Abstract
Bladder cancer (BC) is one of the most common male malignant tumors and the most common urological tumor. However, the molecular mechanism and role of PLK1 on bladder cancer were unclear. Therefore, the study aims to explore the potential part of the overall survival of bladder cancer through bioinformatics analysis. GSE121711 and GSE130598, from the Gene Expression Omnibus database. The GEO2R screened differently expressed genes, and DAVID and Metascape were used for functional annotation. The cytoHubba made hub genes identification and expression. A total of 50 BC participants were recruited. After surgery, 50 BC tumor samples from BC patients and 50 adjacent standard bladder tissue samples were obtained. The RT-qPCR assay was performed to verify the expression of hub genes. The Kaplan–Meier Plotter analyzed the effect of hub gene expression for overall survival of BC. The compulsory module of Molecular Complex Detection tool analysis was shown, which included CDK1, TTK, AURKB, MELK, PLK1, and BUB1. And the six hub genes were up-regulated in the BC compared with the normal tissues. The relative expression levels of CDK1, TTK, AURKB, MELK, PLK1, and BUB1 were significantly higher in BC samples compared with the regular kidney tissue groups. The result demonstrated that CDK1, TTK, AURKB, MELK, PLK1, and BUB1 might be considered biomarkers for BC. Overall survival analysis showed that BC patients with high expression level of PLK1 had poorer overall survival times than those with low expression level (P < .05). The expression levels of CDK1, TTK, AURKB, MELK, and BUB1 was not related to the overall survival of BC patients (P > .05). The PLK1 gene might provide new ideas and evidence for bladder cancer research.
Keywords: bladder cancer, differently expressed genes, PLK1, prognosis, survival times, verification
1. Introduction
Bladder cancer (BC) is the most common tumor of the urinary system. Most tumors come from epithelial tissues, and more than 90% are transitional epithelial tumors. According to the American Cancer Society, bladder cancer accounts for 5% of all new cancer cases and 3% of all cancer deaths in the United States. The incidence of bladder cancer will be the second urinary tract cancer in 2020. The 5-year survival rate for bladder cancer patients was only 14%.[1] Although radical cystectomy and neoadjuvant chemotherapy are currently used to treat bladder cancer, the prognosis is still poor due to its high recurrence rate.[2] Long-term exposure to carcinogens, smoking, and chronic bladder infections and long-term foreign body irritation increase the risk of bladder cancer. But the exact cause of bladder cancer is still unknown. Therefore, it is urgent to find more effective biomarkers to diagnose and treat bladder cancer.
Bioinformatics is widely used to process and analyze large amounts of complex biological data. Microarray data information analysis have been commonly used in studying tumors and other diseases to explore the genetic correlation among them.[3,4] Microarray analysis technology can simultaneously obtain the expression information of tens of thousands of genes and then discover the genomic changes related to the occurrence and development of diseases. Currently, many studies[5,6] have used bioinformatics techniques to analyze differentially expressed genes in tumor progression, then study their roles in biological processes, molecular functions and signaling pathways, and elucidate diseases’ pathogenesis as a theoretical basis for early diagnosis and treatment.
Polo-like kinases (Plks) are a class of structurally highly conserved serine and threonine protein kinases that play an essential regulatory role in the cell cycle process.[7] Polo-like kinase 1 (PLK1) is one of the main subtypes of Plks. Currently, many studies have found that PLK1 is abnormally expressed in various tumors, including melanoma,[8] colorectal cancer,[9] and non-small cell lung cancer.[10] Similarly, Plk1 Overexpression was found in bladder cancer studies, and this Overexpression was associated with higher pathological grade (P = .0024) and multiple tumors (P = .0241).[11] But its specific mechanism of action has not been studied.
Therefore, this paper intends to use bioinformatics analysis to explore the hub genes of bladder cancer and aims to explore the potential role in the overall survival of bladder cancer.
2. Materials and Methods
2.1. Dataset
We present the following article following the PRISMA Checklist. We downloaded two gene profiles, GSE121711 and GSE130598, from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). The GSE121711 includes eight tumoral bladder tissues and ten non-tumoral bladder tissues, while the GSE130598 includes 24 24 non-tumoral bladder tissues.
2.2. Differently expressed genes identification
We applied GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r), an online tool based on GEOquery and limma R packages, to identify Differently expressed genes (DEGs) between esophagus cancer and normal group. The cutoff criteria were that P value < .05 and a log (FC) > 1 or log (FC) < −1.
2.3. DEGs annotation
DAVID (https://david.ncifcrf.gov/home.jsp) and Metascape (http://metascape.org/gp/index.html) are two powerful annotation tools that can perform the biological process (BP), cellular component (CC), molecular function (MF), and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis on genes. We annotated the function of common DEGs through DAVID and Metascape.
2.4. Protein–protein interaction network construction
The Search Tool for the Retrieval of Interacting Genes (STRING) (http://string-db.org) can convert DEGs into expressed proteins and construct a protein–protein interaction (PPI) network. We got a PPI network of common DEGs through STRING and visualized it by Cytoscape (version 3.8.0).
2.5. Hub genes identification and expression
Molecular Complex Detection tool (MCODE) (version 1.6.1), an open plug-in of Cytoscape, was used to identify significant modules from the PPI network. The criteria were that the maximum depth = 100, MCODE scores > 5, cutoff = 2, k-score = 2, and node score cutoff = 0.2.
2.6. RT-qPCR assay
A total of 50 BC participants were recruited. After surgery, 50 BC tumor samples from BC patients and 50 adjacent standard bladder tissue samples were obtained. The research conformed to the Declaration of Helsinki and was authorized by the Human Ethics and Research Ethics Committees of the Fourth Hospital of Hebei Medical University. Informed consent was obtained from all participants.
Total RNA was extracted from 50 BC tumor samples and 50 adjacent standard bladder tissue samples by the RNAiso Plus (Trizol) kit (Thermofisher, Waltham, MA) and reverse transcribed to cDNA. RT-qPCR was performed using a Light Cycler® 4800 System with specific primers for genes. The RQ values (2−ΔΔCt, where Ct is the threshold cycle) of each sample were calculated and are presented as fold changes in gene expression relative to the control group. GAPDH was used as an endogenous control.
2.7. Effect of hub gene expression for overall survival of BC
The Kaplan–Meier Plotter analyzed the effect of hub gene expression for overall survival of BC.
3. Results
3.1. DEGs
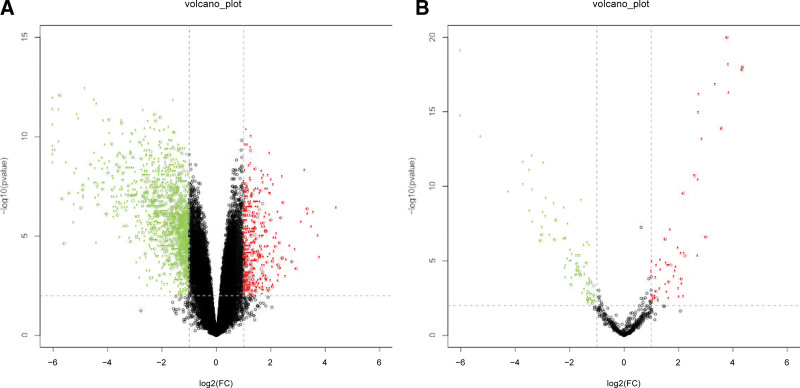
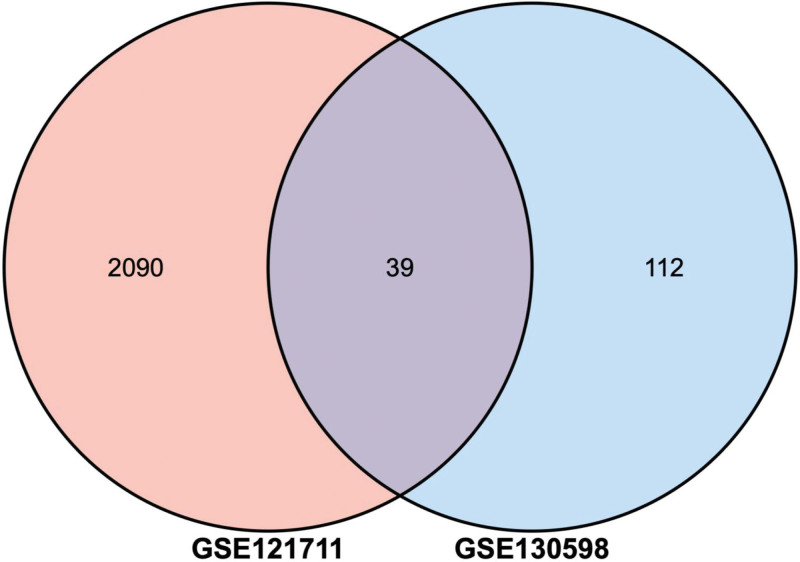
Two volcano plots present the DEGs in the GSE121711 and GSE130598 (Fig. 1A and B). The Venn diagram revealed 39 DEGs shared by the two datasets (Fig. 2).
Figure 1.
Two volcano plots present the DEGs. (A) The DEGs in the GSE121711. (B) The DEGs in the GSE130598. DEGs = differently expressed genes.
Figure 2.
The common DEGs between GSE121711 and GSE130598. DEGs = differently expressed genes.
3.2. DEGs annotation
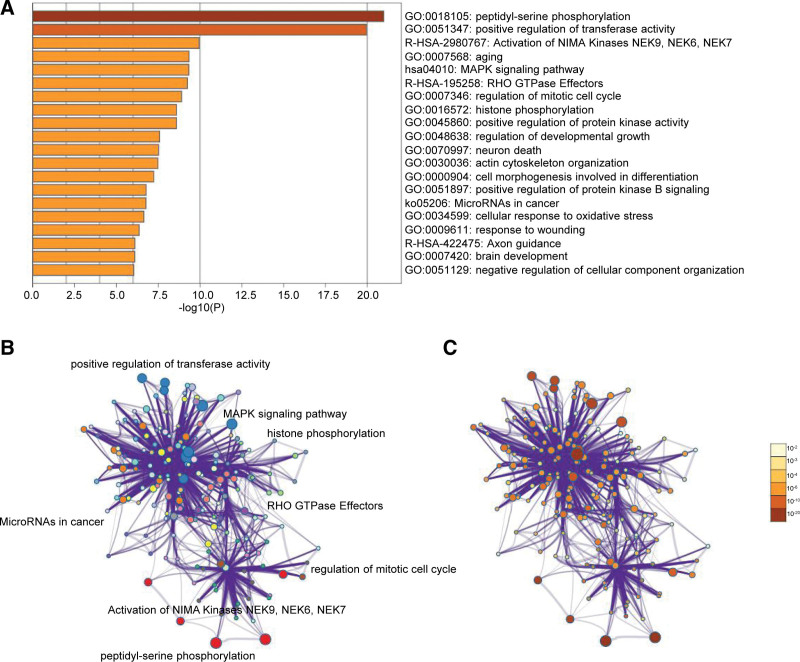
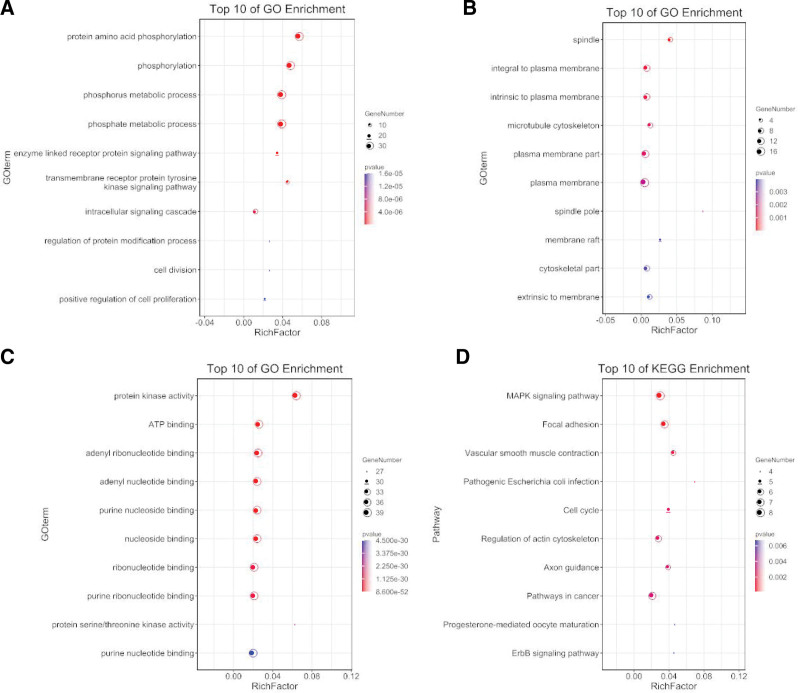
Enrichment analysis by Metascape is displayed in Figure 3. The DEGs related to BP, CC, MF, and KEGG enrichment analysis were shown in bubble diagrams separately (Fig. 4A–D). The variations in DEGs linked with BP were mainly enriched in protein amino acid phosphorylation, phosphorylation, phosphorus metabolic process, phosphate metabolic process, enzyme-linked receptor protein signaling pathway, transmembrane receptor protein tyrosine kinase signaling pathway, intracellular signaling cascade, regulation of protein modification process, cell division, and positive regulation of cell proliferation (Fig. 4A). The spindle, microtubule cytoskeleton, plasma membrane part, plasma membrane, spindle pole, membrane raft, cytoskeletal component, and extrinsic membrane were the critical regions of the plasma membrane where the alterations in DEGs associated with CC were enriched (Fig. 4B). The variations in DEGs linked with MF were mainly enriched in protein kinase activity, ATP binding, adenyl ribonucleotide binding, adenyl nucleotide binding, purine nucleoside binding, nucleoside binding, ribonucleotide binding, purine ribonucleotide binding, protein serine/threonine kinase activity, and purine nucleotide binding (Fig. 4C). The variations in DEGs linked with KEGG were mainly enriched in MAPK signaling pathway, Focal adhesion, Vascular smooth muscle contraction, Pathogenic Escherichia coli infection, Cell cycle, Regulation of actin cytoskeleton, Axon guidance, Pathways in cancer, Progesterone-mediated oocyte maturation, ErbB signaling pathway (Fig. 4D).
Figure 3.
Enrichment analysis for the DEGs by Metascape. (A) Heatmap of enriched terms across input differently expressed gene lists, colored by P values, via the Metascape. (B) Network of enriched terms colored by cluster identity, where nodes that share the same cluster identity are typically close to each other. (C) Network of enriched terms colored by P value, where terms containing more genes tend to have a more significant P value. DEGs = differently expressed genes.
Figure 4.
The functional annotation for the DEGs based on the DAVID. (A) BP, (B) CC, (C) MF, and (D) KEGG. BP = biological process, CC = cellular component, DEGs = differently expressed genes, KEGG = Kyoto Encyclopedia of Genes and Genomes, MF = molecular function.
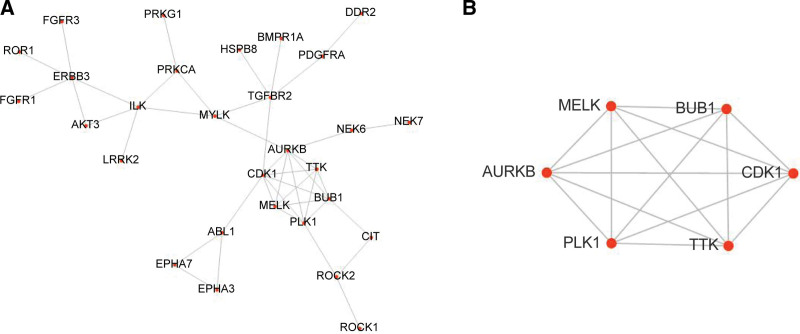
3.3. Protein–protein interaction network and hub genes
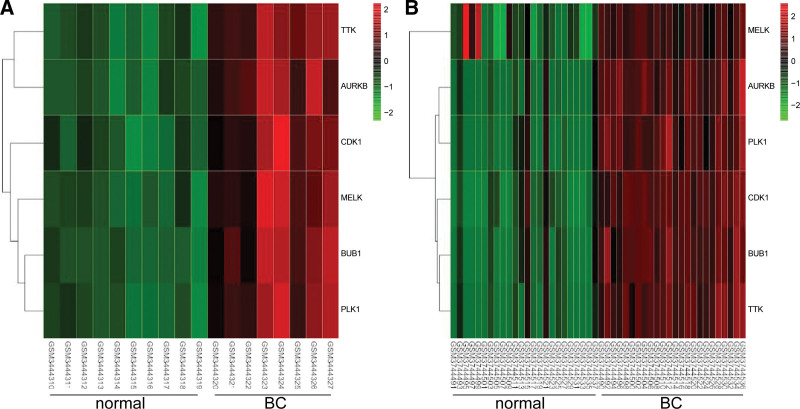
PPI was displayed in Figure 5A. The key MCODE analysis module was shown, including CDK1, TTK, AURKB, MELK, PLK1, and BUB1 (Fig. 5B). Two heat maps showed the expressions of the hub genes in GSE121711 (Fig. 6A) and GSE130598 (Fig. 6B). And the six hub genes were up-regulated in the tumoral compared with the normal tissues.
Figure 5.
The PPI network and hub genes. (A) PPI network. (B) The key module of MCODE analysis with six hub genes CDK1, TTK, AURKB, MELK, PLK1, and BUB1. MCODE = Molecular Complex Detection tool, PPI = protein–protein interaction.
Figure 6.
The expression analysis for the hub genes. (A) The heat map showed the expressions of the hub genes in GSE121711. (B) The heat map showed the expressions of the hub genes in GSE130598.
3.4. Results of RT-qPCR analysis
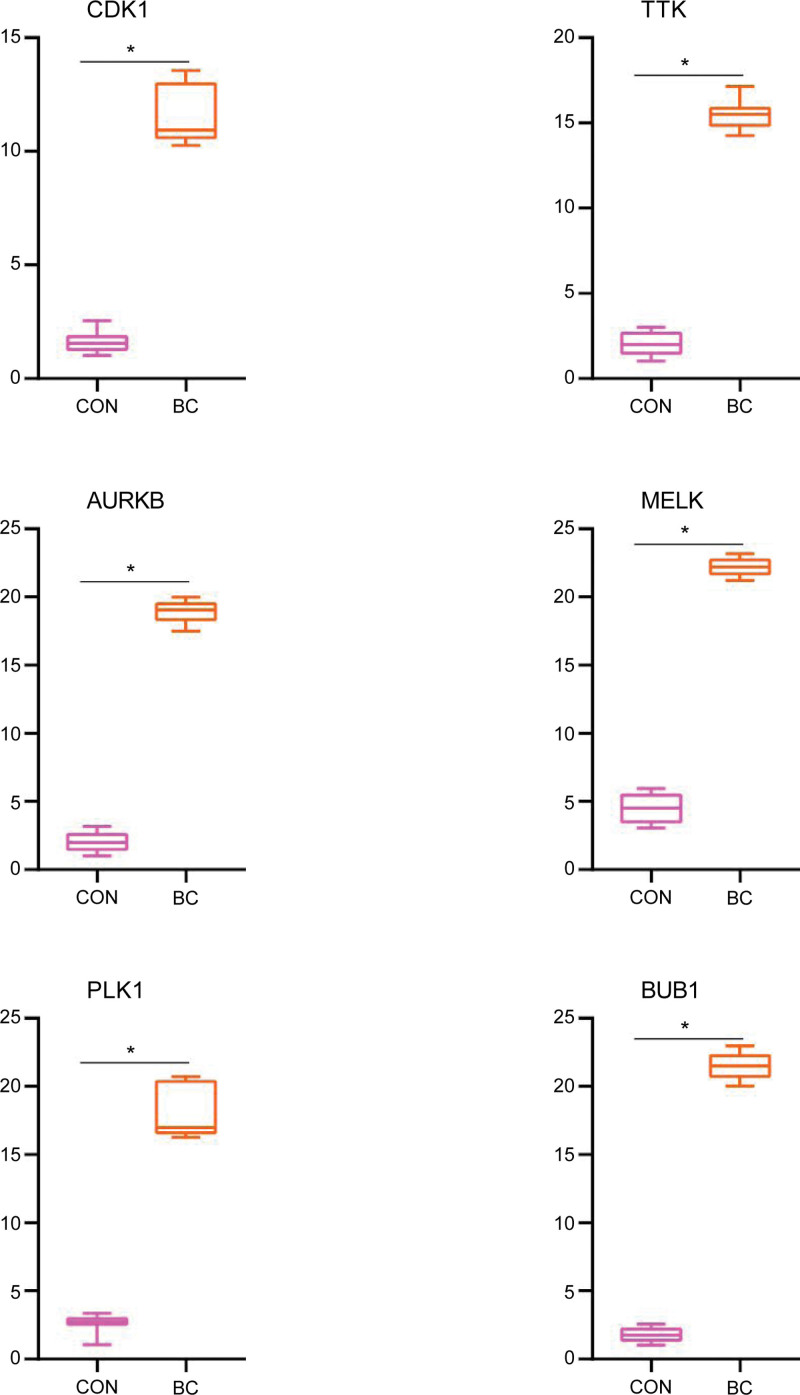
As presented in Figure 7, the relative expression levels of CDK1, TTK, AURKB, MELK, PLK1, and BUB1 were significantly higher in BC samples compared with the regular kidney tissue groups. The result demonstrated that CDK1, TTK, AURKB, MELK, PLK1, and BUB1 might be considered biomarkers for BC.
Figure 7.
Relative expression of CDK1, TTK, AURKB, MELK, PLK1, and BUB1 by RT-qPCR analysis. *P < .05, compared with normal bladder tissues.
3.5. Association between hub gene expression and overall survival
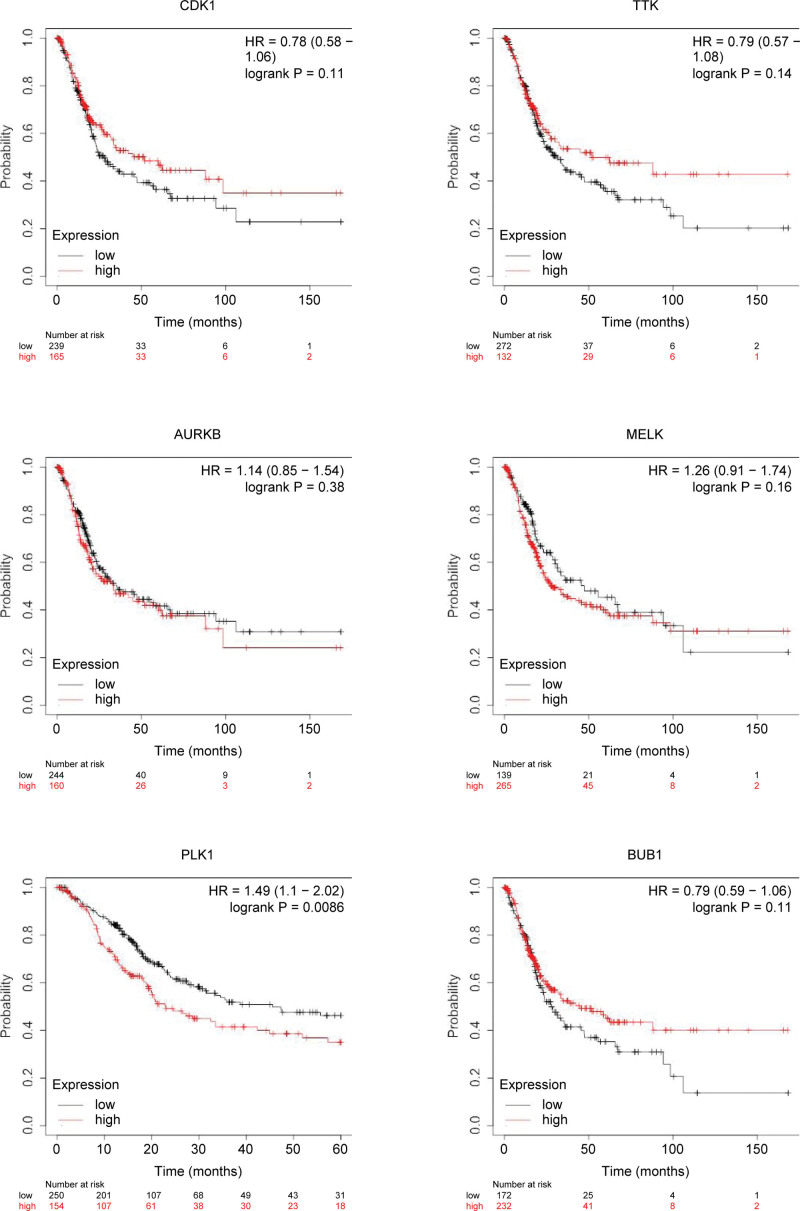
Overall survival analysis showed that BC patients with high expression level of PLK1 had poorer overall survival times than those with low expression level (P < .05). The expression levels of CDK1, TTK, AURKB, MELK, and BUB1 was not related to the overall survival of BC patients (P > .05, Fig. 8).
Figure 8.
The effect of gene expression on overall survival.
4. Discussion
Bladder cancer is one of the most common male malignant tumors and the most common urological tumor.[12] The biological behavior of bladder cancer is complex and has the characteristics of easy recurrence and metastasis. Local progression and distant metastasis occur in approximately 33% of primary bladder cancers,[13] but the specific mechanism of occurrence and development has not been studied. In-depth exploration of the molecular mechanism of bladder cancer is significant for researching targeted drugs. This study used bioinformatics to analyze bladder cancer and normal tissues and screened core genes. The main result of this study was that the PLK1 gene was highly expressed in bladder cancer, and when this gene was highly expressed, the prognosis of patients was poor.
Polo-like kinase 1 (PLK1), a member of the silk/threonine protein kinase family, plays various roles in the cell cycle. PLK1 is critical for precisely regulating cell division and maintaining genomic stability in mitosis, spindle assembly, and DNA damage responses.[14] Studies have found that it is highly expressed in prostate cancer,[15] neuroblastoma cells,[16] acute myeloid leukemia,[17] cervical cancer,[18] and other malignant tumors, plays an essential role in the initiation, maintenance and completion process of mitosis, and is closely related to survival and prognosis. Studies have suggested that blocking PLK1 expression by using antibodies, RNA interference or kinase inhibitors can effectively inhibit tumor cell proliferation and induce cell apoptosis.[19] Inhibition of PLK1 expression can lead to tumor cell death by interfering with multiple stages of mitosis, and PLK1 is expected to be a potential target for cancer therapy.[20,21]
PLK1 is overexpressed in cisplatin-resistant gastric cancer cells, and the sensitivity of tumor cells to chemotherapy is increased after the transfer of SI-PLk1 and BI2536.[20] Li et al[15] showed that Overexpression of PLK1 activates prostatic cancer oncogenesis and affects the cell reprogramming process, leading to increased migration and invasion of prostate cancer cells. PLK1-specific inhibitor BI2536 has been used in phase II clinical trials for some cancers.[22] Studies have shown[23] that PLK1 expression in bladder urothelial carcinoma and highly invasive bladder T24 cells were significantly higher than in normal bladder tissues and superficial bladder BIU-87 cells. After increasing PLK1 inhibitor (Scytonemin) concentration, tumor cell proliferation and invasion activity were significantly decreased, and G2/M phase arrest occurred. PLK1 expression is associated with critical histopathological features (grade and stage) and recurrence and metastasis of urothelial carcinoma of the bladder. In the nude mouse model, PLK1 inhibitor RO3280 inhibited the growth and cell cycle progression of bladder cancer cells and blocked xenograft growth of bladder cancer.[24] Wu et al[25] used four PLK1 inhibitors to inhibit PLK1 expression and found that various bladder cancer cells were more sensitive to radiotherapy and chemotherapy. At the same time, the proliferation ability of tumor cells decreased. The above literature review is consistent with our results, the PLK1 gene is highly expressed in bladder cancer, and when the gene is highly expressed, the prognosis of patients is poor. It is speculated that the PLK1 gene plays an essential role in the growth and development of bladder cancer.
Despite the rigorous bioinformatics analysis in this paper, there are still some deficiencies. In this study, no animal experiments of gene overexpression or knockout were conducted to verify its function further. Therefore, in future research, we should explore this aspect in-depth.
5. Conclusion
PLK1 gene may provide new ideas and evidence for targeted bladder cancer therapy.
Author contributions
Conceptualization: Bin Liu.
Data curation: Ling-Bing Meng.
Formal analysis: Jian-Zhi Su, Bo Fan.
Investigation: Jian-Zhi Su, Bo Fan, Shi-Bin Zhao, Hao-Yuan Wang.
Methodology: Shi-Bin Zhao.
Resources: Shi-Bin Zhao.
Software: Hao-Yuan Wang, Tao Li.
Supervision: Hao-Yuan Wang.
Validation: Tao Li, Tian-Yi Wang.
Visualization: Tian-Yi Wang.
Writing – original draft: Ai-Li Zhang, Xiao-Chen Ni.
Writing – review & editing: Ai-Li Zhang, Xiao-Chen Ni.
Abbreviations:
- BC =
- bladder cancer
- BP =
- biological process
- CC =
- cellular component
- DEGs =
- differently expressed genes
- GEO =
- Gene Expression Omnibus
- MCODE =
- Molecular Complex Detection tool
- MF =
- molecular function
- Plks =
- polo-like kinases
- STRING =
- Search Tool for the Retrieval of Interacting Genes
A-LZ and X-CN contributed equally to this work.
The research conformed to the Declaration of Helsinki and was authorized by the Human Ethics and Research Ethics Committees of the Fourth Hospital of Hebei Medical University. Informed consent was obtained for all participants, and they agreed to publish it.
The authors have no funding and conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
How to cite this article: Liu B, Meng L-B, Su J-Z, Fan B, Zhao S-B, Wang H-Y, Li T, Wang T-Y, Zhang A-L, Ni X-C. PLK1 as one novel target for the poor prognosis of bladder cancer: An observational study. Medicine 2022;101:39(e30723).
Contributor Information
Bin Liu, Email: liubinsy123@163.com.
Ling-Bing Meng, Email: 314597690@qq.com.
Jian-Zhi Su, Email: jianzhi1288@163.com.
Bo Fan, Email: fanbo4587@163.com.
Shi-Bin Zhao, Email: zhaoshibin124@sina.com.
Hao-Yuan Wang, Email: tywang6688@163.com.
Tao Li, Email: 1187638487@qq.com.
Tian-Yi Wang, Email: tywang6688@163.com.
References
- [1].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- [2].Zigeuner R. Bladder cancer in 2016: news in diagnosis, treatment, and risk group assessment. Nat Rev Urol. 2017;14:74–6. [DOI] [PubMed] [Google Scholar]
- [3].Zhou Y, Liepe J, Sheng X, et al. GPU accelerated biochemical network simulation. Bioinformatics. 2011;27:874–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Nobile MS, Cazzaniga P, Tangherloni A, et al. Graphics processing units in bioinformatics, computational biology and systems biology. Brief Bioinform. 2017;18:870–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chaudhary K, Poirion OB, Lu L, et al. Deep learning-based multi-omics integration robustly predicts survival in liver cancer. Clin Cancer Res. 2018;24:1248–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Qiu Z, Li H, Zhang Z, et al. A pharmacogenomic landscape in human liver cancers. Cancer Cell. 2019;36:179–93.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Park JE, Soung NK, Johmura Y, et al. Polo-box domain: a versatile mediator of polo-like kinase function. Cell Mol Life Sci. 2010;67:1957–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Sanchez IM, Purwin TJ, Chervoneva I, et al. In vivo ERK1/2 reporter predictively models response and resistance to combined BRAF and MEK inhibitors in melanoma. Mol Cancer Ther. 2019;18:1637–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ran Z, Chen W, Shang J, et al. Clinicopathological and prognostic implications of polo-like kinase 1 expression in colorectal cancer: a systematic review and meta-analysis. Gene. 2019;721:144097. [DOI] [PubMed] [Google Scholar]
- [10].Reda M, Ngamcherdtrakul W, Gu S, et al. PLK1 and EGFR targeted nanoparticle as a radiation sensitizer for non-small cell lung cancer. Cancer Lett. 2019;467:9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yamamoto Y, Matsuyama H, Kawauchi S, et al. Overexpression of polo-like kinase 1 (PLK1) and chromosomal instability in bladder cancer. Oncology. 2006;70:231–7. [DOI] [PubMed] [Google Scholar]
- [12].Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. [DOI] [PubMed] [Google Scholar]
- [13].Choueiri TK, Raghavan D. Chemotherapy for muscle-invasive bladder cancer treated with definitive radiotherapy: persisting uncertainties. Nat Clin Pract Oncol. 2008;5:444–54. [DOI] [PubMed] [Google Scholar]
- [14].Lens SM, Voest EE, Medema RH. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat Rev Cancer. 2010;10:825–41. [DOI] [PubMed] [Google Scholar]
- [15].Li J, Wang R, Kong Y, et al. Targeting Plk1 to enhance efficacy of olaparib in castration-resistant prostate cancer. Mol Cancer Ther. 2017;16:469–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Pajtler KW, Sadowski N, Ackermann S, et al. The GSK461364 PLK1 inhibitor exhibits strong antitumoral activity in preclinical neuroblastoma models. Oncotarget. 2017;8:6730–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tao YF, Li ZH, Du WW, et al. Inhibiting PLK1 induces autophagy of acute myeloid leukemia cells via mammalian target of rapamycin pathway dephosphorylation. Oncol Rep. 2017;37:1419–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yang X, Chen G, Li W, et al. Cervical cancer growth is regulated by a c-ABL-PLK1 signaling Axis. Cancer Res. 2017;77:1142–54. [DOI] [PubMed] [Google Scholar]
- [19].Bu Y, Yang Z, Li Q, et al. Silencing of polo-like kinase (Plk) 1 via siRNA causes inhibition of growth and induction of apoptosis in human esophageal cancer cells. Oncology. 2008;74:198–206. [DOI] [PubMed] [Google Scholar]
- [20].Chen Z, Chai Y, Zhao T, et al. Effect of PLK1 inhibition on cisplatin-resistant gastric cancer cells. J Cell Physiol. 2019;234:5904–14. [DOI] [PubMed] [Google Scholar]
- [21].Ferrarotto R, Goonatilake R, Yoo SY, et al. Epithelial-mesenchymal transition predicts polo-like Kinase 1 inhibitor-mediated apoptosis in non-small cell lung cancer. Clin Cancer Res. 2016;22:1674–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Liu Z, Sun Q, Wang X. PLK1, a potential target for cancer therapy. Transl Oncol. 2017;10:22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhang Z, Zhang G, Kong C. High expression of polo-like kinase 1 is associated with the metastasis and recurrence in urothelial carcinoma of bladder. Urol Oncol. 2013;31:1222–30. [DOI] [PubMed] [Google Scholar]
- [24].Zhang Z, Zhang G, Kong C. Targeted inhibition of Polo-like kinase 1 by a novel small-molecule inhibitor induces mitotic catastrophe and apoptosis in human bladder cancer cells. J Cell Mol Med. 2017;21:758–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wu J, Ivanov AI, Fisher PB, et al. Polo-like kinase 1 induces epithelial-to-mesenchymal transition and promotes epithelial cell motility by activating CRAF/ERK signaling. Elife. 2016;5:e10734. [DOI] [PMC free article] [PubMed] [Google Scholar]