Abstract
Background
This study was the first human validation of the gram-positive bacterial DNA polymerase IIIC target in patients with Clostridioides difficile infection. The primary objectives were to assess clinical cure rates and adverse events (AEs). Secondary objectives were to evaluate plasma/fecal pharmacokinetics, microbiologic eradication, microbiome and bile acid effects, and sustained clinical cure (SCC) with ibezapolstat.
Methods
This single-arm, open-label, phase 2a study enrolled adults with C. difficile infection at 4 US centers. Patients received ibezapolstat 450 mg orally every 12 hours for 10 days and followed for an additional 28 days to assess study objectives.
Results
Ten patients with a mean (standard deviation [SD]) age of 49 [15] years were enrolled. Seven AEs were reported classified as mild-moderate. Plasma levels of ibezapolstat ranged from 233 to 578 ng/mL while mean (SD) fecal levels were 416 (494) µg/g stool by treatment day 3 and >1000 µg/g stool by days 8–10. A rapid increase in alpha diversity in the fecal microbiome was noted after starting ibezapolstat therapy, which was maintained after completion of therapy. A proportional decrease in Bacteroidetes phylum was observed (mean change [SD], −10.0% [4.8%]; P = .04) with a concomitantly increased proportion of Firmicutes phylum (+14.7% [5.4%]; P = .009). Compared with baseline, total primary bile acids decreased by a mean (SD) of 40.1 (9.6) ng/mg stool during therapy (P < .001) and 40.5 (14.1) ng/mg stool after completion of therapy (P = .007). Rates of both initial clinical cure and SCC at 28 days were 100% (10 of 10 patients).
Conclusions
In this phase 2a study, 10 of 10 patients achieved SCC, demonstrated favorable pharmacokinetics, minimal AEs, and beneficial microbiome and bile acids results. These results support continued clinical development.
Keywords: Clostridioides difficile, DNA polymerase IIIC, humans, Enterocolitis, Male, Female
This single-arm, phase 2a study was the first validation of ibezapolstat in adult patients with Clostridioides difficileinfection. Ten of 10 patients achieved sustained clinical cure and demonstrated favorable pharmacokinetics, minimal adverse events, and beneficial microbiome and bile acids results.
Clostridioides difficile infection (CDI) is the most common cause of healthcare-associated infections in the United States [1], where it is responsible for almost 500 000 incident infections and 29 000 deaths [2]. Current treatment guidelines recommend only 2 antibiotics for initial treatment of CDI, namely, oral vancomycin or fidaxomicin [3]. Vancomycin is associated with unacceptably high recurrence rates and both antibiotics are associated with the emergence of antimicrobial resistance [4–6]. CDI recurrence is due to the continued perturbation of the gut microbiome, most commonly characterized by decreased proportions of Firmicutes, Bacteroidetes, and Actinobacteria phyla with subsequent overgrowth of Proteobacteria [7]. These taxa changes reduce colonization resistance to C. difficile by eliminating the taxa responsible for bile acid metabolism, leading to a higher concentration of primary bile acids which support spore germination and onset of CDI or recurrent CDI [8]. Ideally, new drugs in clinical development should have a unique mechanism of action with similarly potent activity against C. difficile but without activity against key host microbiota [9]. This spectrum of activity is crucial to prevent the treatment-associated dysbiosis that allows for further germination and infection by C. difficile after therapy completion.
Ibezapolstat (formerly ACX-362E) represents a unique class of gram-positive selective spectrum antimicrobials that bind to and inhibit bacterial DNA polymerase IIIC [10]. The DNA polymerase IIIC enzyme is essential for replication of low–G + C content (fewer G and C DNA bases than A and T bases) in gram-positive bacteria, including Firmicutes such as C. difficile, yet it is absent in Actinobacteria and gram-negative host microbiota, including Bacteroidetes species. Ibezapolstat was minimally absorbed in a hamster model, leading to high colonic and low systemic concentrations and was shown to be effective for CDI [11]. A phase 1 healthy volunteer study demonstrated a similarly advantageous pharmacokinetic (PK) profile and a favorable safety profile [12]. In contrast to vancomycin, ibezapolstat did not cause overgrowth of Proteobacteria and preserved a favorable ratio of secondary-to-primary bile acids that would be predictive of an anti-CDI recurrence effect.
This phase 2a study was conducted as the first human validation of the DNA polymerase IIIC target in a diseased population of patients with CDI. The primary objectives of this study were to assess CDI clinical cure rates 2 days after the end of treatment (EOT) and the safety/tolerability of ibezapolstat given to adult patients with CDI. The secondary objectives were to evaluate plasma and fecal PK characteristics, microbiologic eradication, quantitative microbiome changes in relevant fecal bacterial communities and microbial diversity, bile acid effects, and sustained clinical cure (SCC) associated with ibezapolstat in patients with CDI.
METHODS
Study Design
This was a single-arm, open-label, phase 2a segment of a multicenter phase 2 trial, and enrolled adults at 4 centers in the United States in 2019–2020 (protocol no. ACX-362E-201; ClinicalTrials.gov identifier NCT04247542). Patients received three 150-mg ibezapolstat capsules (total dose, 450 mg) orally every 12 hours for 10 days. After the EOT, patients were followed up for an additional 28 days to evaluate clinical response, adverse events (AEs), and status of the fecal microbiome. The study was conducted in accordance with the Declaration of Helsinki. The study protocol and amendments were approved by an institutional review board at each study site, and written informed consent was obtained for each enrolled subject before the study commenced.
Patients
Eligible participants included adults aged 18–90 years with CDI defined as >3 watery bowel movements in the 24 hours before enrollment and classified as nonsevere CDI, as defined by Infectious Diseases Society of America/Society for Healthcare Epidemiology of America guidelines (white blood cell count ≤15 000/mL and serum creatinine level <1.5 mg/dL) [13]. Enrolled patients must have had CDI diagnosed using a positive free toxin–based fecal test (C. DIFF QUIK CHEK COMPLETE [TechLab] or Immunocard Toxin A&B [Meridian Bioscience]). Patients were excluded if they had >24 hours of other CDI-directed antibiotics at the time of enrollment, probiotic or laxative receipt, >3 episodes of CDI in the previous 12 months or >1 prior episode in the last 3 months, immunocompromising conditions or medications, inflammatory bowel disease, pregnancy or lactation, active gastroenteritis due to another microorganism, major gastrointestinal surgery within 3 months of enrollment (appendectomy or cholecystectomy permitted), or elevated liver function values (defined as >2 times the upper limit of normal for alanine aminotransferase or aspartate aminotransferase).
Safety Assessments
Safety evaluations included AE assessment, physical examination, vital signs, clinical laboratory tests (chemistry, hematology, and urinalysis), and electrocardiography. Safety end points for all subjects were recorded including nature, frequency, and severity of AEs. AEs were assessed at each visit beginning from the time of enrollment and classified according to the Medical Dictionary for Regulatory Activities (MedDRA; version 15.0). AE severity (mild, moderate, or severe) and causality (unrelated, possibly related, or probably related to the study medication) were assessed by the investigator at each site.
PK Evaluations
Plasma samples were obtained 2 and 4 hours after the first daily ibezapolstat administration on days 1, 5, and 10. Fecal samples were collected at baseline and daily during days 1–10 of ibezapolstat receipt. Plasma and fecal concentrations were assayed by AltaSciences (Laval), and PK analyses were performed by Learn and Confirm.
Microbiology
Stool samples were cultured for C. difficile growth on a selective cycloserine-cefoxitin fructose agar at 37°C under anaerobic conditions for 48 hours [5]. Isolates were identified as C. difficile based on growth and morphology and confirmed by PCR for C. difficile toxin and tpi genes. C. difficile was strain typed using a PCR-based ribotyping method, as described elsewhere [14]. Minimum inhibitory concentrations were determined for ibezapolstat by broth microdilution in 0.1% sodium taurocholate brain heart infusion media [15].
Microbiome and Bile Acid Evaluations
Fecal samples for microbiome analysis were collected daily during ibezapolstat dosing and on days 2, 10, 20, and 28 after the EOT. Stool DNA extraction was performed using the Qiagen DNeasy PowerSoil Pro Kit (Qiagen; catalog no. 12888-100) in a QIAcube automated DNA extraction system (Qiagen) according to the manufacturer’s instructions. Microbiome characterization was performed by sequencing the V1–V3 region of the 16S ribosomal RNA gene, using the MiSeq system (Illumina) [16, 17]. Each sample was amplified using a barcoded primer, which yielded a unique sequence identifier tagged onto each individual sample library. Genomic DNA was normalized before polymerase chain reaction (PCR) analysis, and PCR products were normalized before pooling. Illumina-based sequencing yielded >15 000 reads per sample. Bile acids were quantified using targeted liquid chromatography mass spectrometric analysis performed on a QTRAP 5500 mass spectrometer (Sciex), adapted from a previously described method [18]. Bile acid levels were normalized by the corresponding stool sample weight.
Efficacy Assessments
The primary efficacy outcome measure was initial clinical cure at the EOT, defined as resolution of diarrhea in the 24-hour period before the EOT and maintained for ≥48 hours after the EOT. SCC was defined as clinical cure with no recurrence of CDI within 28 (±2) days after the EOT.
Statistical Analysis
An intent-to-treat analysis of patients receiving ≥1 dose of ibezapolstat was conducted. Descriptive statistics were calculated for efficacy, safety/tolerability, and PK data generated using SAS version 9.4 software (SAS Institute). Results are expressed as means with standard deviations (SDs) unless otherwise stated. Microbiome summary plots and data visualization was prepared using R software, version 4.1.1 (R Core Team 2021) [19]. Alpha diversity for each sample was assessed with the VeganR package version 2.4-2, using the Shannon diversity index and the inverse Simpson index. Differences in alpha diversity (with both indexes) and bile acids between baseline and during or after therapy were determined using linear regression models. Proportional changes of bacterial taxa over the 10-day dosing interval were calculated using linear regression models for taxa with a ≥1% proportional change during the study time period. Differences were considered significant at P < .05.
RESULTS
Patients
Ten patients aged were enrolled, with a mean (SD) age of 49 (15) years (50% female; 100% white race; 80% Hispanic or Latino ethnicity). All 10 patients received ibezapolstat and completed the study (Supplementary Figure 1). The median number of unformed bowel movements in the 24 hours before the start of therapy was 4 (range, 3–10). Two of 10 patients received <24 hours of antibiotics, either metronidazole or vancomycin, before starting ibezapolstat. No patients were hospitalized before or after enrollment.
Safety
A summary of the AEs is provided in Supplementary Table 1. Seven AEs were reported in 4 of the 10 patients, with 4 occurring in a single subject. None of the AEs were serious AEs. The severity of AEs was mild (n = 2), moderate (n = 4), and severe (n = 1; drug-unrelated migraine headache). The most common AEs were headache (n = 2) or nausea (n = 2); both episodes of nausea were regarded by the investigator as “probably related” to the study drug. No treatment was required for these AEs, and no AE required a change to the study drug schedule or withdrawal of dosing. All AEs were resolved by the end of the study. No significant clinical laboratory test abnormalities were detected
PK Results
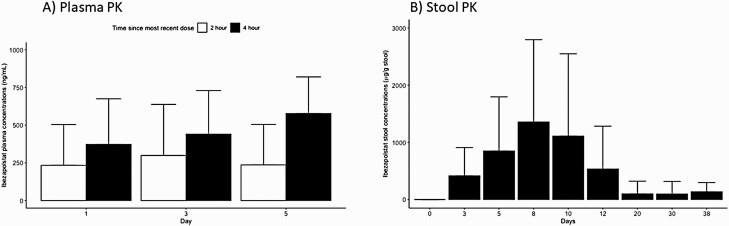
The 2–4-hour postdose ibezapolstat plasma levels ranged from 233 to 578 ng/mL, with higher concentrations observed at 4 hours (range, 373–578 ng/mL) than at 2 hours (234–299 ng/mL). The mean ibezapolstat stool concentration (SD) was 416 (494) µg/g stool by day 3 of therapy, >1000 µg/g stool by days 8–10, and 535 (748) µg/g stool 2 days after the EOT. Three of 4 stool samples collected on day 38 continued to have detectable stool concentrations of ibezapolstat (mean [SD], 136 [161] µg/g stool). Baseline stool and plasma concentrations (before drug administration) were undetectable. Full stool and plasma PK data are shown in Figure 1.
Figure 1.
Ibezapolstat pharmacokinetics in plasma (A) and stool (B) samples. Values represent means with standard deviations.
Microbiology Results
Toxigenic C. difficile grew in 6 of 7 baseline stool samples (86%) available for microbiology studies but not in stool samples from any other sampling day (range, 7–9 samples per day). Identified ribotypes included F078–126 (n = 2), F014–020 (n = 2), F106 (n = 1), and FP435 (n = 1). The minimum inhibitory concentrations of ibezapolstat were 0.25 (n = 1), 0.5 (n = 3), or 1.0 (n = 1) ug/mL.
Microbiome and Bile Acid Results
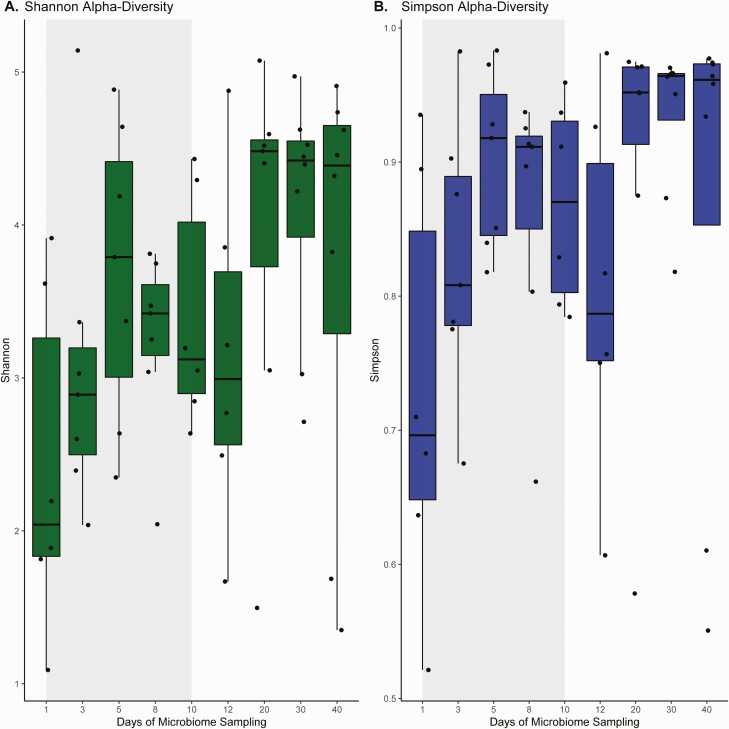
Eight participants provided stool samples for microbiome and bile acid analysis. Although interindividual changes were noted, a rapid increase in alpha diversity was noted from baseline samples using both the inverse Simpson and Shannon diversity indexes (Figure 2). Compared with baseline, inverse Simpson index diversity increased by a mean (SD) of 0.14 (0.06) points during ibezapolstat therapy (P = .02) and by 0.22 (0.10) points after the EOT (P = .003).
Figure 2.
Summary estimates of changes in alpha diversity over time with the Shannon diversity (A) and inverse Simpson (B) indexes.
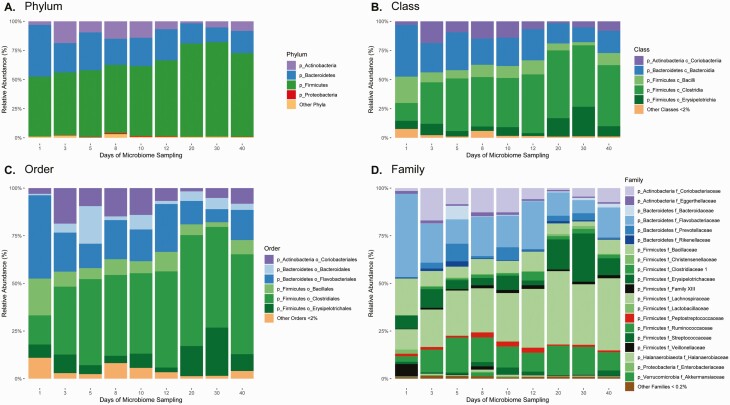
Similar results were observed using the Shannon diversity index; diversity increased by a mean (SD) of 0.98 (0.48) points during ibezapolstat therapy (P = .049) and by 1.7 (0.87) points after the EOT (P = .04), compared with baseline. Taxa changes during and after ibezapolstat therapy are shown in Figure 3. A proportional decrease in Bacteroidetes phylum was observed (mean change [SD], −10.0% [4.8%]; P = .04), most commonly owing to a decreased proportion of Bacteroidia class taxa (−10.0% [4.8%]) and Flavobacteriaceae family taxa (−8.8% [4.8%]). An increased proportion of Firmicutes phylum was observed (mean change [SD], +14.7% [5.4%]; P = .009), most commonly owing to an increased proportion of Lachnospiraceae (+12.7% [6.0%]) and Ruminococcaceae (+2.8% [2.7%]). Other Firmicutes had decreased proportions, most notably Bacillales (mean change [SD], −4.4% [2.3%]) and Lactobacillales (−3.7% [2.2%]) order taxa. Abundance tables for individual patients are shown in Supplementary Figure 2.
Figure 3.
Effects of ibezapolstat on relative abundance of taxa by phylum (A), class (B), order (C), and family (D).
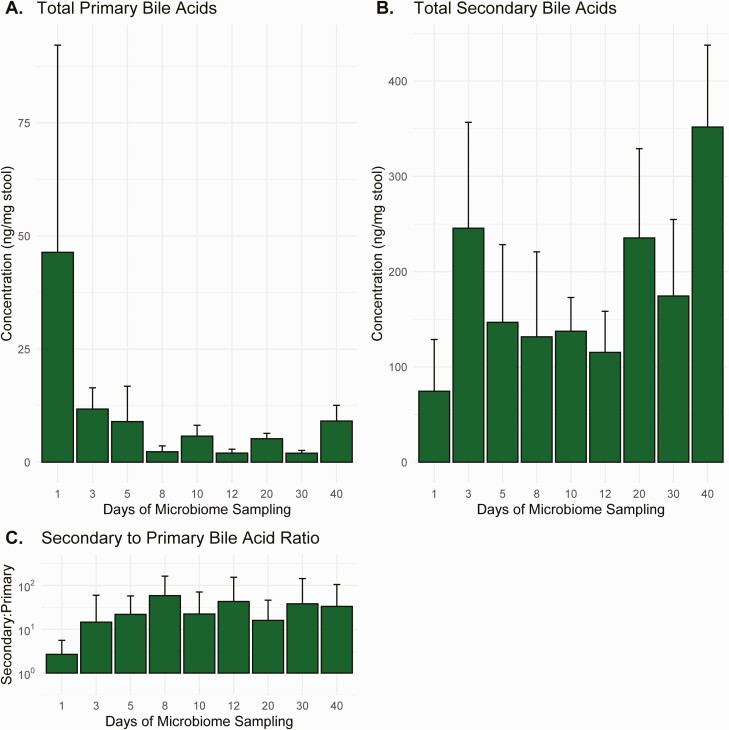
Results of the bile acid analysis are shown in Figure 4. Compared with baseline, total primary acid concentrations in stool samples decreased by a mean (SD) of 40.1 (9.6) ng/mg stool during therapy (P < .001) and 40.5 (14.1) ng/mg stool after the EOT (P = .007). Compared with baseline, total secondary bile acid concentrations increased by a mean (SD) of 65.6 (146.7) ng/mg stool during therapy (P = .66) and 97.5 (215.4) ng/mg stool after the EOT (P = .65).
Figure 4.
Changes over time in primary (A) and secondary (B) bile acid concentrations and the ratio of secondary to primary bile acid concentrations (C). Values represent means with standard errors.
Efficacy Outcomes
The initial clinical cure rate at the EOT was 100% (10 of 10 patients). The mean time to resolution of diarrhea was 5 days (range, 3–7 days). The SCC rate at 28 ± 2 days after the EOT was also 100% (10 of 10 patients).
DISCUSSION
In this open-label, phase 2a study, ibezapolstat was well tolerated and had a safety profile consistent with results from the phase I study [12]. PK findings were also similar to those seen in the healthy volunteer study. Ibezapolstat achieved high stool concentrations and plasma concentrations that did not exceed 1 ug/mL. Favorable changes to the microbiome were observed, most notably C. difficile eradication by day 3 and an increased proportion of healthy microbiota, including Clostridiales order taxa known to metabolize primary bile acids to secondary bile acids via the 7α-dehydroxylation pathway [20]. These proportional changes were associated with bile acid changes, including a reduction in primary and an increase in secondary bile acids during ibezapolstat therapy, predictive biomarkers of a lower chance of CDI recurrence. Finally, clinical efficacy evaluations demonstrated that 100% of the 10 patients experienced initial clinical cure and SCC evaluated at 28 days.
There are currently only 2 Food and Drug Administration–indicated antibiotics for the treatment of CDI: vancomycin and fidaxomicin [3]. As resistance to both these antibiotics has been noted, new CDI-directed antibiotics are urgently needed [4, 5]. The first-in-class gram-positive selective-spectrum antimicrobial ibezapolstat is a novel DNA polymerase IIIC inhibitor with a unique mechanism of action that targets low–G + C content gram-positive bacteria [21].
The microbiome studies in this phase 2a study provide additional insight into the effect of ibezapolstat on a mixed bacterial community such as the gut microbiome. Because ibezapolstat has no direct activity on gram-negative organisms, the decrease in Bacteroidetes phylum was perhaps a secondary result of ibezapolstat’s effect on other targeted gram-positive bacteria. Likewise, the increased proportion of the favorable Clostridiales order taxa demonstrates ibezapolstat selectivity within low–G + C content bacteria. DNA polymerase IIIC is thought to be present in most Firmicutes, and the reasons for this selectivity will require further study.
Although the current study is underpowered to statistically evaluate secondary bile acid changes, an increased concentration of secondary bile acids was observed during and after ibezapolstat therapy, which is known to be correlated with colonization resistance against C. difficile [22]. In addition, the decrease in primary bile acids and the favorable increase in the ratio of secondary to primary bile acids suggest that ibezapolstat may reduce the likelihood of CDI recurrence compared with vancomycin. Phase 2b and 3 studies will allow further investigations of these mechanistic findings. Finally, although favorable efficacy results were demonstrated, these will need to be validated in a larger population using a double-blind study design.
In conclusion, in the current study, ibezapolstat was well tolerated in adults with CDI and demonstrated a PK profile ideal for a CDI antibiotic with low systemic absorption and high colonic concentrations. Advantageous microbiome abundance and bile acids changes coupled with successful efficacy data support the continued development of ibezapolstat for use in the adult CDI population.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. Conceptualization: K. W. G., A. J. G. L., K. B., M. J. A., and M. H. S.
Methodology and investigation: All authors. Visualization: K. W. G., J. M., C. H., and J. J. Funding acquisition: K. W. G. Project administration: K. W. G., W. W., C. K. L., A. J. G. L., K. B., M. J. A., and B. H. Supervision: K. W. G., W. W., C. K. L., A. J. G. L., K. B., M. J. A., and B. H. Writing (original draft): K. W. G., J. M., J. J., and A. J. G. L. Writing (review and editing): All authors.
Financial support. This work was supported by Acurx Pharmaceuticals (research grant to the University of Houston).
Contributor Information
Kevin W Garey, University of Houston College of Pharmacy, Houston, Texas, USA; The University of Texas Health Science Center at Houston School of Public Health, Houston, Texas, USA.
Jacob McPherson, University of Houston College of Pharmacy, Houston, Texas, USA.
An Q Dinh, The University of Texas Health Science Center at Houston School of Public Health, Houston, Texas, USA.
Chenlin Hu, University of Houston College of Pharmacy, Houston, Texas, USA.
Jinhee Jo, University of Houston College of Pharmacy, Houston, Texas, USA.
Weiqun Wang, University of Houston College of Pharmacy, Houston, Texas, USA.
Chris K Lancaster, University of Houston College of Pharmacy, Houston, Texas, USA.
Anne J Gonzales-Luna, University of Houston College of Pharmacy, Houston, Texas, USA.
Caroline Loveall, University of Houston College of Pharmacy, Houston, Texas, USA.
Khurshida Begum, University of Houston College of Pharmacy, Houston, Texas, USA.
M Jahangir Alam, University of Houston College of Pharmacy, Houston, Texas, USA.
Michael H Silverman, Acurx Pharmaceuticals, Inc, Staten Island, New York, USA.
Blake M Hanson, The University of Texas Health Science Center at Houston School of Public Health, Houston, Texas, USA.
References
- 1. Magill SS, O’Leary E, Janelle SJ, et al. Changes in prevalence of health care-associated infections in U.S. Hospitals. N Engl J Med 2018; 379:1732–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lessa FC, Winston LG, McDonald LC; Emerging Infections Program C. difficile Surveillance Team. Burden of Clostridium difficile infection in the United States. N Engl J Med 2015; 372:2369–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Johnson S, Lavergne V, Skinner AM, et al. Clinical practice guideline by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA): 2021 focused update guidelines on management of Clostridioides difficile infection in adults. Clin Infect Dis 2021; 73:755–7. [DOI] [PubMed] [Google Scholar]
- 4. Shen WJ, Deshpande A, Hevener KE, et al. Constitutive expression of the cryptic vanGCd operon promotes vancomycin resistance in Clostridioides difficile clinical isolates. J Antimicrob Chemother 2020; 75:859–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gonzales-Luna AJ, Spinler JK, Oezguen N, et al. Systems biology evaluation of refractory Clostridioides difficile infection including multiple failures of fecal microbiota transplantation. Anaerobe 2021; 70:102387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oka D, Yamaya N, Kuno T, et al. In vitro and in vivo antibacterial activities of a novel quinolone compound, OPS-2071, against Clostridioides difficile. Antimicrob Agents Chemother 2021; 65:e01170-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ.. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J Clin Gastroenterol 2010; 44:354–60. [DOI] [PubMed] [Google Scholar]
- 8. Alam MJ, Walk ST, Endres BT, et al. Community environmental contamination of toxigenic Clostridium difficile. Open Forum Infect Dis 2017; 4:ofx018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Basseres E, Endres BT, Dotson KM, Alam MJ, Garey KW.. Novel antibiotics in development to treat Clostridium difficile infection. Curr Opin Gastroenterol 2017; 33:1–7. [DOI] [PubMed] [Google Scholar]
- 10. Xu WC, Silverman MH, Yu XY, Wright G, Brown N.. Discovery and development of DNA polymerase IIIC inhibitors to treat gram-positive infections. Bioorg Med Chem 2019; 27:3209–17. [DOI] [PubMed] [Google Scholar]
- 11. van Eijk E, Boekhoud IM, Kuijper EJ, Bos-Sanders I, Wright G, Smits WK.. Genome location dictates the transcriptional response to PolC inhibition in Clostridium difficile. Antimicrob Agents Chemother 2019; 63:e01263-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Garey KW, Begum K, Lancaster C, et al. A randomized, double-blind, placebo-controlled, single and multiple ascending dose phase 1 study to determine the safety, pharmacokinetics and food and faecal microbiome effects of ibezapolstat administered orally to healthy subjects. J Antimicrob Chemother 2020; 75:3635–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McDonald LC, Gerding DN, Johnson S, et al. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis 2018; 66:e1–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gonzales-Luna AJ, Carlson TJ, Dotson KM, et al. PCR ribotypes of Clostridioides difficile across Texas from 2011 to 2018 including emergence of ribotype 255. Emerg Microbes Infect 2020; 9:341–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Begum K, Basseres E, Miranda J, et al. In vitro activity of omadacycline, a new tetracycline analog, and comparators against Clostridioides difficile. Antimicrob Agents Chemother 2020; 64:e00522-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fadrosh DW, Ma B, Gajer P, et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014; 2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Walker JN, Hanson BM, Pinkner CL, et al. Insights into the microbiome of breast implants and periprosthetic tissue in breast implant-associated anaplastic large cell lymphoma. Sci Rep 2019; 9:10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Scherer M, Gnewuch C, Schmitz G, Liebisch G.. Rapid quantification of bile acids and their conjugates in serum by liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 2009; 877:3920–5. [DOI] [PubMed] [Google Scholar]
- 19. R Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing,2013. Available at: http://www.R-project.org/. Accessed 22 February 2022. [Google Scholar]
- 20. Ridlon JM, Kang DJ, Hylemon PB.. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 2006; 47:241–59. [DOI] [PubMed] [Google Scholar]
- 21. Kullar R, Tran MN, Goldstein EJC.. Investigational treatment agents for recurrent Clostridioides difficile infection (rCDI). J Exp Pharmacol 2020; 12:371–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Winston JA, Theriot CM.. Impact of microbial derived secondary bile acids on colonization resistance against Clostridium difficile in the gastrointestinal tract. Anaerobe 2016; 41:44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.