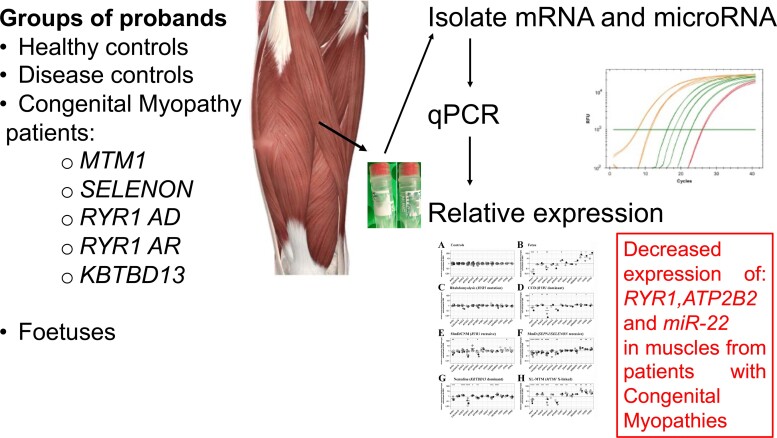
Abstract
Congenital myopathies are a group of early onset muscle diseases of variable severity often with characteristic muscle biopsy findings and involvement of specific muscle types. The clinical diagnosis of patients typically relies on histopathological findings and is confirmed by genetic analysis. The most commonly mutated genes encode proteins involved in skeletal muscle excitation–contraction coupling, calcium regulation, sarcomeric proteins and thin–thick filament interaction. However, mutations in genes encoding proteins involved in other physiological functions (for example mutations in SELENON and MTM1, which encode for ubiquitously expressed proteins of low tissue specificity) have also been identified. This intriguing observation indicates that the presence of a genetic mutation impacts the expression of other genes whose product is important for skeletal muscle function. The aim of the present investigation was to verify if there are common changes in transcript and microRNA expression in muscles from patients with genetically heterogeneous congenital myopathies, focusing on genes encoding proteins involved in excitation–contraction coupling and calcium homeostasis, sarcomeric proteins, transcription factors and epigenetic enzymes. Our results identify RYR1, ATPB2B and miRNA-22 as common transcripts whose expression is decreased in muscles from congenital myopathy patients. The resulting protein deficiency may contribute to the muscle weakness observed in these patients. This study also provides information regarding potential biomarkers for monitoring disease progression and response to pharmacological treatments in patients with congenital myopathies.
Keywords: congenital myopathies, mutations, muscle biopsies, transcripts miRNAs, expression
In this study, Bachmann et al. investigate changes in transcript and microRNA expression in muscles from patients with genetically heterogeneous congenital myopathies (CMs). Their results identify RYR1, ATPB2B and miRNA-22 as common transcripts whose expression is decreased in muscles from patients harbouring RYR1, SELENON, MTM1 and KBTBD13-related CMs
Graphical Abstract
Graphical abstract.
Introduction
Congenital myopathies (CMs) are a group of generally non-progressive or slowly progressive early onset muscle disorders characterized by muscle weakness and reduced muscle tone. They preferentially affect proximal limb and axial muscles but, depending on the genetic mutation, CMs can impact the function of cardiac, facial, extraocular and respiratory muscles.1 Some patients, especially those affected by the more severe forms, may have feeding difficulties and skeletal abnormalities including scoliosis and foot deformities. The overall incidence of CMs as a group is estimated at 1:26 000–1:50 000. Mutations in >20 genes have been reported to date, the most commonly affected genes being those encoding proteins involved in skeletal muscle excitation–contraction coupling (ECC) and calcium homeostasis, thin–thick filament assembly and titin, though mutations in genes encoding ubiquitously expressed proteins are also common.1–4 A striking aspect of CMs is their pleiotropy (different clinical and histopathological phenotypes caused by different mutations in one gene) and their genetic heterogeneity (one phenotype caused by mutations in different genes).
Mutations in RYR1 provide an excellent example of pleiotropy. This gene encodes the ryanodine receptor 1 calcium-release channel (RyR1) of the sarcoplasmic reticulum and mutations therein have been identified in ∼30% of all CM patients.2,5 Interestingly, mutations in RYR1 are linked to various disease phenotypes, including those with dominantly inherited central core disease (CCD, MIM#11700) and mainly recessively inherited subgroups of multiminicore disease (MmD, MIM#255320), centronuclear myopathy (CNM) and congenital fibre type disproportion. Additionally, RYR1 mutations are the underlying cause of the majority of cases of malignant hyperthermia (MH) susceptibility (MHS, MIM #145600) a pharmacogenetic disorder triggered by depolarizing muscle relaxants or volatile anaesthetics in genetically predisposed individuals. MH-associated RYR1 mutations have also been implicated in some forms of rhabdomyolysis/exercise induced/heat intolerance,1,6,7 King Denborough syndrome a myopathic syndrome characterized by skeletal abnormalities, dysmorphic features and MH susceptibility,8 as well as some forms of periodic paralysis.9 These results highlight the fact that patients with mutations in one gene may present with a variety of different phenotypes.
MmD, CNM and nemaline myopathy illustrate the genetic heterogeneity of CMs. Indeed, some forms of MmD and particularly the rigid spine muscular dystrophy subtype are caused by recessive mutations in SELENON (formerly known as SEPN1; MIM#606210). SELENON encodes for a ubiquitous selenocysteine-containing protein expressed in the ER of several tissues including brain, lung, spleen and skeletal muscle.10,11 CNM can be caused by mutations in a number of genes including RYR1 (autosomal recessive, AR), TTN (AR), DNM2 (autosomal dominant, AD), BIN1 (AD and AR), SPEG (AR) and MTM1 (X-linked).12,13 The X-linked form (MIM#310400) also known as XL-MTM is by far the most severe subtype of CNMs and is caused by mutations in the lipid phosphatase myotubularin 1 a protein that is expressed in many tissues including brain, liver, endocrine tissues, gastrointestinal tract, kidney and skeletal muscle. Myotubularin de-phosphorylates phosphatidylinositol 3 monophosphate and phosphatidylinositol 3,5-bisphosphate, lipids playing a role in intracellular vesicle trafficking and autophagy.14 Interestingly, MTM1-related CNM is accompanied by an up-regulation of DNM2 and this negatively affects the structure and function of triads, suggesting that regulating DNM2 expression may be a potential target for therapeutic intervention.15,16
Finally, nemaline myopathy is linked to mutations in several genes including NEB, ACTA1, TPM3, KLHL40, KLHL41, LMOD3, KBTBD13, MYPN and more. Many of these genes are implicated in sarcomeric assembly and maintenance and are associated with variable degrees of muscle weakness.17 The KBTBD13-dominant NEM6 form (MIM#609273) is characterized by progressive proximal and neck weakness, gait abnormalities and exercise intolerance. KBTBD13 was recently reported to be an actin-binding protein and mutations therein were shown to impair muscle relaxation.18
In the present study, we investigated whether the expression of different groups of transcripts are similarly impacted in muscles from CM patients with AD and AR RYR1-related myopathies, SELENON-related MmD, KBTBD13-related nemaline myopathy and MTM1-related XL-MTM. The underlying hypothesis being that the primary genetic defects may cause downstream changes in the expression of genes important for muscle function, leading to common biochemical or functional changes. In other words, muscles from patients with different CMs may show similar changes in the expression of transcripts, leading to partially overlapping phenotypes.
In this study, we focus on the expression of transcripts involved in muscle function, ECC, calcium homeostasis, contractile and sarcomeric proteins were impacted. This selection is based on the fact that proteins involved in ECC, calcium regulation and contraction are essential for skeletal muscle contraction and changes in their expression may explain the diminished muscle function. Additionally, changes in the expression of epigenetic enzymes, of transcription factors and of microRNAs (miRNAs), may be causally linked to the altered gene expression which has been observed in some CM.19–23
The results show that the expression levels of RYR1, ATPB2B and miRNA-22 are decreased in muscles from all CM groups. In addition, distinct CMs are characterized by specific transcriptional alterations. The results of this study provide a profile of the changes occurring in muscles from patients affected by different CMs and may be useful in the future to help classify patients, monitor disease progression and potentially evaluate the response to pharmacological treatment.
Materials and methods
Compliance with ethical standards
All procedures were performed in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This study was approved by the Ethikkommission Nordwest- und Zentralschweiz (permit N° EKNZ 2014-065); all subjects gave written informed consent to carry out this work.
Muscle biopsies
Quadriceps muscle biopsies from patients with genetically confirmed variants and healthy non-affected individuals undergoing the in vitro contracture test were used. For some patients, residual material of needle biopsies for other diagnostic purposes was used. The patients with the rhabdomyolysis/exercise intolerance phenotype had previously presented elevated creatine kinase (CK) levels (CK > 10 000 IU/l). All healthy controls underwent RYR1 sequencing and none were found to carry any RYR1 variants.
Quantitative polymerase chain reaction
Was performed as previously described.19 Briefly, RNA was isolated from muscle biopsies using TRIzol™ reagent (Thermo Fischer; 15596026) following the manufacturer’s protocol. For subsequent quantitative real-time polymerase chain reaction (qPCR), 1000 ng of RNA were reverse-transcribed to cDNA using the high-capacity cDNA reverse transcription kit (Applied Biosystems; 4368814) on an Applied Biosystems 2720 Thermal Cycler. The cDNA was amplified using PowerUP™ Sybr™ Green Master Mix (Applied Biosystems; A25742) for regular DNA quantification on an Applied Biosystems 7500 Fast Real-time PCR System running 7500 software version 2.3. A complete list of the transcripts whose expression was investigated is presented in Supplementary Table 1.
Each reaction was performed in duplicate and results are expressed as log2-fold change relative to controls; gene expression was normalized to the muscle-specific housekeeping gene DESMIN (DES). DES was selected as a housekeeping gene, since desmin is expressed at high levels in all vertebrate muscle cells, as opposed to GAPDH which is expressed in all mammalian cells. A table comparing the Ct values for DES and GAPDH in muscles from controls and patients with AD-RYR1 mutations and SEPN1 mutations is shown in Supplementary Table 2. The sequences of the primers used are listed in Supplementary Table 3.
Because of the large number of samples, each 96-well PCR plate contained only samples from healthy controls and from a specific disease group (or foetal muscles). Furthermore, since the amount of biological material was very limited, for some patients the expression levels of a restricted number of transcripts were evaluated. Data were centred using average ΔCt value of the controls which were included in each 96-well PCR plate. Results are presented as log2-fold change in patients/foetal muscles relative to healthy controls.
MiRNA quantification
For quantification of miRNAs, 450 ng of RNA isolated from biopsies using TRIzol™ were treated with TaqMan™ miRNA reverse transcription kit (Applied Biosystems; 4366596) on an Applied Biosystems 2720 Thermal Cycler using a miRNA primer mix, as previously described.19 Subsequent qPCR was performed using TaqMan™ Universal Master Mix II, no UNG (Applied Biosystems; 4440040) according to the manufacturer’s protocol using specific miRNA probes listed in Supplementary Table 4. Quantification was performed on an Applied Biosystems 7500 Fast Real-time PCR System running 7500 software version 2.3 and each reaction was performed in duplicate. Results were normalized to the human housekeeping snRNA U6. Each 96-well PCR plate contained only samples from healthy controls and from a specific disease group (or foetal muscles). Data were centred using average ΔCt value of the controls included in each 96-well PCR plate. Results are presented as log2-fold change in patients/foetal muscles, relative to healthy controls.
Statistical analysis
Since each test plate contained only samples from the control group and a specific disease group (or foetuses), statistical analysis was only made between these two groups. Statistical analysis was performed using ‘R’ version 4.2.0 running on platform x86_64-apple-darwin13.4.0 (64 bits). Log2-fold changes and standard errors were estimated using the linear model fitted to every gene, with disease as an explanatory variable. Moderated t-statistics was calculated using the limma package.24 Obtained P-values were adjusted for multiple testing using Benjamini–Hochberg method to control the false discovery rate.25 All quantitative PCR and miRNA figures were created using R Studio (version 1.4.1106) and assembled to panels using Adobe Photoshop CS6.
Data availability
Data sharing is not applicable, since all results are included in the manuscript.
Results
Selection of patients and mutations
Previous investigations have demonstrated that muscles from patients with CMs carrying mutations in RYR1, SELENON and MTM1 have reduced levels of RyR1.19–23 In the present investigation, we expanded the study and directly compared the expression of transcripts encoding other proteins involved in ECC and calcium homeostasis, myofilaments, transcription factors and regulators, epigenetic enzymes (see Supplementary Table 1 for the transcripts that were analysed and summary of the function of the encoded proteins) as well as selected families of miRNAs, in muscle biopsies from patients who had previously undergone diagnostic genetic testing. We also included disease controls, namely samples from patients carrying dominant RYR1 mutations associated with exertional rhabdomyolysis/exercise intolerance and foetal muscles. Supplementary Table 5 shows the phenotypes and genotypes of the patients we investigated. A total of 50 patients were included and pathogenic mutations were identified in 96% (48/50) of them. In particular, 13 probands were diagnosed as having rhabdomyolysis/exercise intolerance having suffered one or more episodes of exertional rhabdomyolysis (CK > 10 000 IU/l). All carried RYR1 mutations and were also classified as MHS by the in vitro contracture test. Three probands were diagnosed as having AD CCD of which two carried pathogenic RYR1 mutations. Eight probands were diagnosed as having AR MmD/CNM and carried RYR1 mutations (in one patient, the second mutation was not identified). Fourteen probands were diagnosed as having AR MmD and carried SELENON mutations. Six probands originating from two families were diagnosed as having AD NEM6 (KBTBD13 nemaline myopathy). Six probands were diagnosed as having XL-MTM and carried MTM1 mutations. Classification of the variants identified in the patients and reference sequence of the mutated genes are presented in Supplementary Table 6. Muscle biopsies from 12 healthy non-affected individuals were used as control. Transcript expression levels in muscles of patients/foetal muscles were compared with those observed in muscles from healthy controls, which were set to 1. Gene expression could not be evaluated for all genes in all probands because of the lack of a sufficient amount of biological material.
Expression of transcripts encoding proteins involved in ECC and calcium homeostasis
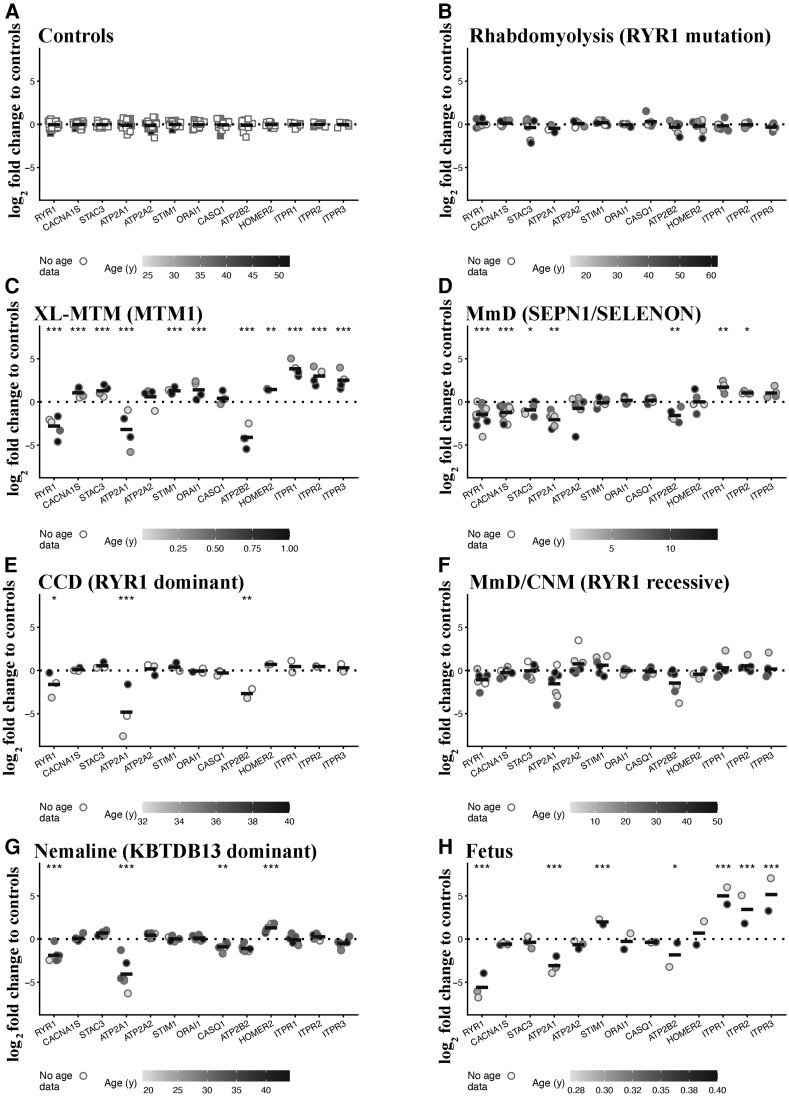
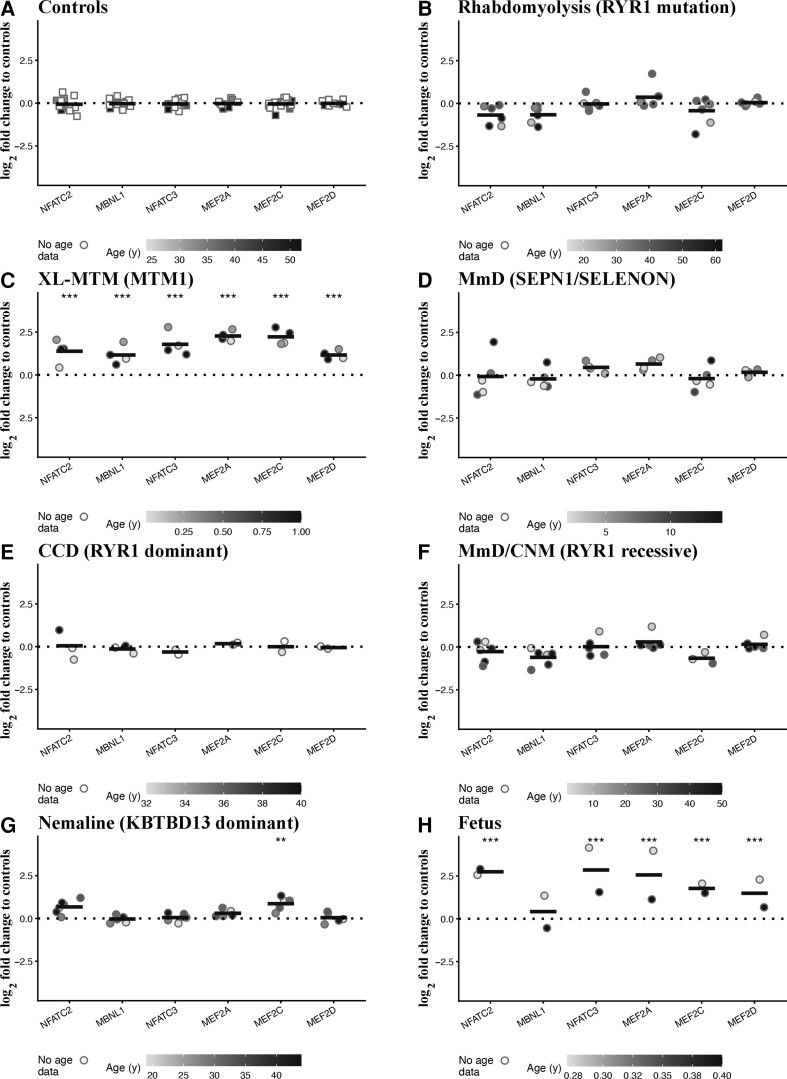
Figure 1 shows the results obtained when analysing transcripts encoding the main proteins involved in skeletal muscle ECC and calcium homeostasis in each group of patients with the different panels divided by disease and genotype. Each symbol represents the average value (of duplicate runs) obtained from a single patient plotted on a log2 scale with the horizontal line representing the mean value.
Figure 1.
Muscles from patients with CM show significant changes in the expression levels of transcripts encoding proteins involved in ECC and calcium homeostasis. Expression levels of the indicated transcripts were determined by qPCR and normalized to the expression of DES. Muscle biopsies were from: (A) healthy controls; (B) patients with exertional rhabdomyolysis/heat stroke/exercise intolerance carrying RYR1 mutations; (C) patients with MTM1-related XL-MTM; (D) patients with AR SELENON-related MmD; (E) patients with AD-RYR1-related CCD; (F) patients with AR RYR1-related MmD/CNM; (G) patients with AD KBTBD13-related nemaline myopathy; (H) foetuses. The greyscale given to the symbols reflects the age range of the patients and the scale bar at the bottom of each panel correlates greyscale to age. Empty symbols represent patients or probands whose age was not known. Square symbols represent results from controls; circles represent results from disease patients and foetuses. The relative transcript expression in patient muscles was compared with that in muscles from healthy controls that was set to 1. Statistical analysis was performed using the ‘R’ version 4.2.0 running on platform x86_64-apple-darwin13.4.0 (64 bits). Comparisons of each disease group (or foetus) to controls were calculated using the limma package24 of ‘R’. Obtained P-values were adjusted for multiple testing using Benjamini–Hochberg method to control the false discovery rate. Means were considered statistically significant, when the adjusted P-values were <0.05. The horizontal black bar represents the mean content levels in patient muscles. *P < 0.05; **P < 0.01; ***P < 0.001.
None of the transcripts examined encoding proteins involved in ECC and calcium homeostasis26–35 varied between muscles from healthy controls (Fig. 1A) and disease controls, i.e. patients with RYR1-related rhabdomyolysis/exercise intolerance (Fig. 1B). Muscles from patients with MTM1 mutations showed the greatest changes in transcript levels, with all transcripts except ATP2A2 and CASQ1, being significantly altered (Fig. 1C). In particular, RYR1 transcript levels were reduced by almost 10-fold (the mean log2-fold change was −2.75 adjusted P = 2.89E − 09), whereas the transcript levels encoding the three InsP3R isoforms were significantly increased (the mean log2-fold changes for ITPR1, ITPR2 and ITPR3 were 3.86 adjusted P = 1.35E − 09, 3.00 adjusted P = 1.18E − 08 and 2.53 adjusted P = 1.62E − 04, respectively). The expression levels of CACNA1S, STAC3, STIM1, ORAI1 and HOMER were significantly increased (the mean log2-fold changes were 1.06 adjusted P = 2.22E − 04, 1.32 adjusted P = 4.33E − 04, 1.30 adjusted P = 3.07E − 05, 1.44 adjusted P = 8.82E − 06 and 1.46 adjusted P = 0.001, respectively). Muscles from patients with AR SELENON MmD also showed significantly altered expression levels of several transcripts (Fig. 1D), including lower levels of transcripts encoding RYR1, CACNA1S, STAC3, ATP2A1 and ATP2B2 (the mean log2-fold changes were −1.39 adjusted P = 6.61E − 05, −1.20 adjusted P = 2.35E − 08, −0.89 adjusted P = 0.016, −1.95 adjusted P = 0.0034 and −1.45 adjusted P = 0.0077, respectively) and increased levels of ITPR1 and ITPR2 (the mean log2-fold changes were 1.73 adjusted P = 0.0013 and 1.05 adjusted P = 0.021, respectively).
Importantly, in muscles from all other CM patients the levels of RYR1 and ATP2B2 were reduced compared with healthy controls though they did not always reach significance when the adjusted P-values were used. Their mean log2-fold change levels in patients with AD-RYR1-related CCD were −1.58 (adjusted P = 0.081) and −2.56 (adjusted P = 0.0025; Fig. 1E), in patients with AR RYR1-related MmD/CNM, they were reduced to −1.03 (adjusted P = 0.063) and −1.34 (adjusted P = 0.052; Fig. 1F), and in AD nemaline myopathy patients, they were −1.85 (adjusted P = 8.74E − 05) and −0.98 adjusted (P = 0.076; Fig. 1G). Foetal muscles have a distinct transcript expression pattern (Fig. 1H), showing a more than 10-fold reduction of RYR1 (the mean log2-fold change was −5.56 adjusted P = 1.10E − 14) and ATP2A1 (the mean log2-fold change was −2.97 adjusted P = −8.41E − 04) and significantly higher levels of STIM1, ITPR1, ITPR2 and ITPR3 (mean log2-fold changes were 1.99 adjusted P = 4.34E − 06, 5.02 adjusted P = 1.67E − 09, 3.44 adjusted P = 1.61E − 07 and 5.17 adjusted P = 1.45E − 07, respectively; see Supplementary Table 7 for complete data set and statistical analysis).
In conclusion, these results show that there are specific changes in the expression levels of several transcripts in muscles from all patients with CM, in particular: (i) decreased levels of RYR1 and ATP2B2; (ii) increased levels of transcripts encoding for one or more of ITPR isoforms; and (iii) diseases which more strongly impact muscle function such as XL-MTM1 were accompanied by the greatest changes in the expression of transcripts encoding proteins involved in calcium homeostasis. Importantly, muscles from patients carrying dominant RYR1 mutations linked to rhabdomyolysis did not exhibit any changes in the expression of the investigated transcripts.
Expression of transcripts encoding contractile and sarcomeric proteins
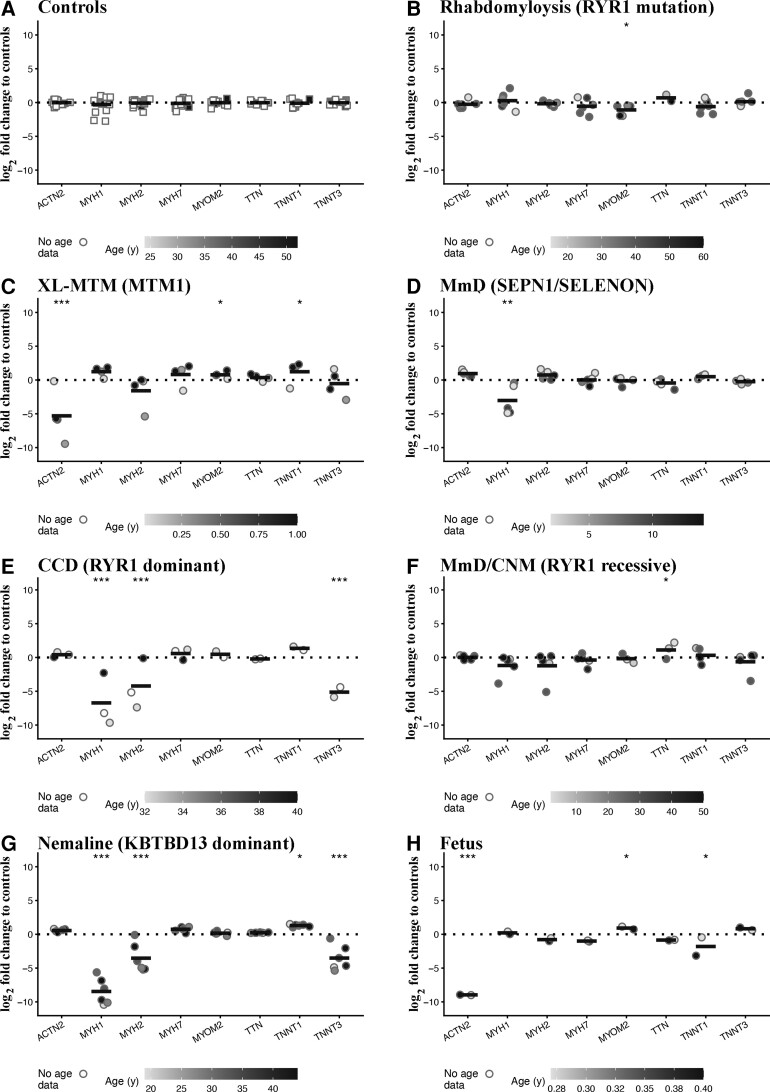
Muscle groups specialized for different tasks express different myofilament isoforms whose shortening velocity is determined by the expression of specific myosin heavy chain (MyHC) isoforms.36 Additionally, there are many proteins involved in the assembly and organization of the contractile apparatus with mutations in genes encoding some sarcomeric proteins are linked to specific forms of CMs.1 In this part of the study, we investigated changes in the expression of a subset of these transcripts, focusing on the transcripts encoding MyHC isoforms, α-actinin, myomesin, titin and troponin T.36,37 Interestingly, muscle biopsies from disease controls, i.e. patients with RYR1-related rhabdomyolysis/exercise intolerance, showed significantly lower expression levels of MYOM2 (the mean log2-fold change was −1.07 adjusted P = 0.01; Fig. 2B) compared with healthy controls. On the other hand, muscles from patients with MTM1 mutation showed increased expression levels of MYOM2 and TNNT1 (the mean log2-fold changes were 0.79 adjusted P = 0.01 and 1.32 adjusted P = 0.01, respectively) and decreased levels of ACTN2 (the mean log2-fold change was −5.26 adjusted P = 1.03E − 10; Fig. 2C). Muscles from patients with AR SELENON-related MmD showed decreased levels of MYH1 (the mean log2-fold change was −2.74 adjusted P = 0.003; Fig. 2D). Compatible with a loss of fast twitch fibres, muscle biopsies from patients with AD-RYR1-related CCD showed a significant decrease in MYH1, MYH2 and TNNT3 (mean log2-fold changes were −6.43 adjusted P = 9.09E − 07, −4.13 adjusted P = 7.29E − 04 and −5.10 adjusted P = 2.36E − 05, respectively; Fig. 2E), whereas those from AR RYR1-related MmD/CNM showed a significant increased expression of TTN (the mean log2-fold change was 1.13 adjusted P = 0.03; Fig. 2F). Interestingly, muscles from patients with AD KBTBD13-related nemaline myopathy who also have a loss of fast twitch fibres, showed reduced expression levels of all MYH1, MYH2 and TNNT3 (the mean log2-fold changes were −8.17 adjusted P = 2.78E − 13, −3.44 adjusted P = 1.55E − 04 and −3.48 adjusted P = 1.55E − 05, respectively) and increased expression of TNNT1 (the mean log2-fold change was 1.39 adjusted P = 0.01; Fig. 2G). Foetal muscles (Fig. 2H) showed a >200-fold lower expression level of ACTN2 (the mean log2-fold change was −8.95 adjusted P = 7.69E − 14), increased levels of MYOM2 (the mean log2-fold change was 0.944 adjusted P = 0.03) and decreased levels of TNNT1 (the mean log2-fold change was −1.70 adjusted P = 0.02; see Supplementary Table 8 for the complete data set and statistical analysis).
Figure 2.
Muscles from patients with rhabdomyolysis and congenital myopathies show significant changes in the expression levels of transcripts encoding contractile and sarcomeric proteins. Expression levels of the indicated transcripts were determined by qPCR and normalized to the expression of DES. Muscle biopsies were from: (A) healthy controls; (B) patients with exertional rhabdomyolysis/heat stroke/exercise intolerance carrying RYR1 mutations; (C) patients with MTM1-related XL-MTM; (D) patients with AR SELENON-related MmD; (E) patients with AD-RYR1-related CCD; (F) patients with AR RYR1-related MmD/CNM; (G) patients with AD KBTBD13-related nemaline myopathy; (H) foetuses. The greyscale given to the symbols reflects the age range of the patients and the scale bar at the bottom of each panel correlates greyscale to age. Empty symbols represent patients or probands whose age was not known. Square symbols represent results from controls; circles represent results from disease patients and foetuses. The relative transcript expression in patient muscles was compared with that in muscles from healthy controls that was set to 1. Statistical analysis was performed using ‘R’ version 4.2.0 running on platform x86_64-apple-darwin13.4.0 (64 bits). Comparisons of each disease group (or foetus) to controls were calculated using the limma package24 of ‘R’. Obtained P-values were adjusted for multiple testing using Benjamini–Hochberg method to control the false discovery rate. Means were considered statistically significant when the adjusted P-values were <0.05. The horizontal black bar represents the mean content levels in patient muscles. *P < 0.05; **P < 0.01; ***P < 0.001.
In short, (i) muscles from patients with MTM1 mutations showed increased levels of TNNT1 and, similar to foetal muscles, very low expression levels of ACTN2; (ii) muscles from patients with SELENON-related MmD showed a decrease of MYH1 expression; (iii) compatible with the loss of fast twitch Type 2 fibres, muscles from patients with CCD and nemaline myopathy exhibit decreased expression of genes encoding MYH1, MYH2 and TNNT3 and increased expression levels of TNNT1; and (iv) muscles from rhabdomyolysis/exercise intolerance individuals with dominant RYR1 mutations showed decreased levels of MYOM2.
Expression of transcripts encoding enzymes involved in epigenetic modifications
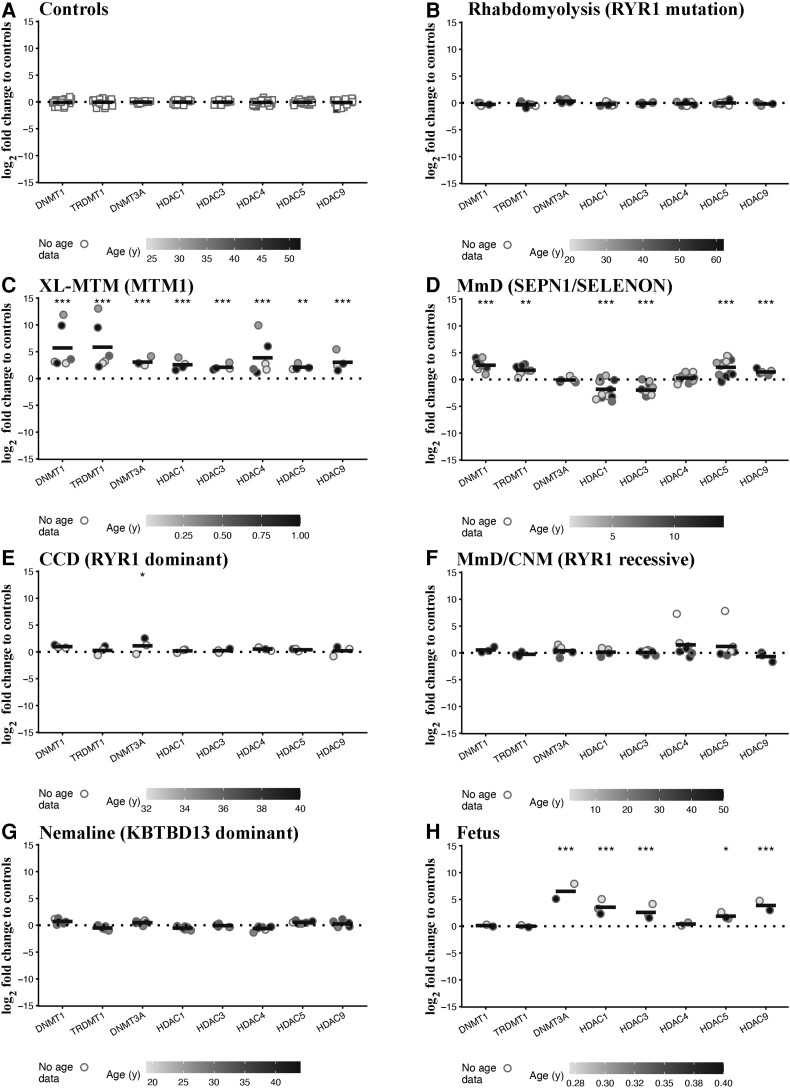
We subsequently investigated the levels of transcripts encoding epigenetic enzymes including DNA methyl transferases (DNMTs) and Classes I and II histone deacetylases (HDACs). We hypothesize that the activity of these enzymes may be responsible in part, for the altered expression levels of muscle transcripts since they strongly influence the structure of chromatin, by rendering it more accessible (euchromatin) or less accessible (heterochromatin) to transcription.38,39 Our results show that the expression levels of DNMTs and HDACs were similar in muscles from healthy controls (Fig. 3A) and disease controls, i.e. patients with RYR1-related exertional rhabdomyolysis (Fig. 3B). On the other hand, muscles from XL-MTM patients showed a significant increase in all DNMT and HDAC isoforms examined (Fig. 3C). The mean log2-fold changes of transcripts encoding DNMT1, TRDMT1 (DNMT2) and DNMT3A were 5.79 adjusted (P = 6.50E − 11), 5.91 adjusted (P = 6.96E − 11) and 3.09 (adjusted P = 3.71E − 10), respectively. The levels of Class I HDACs including HDAC1 and HDAC3 were significantly increased (the mean log2-fold changes were 2.61 adjusted P = 4.16E − 06 and 2.14 adjusted P = 2.66E − 07, respectively) as were those of Class IIa HDACs, including HDAC4, HDAC5 and HDAC9 (the mean log2-fold changes were 3.95 adjusted P = 6.83E − 08, 2.16 adjusted P = 0.001 and 3.14 adjusted P = 1.18E − 09, respectively). Muscles from patients with AR SELENON also showed significantly increased expression levels of DNMT1 and TRDMT1 (the mean log2-fold changes were 2.75 adjusted P = 6.61E − 05 and 1.79 adjusted P = 0.006, respectively), of HDAC5 and HDAC9 (the mean log2-fold changes were 2.33 adjusted P = 5.59E − 06 and 1.51 adjusted P = 9.92E − 04, respectively) and decreased levels of Class I HDACs (the mean log2-fold change of HDAC1 and HDAC3, were −1.78 adjusted P = 7.78E − 06 and −1.94 adjusted P = 6.83E − 10, respectively; Fig. 3D). In muscles from patients with AD-RYR1-related CCD DNMT3A expression levels were increased (the mean log2-fold change was 1.16 adjusted P = 0.03), whereas in AR RYR1-related MmD/CNM, HDAC4 expression levels were increased though they did not achieve statistical significance (the mean log2-fold change was 1.58 adjusted P = 0.06; Fig. 3F). Interestingly, the levels (fold change) of transcript encoding epigenetic enzymes were not impacted in muscles from patients with AD nemaline myopathy (Fig. 3G) and in AD-RYR1-related CCD only, the expression level of DNMT3A was increased (the mean log2-fold change was 1.16 adjusted P = 0.03; Fig. 3E). Foetal muscles showed a significant increase in DNMT3A (the mean log2-fold change was 6.53 adjusted P = 7.28E − 16) as well as in HDAC1, HDAC3, HDAC5 and HDAC9 expression (the mean log2-fold changes were 3.58 adjusted P = 1.26E − 07, 2.62 adjusted P = 8.71E − 08, 1.92 adjusted P = 0.01 and 3.98 adjusted P = 4.56E − 08, respectively; Fig. 3H, see Supplementary Table 9 for the complete data set and statistical analysis).
Figure 3.
Muscles from patients with congenital myopathies show significant changes in the expression levels of transcripts encoding enzymes involved in epigenetic modifications. Expression levels of the indicated transcripts were determined by qPCR and normalized to the expression of DES. Muscle biopsies were from: (A) healthy controls; (B) patients with exertional rhabdomyolysis/heat stroke/exercise intolerance carrying RYR1 mutations; (C) patients with MTM1-related XL-MTM; (D) patients with AR SELENON-related MmD; (E) patients with AD RYR1-related CCD; (F) patients with AR RYR1-related MmD/CNM; (G) patients with AD KBTBD13-related nemaline myopathy; (H) foetuses. The greyscale given to the symbols reflects the age range of the patients and the scale bar at the bottom of each panel correlates greyscale to age. Empty symbols represent patients or probands whose age was not known. Square symbols represent results from controls; circles represent results from disease patients and foetuses. The relative transcript expression in patient muscles was compared with that in muscles from healthy controls that was set to 1. Statistical analysis was performed using ‘R’ version 4.2.0 running on platform x86_64-apple-darwin13.4.0 (64 bits). Comparisons of each disease group (or foetus) to controls were calculated using the limma package24 of ‘R’. Obtained P-values were adjusted for multiple testing using Benjamini–Hochberg method to control the false discovery rate. Means were considered statistically significant when the adjusted P-values were <0.05. The horizontal black bar represents the mean content levels in patient muscles. *P < 0.05; **P < 0.01; ***P < 0.001.
Taken together, these results show that muscles of CM patients (excluding KBTBD13-related nemaline myopathy patients) as well as foetal muscles exhibit increased levels of transcripts encoding DNMTs and/or mainly, Class II HDACs. Patients carrying RYR1 mutations linked to exertional rhabdomyolysis/exercise intolerance did not show changes in the expression of any of the investigated transcripts encoding epigenetic enzymes.
Expression of transcripts encoding skeletal muscle transcription factors and splicing regulators
We next examined the levels of a subset of skeletal muscle transcription factors, including NFAT and MEF2 family members,40–42 and of muscleblind (MBNL1).43 The transcript expression levels in muscles from disease controls, i.e. patients with RYR1-related rhabdomyolysis/exercise intolerance were similar to those found in muscles from controls (Fig. 4B). Muscles of XL-MTM patients showed the greatest changes in transcript expression levels and importantly, the expression of all investigated transcripts was significantly increased (Fig. 4C). The expression (the mean log2-fold changes were: 1.44 adjusted P = 2.22E − 04 for NFATC2, 1.83 adjusted P = 3.48E − 0.6 for NFATC3, 2.29 adjusted P = 1.17E − 07 for MEF2A, 2.26 adjusted P = 8.43E − 09 MEF2C, 1.17 adjusted P = 8.82E − 06 for MEF2D and 1.19 adjusted P = 4.84E − 05 for MBNL1). None of the investigated transcripts were changed in muscles from patients with SELENON-related MmD (Fig. 4D) nor from patients with AD-RYR1-related CCD (Fig. 4E) though in AR RYR1 MmD/CNM MBNL1 levels were reduced though the fold change did not achieve significance (the mean log2-fold change was −0.57 adjusted P = 0.06; Fig. 4F). MEF2C levels were significantly increased in muscles of patients with AD nemaline myopathy (the mean log2-fold change was 0.90 adjusted P = 0.006; Fig. 4G). Foetal muscles also showed significant increased expression levels of all the transcripts examined except for MBNL1 whose levels were unchanged (Fig. 4H; see Supplementary Table 10 for the complete data set and statistical analysis).
Figure 4.
Muscles from patients with congenital myopathies show significant changes in the expression levels of transcripts encoding transcription factors and splicing regulators. Expression levels of the indicated transcripts were determined by qPCR and normalized to the expression of DES. Muscle biopsies were from: (A) healthy controls; (B) patients with exertional rhabdomyolysis/heat stroke/exercise intolerance carrying RYR1 mutations; (C) patients with MTM1-related XL-MTM; (D) patients with AR SELENON-related MmD; (E) patients with AD RYR1-related CCD; (F) patients with AR RYR1-related MmD/CNM; (G) patients with AD KBTBD13-related nemaline myopathy; (H) foetuses. The greyscale given to the symbols reflects the age range of the patients and the scale bar at the bottom of each panel correlates greyscale to age. Empty symbols represent patients or probands whose age was not known. Square symbols represent results from controls; circles represent results from disease patients and foetuses. The relative transcript expression in patient muscles was compared with that in muscles from healthy controls that was set to 1. Statistical analysis was performed using ‘R’ version 4.2.0 running on platform x86_64-apple-darwin13.4.0 (64 bits). Comparisons of each disease group (or foetus) to controls were calculated using the limma package24 of ‘R’. Obtained P-values were adjusted for multiple testing using Benjamini–Hochberg method to control the false discovery rate. Means were considered statistically significant when the adjusted P-values were <0.05. The horizontal black bar represents the mean content levels in patient muscles. *P < 0.05; **P < 0.01; ***P < 0.001.
These results indicate that in the muscles from CM patients: (i) NFATC2 and NFATC3 levels do not correlate with the expression of specific slow twitch muscle genes and (ii) the expression level of different MEF2 isoforms increases in muscles from XL-MTM and AD nemaline myopathy patients.
Expression of miRNAs in muscles from patients with different muscle diseases
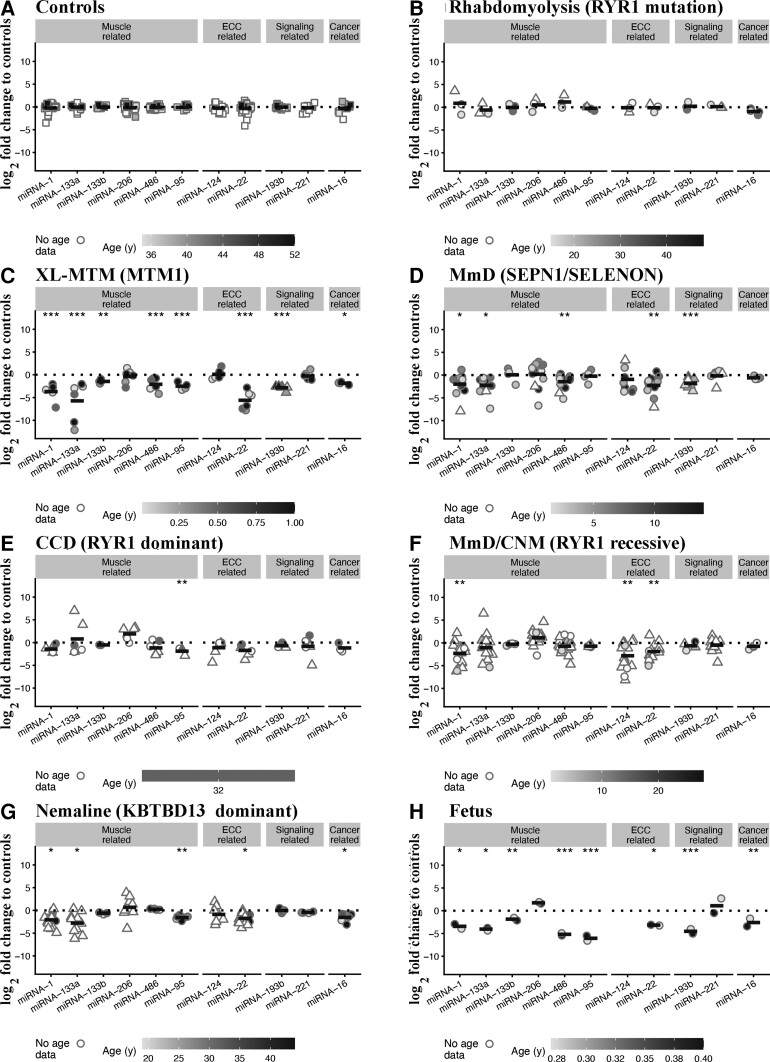
To date more than 2000 miRNAs that have been identified in humans and their expression levels are dysregulated in disorders including cancer, ALS, MmD and muscular dystrophies.23,44–47 We next investigated miRNA expression levels in muscles from foetuses, disease controls and CM patients and compared them to the expression in muscles from healthy controls (the latter were set to 1, Fig. 5A). We chose to quantify the expression of myo-miRNAs, i.e. those that are specifically expressed in muscles including miRNA-1, -95, -133, -206 and -486,48 miRNAs that are predicted to bind to the 3′-UTR of the RYR1 (miRNA-22 and -124) and to bind to HDACs (epigenetic related), and miRNAs binding to transcripts whose products are involved in signalling (miRNA-19 and 221) and tumour suppression/proliferation (cancer related, miRNA-16). Because of the lack of sufficient biological material for the groups of MmD/CNM, CCD, XL-MTM, nemaline and SEPN1 patients, we added data from our previously published results,19,21,23 in order to increase sample sizes (data from previously published results are shown as triangles in Fig. 5). Similar to gene expression results, muscles from disease controls, i.e. patients with RYR1-related rhabdomyolysis/exercise intolerance showed no changes in expression of any of the investigated miRNAs (Fig. 5B). On the other hand, miRNA expression levels were disrupted in muscles from all CM patients. Indeed, muscles from XL-MTM patients exhibited the greatest changes in miRNA expression levels, showing significantly lower expression levels of miRNA-1, -95, -133a, -133b, -486, -22, -193b and -16 (the mean log2-fold changes were −3.44 adjusted P = 9.03E − 05, −2.42 adjusted P = 6.53E − 06, −5.66 adjusted P = 6.53E − 06, −1.42 adjusted P = 0.002, −2.42 adjusted P = 9.92E − 04, −5.27 adjusted P = 1.24E − 10, −2.81 adjusted P = 1.34E − 12 and −1.50 adjusted P = 0.01, respectively; Fig. 5C). Muscles from AR SELENON-related MmD patients showed significantly decreased expression of miRNA-1, -133a, -486, -22 and -193b (the mean log2-fold changes were −1.73 adjusted P = 0.01, −2.17 adjusted P = 0.02, −1.39 adjusted P = 0.005, −1.93 adjusted P = 0.003 and −1.74 adjusted P = 9.79E − 09, respectively; Fig. 5D). In muscles from AD-RYR1-related CCD patients, expression levels of miRNA-95 were significantly decreased (the mean log2-fold change was −1.80 adjusted P = 0.006; Fig. 5E). Muscles from AR RYR1-related MmD/CNM patients showed significantly decreased expression levels of miRNA-1, -22 and -124 (the mean log2-fold changes were −2.06 adjusted P = 0.003, −1.58 adjusted P = 0.04 and −2.57 adjusted P = 0.003, respectively; Fig. 5F). Muscles from AD nemaline myopathy patients showed significantly decreased expression levels of miRNA-1, -95, 133a, -22 and -16 (the mean log2-fold changes were −1.82 adjusted P = 0.01, −1.49 adjusted P = 0.003, −2.73 adjusted P = 0.01, −1.41 adjusted P = 0.01 and −1.17 adjusted P = 0.01, respectively; Fig. 5G). Foetal muscles showed a significant decrease in the expression of all miRNA investigated except miRNA-206 and miRNA-221 that were unchanged (Fig. 5H). MiRNA-124 was not detected in foetal muscles (see Supplementary Table 11 for the complete data set and statistical analysis).
Figure 5.
Muscles from patients with congenital myopathies show significant changes in the expression levels of miRNAs targeting transcripts encoding proteins involved in ECC, muscle, epigenetic and signalling-related transcripts. Expression levels of the indicated transcripts were determined by qPCR and normalized to the expression of U6. Muscle biopsies were from: (A) healthy controls; (B) patients with exertional rhabdomyolysis/heat stroke/exercise intolerance carrying RYR1 mutations; (C) patients with MTM1-related XL-MTM; (D) patients with AR SELENON-related MmD; (E) patients with AD RYR1-related CCD; (F) patients with AR RYR1-related MmD/CNM; (G) patients with AD KBTBD13-related nemaline myopathy; (H) foetuses. The greyscale given to the symbols reflects the age range of the patients and the scale bar at the bottom of each panel correlates greyscale to age. Empty symbols represent patients or probands whose age was not known. Square symbols represent results from controls; circles represent results from disease patients and foetuses obtained in the present study; triangles represent data obtained from previous investigations.19,21,23 The relative transcript expression in patient muscles was compared with that in muscles from healthy controls that was set to 1. Statistical analysis was performed using ‘R’ version 4.21.0 running on platform x86_64-apple-darwin13.4.0 (64 bits). Comparisons of each disease group (or foetus) to controls were calculated using the limma package24 of ‘R’. Obtained P-values were adjusted for multiple testing using Benjamini–Hochberg method to control the false discovery rate. Means were considered statistically significant when the adjusted P-values were <0.05. The horizontal black bar represents the mean content levels in patient muscles. *P < 0.05; **P < 0.01; ***P < 0.001.
In conclusion, (i) the expression levels of most myo-miRNAs and miRNA-22 are downregulated in muscles of all CM patients; (ii) miRNA-95 expression levels are significantly decreased in CM patients carrying dominant RYR1 mutations; and (iii) foetal muscles show altered expression levels of most of the miRNAs that were investigated.
Discussion
In this study, we performed a targeted transcriptome analysis on muscle biopsies from 50 patients with genetically diverse muscle disorders. We focused on the expression levels of transcript encoding proteins involved in important aspects of skeletal muscle function including ECC, Ca2+ homeostasis, contractile and sarcomeric proteins and regulation of transcription, in order to identify (i) variations in gene expression that may contribute to the development of muscle weakness observed in all these conditions (i.e. generic targets) and (ii) disease-specific transcriptional changes (i.e. disease-specific targets). Our results show that those disorders which impact muscle function more strongly are accompanied by an overall change in expression of a multitude of transcripts. On the other hand, in muscles from mildly affected individuals such as the disease control group, the expression levels of transcripts and miRNAs were almost indistinguishable from those of the healthy control group. This is an important finding since disease controls carry mutations in the RYR1 gene resulting in hypersensitivity to RyR1 agonists, but are otherwise healthy and do not exhibit a weak muscle phenotype.
Our results show that the expression levels of transcripts encoding many proteins important for muscle function, development and miRNAs (generic targets) are significantly altered in muscles from all CM patients. In particular, muscles from all CM patients investigated showed some commonly mis-regulated transcripts, including reduced levels of RYR1, ATP2B2, miRNA-1, miRNA-22, increased levels of DNMT and Class II HDACs. RYR1 encodes the RyR1 sarcoplasmic reticulum Ca2+ channel one of the key proteins involved in skeletal muscle ECC. Decreased RyR1 protein content has been reported in muscles from patients with AR RYR1 and SELENON-related MmD and MTM1-related XL-MTM19–23 but not in patients with nemaline myopathy. In line with previous findings, RYR1 expression was either unchanged or reduced in muscles of CCD patients carrying dominant RYR1 mutations.49,50 This variable expression may be linked to the type of mutation present in the proband, which may influence the stability of the transcript. It should also be mentioned that most CCD-linked RYR1 mutations strongly influence the biophysical properties of the RyR1 calcium channel resulting in less calcium released during ECC.51,52 The reduced amount of RyR1 protein and/or the presence of mutant channels is expected to induce lower amounts of calcium released during ECC and consequently to weaker muscles.
ATP2B2 encodes for the plasma membrane Ca2+ATPase (PMCA) a pump involved in extruding Ca2+ ions from the myoplasm to the extracellular environment.31 The observed decreased expression of ATP2B2 may be a compensatory mechanism of the muscle to the lower levels of myoplasmic calcium achieved during ECC consequent to the reduced RyR1 calcium channels. In line, Cully et al.53 reported that muscles from mdx mice that show increased levels of cytosolic calcium, exhibit increased levels of PMCA levels. Nevertheless, the overall impact of reduced PMCA expression is difficult to decipher, especially since skeletal muscles express other calcium pumps (SERCAs) and transporters (Na/Ca exchanger) involved in regulating the myoplasmic calcium concentration. A reduction in PMCA content would decrease the extrusion rate of calcium from the myoplasm into the extracellular environment and this may affect the open probability of the RyR1, since its Po is decreased at high-calcium concentrations.54
Interestingly, transcripts encoding the three InsP3R calcium channel isoforms namely ITPR1, ITPR2 and ITPR3 were elevated in muscle biopsies from many CM patients. Increased ITPR expression was reported in muscles from patients with AD and AR RYR1 CM.49,55 Even though InsP3Rs are calcium channels, they cannot functionally replace the RyR1 because of their different biophysical properties, mode of activation (they are ligand-gated channels and are physiologically activated by the second messenger inositol 1,4,5-trisphosphate), the fact that they are not coupled to the voltage sensing dihydropyridine receptor and most likely not located in the junctional face membrane. Remarkably, Suman et al.55 showed that in myotubes from patients carrying RYR1 mutations, there is an inverse correlation between RyR1 content and (pan)InsP3R content. However, no insight into the mechanism leading to the switch in expression of these intracellular Ca2+ channels was provided and it is reasonable to hypothesize that it may be partially caused by the presence of a greater number of immature muscle cells in patient-derived myotube cultures.
On the other hand, the underlying cause leading to the expression of InsP3R may be linked to the lack of Ca2+ signals originating from fewer or mutated RyR1 Ca2+ channels which in turn may impact the muscle proteostatic machinery, leading to the aberrant expression of many proteins. Indeed, (i) in the absence of RyR1, mouse muscles aberrantly express >300 genes, including atypical expression of transcription factors, contractile proteins, muscle-specific structural proteins, and muscle regulatory factors56; (ii) RYR1 silencing induces a decrease in myotube area and fusion index57; (iii) pharmacologically blocking RyR1 activity inhibits in vitro differentiation of foetal myoblasts58; (iv) muscles from XL-MTM patients express the lowest levels of RYR1 and show the greatest changes in overall transcript expression; and (v) expression of the MyHC-EO isoform during development is dependent on the function of the vestibular system and when reduced amounts of mutant RyR1 channels are present, extraocular muscles fail to express MyHC-EO.59,60 Hence, it is reasonable to assume that calcium release mediated by RyR1 channels also regulates gene expression. Aside its indirect influence on the expression of myogenic factors such as Pax7, MyoD, MyoG and Mrf4, transcription factors and signalling molecules,56 we hypothesize that RyR1-mediated calcium signals impact the accessibility of chromatin to transcription by influencing epigenetic enzymes such as HDACs and DNMT. HDACs can be subdivided into three main groups of which Subgroups IIa (HDAC-4, -5, -7 and -9) and IIb (HDAC-6 and -10) exhibit tissue specificity and are highly expressed in brain, heart and skeletal muscle.39,61 When present in the nucleus, HDAC-4 and -5 directly bind and sequester MEF2 thereby repressing its transcriptional activity.41,42 Phosphorylation of HDAC-4 by calcium calmodulin–dependent protein kinase (CamK) promotes its export into the cytosol thereby de-repressing the transcription of MEF2-dependent genes.41 The physiological validity of this pathway which provides a direct link between calcium homeostasis and HDAC function was demonstrated in mouse cardiomyocytes. In this cellular model, KCl-induced depolarization (which causes a cytoplasmic calcium transient) leads to the export of HDAC-4 and -5 from the nucleus into the cytoplasm and to increased acetylation of histone H3 and H4.62 The transcriptional repressing activity of HDACs is strengthened by DNA methylating enzyme (DNMT1, DNMT2/TRDMT1 and DNMT3) such that de-acetylated histones preferentially accumulate on hypermethylated DNA.63
Thus, gene transcription is finely controlled within tissues and cells and its regulation occurs at different hierarchical levels, including chromatin accessibility and the presence of specific transcription factors. An additional level of post-transcriptional control is provided by miRNAs which preferentially bind to 3′-UTR of a transcript leading to its degradation. One miRNA can bind to several targets and the 3′-UTR of one transcript can bind several miRNAs thereby creating an intracellular epigenetic circuit.44 DNA methylation and histone modifications regulate the expression of various miRNAs64 (including miRNA-124, -137 and -148) and a multitude of miRNAs target HDACs and DNMTs (including miRNA-206, -1 and -95). The potential role of such an epigenetic circuit in CM is exemplified by the fact that 26 miRNAs are predicted to target RYR1 including miRNA-124 and -22 and miRNA-22 also binds to the 3′-UTR of HDAC4 (http://www.mirdb.org/cgi-bin/search.cgi). Thus, decreased content of miRNA-22 may lead to increased levels of HDAC4. Furthermore, miRNA-206 targets >100 transcripts including DNMT3a, MEF2D and HDAC465 (http://www.mirdb.org/mirdb/index.html). Our results clearly show that miRNA expression is altered in human muscles carrying genetically diverse mutations, but the functional significance of these changes is difficult if not impossible to determine and will require more in depth investigations.
In the present study, we also analysed transcript expression in foetal muscles and our results reveal similarities in RNA expression between foetal muscles and muscles from patients affected by CM, especially younger patients affected by XL-MTM. This observation deserves to be investigated in the future and may shed important clues as to developmental abnormalities of muscles caused by the primary genetic defects. Of interest, compared with mature healthy muscles, foetal muscles exhibit low expression levels of RYR1 and high levels of ITPR1, ITPR2, ITPR3, STIM1, DNMT3A and of all HDACs (except HDAC4) as well of transcription factors belonging to the NFAT and MEF2 families.
Conclusion
This study provides important insight into the changes occurring in muscles of patients with genetically diverse CM. From a global point of view, muscles from patients with RYR1 mutations associated with rhabdomyolysis were similar to controls, whereas muscles from XL-MTM patients showed the greatest changes in gene expression, affecting almost all of the transcripts and miRNAs investigated. Thus, the lack of the lipid phosphatase myotubularin 1 dramatically influences the overall composition of mature skeletal muscles. We are aware of the limitations of qPCR-based approaches, and particularly that they do not provide a global overview of all transcriptional changes occurring in a tissue. In addition, the utility of our data in a clinical setting needs to be further developed before it could be used to extrapolate clinical biomarkers. Nevertheless, our study provides a snapshot of the changes occurring at a specific timepoint during disease and presents important information as to the correlation between particular types of mutations and gene expression. The results of this study support the idea for developing cross therapies for patients with genetically diverse CM. Furthermore, our results may provide useful information in the future, for clinicians monitoring transcript expression in patients enrolled in clinical trials using drugs to improve muscle function.
Supplementary Material
Acknowledgements
The authors thank Pierpaolo Ala for his expert technical support, as well as the staff of the Bioinformatics core facility of the Department of Biomedicine, Basel University Hospital for support with statistical analysis.
Abbreviations
- AD =
autosomal dominant
- AR =
autosomal recessive
- CCD =
central core disease
- CM =
congenital myopathies
- CNM =
centronuclear myopathy
- DNMT =
DNA methyl transferase
- ECC =
excitation–contraction coupling
- HDAC =
histone deacetylase
- MmD =
multiminicore disease
- MH (S) =
malignant hyperthermia (susceptibility)
- miRNA =
microRNA
- MyHC =
myosin heavy chain
- PMCA =
plasma membrane Ca2+ATPase
- RyR1 =
ryanodine receptor 1
- SERCA =
sarcoplasmic reticulum Ca2+ATPase
Contributor Information
Christoph Bachmann, Department of Biomedicine, Basel University Hospital, Hebelstrasse 20, Basel 4031, Switzerland; Department of Neurology, Basel University Hospital, Hebelstrasse 20, Basel 4031, Switzerland.
Martina Franchini, Department of Biomedicine, Basel University Hospital, Hebelstrasse 20, Basel 4031, Switzerland; Department of Neurology, Basel University Hospital, Hebelstrasse 20, Basel 4031, Switzerland.
Luuk R Van den Bersselaar, Department of Anesthesiology, Malignant Hyperthermia Investigation Unit, Canisius Wilhelmina Hospital, Nijmegen, The Netherlands; Department of Neurology, Donders Institute for Brain, Cognition and Behaviour, Radboud University Medical Center, Nijmegen, The Netherlands.
Nick Kruijt, Department of Neurology, Donders Institute for Brain, Cognition and Behaviour, Radboud University Medical Center, Nijmegen, The Netherlands.
Nicol C Voermans, Department of Neurology, Donders Institute for Brain, Cognition and Behaviour, Radboud University Medical Center, Nijmegen, The Netherlands.
Karlijn Bouman, Department of Neurology, Donders Institute for Brain, Cognition and Behaviour, Radboud University Medical Center, Nijmegen, The Netherlands; Department of Pediatric Neurology, Donders Institute for Brain, Cognition and Behaviour, Amalia Children’s Hospital, Radboud University Medical Center, Nijmegen, The Netherlands.
Erik-Jan Kamsteeg, Department of Clinical Genetics, Radboud Institute for Molecular Life Sciences, Radboud University, Nijmegen Medical Centre, Nijmegen, The Netherlands.
Karl Christian Knop, Muskelhistologisches Labor, Neurologische Abteilung, Asklepios Klinik St. Georg, Lohmuehlenstraße 5, Hamburg 20099, Germany.
Lucia Ruggiero, Dipartimento di Neuroscienze, Scienze Riproduttive ed Odontostomatologiche, Università degli Studi di Napoli Federico II, Via Pansini 5, Napoli 80131, Italy.
Lucio Santoro, Dipartimento di Neuroscienze, Scienze Riproduttive ed Odontostomatologiche, Università degli Studi di Napoli Federico II, Via Pansini 5, Napoli 80131, Italy.
Yoram Nevo, Institute of Neurology, Schneider Children’s Medical Center of Israel, Petah Tiqva, Israel.
Jo Wilmshurst, Paediatric Neurology, Red Cross War Memorial Children’s Hospital, Neuroscience Institute, University of Cape Town, Cape Town, South Africa.
John Vissing, Department of Neurology, section 8077, Rigshospitalet, University of Copenhagen, Blegdamsvej 9, Copenhagen DK-2100, Denmark.
Michael Sinnreich, Department of Biomedicine, Basel University Hospital, Hebelstrasse 20, Basel 4031, Switzerland; Department of Neurology, Basel University Hospital, Hebelstrasse 20, Basel 4031, Switzerland.
Daniele Zorzato, GKT School of Medical Education, King’s College London, Hodgkin Building, Newcomen Street, London SE1 1UL, UK.
Francesco Muntoni, Dubowitz Neuromuscular Centre and MRC Centre for Neuromuscular Diseases, UCL, Institute of Child Health, London, UK; NIHR Great Ormond Street Hospital Biomedical Research Centre, London, UK.
Heinz Jungbluth, Department of Paediatric Neurology, Neuromuscular Service, Evelina Children’s Hospital, St. Thomas’ Hospital, London, UK; Department of Basic and Clinical Neuroscience, Institute of Psychiatry, Psychology and Neuroscience (IoPPN), King’s College London, London, UK; Randall Center for Cell and Molecular Biophysics, Muscle Signalling Section, Faculty of Life Sciences and Medicine, King’s College, London, UK.
Francesco Zorzato, Department of Biomedicine, Basel University Hospital, Hebelstrasse 20, Basel 4031, Switzerland; Department of Neurology, Basel University Hospital, Hebelstrasse 20, Basel 4031, Switzerland; Department of Life Science and Biotechnology, University of Ferrara, Via Borsari 46, Ferrara 44100, Italy.
Susan Treves, Department of Biomedicine, Basel University Hospital, Hebelstrasse 20, Basel 4031, Switzerland; Department of Neurology, Basel University Hospital, Hebelstrasse 20, Basel 4031, Switzerland; Department of Life Science and Biotechnology, University of Ferrara, Via Borsari 46, Ferrara 44100, Italy.
Funding
This study was supported by a grant from the Swiss National Science Foundation (SNF N° 31003A-184765). The research leading to these results has received funding from the Muscular Dystrophy Campaign grant ‘Understanding the function of novel congenital muscular dystrophies and congenital myopathies genes’ [Grant reference number: 18GRO-PG24-0271]. The Neuromuscular Biobank of the Dubowitz Neuromuscular Centre at University College London (UCL) receives funding from the Medical Research Council and from the National Institute of Health and Care Research (NIHR) Biomedical Research Centre at Great Ormond Street Hospital, Great Ormond Street, London, UK..
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain Communications online.
References
- 1. Jungbluth H, Treves S, Zorzato F, et al. . Congenital myopathies: disorders of excitation–contraction coupling and muscle contraction. Nat Rev Neurol 2018;14:151–167. [DOI] [PubMed] [Google Scholar]
- 2. Amburgey K, McNamara N, Bennett LR, McCormick ME, Acsadi G, Dowling JJ. Prevalence of congenital myopathies in a representative pediatric United States population. Ann Neurol 2011;70:662–665. [DOI] [PubMed] [Google Scholar]
- 3. Gonorazky HD, Dowling JJ, Volpatti JR, Vajsar J. Signs and symptoms in congenital myopathies. Semin Pediatr Neurol 2019;29:3–11. [DOI] [PubMed] [Google Scholar]
- 4. Westra D, Schouten MI, Stunnenberg BC, et al. . Panel-based exome sequencing of neuromuscular disorders as a diagnostic service. J Neuromuscul Dis 2019;6:241–258. [DOI] [PubMed] [Google Scholar]
- 5. Snoeck M, van Engelen BGM, Küsters B, et al. . RYR1-related myopathies: A wide spectrum of phenotypes throughout life. Eur J Neurol 2015;22:1094–1112. [DOI] [PubMed] [Google Scholar]
- 6. Jungbluth H, Ochala J, Treves S, Gautel M. Current and future therapeutic approaches to the congenital myopathies. Semin Cell Dev Biol 2017;64:191–200. [DOI] [PubMed] [Google Scholar]
- 7. Treves S, Jungbluth H, Voermans N, Muntoni F, Zorzato F. Ca2+ handling abnormalities in early-onset muscle diseases: Novel concepts and perspectives. Semin Cell Dev Biol 2017;64:201–212. [DOI] [PubMed] [Google Scholar]
- 8. Dowling JJ, Lillis S, Amburgey K, et al. . King–Denborough syndrome with and without mutations in the skeletal muscle ryanodine receptor (RYR1) gene. Neuromuscul Disord 2011;21:420–427. [DOI] [PubMed] [Google Scholar]
- 9. Matthews E, Neuwirth C, Jaffer F, et al. . Atypical periodic paralysis and myalgia. Neurology 2018;90:e412–e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Petit N, Lescure A, Rederstorff M, et al. . Selenoprotein N: An endoplasmic reticulum glycoprotein with an early developmental expression pattern. Hum Mol Genet 2003;12:1045–1053. [DOI] [PubMed] [Google Scholar]
- 11. Castets P, Lescure A, Guicheney P, Allamand V. Selenoprotein N in skeletal muscle: From diseases to function. J Mol Med 2012;90:1095–1107. [DOI] [PubMed] [Google Scholar]
- 12. Jungbluth H, Wallgren-Pettersson C, Laporte J. Centronuclear (myotubular) myopathy. Orphanet J Rare Dis 2008;3:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Romero NB. Centronuclear myopathies: A widening concept. Neuromuscul Disord 2010;20:223–228. [DOI] [PubMed] [Google Scholar]
- 14. Hnia K, Vaccari I, Bolino A, Laporte J. Myotubularin phosphoinositide phosphatases: Cellular functions and disease pathophysiology. Trends Mol Med 2012;18:317–327. [DOI] [PubMed] [Google Scholar]
- 15. Cowling BS, Chevremont T, Prokic I, et al. . Reducing dynamin 2 expression rescues X-linked centronuclear myopathy. J Clin Invest 2014;124:1350–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maani N, Sabha N, Rezai K, et al. . Tamoxifen therapy in a murine model of myotubular myopathy. Nat Commun 2018;9:4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sewry CA, Laitila JM, Wallgren-Pettersson C. Nemaline myopathies: A current view. J Muscle Res Cell Motil 2019;40:111–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de Winter JM, Molenaar JP, Yuen M, et al. . KBTBD13 is an actin-binding protein that modulates muscle kinetics. J Clin Invest 2020;130:754–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bachmann C, Noreen F, Voermans NC, et al. . Aberrant regulation of epigenetic modifiers contributes to the pathogenesis in patients with selenoprotein N-related myopathies. Hum Mutat 2019;40:962–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou H, Jungbluth H, Sewry CA, et al. . Molecular mechanisms and phenotypic variation in RYR1-related congenital myopathies. Brain 2007;130:2024–2036. [DOI] [PubMed] [Google Scholar]
- 21. Bachmann C, Jungbluth H, Muntoni F, Manzur AY, Zorzato F, Treves S. Cellular, biochemical and molecular changes in muscles from patients with X-linked myotubular myopathy due to MTM1 mutations. Hum Mol Genet 2017;26:320–332. [DOI] [PubMed] [Google Scholar]
- 22. Al-Qusairi L, Weiss N, Toussaint A, et al. . T-tubule disorganization and defective excitation-contraction coupling in muscle fibers lacking myotubularin lipid phosphatase. Proc Natl Acad Sci USA 2009;106:18763–18768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rokach O, Sekulic-Jablanovic M, Voermans N, et al. . Epigenetic changes as a common trigger of muscle weakness in congenital myopathies. Hum Mol Genet 2015;24:4636–4647. [DOI] [PubMed] [Google Scholar]
- 24. Ritchie ME, Phipson B, Wu D, et al. . Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Statist Soc Ser B 2015;57:289–300. [Google Scholar]
- 26. MacLennan DH, Duff C, Zorzato F, et al. . Ryanodine receptor gene is a candidate for predisposition to malignant hyperthermia. Nature 1990;343:559–561. [DOI] [PubMed] [Google Scholar]
- 27. Tanabe T, Takeshima H, Mikami A, et al. . Primary structure of the receptor for calcium channel blockers from skeletal muscle. Nature 1987;328:313–318. [DOI] [PubMed] [Google Scholar]
- 28. Nelson BR, Wu F, Liu Y, et al. . Skeletal muscle-specific T-tubule protein STAC3 mediates voltage-induced Ca2+ release and contractility. Proc Natl Acad Sci USA 2013;110:11881–11886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Polster A, Perni S, Bichraoui H, Beam KG. Stac adaptor proteins regulate trafficking and function of muscle and neuronal L-type Ca2+ channels. Proc Natl Acad Sci USA 2015;112:602–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. MacLennan DH. Ca2+ signalling and muscle disease. Eur J Biochem 2000;267:5291–5297. [DOI] [PubMed] [Google Scholar]
- 31. Carafoli E. Calcium pump of the plasma membrane. Physiol Rev 1991;71:129–153. [DOI] [PubMed] [Google Scholar]
- 32. Qiu R, Lewis RS. Structural features of STIM and Orai underlying store-operated calcium entry. Curr Opin Cell Biol 2019;57:90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Feng W, Tu J, Yang T, et al. . Homer regulates gain of ryanodine receptor type 1 channel complex. J Biol Chem 2002;277:44722–44730. [DOI] [PubMed] [Google Scholar]
- 34. Shin DM, Dehoff M, Luo X, et al. . Homer 2 tunes G protein–coupled receptors stimulus intensity by regulating RGS proteins and PLCβ GAP activities. J Cell Biol 2003;162:293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Michikawa T, Miyawaki A, Furuichi T, Mikoshiba K. Inositol 1,4,5-trisphosphate receptors and calcium signaling. Crit Rev Neurobiol 1996;10:39–55. [DOI] [PubMed] [Google Scholar]
- 36. Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev 2011;91:1447–1531. [DOI] [PubMed] [Google Scholar]
- 37. Obermann WMJ. Molecular structure of the sarcomeric M band: Mapping of titin and myosin binding domains in myomesin and the identification of a potential regulatory phosphorylation site in myomesin. EMBO J 1997;16:211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lyko F. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat Rev Genet 2018;19:81–92. [DOI] [PubMed] [Google Scholar]
- 39. Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat Rev Genet 2009;10:32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chin ER, Olson EN, Richardson JA, et al. . A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev 1998;12:2499–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McKinsey TA, Zhang CL, Lu J, Olson EN. Signal dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 2000;408:106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lu J, McKinsey TA, Nicol RL, Olson EN. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc Natl Acad Sci USA 2000;97:4070–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang ET, Cody NAL, Jog S, et al. . Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell 2012;150:710–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev 2015;87:3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Morton SU, Sefton CR, Zhang H, et al. . microRNA-mRNA profile of skeletal muscle differentiation and relevance to congenital myotonic dystrophy. Int J Mol Sci 2021;22(5):2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ma G, Wang Y, Li Y, et al. . MiR-206, a key modulator of skeletal muscle development and disease. Int J Biol Sci 2015;11:345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zaharieva IT, Calissano M, Scoto M, et al. . Dystromirs as serum biomarkers for monitoring the disease severity in Duchenne muscular dystrophy. PLoS One 2013;8(11):e80263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Güller I, Russell AP. MicroRNAs in skeletal muscle: Their role and regulation in development, disease and function. J Physiol 2010;588:4075–4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhou H, Rokach O, Feng L, et al. . Ryr1 deficiency in congenital myopathies disrupts excitation-contraction coupling. Hum Mutat 2013;34:986–996. [DOI] [PubMed] [Google Scholar]
- 50. Cacheux M, Blum A, Muriel S, et al. . Functional characterization of a central core disease RyR1 mutation (p.Y4861H) associated with quantitative deficit in RyR1 protein. J Neuromuscul Dis 2015;2:421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Treves S, Jungbluth H, Muntoni F, Zorzato F. Congenital muscle disorders with cores: The ryanodine receptor calcium channel paradigm. Curr Opin Pharmacol 2008;8:319–326. [DOI] [PubMed] [Google Scholar]
- 52. Dirksen RT, Avila G. Altered ryanodine receptor function in central core disease: Leaky or uncoupled Ca2+ release channels? Trends Cardiovasc Med 2002;12:189–197. [DOI] [PubMed] [Google Scholar]
- 53. Cully TR, Edwards JN, Friedrich O, et al. . Changes in plasma membrane Ca-ATPase and stromal interacting molecule 1 expression levels for Ca2+ signaling in dystrophic mdx mouse muscle. Am J Physiol Cell Physiol 2012;303:C567–C576. [DOI] [PubMed] [Google Scholar]
- 54. Sutko JL, Airey JA. Ryanodine receptor Ca2+ release channels: Does diversity in form equal diversity in function? Physiol Rev 1996;76:1027–1071. [DOI] [PubMed] [Google Scholar]
- 55. Suman M, Sharpe JA, Bentham RB, et al. . Inositol trisphosphate receptor-mediated Ca2+ signalling stimulates mitochondrial function and gene expression in core myopathy patients. Hum Mol Genet 2018;27:2367–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Filipova D, Walter AM, Gaspar JA, et al. . Gene profiling of embryonic skeletal muscle lacking type 1 ryanodine receptor Ca2+ release channel. Sci Rep 2016;6:20050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Meyer P, Notarnicola C, Meli AC, et al. . Skeletal ryanodine receptors are involved in impaired myogenic differentiation in Duchenne muscular dystrophy patients. Int J Mol Sci 2021;22:12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pisaniello A, Serra C, Rossi D, et al. . The block of ryanodine receptors selectively inhibits fetal myoblast differentiation. J Cell Sci 2003;116:1589–1597. [DOI] [PubMed] [Google Scholar]
- 59. Brueckner JK, Ashby LP, Prichard JR, Porter JD. Vestibular-ocular pathways modulate extraocular muscle myosin expression patterns. Cell Tissue Res 1999;295:477–484. [DOI] [PubMed] [Google Scholar]
- 60. Eckhardt J, Bachmann C, Benucci S, et al. . Molecular basis of impaired extraocula muscle function in a mouse model of congenital myopathy due to compound heterozygous Ryr1 mutations. Hum Mol Genet 2020;29:1330–1339. [DOI] [PubMed] [Google Scholar]
- 61. Seto E, Yoshida M. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb Perspect Biol 2014;6:a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sharma A, Nguyen H, Geng C, Hinman MN, Luo G, Lou H. Calcium-mediates histone modifications regulate alternative splicing in cardiomyocytes. Proc Natl Acad Sci USA 2014;111:E4920–E4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schübeler D, Lorincz MC, Cimbora DM, et al. . Genomic targeting of methylated DNA: Influence of methylation on transcription, replication, chromatin structure, and histone acetylation. Mol Cell Biol 2000;20:9103–9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yao Q, Chen Y, Zhou X. The roles of microRNAs in epigenetic regulation. Curr Opin Chem Biol 2019;51:11–17. [DOI] [PubMed] [Google Scholar]
- 65. Salant GM, Tat KL, Goodrich JA, Kugel JF. miR-206 knockout shows it is critical for myogenesis and directly regulates newly identified target mRNAs. RNA Biol 2020;17:956–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data sharing is not applicable, since all results are included in the manuscript.