metastasis. Due to its aberrant expression in cancer, the mechanism of action of CLIC1 in cancer has been carefully studied. In this study, we tried to investigate the relationship between CLIC1 and lung adenocarcinoma (LUAD).
Methods:
The RNA-sequencing data and clinical information of CLIC1 in lung adenocarcinoma were collected from the the cancer genome altas (TCGA) database and analyzed with R software. Paired t test and Mann–Whitney U test were used to detect differences between LUAD tissue and adjacent normal tissue, and the pROC software package performed reactive oxygen species (ROC) curves to detect cutoff values for CLIC1. The expression of CLIC1 in normal human tissues was extracted from the human protein altas (HPA) database, and analyzed clinical proteomic tumor analysis consortium by using UALCAN programme. The relationship between CLIC1 and LUAD was explored by enrichment analysis using gene oncology and Kyoto encyclopedia of genes and genomes. The tumor immunity estimation resource (TIMER) and integrated repository portal for tumor-immune system interactions (TISIDB) databases were used to analyze the correlation between CLIC1 and LUAD immune cell infiltration. Survival analysis of CLIC1 in LUAD was assessed by the PrognoScan database.
Results:
Compared with normal tissues, both mRNA (messenger Ribose Nucleic Acid) and protein of CLIC1 were overexpressed in LUAD, which was associated with shorter overall survial (OS). In addition, CLIC1 expression was in connection with some clinical-pathological characteristics like tumor node metatasis stages and lymph node metastases. What’s more, CLIC1 may play a role in the immune infiltration of LUAD.
Conclusion:
In summary, CLIC1 is up-regulated in LUAD and is associated with tumor metastasis, tumor staging, and OS. It may be regarded as a novel marker for prognostic judgement in LUAD.
Keywords: biomarker, CLIC1, immune infiltration, lung adenocarcinoma, prognosis
1. Introduction
Lung cancer is the leading cause of cancer death in China and worldwide, and nearly 50% of lung cancer patients are diagnosed with lung adenocarcinoma (LUAD).[1] Complete surgical resection is recommended for stage Ⅰ to Ⅱ lung cancer and the 5-year survival rate is more than 50%.[2] While the advanced LUAD patients should be considered for conventional radiotherapy and chemoradiotherapy, molecular-targeted therapy and immunotherapy. The 5-year survival rate is low at 15%.[3] With the wide application of the low-dose computer tomography (CT) in lung cancer screening, the diagnosis rate of early lung cancer has been greatly elevated, which creating an advantage for the treatment of the disease.[4] However, in China, two-thirds of patients have lost their chance of surgery when they see the doctor.[5] Therefore, the annual mortality rate of lung cancer is still rising rapidly in China, bringing a huge burden to the society and economy.[6,7]
Chloride intracellular channel 1 (CLIC1) is a transmembrane protein that plays an important role in regulating cell volume, acidifying organelles, performing epithelial transport and regulating electrical excitation.[8–10] Recently, it has been confirmed that its expression is up-regulated in gastric cancer, colon cancer and liver cancer.[11–13] The exact mechanism of this phenomenon needs further study. Gurski et al found that CLIC1 plays a key role in maintaining the stability of invadopodia in endothelial and tumor cells embedded in a 3-dimensional (3D) matrix of fibrin.[14] Peng et al showed that CLIC1 might influence cell migration, tumor invasion and metastasis by regulating the formation of cell-matrix adhesions and membrane protrusions through the recruitment of PIP5Ks to the plasma membrane.[15] Overexpression of CLIC1 in tumor cells makes it a novel prognostic factor for various cancers. However, the association between CLIC1 and lung adenocarcinoma remains unclear.
2. Materials and Methods
In the present study, ethical approval was unnecessary because all analytical data were derived from publicly available database (https://portal.gdc.cancer.gov/).
2.1. Data acquisition and processing
We downloaded from the cancer genome altas (TCGA) official website (https://portal.gdc.cancer.gov/) several kinds of cancers, including LUAD CLIC1 transcriptome data and corresponding clinical information.[16] The 30 enrolled cancer types contained at least 5 samples in the normal group. The initial downloaded data with FPKM format was transformed into TPM format and log2 format for further study. RNA-Seq data of 535 lung adenocarcinoma and 59 adjacent normal tissue data were retained. The selected samples contained CLIC1 gene expression data and relevant clinical information, including age, sex, smoker status, T, N, M stage, pathologic stage, tumor location and survival condition, etc. As all data were downloaded from the public database, the study did not require the approval of the ethics committee.
2.2. Expression analysis of CLIC1
The mRNA expression data were characterized by mean ± SD. All statistical analyses were performed by R (v3.6.3) (https://www.r-project.org/), and differences were visualized by R package ggplot2. Paired t test and Mann–Whitney U test were applied to detect the differences between LUAD tissue and adjacent normal tissues. Reactive oxygen species (ROC) curve was performed to detect the cutoff of CLIC1 by using the pROC software package.[17]
In this study, a comprehensive analysis of CLIC1 protein expression was presented by UALCAN (http://ualcan.path.uab.edu/).[18] And we extracted the expression of CLIC1 in normal human tissues from the human protein atlas (HPA) database.[19]
2.3. Protein–protein interaction (PPI) and enrichment analysis of CLIC1
The PPI network was constructed on the basis of the public database STRING (http://string-db.org) to retrieve the co-expressed genes. The functional enrichment analysis of the co-expressed genes using gene ontology and Kyoto encyclopedia of genes and genomes which were performed by the “ClusterProfiler” package and visualized by the “ggplot2” package.[20,21]
2.4. Tumor immune estimation resource (TIMER) database
The association of CLIC1 expression in LUAD and six immune infiltrates, including B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells, are performed base on TIMER database.[22]
2.5. Tumor-immune system interaction database (TISIDB) analysis
TISIDB (http://cis.hku.hk/TISIDB/) was used to explore the association between CLIC1 and tumor-infiltrating lymphocytes (TILs) in human tumors.[22] On the basis of the gene expression profile, gene set variation analysis is used to infer the relative abundance of TILs. The correlation between CLIC1 and TILs was detected by Spearman’s test.
2.6. Survival analysis
The correlation between CLIC1 expression and overall survival in LUAD with two different datasets (jacob-00182-HLM, GSE31210) was performed by the Prognostic Scan database.[23]
3. Results
3.1. Elevated expression of CLIC1 in pan-cancer perspective
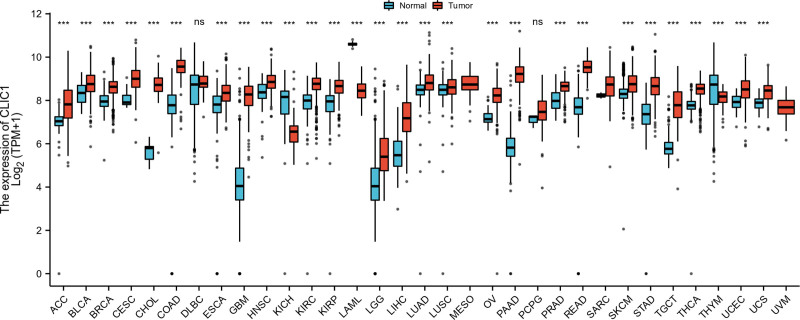
Three tumor types (MESO, SARC, UVM) with a number less than 5 in the normal groups were excluded. So our final work covered 30 cancer types. As shown in Figure 1, compared with normal tissues, CLIC1 expression was up-regulated in 28 tumors in the complete set of analyses. This data provided a result that the mRNA expression of CLIC1 is abnormal in most cancers.
Figure 1.
Expression pattern of CLIC1 in Pan-cancer perspective. CLIC1 = chloride intracellular channel 1.
3.2. Expression of CLIC1 in patients with LUAD
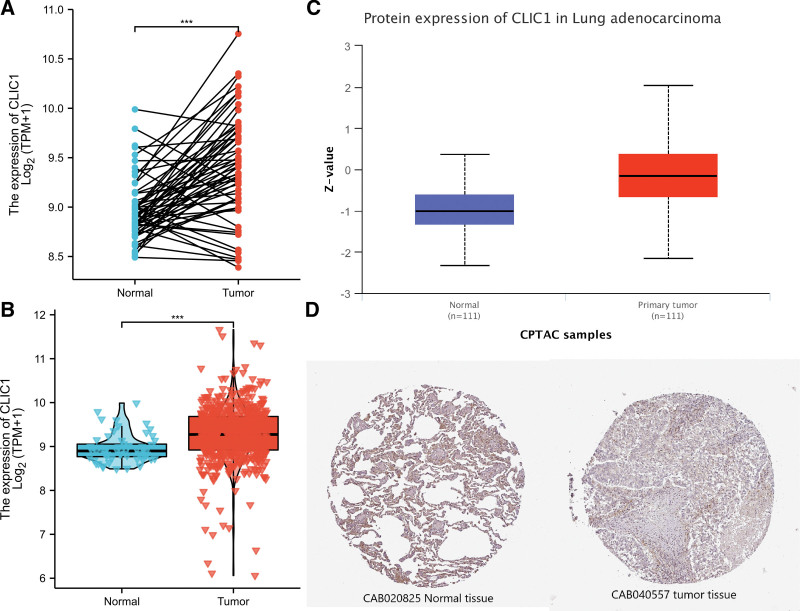
By analyzing the expression data of CLIC1 in TCGA and HPA, we determined the mRNA and protein expression of CLIC1 in LUAD. As shown in Figure 2A, paired data analysis showed that the expression level of CLIC1 mRNA in LUAD tissues (n = 57) was significantly higher than that in adjacent normal tissues (n = 57) (8.973 ± 0.319 vs 9.405 ± 0.538 P < .001). In Figure 2B, the result of unpaired data analysis also showed that the expression level of CLIC1 mRNA in LUAD tissues (n = 535) was significantly higher than that in adjacent normal tissues (n = 59). (8.961 ± 0.320 vs 9.275 ± 0.642 P < .001).
Figure 2.
The mRNA and protein expression of CLIC1 in lung adenocarcinoma. (A) The mRNA expression levels of CLIC1 in 57 lung adenocarcinoma and matched-adjacent normal samples. (B) The mRNA expression levels of CLIC1 in 535 lung adenocarcinoma samples and 59 normal samples. (C) The protein expression levels of CLIC1 based on CPTAC. (D) The protein levels of CLIC1 based on Human Protein Atlas. CPTAC = clinical proteomic tumor analysis consortium, CLIC1 = chloride intracellular channel 1.
The results of the comprehensive analysis of CLIC1 protein expression were as follows. Clinical proteomic tumor analysis consortium analysis showed that the expression of CLIC1 protein in LUAD tissues was significantly higher than that in normal tissues (Fig. 2C). HPA immunohistochemical staining showed that CLIC1 protein expression was up-regulated in LUAD tissues (Fig. 2D). These results indicate that both mRNA and CLIC1 protein expression are up-regulated in LUAD tissue.
3.3. Relationships between CLIC1 mRNA levels and clinical pathological characteristics of LUAD patients
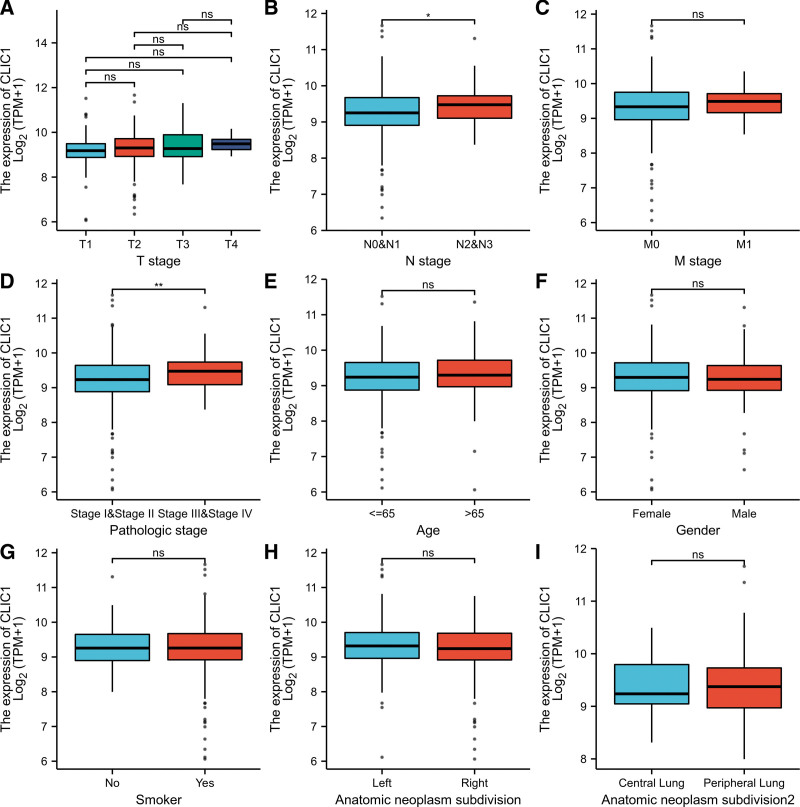
Baseline characteristics of LUAD patients were shown in Table 1. Using TCGA data, Mann–Whitney U test and logistic regression analysis were performed to investigate the relationship between clinical-pathological features and the CLIC1 mRNA expression. As can be seen from Table 1 and Figure 3A–I, the expression level was significantly correlated with stage N disease (P = .006), pathological stage (P = .036) and overall survival (P = .026). CLIC1 expression was higher in patients with lymph node metastases (P = .016), and in patients with high pathological stage (P = .001). However, no statistically significant correlation between expression levels was found between CLIC1 and other clinical-pathological features such as T stage (P = .097), M stage (P = .729), age (P = .658), smokers conditions (P = .901), and anatomic subdivision (right vs. left, P = .350; peripheral versus central, P = .305). In summary, these results suggested that CLIC1 is highly correlated with lymph node metastases and tumor node metatasis staging, further suggesting that CLIC1 may be a biomarker of poor prognosis in LUAD.
Table 1.
Clinical characteristics of the lung adenocarcinoma patients (TCGA).
| Characteristic | Low expression of CLIC1 | High expression of CLIC1 | P |
|---|---|---|---|
| n | 267 | 268 | |
| T stage, n (%) | .097 | ||
| T1 | 96 (18%) | 79 (14.8%) | |
| T2 | 139 (26.1%) | 150 (28.2%) | |
| T3 | 24 (4.5%) | 25 (4.7%) | |
| T4 | 5 (0.9%) | 14 (2.6%) | |
| N stage, n (%) | .006* | ||
| N0 | 188 (36.2%) | 160 (30.8%) | |
| N1 | 36 (6.9%) | 59 (11.4%) | |
| N2 | 29 (5.6%) | 45 (8.7%) | |
| N3 | 1 (0.2%) | 1 (0.2%) | |
| M stage, n (%) | .729 | ||
| M0 | 165 (42.7%) | 196 (50.8%) | |
| M1 | 10 (2.6%) | 15 (3.9%) | |
| Pathologic stage, n (%) | .036* | ||
| Stage I | 163 (30.9%) | 131 (24.9%) | |
| Stage II | 55 (10.4%) | 68 (12.9%) | |
| Stage III | 34 (6.5%) | 50 (9.5%) | |
| Stage IV | 11 (2.1%) | 15 (2.8%) | |
| Gender, n (%) | .575 | ||
| Female | 139 (26%) | 147 (27.5%) | |
| Male | 128 (23.9%) | 121 (22.6%) | |
| Age, n (%) | .658 | ||
| ≤65 | 132 (25.6%) | 123 (23.8%) | |
| >65 | 129 (25%) | 132 (25.6%) | |
| Smoker, n (%) | .901 | ||
| No | 39 (7.5%) | 36 (6.9%) | |
| Yes | 225 (43.2%) | 221 (42.4%) | |
| Anatomic neoplasm subdivision, n (%) | .350 | ||
| Left | 96 (18.5%) | 109 (21%) | |
| Right | 162 (31.2%) | 153 (29.4%) | |
| Anatomic neoplasm subdivision 2, n (%) | .305 | ||
| Central lung | 33 (17.5%) | 29 (15.3%) | |
| Peripheral Lung | 56 (29.6%) | 71 (37.6%) | |
| OS event, n (%) | .026* | ||
| Alive | 184 (34.4%) | 159 (29.7%) | |
| Dead | 83 (15.5%) | 109 (20.4%) |
CLIC1 = Chloride intracellular channel 1, OS = overall survival.
p < .05.
Figure 3.
Relationships between CLIC1 mRNA levels and clinical pathological characteristics. CLIC1 = chloride intracellular channel 1.
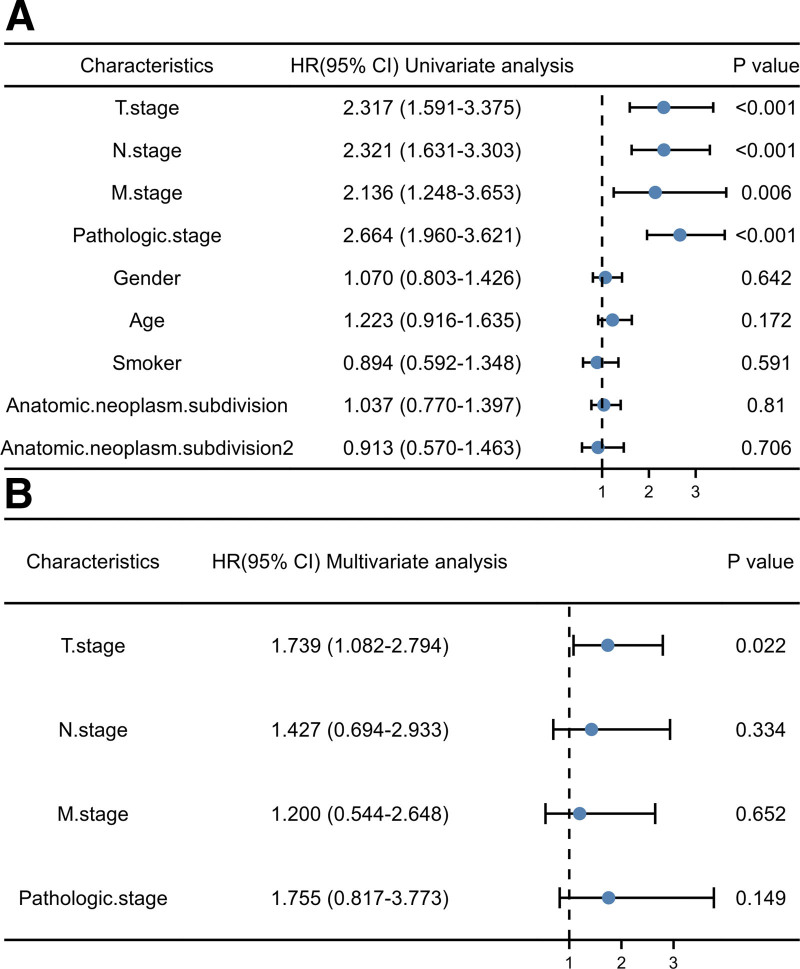
Additionally, Cox univariate survival analysis showed that T, N, M stage and pathological stage were poorly correlated with overall survival (Fig. 4A). Cox multivariate survival analysis further showed that T stage (P = .022), N stage (P = .334), M stage (P = .652) and pathological stages (P = .149) were the factors influencing the survival time of LUAD patients (Fig. 4B), confirming the previous result. These results indicated that CLIC1 expression can not only participate in guiding clinical work as common clinical phenotypes, but also play a superior role in evaluating clinical outcomes in patients with T, N, and M stages (Table 2).
Figure 4.
Cox regression analysis of forest map.
Table 2.
Cox regression analysis of clinical prognosis.
| Characteristics | Total (N) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | ||
| T. stage (T1&T2 vs T3&T4) | 523 | 2.317 (1.591–3.375) | <.001 * | 1.739 (1.082–2.794) | .022 |
| N. stage (N0&N1 vs N2&N3) | 510 | 2.321 (1.631–3.303) | <.001 * | 1.427 (0.694–2.933) | .334 |
| M. stage (M0 vs M1) | 377 | 2.136 (1.248–3.653) | .006 * | 1.200 (0.544–2.648) | .652 |
| Pathologic.stage (Stage I& II vs Stage III&IV) | 518 | 2.664 (1.960–3.621) | <.001 * | 1.755 (0.817–3.773) | .149 |
| Gender (Female vs Male) | 526 | 1.070 (0.803–1.426) | .642 | ||
| Age (≤65 vs > 65) | 516 | 1.223 (0.916–1.635) | .172 | ||
| Smoker (No vs Yes) | 512 | 0.894 (0.592–1.348) | .591 | ||
| Anatomic. neoplasm. subdivision (Left vs Right) | 512 | 1.037 (0.770–1.397) | .81 | ||
| Anatomic.neoplasm.subdivision2 (Central Lung vs Peripheral Lung) | 182 | 0.913 (0.570–1.463) | .706 | ||
CI = confidence interval, HR = hazard ratio.
P < .05.
3.4. Differential RNA-seq levels of CLIC1 as a prospective biomarker to distinguish LUAD samples from normal samples
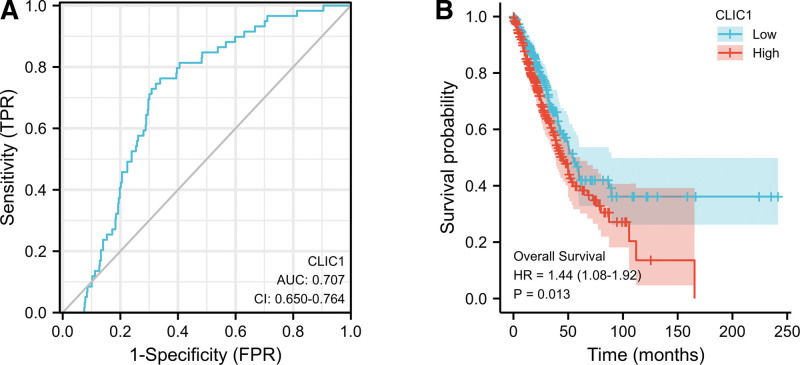
In order to study the value of CLIC1 in distinguishing LUAD samples from normal samples, ROC curve analysis was conducted and it was found that the AUC value of CLIC1 was 0.707 (95%CI: 0.650–0.764) (Fig. 5A). When the cutoff value was 9.060, the sensitivity, specificity and accuracy of CLIC1 were 66.2%, 76.3%, and 74.6%, respectively. The positive predictive value was 19.9%, and the negative predictive value was 96.2%. These findings indicated that CLIC1 may be a promising biomarker for distinguishing LUAD tissue from normal tissue.
Figure 5.
ROC and Kaplan–Meier curves for CLIC1. CLIC1 = chloride intracellular channel 1, ROC = reactive oxygen species.
3.5. High mRNA expression of CLIC1 was associated with short OS
Kaplan–Meier curve and PrognoScan database were used to investigate the relationship between CLIC1 mRNA expression and overall survival (OS) in patients with LUAD. As shown in Figure 5B, the OS of patients with high CLIC1 expression was significantly shorter than low CLIC1 expression (45.2 vs 54.1 months, HR = 1.44, P = .013). These data indicated that high CLIC1 mRNA expression is a biomarker of poor prognosis in LUAD.
3.6. PPI network and functional annotations
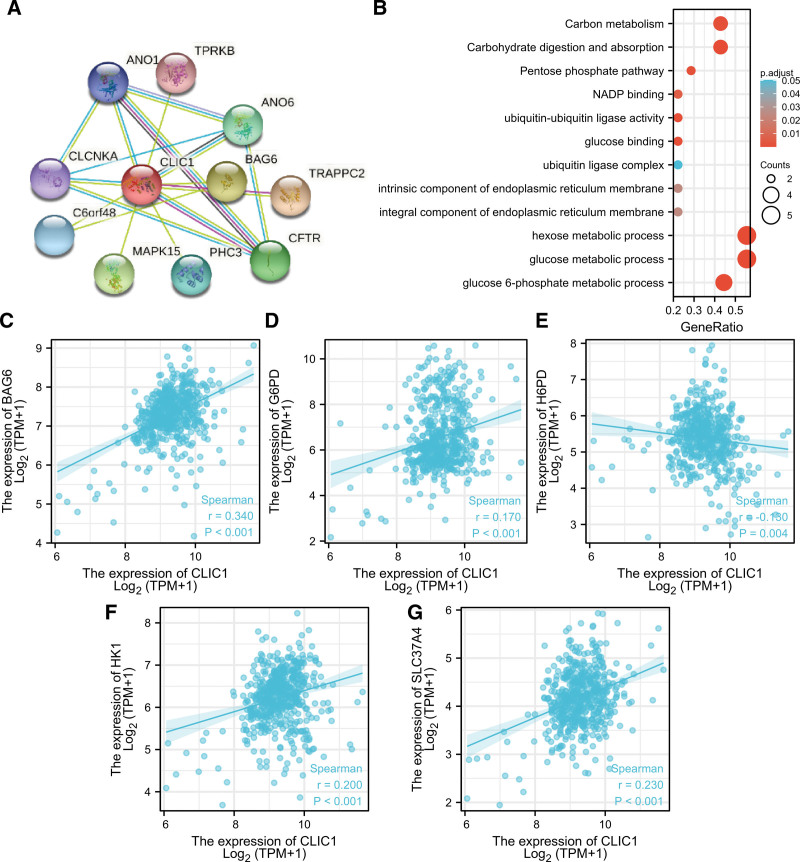
A PPI network and functional annotations were constructed by using the STRING database for gene ontology and Kyoto encyclopedia of genes and genomes analysis. Figure 6A shows CLIC1 and its network of 11 co-expressed genes. As shown in Figure 6B, changes in CLIC1 bio-processes are related to the tissues of hexose and glucose metabolism. Functional annotations indicated that these genes are involved in ubiquitin ligase complexes. The correlation analyses of CLIC1 and co-expressed gene expression in LUAD from TCGA was shown in Figure 6C–G.
Figure 6.
PPI networks and functional enrichment analyses. (A) A network of CLIC1 and its co-expression genes. (B) Functional enrichment analyses of 11 involved genes. (C-G) The correlation analyses between the expression of CLIC1 and co-expressed genes in lung adenocarcinoma. PPI = protein–protein interaction.
3.7. Correlation analysis between CLIC1 expression and immune cell infiltration in LUAD
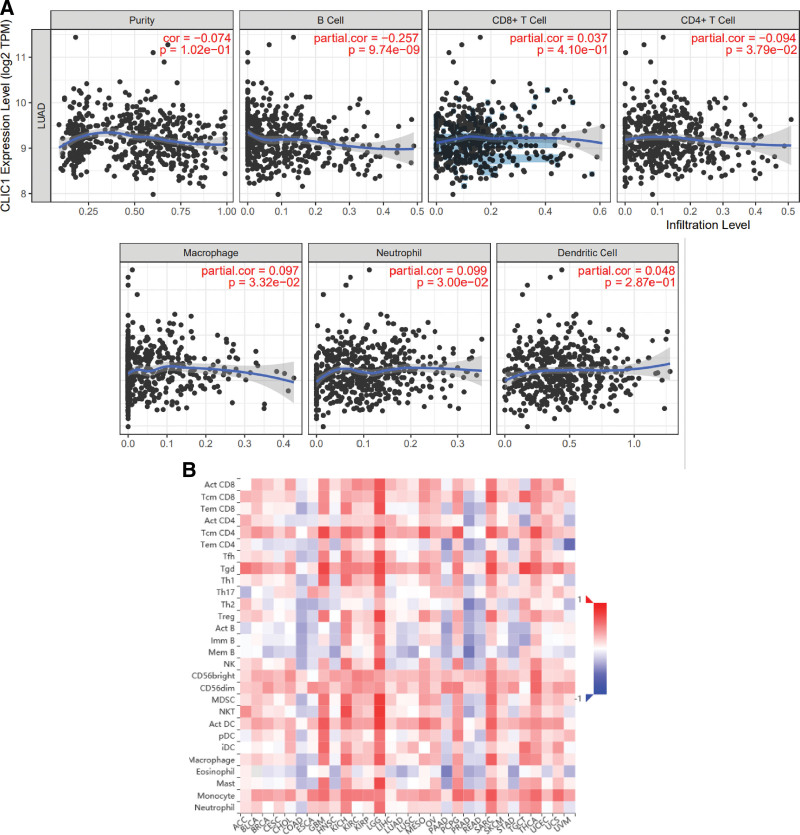
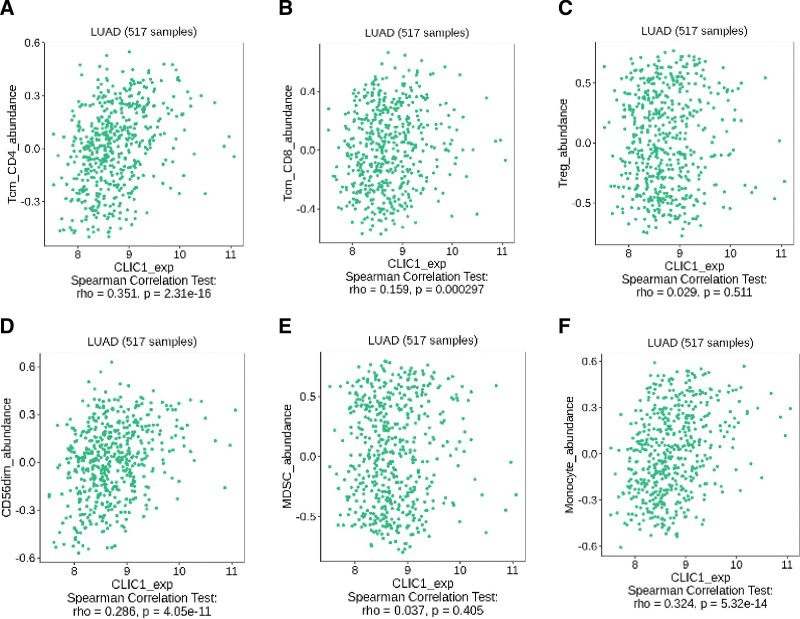
The correlation between CLIC1 expression and the six types of tumor-infiltrating immune cells was studied by using TIMER database and TISIDB database. The result suggested that CLIC1 expression was correlationed with B Cell (r = −0.257, P = 9.74e-09), CD4+ T cell (r = −0.094, P = 3.79e-02), macrophage (R = 0.097, P = 3.32e-02), neutrophil (R = 0.099, P = 3.00e-02) (Fig. 7A). Figure 7B shows the relationship between CLIC1 expression and 28 TILs in human cancers. As shown in Figure 8, CLIC1 expression was correlated with abundant expression of CD8+ T cells (R = 0.159, P = .000297), CD4+ T cells (R = 0.351, P = 2.31 e-16), monocytes (R = 0.324, P = 5.32 e-14) and CD56dim cells (R = 0.286, P = 4.05 e-11). These data suggest that CLIC1 may play a specific role in immune infiltration of LUAD.
Figure 7.
Correlations of CLIC1 expression with immune infiltration level. (A) CLIC1 expression has correlations with B Cell, CD4+ T cell, macrophage, and neutrophil in lung adenocarcinoma. (B) Relations between the expression of CLIC1 and 28 types of TILs across human cancers. CLIC1 = chloride intracellular channel 1, TILs = tumor-infiltrating lymphocytes.
Figure 8.
CLIC1 was correlated with abundance of CD8+ T cells, CD4+ T cells, monocyte cells, and CD56dim cells. CLIC1 = chloride intracellular channel 1.
4. Discussion
Compared with normal lung tissues, mRNA expression of CLIC1 was up-regulated in LUAD tissues in the study. Elevated CLIC1 expression was closely associated with lymph node metastasis. ROC curve analysis indicated that CLIC1 might be a promising biomarker of LUAD. Kaplan–Meier curve analysis showed that high CLIC1 expression was associated with poor OS, suggesting that abnormal CLIC1 expression may also be a potential biomarker for poor prognosis of LUAD. In addition,the correlation analysis between CLIC1 expression and immune cell infiltration in LUAD tissue predicted that CLIC1 may play a specific role in the process of cellular immunity, which may be a novel direction of immunotherapy for LUAD patients.
While the rate of early diagnosis has increased, the high false-positive rate of low-dose CT screening for lung cancer has also brought about the problem of over-diagnosis and over-treatment.[24] Therefore, the development of noninvasive adjuvant biomarkers may be of great help in reducing the false positive rate of CT. Biomarkers may be produced in cancer cells, tumor microenvironment, or host response to cancer.[25] As cancer progresses, certain proteins released by tumor cells can enter the bloodstream.[9] These proteins can be detected in serum or plasma and used as tumor screening markers. For instance, CA-125 and CA19-9 are currently the most widely used clinical tumor markers, and they are sensitive to ovarian and pancreatic cancer, respectively.[26–29] CLIC1 also is another protein that may play a similar role.
CLIC1, also known as NCC27, is a highly conserved chloride ion channel that exists in both soluble and integral membrane forms.[30] Like other members of the CLIC family, CLIC1 has obvious chloride channel function and biological activity of regulating cell volume, intracellular organelle acidity, ionic homeostasis and pH.[30,31] CLIC1 has been strongly associated with gastrointestinal tumors, including pancreatic duct adenocarcinoma, liver tumor, gallbladder cancer, colorectal cancer, and gastric cancer in many studies.[11,12,32–34] However, there are few studies on the relationship between CLIC1 and gastrointestinal tumors. Statistical analysis of plasma CLIC1 levels indicated that CLIC1 could be used as a marker for early detection of nasopharyngeal carcinoma.[9] However,a retrospective analysis of a small sample showed that CLIC1 may be closely related to the occurrence and development of LUAD, and can be used as an effective marker to predict the prognosis of the disease.[10] Therefore, ROC curve analysis was used to further determine whether CLIC1 has clinical significance in the diagnosis of LUAD. The results showed that CLIC1 had a significant AUC value in LUAD detection, with a sensitivity of 69.5%, specificity of 93.2% and an accuracy of 71.9%. Based on our findings, we suggest that CLIC1 may serve as a potential diagnostic biomarker.
The relationship between CLIC1 and tumor metastasis and development has been concerned for a long time. With the deepening of research, the mechanism between the two is gradually clarified. It has been confirmed that tumor metastasis to the lung is related to the ability of tumor cells to produce aggressive feet in coagulated plasma. This mechanism relies on integrin αvβ3 and fibronectin.[35] In another paper, Gurski et al determined that CLIC1 cooperates with integrin αvβ3 and fibronectin to support fibrin invasion and colony formation in vitro.[14] Inspired by the relationship between CLIC1 and Alzheimer’s disease, Chang et al proposed that CLIC1 may also induce the production of reactive oxygen species (ROS) during carcinogenesis, thereby promoting cell proliferation, cell movement, invasion, metastasis, and angiogenesis.[9] Membrane protrusion and extracellular matrix adhesion are two basic processes of cell migration, which are essential for embryonic development, wound healing, immune response and tumor invasion and metastasis.[15,36,37] And recently Peng et al have found that by recruiting PIP5Ks to the plasma membrane, CLIC1 regulates cell-matrix adhesion and membrane protrusion formation spatiotemporally.[15]
Moreover, studies have shown that the high expression of CLIC1 is closely related to lymph node metastasis, lymphatic infiltration, perineural infiltration, pathological stage and poor survival.[11,34,38] This is not completely consistent with our findings. In our study, the increase in CLIC1 was statistically significant only in patients with lymph node metastasis (P = .006) and patients with high pathological stages (P = .001). And the Kaplan–Meier curve analysis result (HR = 1.44, P = .013) further indicated that CLIC1 could be a promising biomarker for predicting the prognosis of LUAD.
The functions of CLICs in innate immunity and inflammasome is unclear.[39] Previous studies have suggested that CLIC4 has an innate immune function because CLIC4-deficient mice are resistant to LPS-induced septic shock, although the mechanism is still unclear.[40] A study has shown that CLICs acts on the downstream of the potassium efflux-mitochondrial ROS axis to promote NLRP3 inflammasome activation, whose dysregulation is related to tumor pathogenesis.[39,41] CLIC1 is known to be involved in inflammatory processes by regulating macrophage phagosome function such as pH and proteolysis.[30] Salon et al have proved that CLIC1 regulates the pH of dendritic cells phagosomes to ensure optimal processing of antigen presentation to antigen-specific T cells in vivo and in vitro.[42] Yu et al used CLIC1 and Mycobacterium tuberculosis heat shock protein 70 to synthesize fusion protein (MtHsp70-CLIC1) and confirmed that dendritic cells pulsed by MtHsp70-CLIC1 could enhance anti-tumor immunity against ovarian cancer.[43] All this seems to suggest that CLIC1 may play a crucial role in anti-tumor immunotherapy. Here, our results also support this assumption, found that high expression of CLIC1 is associated with multiple immune cells (B Cell, CD4+ T cell, macrophage, neutrophil, CD8+ T cells and CD56dim cells). However, further studies are needed to confirm this association.
Our study has several limitations. Firstly, all analyses were based on online databases, so our conclusions lack further clinical sample studies to confirm them. Secondly, in vitro and in vivo experiments should be designed to further study the detailed mechanism of the effect of CLIC1 on the immune infiltration of lung adenocarcinoma.
In summary, this study confirmed that CLIC1 is highly expressed in LUAD tissues, which is different from ordinary tissues. It is highly correlated with lymph node metastasis, tumor node metatasis stage and OS, and may be used as a prognostic indicator of LUAD. Meanwhile, CLIC1 may play a special role in the immune infiltration of LUAD.
Author contributions
Conceptualization: Zhiqiang Chen, Wenmin Chen.
Data curation: Ruilan Huang, Daman Chen, Zhuoyao Li, Xiangjun Qi.
Methodology: Ruilan Huang, Daman Chen, Zhuoyao Li, Xiangjun Qi.
Visualization: Lingling Sun, Lizhu Lin, Zhiquan Zhang.
Writing—original draft: Zhiqiang Chen, Wenmin Chen.
Writing—review & editing: Zhiqiang Chen, Lingling Sun, Lizhu Lin, Zhiquan Zhang.
Abbreviations:
- CLIC1 =
- chloride intracellular channel 1
- HPA =
- human protein atlas
- LUAD =
- lung adenocarcinoma
- OS =
- overall survival
- PPI =
- protein–protein interaction
- ROC =
- reactive oxygen species
- TCGA =
- the cancer genome altas
- TILs =
- tumor-infiltrating lymphocytes
- TIMER =
- tumour immune estimation resource
- TISIDB =
- Tumor-immune system interaction database
The work was supported by National Natural Science Foundation of China (No. 81973775 and 81573780) and Natural Science Foundation of Guangdong Province (No. 2020A1515011176 and 2018B030311023).
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are publicly available.
How to cite this article: Chen Z, Chen W, Huang R, Chen D, Li Z, Qi X, Sun L, Lin L, Zhang Z. Comprehensive analysis of clinical prognosis and CLIC1 immune invasion in lung adenocarcinoma. Medicine 2022;101:39(e30760).
Contributor Information
Zhiqiang Chen, Email: 1875806777@qq.com.
Wenmin Chen, Email: 1875806777@qq.com.
Ruilan Huang, Email: 906469305@qq.com.
Daman Chen, Email: 1875806777@qq.com.
Zhuoyao Li, Email: 1169046200@qq.com.
Xiangjun Qi, Email: 20201120220@stu.gzucm.edu.cn.
Lingling Sun, Email: lingling.sun2017@gmail.com.
Zhiquan Zhang, Email: zhang-zq-jy@163.com.
References
- [1].Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2016;389:299–311. [DOI] [PubMed] [Google Scholar]
- [3].Lu J, Zhong H, Chu T, et al. Role of anlotinib-induced CCL2 decrease in anti-angiogenesis and response prediction for nonsmall cell lung cancer therapy. Eur Respir J. 2019;53:1801562. [DOI] [PubMed] [Google Scholar]
- [4].Dai C, Shen J, Ren Y, et al. Choice of surgical procedure for patients with non-small-cell lung cancer ≤ 1 cm or > 1 to 2 cm among lobectomy, segmentectomy, and wedge resection: a population-based study. J Clin Oncol. 2016;34:3175–82. [DOI] [PubMed] [Google Scholar]
- [5].Hong QY, Wu GM, Qian GS, et al. Prevention and management of lung cancer in China. Cancer. 2015;121:3080–8. [DOI] [PubMed] [Google Scholar]
- [6].Yang D, Liu Y, Bai C, et al. Epidemiology of lung cancer and lung cancer screening programs in China and the United States. Cancer Lett. 2020;468:82–7. [DOI] [PubMed] [Google Scholar]
- [7].Cao M, Chen W. Epidemiology of lung cancer in China. Thorac Cancer. 2019;10:3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Shi ZH, Zhao C, Wu H, et al. CLIC1 Protein a candidate prognostic biomarker for malignant-transformed hydatidiform moles. Int J Gynecol Cancer. 2011;21:153–60. [DOI] [PubMed] [Google Scholar]
- [9].Chang YH, Wu CC, Chang KP, et al. Cell secretome analysis using hollow fiber culture system leads to the discovery of CLIC1 protein as a novel plasma marker for nasopharyngeal carcinoma. J Proteome Res. 2009;8:5465–74. [DOI] [PubMed] [Google Scholar]
- [10].Wang W, Xu X, Wang W, et al. The expression and clinical significance of CLIC1 and HSP27 in lung adenocarcinoma. Tumor Biol. 2011;32:1199–208. [DOI] [PubMed] [Google Scholar]
- [11].Chen CD, Wang CS, Huang YH, et al. Overexpression of CLIC1 in human gastric carcinoma and its clinicopathological significance. Proteomics. 2007;7:155–67. [DOI] [PubMed] [Google Scholar]
- [12].Petrova DT, Asif AR, Armstrong VW, et al. Expression of chloride intracellular channel protein 1 (CLIC1) and tumor protein D52 (TPD52) as potential biomarkers for colorectal cancer. Clin Biochem. 2008;41:1224–36. [DOI] [PubMed] [Google Scholar]
- [13].Huang JS, Chao CC, Su TL, et al. Diverse cellular transformation capability of overexpressed genes in human hepatocellular carcinoma. Biochem Biophys Res Commun. 2004;315:950–8. [DOI] [PubMed] [Google Scholar]
- [14].Lisa AG, Knowles LM, Basse PH, et al. Relocation of CLIC1 promotes tumor cell invasion and colonization of fibrin. Mol Cancer Res. 2015;13:273–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Peng JM, Lin SH, Yu MC, et al. CLIC1 recruits PIP5K1A/C to induce cell-matrix adhesions for tumor metastasis. J Clin Invest. 2021;131:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang Z, Jensen MA, Zenklusen JC. A practical guide to The Cancer Genome Atlas (TCGA). Methods Mol Biol. 2016;1418:111–41. [DOI] [PubMed] [Google Scholar]
- [17].Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinfor. 2011;12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ellis MJ, Gillette M, Carr SA, et al. Connecting genomic alterations to cancer biology with proteomics: the NCI Clinical Proteomic Tumor Analysis Consortium. Cancer Discov. 2013;3:1108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Navani S. Manual evaluation of tissue microarrays in a high-throughput research project: the contribution of Indian surgical pathology to the Human Protein Atlas (HPA) project. Proteomics. 2016;16:1266–70. [DOI] [PubMed] [Google Scholar]
- [20].Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kanehisa M, Sato Y, Kawashima M, et al. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].He M, Han Y, Cai C, et al. CLEC10A is a prognostic biomarker and correlated with clinical pathologic features and immune infiltrates in lung adenocarcinoma. J Cell Mol Med. 2021;25:3391–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lu G, Zhou Y, Zhang C, et al. Upregulation of LIMK1 Is correlated with poor prognosis and immune infiltrates in lung adenocarcinoma. Front Genet. 2021;12:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ten Haaf K, van der Aalst CM, de Koning HJ. Clinically detected non-aggressive lung cancers: implications for overdiagnosis and overtreatment in lung cancer screening. Thorax. 2018;73:407–8. [DOI] [PubMed] [Google Scholar]
- [25].Sozzi G, Boeri M. Potential biomarkers for lung cancer screening. Transl Lung Cancer Res. 2014;3:139–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Luo G, Jin K, Deng S, et al. Roles of CA19-9 in pancreatic cancer: biomarker, predictor and promoter. Biochim Biophys Acta Rev Cancer. 2021;1875:188409. [DOI] [PubMed] [Google Scholar]
- [27].Dunn PM. CA19-9 and pancreatic cancer. Ann Intern Med. 1989;111:343. [DOI] [PubMed] [Google Scholar]
- [28].Duffy MJ, Bonfrer JM, Kulpa J, et al. CA125 in ovarian cancer: European Group on tumor markers guidelines for clinical use. Int J Gynecol Cancer. 2005;15:679–91. [DOI] [PubMed] [Google Scholar]
- [29].Bast RC, Xu FJ, Yu YH, et al. CA 125: the past and the future. Int J Biol Markers. 1998;13(4):179–87. [DOI] [PubMed] [Google Scholar]
- [30].Harrop SJ, DeMaere MZ, Fairlie WD, et al. Crystal structure of a soluble form of the intracellular chloride ion channel CLIC1 (NCC27) at 1.4-Å resolution. J Biol Chem. 2001;276:44993–5000. [DOI] [PubMed] [Google Scholar]
- [31].Jentsch TJ. Chloride channels: a molecular perspective. Curr Opin Neurobiol. 1996;6:303–10. [DOI] [PubMed] [Google Scholar]
- [32].Jia N, Dong S, Zhao G, et al. CLIC1 overexpression is associated with poor prognosis in pancreatic ductal adenocarcinomas. J Cancer Res Ther. 2016;12:892–6. [DOI] [PubMed] [Google Scholar]
- [33].Wang JW, Peng SY, Li JT, et al. Identification of metastasis-associated proteins involved in gallbladder carcinoma metastasis by proteomic analysis and functional exploration of chloride intracellular channel 1. Cancer Lett. 2009;281:71–81. [DOI] [PubMed] [Google Scholar]
- [34].Zhang S, Wang XM, Yin ZY, et al. Chloride intracellular channel 1 is overexpression in hepatic tumor and correlates with a poor prognosis. APMIS. 2013;121:1047–53. [DOI] [PubMed] [Google Scholar]
- [35].Knowles LM, Gurski LA, Engel C, et al. Integrin αvβ3 and fibronectin upregulate Slug in cancer cells to promote clot invasion and metastasis. Cancer Res. 2013;73:6175–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Murai T, Kawashima H, Naor D. Editorial: cell-cell and cell-matrix adhesion in immunobiology and cancer. Front Immunol. 2020;10:3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Schaks M, Rottner K. Actin dynamics in cell migration. Essays Biochem. 2019;63:483–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ding Q, Li M, Wu X, et al. CLIC1 overexpression is associated with poor prognosis in gallbladder cancer. Tumour Biol. 2015;36:193–8. [DOI] [PubMed] [Google Scholar]
- [39].Tang T, Lang X, Xu C, et al. CLICs-dependent chloride efflux is an essential and proximal upstream event for NLRP3 inflammasome activation. Nat Commun. 2017;8:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].He G, Ma Y, Chou XY, et al. Role of CLIC4 in the host innate responses to bacterial lipopolysaccharide. Eur J Immunol. 2011;41:1221–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Moossavi M, Parsamanesh N, Bahrami A. Role of the NLRP3 inflammasome in cancer. Mol Cancer. 2018;17:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Salao K, Jiang L, Li H, et al. CLIC1 regulates dendritic cell antigen processing and presentation by modulating phagosome acidification and proteolysis. Biol Open. 2016;5:620–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Yu W, Qu H, Cao G. MtHsp70-CLIC1-pulsed dendritic cells enhance the immune response against ovarian cancer. Biochem Biophys Res Commun. 2017;494:13–9. [DOI] [PubMed] [Google Scholar]