Abstract
Objectives
Antimicrobial drugs are frequently administered in veal calves, but investigations on associations with antimicrobial susceptibility of bacteria are scarce and convey partly contradictory findings. The aim of this study was to investigate associations of antimicrobial use (AMU) during the fattening period with antimicrobial susceptibility shortly before slaughter.
Methods
Detailed treatment data of 1905 veal calves from 38 farms were collected prospectively during monthly farm visits for 1 year (n = 1864 treatments, n = 535 visits); 1582 Escherichia coli, 1059 Pasteurella multocida and 315 Mannheimia haemolytica were isolated from rectal and nasopharyngeal swabs collected before slaughter and subjected to antimicrobial susceptibility testing by microdilution. Associations of antimicrobial treatments with resistant isolates were investigated at the calf level.
Results
Associations of AMU with antimicrobial resistance were observed using generalized linear models. For E. coli, the odds of being resistant were increased with increased AMU (OR 1.36 when number of treatments >1, P = 0.066). Use of tetracyclines was associated with resistance to tetracycline (OR 1.86, P < 0.001) and use of penicillins was associated with resistance to ampicillin (OR 1.66, P = 0.014). No significant associations were observed for P. multocida (use of aminoglycosides: OR 3.66 for resistance to spectinomycin, P = 0.074). For M. haemolytica, the odds of being resistant were increased with increased AMU (OR 4.63, P < 0.001), and use of tetracyclines was associated with resistance to tetracycline (OR 6.49, P < 0.001).
Conclusions
Occurrence of resistant bacteria shortly before slaughter was associated with AMU in veal calves. Prudent and appropriate use may contribute to limit the selection of resistant bacteria on veal farms.
Introduction
The administration of antimicrobials has been extensively reported to lead to selection of resistant bacteria.1 Veal calf production is known for high metaphylactic and therapeutic antimicrobial use (AMU).2–4 High antimicrobial resistance (AMR) has also been reported in this sector,5,6 thus the development of AMR may be a result of this selection process. However, in investigations at the farm level and in nationwide investigations in European countries, associations of AMU with AMR have been detected by some authors, but not by others.5,7–11 Therefore, further research is needed for a deeper understanding of the interplay of antimicrobial treatments and AMR in veal production in order to adapt treatment strategies, improve biosecurity concepts, and eventually reduce AMU and AMR in veal calf farms.
In the present study, we investigated potential associations between AMU and AMR in a comprehensive dataset at the individual veal calf level. The three main aims of the study were: (i) to analyse whether an increasing number of antimicrobial treatments is associated with AMR at the end of the fattening period; (ii) whether treatments with antimicrobials of a specific antimicrobial class are associated with resistance to this class (homologous resistance) or other classes (heterologous resistance); and (iii) whether susceptibility patterns of indicator bacteria Escherichia coli, Pasteurella multocida and Mannheimia haemolytica isolated shortly before slaughter allow for retrospective assumptions on antimicrobial treatment intensity during the fattening period.
Materials and methods
Data were obtained in the frame of a previous study where veal calves were closely followed from birth or purchase until slaughter.3 Briefly, 1905 calves were enrolled in 38 veal farms in Switzerland. Farms were visited once a month for a minimum of 12 consecutive months. On each visit, calves for which slaughter was imminent were swabbed as described below. Data regarding antimicrobial treatments, production parameters, animal welfare and economics were collected during the visits.3,12,13 The calves were fattened according to improved welfare standards (IP-SUISSE) alongside milk production in all farms.14 Farmers worked according to one of two management concepts: the ‘outdoor veal calf’ or ‘conventional’ (19 farms each), as described previously.3 All components and procedures of the study were approved by the competent authority under authorization number BE 71/16.
Treatment records
Antimicrobial treatments were recorded by the farmers or their herd veterinarians at the time of treatment, and the records were checked for clarity and completeness during the farm visits. Treatments were recorded for each calf from the beginning until the end of the fattening period. The beginning was defined as the day of purchase (if the calf was purchased) or the mean age of purchased calves upon arrival at the respective farm (if the calf was born on-farm). The end was defined as the day of slaughter. A total of 535 farm visits was conducted. Treatments were recorded on each farm according to the statutory requirements (identification number of the treated calf, first and last administration date, name of therapeutic product, dosage, indication for treatment, administration route and withdrawal period) using a customized booklet. In addition, information on administration type (individual or group treatment), signs of disease and treatment outcome were recorded. Only treatments with therapeutic products containing antimicrobials were recorded. The study team was not involved in the choice of treatment modalities, which were designed and applied by the farm veterinarians.
Nasopharyngeal and rectal swab sample collection
The sampling procedure included rectal and nasopharyngeal swabbing of each calf and was performed regardless of the presence or absence of clinical signs of disease during the last farm visit before the calves were brought to slaughter. The calves were restrained manually or in a headlock for sampling. For practical reasons, such as sudden death, unforeseen early slaughter without notification of the study team and euthanasia, not all calves enrolled in the study were sampled shortly before leaving for slaughter. Of the 1905 calves enrolled in the study, rectal swabs were not taken from 236 calves and nasopharyngeal swabs were not taken from 235 calves. Correspondingly, rectal samples of 1669 calves and nasopharyngeal samples of 1670 calves were available. For E. coli isolation, a sterile swab (BD BBL CultureSwab, Becton Dickinson AG, Basel, Switzerland) was taken from the rectum and immediately placed into transportation medium for Enterobacteriaceae (DeltaSwab Cary Blair, deltalab, Barcelona, Spain). For isolation of P. multocida and M. haemolytica, one nostril was first disinfected using gauze swabs (Provet AG/Henry Schein Animal Health, Lyssach, Switzerland) soaked in 70% propyl alcohol (F25-A Feinsprit 2% MEK, Alcosuisse AG, Bern, Switzerland). Sterile swabs (COPAN Italia SpA, Brescia, Italy) were inserted through the ventral nasal meatus to swab the nasopharyngeal epithelium, and the samples were then placed in liquid Amies transportation medium (Axonlab SwabAX, Axon Lab AG, Baden, Switzerland).5,15 The samples were transported to the laboratory at room temperature on the day of collection and processed within 24 h after overnight conservation at 4°C if necessary.
Isolation and identification of E. coli and Pasteurellaceae
For E. coli isolation, rectal swabs were spread onto selective agar BROLAC (Thermo Fisher Scientific, Waltham, MA, USA). For Pasteurellaceae isolation, nasopharyngeal swabs were spread onto Pasteurella selective agar (Thermo Scientific Oxoid, Reinach, Switzerland). The plates were incubated at 37°C for 24 h, and one single colony was picked for species identification with MALDI-TOF (Microflex LT, Bruker Daltonics GmbH, Bremen, Germany). Colonies identified as E. coli, P. multocida and M. haemolytica were purified on trypticase soy agar plates containing 5% sheep blood (TSA-SB; Becton, Dickinson and Company, Franklin Lakes, NJ, USA) and incubated at 37°C for 24 h, followed by reconfirmation of species identification using MALDI-TOF. Isolates were then cryopreserved until further analyses in 30% glycerol stocks at –80°C.
Antimicrobial susceptibility testing
If isolation of the target bacterial species from a given sample had been successful, one single isolate was tested for antimicrobial susceptibility. For E. coli, antimicrobial susceptibility was tested by determination of MICs of antimicrobials in CAMHB using EUVSEC Sensititre® plates (Thermo Fisher Scientific). For Pasteurellaceae, MIC was determined in CAMHB supplemented with 5% of laked horse blood and using BOPOF6 Sensititre® plates, according to the manufacturer’s instructions (Thermo Fisher Scientific). Clinical breakpoints (EUCAST, 2018) were used as an indicator to classify E. coli isolates as ‘resistant’.16 Isolates of P. multocida and M. haemolytica were classified as ‘resistant’ based on clinical breakpoints published by the CLSI.17 In this study, the term ‘AMR’ refers to resistant isolates. All antimicrobials used for testing and corresponding breakpoints are listed in Tables S1–S3 (available as Supplementary data at JAC Online).
Statistical analyses
For each calf, the class- or drug-specific treatment incidence was calculated in defined daily doses (TIDDD) from enrolment until swabbing, according to the method of the EMA.18,19 The following formula was used to calculate the TIDDD at the calf level for each antimicrobial treatment before summarizing treatments belonging to the same antimicrobial class or drug:
| (1) |
The total amount of drug used was extracted from the treatment journals. The DDD values and the standard weight were used as indicated by the EMA. Multiplication times 365 provides corrected TIDDD values for a year’s period (with the unit ‘number of days under treatment per calf-year’).20–22 AMU of treated calves is reported as number of treatments administered with the respective antimicrobial class or drug, the total TI of all treatments and the median TI of treatments of the respective class or drug (including 25th/75th quantiles).
Associations between susceptibility status of the isolates (resistant or not) and the administered antimicrobial treatments were investigated. This was done separately for E. coli, P. multocida and M. haemolytica. Calf-level susceptibility data and calf-level treatment data were used to build generalized linear models with a logit link function using ‘R’ Version 3.5.1 (R Core Team, Vienna, Austria; package lme4). The dependent variable was specified as the result of susceptibility testing (resistant to any of the tested antimicrobial drugs or not). The independent variable was specified as number of antimicrobial treatments applied (categorized into ‘untreated’, ‘one treatment’, ‘≥one treatment’). The category ‘untreated’ was used as the reference category. In each model, farm was included as a random effect.
In addition, for each bacterial species, separate generalized linear models with a logit link function were built for the most frequently used antimicrobial drugs and the drugs to which bacteria were most frequently resistant to. This was done in order to investigate homologous (i.e. if resistance to antimicrobial class A is associated with use of class A at the calf level) and heterologous association (i.e. if resistance to antimicrobial class A is associated with use of classes ≠A at the calf level). To avoid type I error due to multiple comparison, a limited number of models were built. Farm was included as a random effect in these models.
Additionally, a machine learning approach was used to test for the predictability of treatment modalities on the presence/absence of resistant bacteria (‘R’ packages lme4, lmerTest, bestNormalize, caret). Three different machine learning algorithms (random forest/elastic net/support vector machine) were trained on 75% of the data to predict whether an animal had been treated in any way (all drugs combined) prior to swabbing, based on the presence/absence of each resistance. The predictive accuracy of the trained models was then tested against the remaining 25% of the observations. For each model, the achieved predictive accuracy was statistically compared with the no-information rate, to indicate whether the constructed model achieved higher predictive power than random chance. Given the low prevalence of P. multocida and M. haemolytica, this analysis was conducted for E. coli isolates only. Clustering of farms was taken into account. The level of significance was set at α = 0.05.
Results
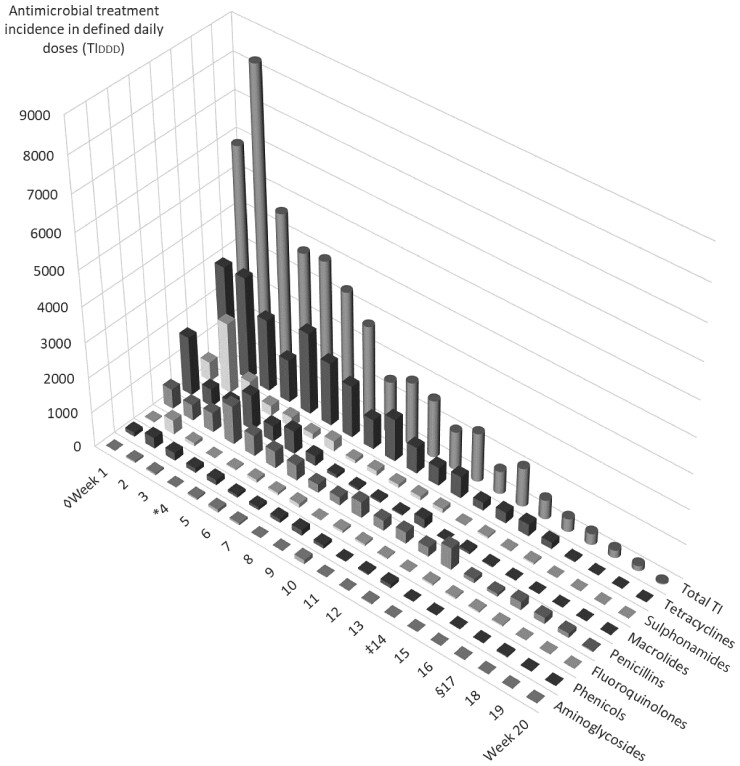
The age of the calves at the beginning of the fattening period (beginning of Week 1) was 4.6 (3.7/5.4) weeks (median, 25th/75th quantiles). In the case of antimicrobial treatment, calves were treated in Week 4.4 (3.3/5.4) of the fattening period. Calves were swabbed in Week 14.3 (12.3/16.9). The duration of the fattening period was 17.0 (15.0/19.0) weeks. Of 1905 calves enrolled at the beginning of the study, 731 calves received antimicrobial treatment during the fattening period (38.4%), through application of a total of 1864 treatments. The TIDDD of each antimicrobial class and antimicrobial drug used are sorted in decreasing order and listed in Tables 1 and 2, respectively. Additionally, the median and 25th/75th quantiles of TIDDD are shown for treatments of the mentioned class or drug. Figure 1 shows overall AMU and AMU grouped per week of the fattening period for the different antimicrobial classes.
Table 1.
Antimicrobial treatment incidence (TI) of 1864 treatments in defined daily doses (DDDs) per treated calf and year (TIDDD)a grouped by antimicrobial classes according to the method of the EMA
| Antimicrobial class | n | Total TIDDD | Calf level TIDDD per year | ||
|---|---|---|---|---|---|
| 25th percentile | median | 75th percentile | |||
| Tetracyclines | 593 | 19846.1 | 13.0 | 29.7 | 38.7 |
| Penicillins | 292 | 7481.3 | 17.1 | 25.0 | 32.1 |
| Macrolides | 372 | 5659.3 | 3.0 | 6.5 | 18.1 |
| Sulphonamides | 343 | 4947.3 | 8.2 | 12.8 | 17.8 |
| Diaminopyrimidines | 60 | 2462.6 | 29.8 | 38.9 | 45.3 |
| Phenicols | 111 | 1659.9 | 6.7 | 11.3 | 17.9 |
| Fluoroquinolones | 51 | 1181.7 | 3.7 | 13.2 | 26.4 |
| Aminoglycosides | 32 | 466.9 | 7.3 | 10.2 | 20.6 |
| Cephalosporins of third generation | 7 | 223.5 | 27.0 | 34.8 | 38.4 |
| Cephalosporins of fourth generation | 2 | 48.7 | 17.8 | 24.3 | 30.9 |
| Lincosamides | 1 | 3.8 | – | – | – |
For each antimicrobial class, the total number of treatments applied during the study period in the 38 study farms is presented (n), as well as the total TI of all treatments with a drug of the respective class (in decreasing order of total TIDDD).
Treatment incidences for individual calf treatments with antimicrobials of the respective class are provided including their 25th percentile, median and 75th percentile.
DDDs and standard weight extracted from: ‘Defined daily doses for animals (DDDvet) and defined course doses for animals (DCDvet): European Surveillance of Veterinary Antimicrobial Consumption (ESVAC), and Revised ESVAC reflection paper on collecting data on consumption of antimicrobial agents per animal species, on technical units of measurement and indicators for reporting consumption of antimicrobial agents in animals’ (www.ema.europa.eu).
Table 2.
Antimicrobial treatment incidence (TI) for 1864 treatments in defined daily doses (DDDs) per treated calf and year (TIDDD)a grouped by antimicrobial drug according to the method of the EMA
| Antimicrobial drug | n | Total TIDDD | Calf level TIDDD per year | ||
|---|---|---|---|---|---|
| 25th percentile | median | 75th percentile | |||
| Chlortetracycline | 309 | 12327.6 | 29.7 | 35.2 | 43.7 |
| Amoxicillin | 223 | 5992.3 | 19.0 | 27.1 | 32.6 |
| Doxycycline | 77 | 4138.1 | 25.0 | 30.4 | 75.7 |
| Sulfadimidine | 278 | 3626.0 | 7.8 | 11.9 | 16.1 |
| Oxytetracycline | 207 | 3380.4 | 6.7 | 9.3 | 20.8 |
| Trimethoprim | 60 | 2462.6 | 29.8 | 38.9 | 45.3 |
| Tulathromycin | 46 | 2215.9 | 25.1 | 27.2 | 33.0 |
| Spiramycin | 91 | 2014.4 | 15.6 | 17.0 | 20.3 |
| Florfenicol | 111 | 1659.9 | 6.7 | 11.3 | 17.9 |
| Procaine benzylpenicillin | 64 | 1392.1 | 12.0 | 17.9 | 28.7 |
| Tylosin | 222 | 1331.4 | 2.4 | 3.3 | 5.2 |
| Phthalylsulfathiazole | 56 | 1152.8 | 14.3 | 19.3 | 21.7 |
| Danofloxacin | 19 | 741.9 | 11.9 | 23.2 | 35.4 |
| Dehydrostreptomycin | 30 | 460.8 | 8.0 | 10.3 | 21.2 |
| Marbofloxacin | 31 | 424.7 | 1.9 | 7.4 | 20.3 |
| Ceftiofur | 7 | 223.5 | 27.0 | 34.8 | 38.4 |
| Tilmicosin | 13 | 97.6 | 6.2 | 7.4 | 8.6 |
| Sulfaguanidine | 3 | 81.2 | 0.5 | 0.5 | 40.4 |
| Amoxicillin + clavulanic acid | 3 | 79.9 | 7.9 | 8.9 | 36.5 |
| Cefquinome | 2 | 48.7 | 17.8 | 24.3 | 30.9 |
| Sulfadiazine | 3 | 40.2 | 4.1 | 5.0 | 18.4 |
| Sulfadoxine | 2 | 37.2 | 12.5 | 18.6 | 24.7 |
| Benzathine benzylpenicillin | 2 | 16.9 | 7.1 | 8.5 | 9.8 |
| Enrofloxacin | 1 | 15.1 | – | – | – |
| Sulfamethoxypyridazin | 1 | 9.9 | – | – | – |
| Lincomycin | 1 | 3.8 | – | – | – |
| Spectinomycin | 1 | 3.5 | – | – | – |
| Neomycin | 1 | 2.6 | – | – | – |
For each antimicrobial drug, the total number of treatments applied during the study period in the 38 study farms is presented (n), as well as the total TI of all treatments with a respective drug (in decreasing order of total TIDDD).
Treatment incidences for individual calf treatments with the respective drug are provided including their 25th percentile, median and 75th percentile.
See Table 1 for footnotes.
Figure 1.
AMU in 38 veal-fattening operations, calculated as treatment incidence in defined daily doses (TIDDD) according to the method of the EMAa. Treatments were grouped by week of the fattening period, for antimicrobial classesb and overall use. aSee Table 1 for footnotes. bClasses with <10 treatments are not shown separately but appear in total TI. ◊Beginning of the fattening period (Week 1). Age of the calves was 4.6 (3.7/5.4) weeks (median, 25th/75th quantiles). *Timepoint of treatment: calves were treated at Week 4.4 (3.3/5.4) of the fattening period. ‡Timepoint of swabbing: calves were swabbed at Week 14.3 (12.3/16.9) of the fattening period. §Duration of the fattening period: 17.0 (15.0/19.0) weeks.
A total of 1582 E. coli, 1059 P. multocida and 315 M. haemolytica were isolated. The most frequent drugs bacteria were resistant to were tetracycline (51.4%), sulfamethoxazole (50.5%) and ampicillin (38.9%) for E. coli; oxytetracycline (74.2%) and spectinomycin (58.1%) for P. multocida; and tilmicosin (16.7%), spectinomycin (3.5%) and danofloxacin (2.9%) for M. haemolytica. Detailed information on clinical breakpoints and susceptibility testing results are listed in Tables S1–S3. Proportions of resistant isolates plotted against the number of antimicrobial treatments are given in Table 3.
Table 3.
Proportion of isolates exhibiting a minimum of one resistance to an antimicrobial drug (=resistant isolate) plotted against the number of antimicrobial treatments for E. coli isolated from rectal swabs, as well as P. multocida and M. haemolytica isolated from nasopharyngeal swabs
| Number of antimicrobial treatments | Proportion of resistant isolates (%) | ||
|---|---|---|---|
| E. coli | P. multocida | M. haemolytica | |
| 0 | 57.7 | 69.6 | 11.8 |
| 1 | 62.1 | 77.0 | 18.9 |
| >1 | 71.7 | 84.9 | 35.6 |
Number of antimicrobial treatments were categorized as none, one, or more than one antimicrobial treatment.
Associations of AMU with AMR were observed for E. coli, P. multocida and M. haemolytica. For E. coli at the level of the individual calf, the odds of being resistant were increased, albeit not significantly so, with increased use of antimicrobials (as number of treatments >1, OR 1.36, P = 0.066; Table 4). Use of tetracyclines was associated with resistance to tetracycline (OR 1.86, P < 0.001) and use of penicillins was associated with resistance to ampicillin (OR 1.66, P = 0.014; Table 5). For P. multocida, use of aminoglycosides was non-significantly associated with resistance to spectinomycin (OR 3.66, P = 0.074). For M. haemolytica, as for E. coli, the odds of being resistant were increased with increased use of antimicrobials (as number of treatments >1, OR 4.63, P < 0.001; Table 4). Animals treated with tetracyclines had a significantly higher odds of carrying resistant isolates of M. haemolytica (OR = 6.49, P < 0.001), but it was not possible to estimate an effect for heterologous treatment, because none of the animals that had received heterologous treatment carried isolates resistant to tetracycline.
Table 4.
Results of the generalized linear models with a logit link function on individual isolates of E. coli isolated from rectal swabs and M. haemolytica isolated from nasopharyngeal swabs exhibiting at least one resistance to an antimicrobial in Swiss veal calves on 38 farms
| Predictors | OR | CI | P |
|---|---|---|---|
| E. coli | |||
| (Intercept) | 1.41 | 1.04–1.93 | 0.029 |
| Number of treatments = 1 | 1.24 | 0.88–1.77 | 0.223 |
| Number of treatments >1 | 1.36 | 0.98–1.90 | 0.066 |
| P. multocida | |||
| (Intercept) | 4.91 | 2.12–11.36 | <0.001 |
| Number of treatments = 1 | 1.15 | 0.62–2.13 | 0.647 |
| Number of treatments >1 | 1.12 | 0.62–2.00 | 0.713 |
| M. haemolytica | |||
| (Intercept) | 0.12 | 0.07–0.22 | <0.001 |
| Number of treatments = 1 | 1.82 | 0.70–4.69 | 0.217 |
| Number of treatments >1 | 4.63 | 2.40–8.91 | <0.001 |
Number of antimicrobial treatments of calves carrying the respective isolate were categorized as no treatment (reference), one, or more than one antimicrobial treatment.
Table 5.
Results of the generalized linear models with a logit link function on E. coli isolated from rectal swabs and P. multocida isolated from nasopharyngeal swabs exhibiting resistance to a given antimicrobial drug when treated with a drug from the same antimicrobial class (homologous treatment) or not (heterologous treatment)
| Predictors | OR | CI | P |
|---|---|---|---|
| E. coli resistance to tetracycline | |||
| (Intercept) | 0.81 | 0.58–1.15 | 0.236 |
| Treatment(s) not containing tetracyclines | 1.05 | 0.74–1.48 | 0.803 |
| Treatment(s) containing tetracyclines | 1.86 | 1.34–2.58 | <0.001 |
| E. coli resistance to ampicillin | |||
| (Intercept) | 0.46 | 0.31–0.66 | <0.001 |
| Treatment(s) not containing penicillins | 1.14 | 0.83–1.57 | 0.420 |
| Treatment(s) containing penicillins | 1.66 | 1.11–2.48 | 0.014 |
| P. multocida resistance to spectinomycin | |||
| (Intercept) | 0.81 | 0.35–1.86 | 0.614 |
| Treatment(s) not containing aminoglycosides | 0.91 | 0.59–1.39 | 0.663 |
| Treatment(s) containing aminoglycosides | 3.66 | 0.88–15.21 | 0.074 |
Bacteria were isolated and treatments were recorded at the calf level on 38 Swiss veal calf farms.
Reference: ‘untreated’.
None of the machine learning models resulted in a high predictability whether an animal had been treated or not, based on the resistance pattern at swabbing (No-Information-Rate = 0.616; AccuracyRandomForest = 0.626, P value = 0.360; AccuracyElasticNet = 0.608, P value = 0.643; AccuracySupportVectorMachine = 0.623, P value = 0.399). In addition, the variable ‘group’ (i.e. management system) was not significantly associated with the outcome.
Discussion
In the present study, AMU and AMR data from 38 farms and 2956 bacterial isolates, respectively, were analysed using an innovative approach for associations at the individual animal level. Bacterial resistance in the commensal flora of veal calves has only been assessed in a few studies, at the level of the farm or the country.5,8–10 Two further studies where associations of AMU with AMR were investigated were conducted in dairy calves and fattening steers.23,24 In contrast to the latter studies, a modified approach was chosen in the present study. Here, antimicrobial treatment modalities were not defined by an experimental protocol, but treatment decisions were at the farm veterinarian’s discretion. In this regard, the experimental setting was unregulated and antimicrobial treatments were administered in real life, independently of the study. This methodological difference is of importance for the discussion of our results. Robust homologous associations of AMU and AMR at the animal level were detected in dairy calves and fattening steers when treatment regimens were restricted to a single drug or antimicrobial class (31 calves and 370 steers, respectively).23,24 In the present study, swabbing was performed at the end of the fattening period to investigate the prevalence of resistant bacteria before the calves entered the downstream food supply chain, where such bacteria may represent a threat to public health. Most calves, however, received treatment considerably earlier in the fattening period than before slaughter. Presence of resistant bacteria is known to be transient in many cases and best detectable a few days to weeks post-treatment.24 As the calves were swabbed after approximatively 10 weeks post-treatment, the selection pressure may already have decreased and the bacterial community almost re-established pre-treatment equilibrium at that time.
In one of the studies mentioned above,23 dairy calves had been randomly assigned to milk containing penicillin G or to a negative control, and a highly significantly reduced susceptibility was observed during treatment in commensal gut bacteria of the treated calves. Similarly, steers having received florfenicol subcutaneously carried more faecal E. coli with resistance to chloramphenicol than controls for several weeks post-treatment.24 In contrast, in a study at the animal level in steers, no associations were detected between AMU and AMR when using a treatment protocol including long-acting oxytetracycline or tilmicosin (depending on the body temperature at arrival at the feedlot), and ceftiofur in case of disease during the fattening process.25 Nonetheless, the authors still suspected an association of AMU with AMR but hold the study design responsible for the fact that no such effect could be detected, as the total TI, not TI at the antimicrobial class or drug level, was used as a predictor variable. In the present study, TI was recorded in more detail, namely at the level of the individual calf. Contradictory findings, i.e. presence or absence of associations between AMU and AMR, have been reported in studies at the farm and national levels in veal calves.8–10,26 This underlines the importance of further research in veal calf farms.
Between 185 and 979 samples were included in previous studies,6,8,9,15,23,27,28 whereas considerably higher numbers of antimicrobial-susceptible and -resistant isolates from treated and non-treated calves were available in the present study (1582 E. coli isolates, 1059 P. multocida and 315 M. haemolytica). Approximatively 12% of the enrolled calves were not sampled due to practical constraints. Calves that died suddenly could have died due to an infection with resistant bacteria and therefore differ systematically from the rest of the population. However, others described pneumonia to be responsible for less than a third of deaths.29 Also, calves that were not swabbed due to sudden death were present on most farms (34 out of 39 farms).
Our findings suggest that different selection mechanisms were present in the veal fattening operations under study. This is in line with the results of analyses of another dataset from the same veal calves, where associations of age and management factors with AMR were investigated at the herd level; a lower AMR prevalence was observed in farms where the effects of risk factors for high AMR were mitigated through thorough changes in management procedures.30 In the present study, investigating associations in logistic regression models allowed for detection of several significant associations between AMU and AMR, as well as for associations close to the significance level at the individual calf level. Interdependency of the calves belonging to the same farm was corrected for by adding a random farm effect to the models. A small to substantial intraclass correlation coefficient of 0.05–0.65 was observed in the models. Likely, besides varying treatment modalities, direct transfer of bacteria between calves and ingestion from the inanimate environment also occurred to varying degrees on the different farms. Models where farm was not added as a random effect showed more significant association for homologous association of AMU–AMR in E. coli (for sulphonamides, OR 2.63, P < 0.001; for diaminopyrimidines, OR 0.05, P < 0.001, data not shown). Similarly, in P. multocida, models without random farm effect showed significant association of AMR with the number of antimicrobial treatments (1 treatment, OR 1.46, P = 0.09; >1 treatment, OR 2.45; P < 0.001, reference is ‘untreated’). Homologous association was significant for tetracyclines (OR 2.90, P < 0.001). Negative associations of AMU and AMR have been described in humans and different animal species, where a complex interplay of resistance determinants has been suspected.31–33
Retrospective estimation of treatment intensity through swabbing and bacterial susceptibility testing at the end of the fattening period would have been an easy way to monitor AMU and AMR in veal farms, which made this hypothesis worth testing. None of the tested machine learning methods was able to provide a reliable prediction for whether an animal had been treated or not based on the pattern of resistances at the end of the fattening period. The most likely cause for this result lies with the fact that the time interval from treatment to sampling was too long and, as mentioned above, selection pressure had already abated by the time of swabbing. Unfortunately, despite the relatively large sample size in comparison with other studies, the chosen machine learning approaches were ineffective. The data were also checked for evidence of clustering of resistant bacteria within farms, but this was not the case (data not shown).
In studies where associations of AMU with AMR in E. coli are investigated, different techniques for species identification can be used. In the past, the most commonly used procedures were based on biochemical tests.6,8,15,23,27,28,34 Species identification by MALDI-TOF including a second MALDI-TOF for species confirmation after purification as performed in the present study led to reliable identification of the bacteria under study. Also, susceptibility test procedures were frequently conducted using disc diffusion in the past.8,23,28,35 In the present study, microdilution was chosen to obtain more precise results. Detailed data on AMU were collected prospectively in a labour-intensive observational setting, in contrast to other studies where AMU data were retrieved through an interviewed person.6,9,10 Analysing data at national level may allow for including more farms; however, the quality of the data may be lower, especially regarding attribution of single treatments to single animals. In the present study, farm visits were performed in a considerably higher frequency than in other veal calf studies, which may have resulted in higher data quality.11,36,37
In conclusion, these results show that associations of AMU and AMR can be detected on veal calf farms at the level of the individual animal using AMU and AMR data at the animal level. The presence of resistant bacteria at the end of the fattening period is associated with AMU during that period. Implications deriving from selection of antimicrobial-resistant bacteria must be considered whenever antimicrobial treatment is applied to animals. These selection processes underline the importance of alternative strategies to improve management practices in veal farms, in order to reinforce animal health, decrease disease incidence and thereby reduce the need for antimicrobial treatments, as it may allow for reducing the selection of resistant bacteria.
Supplementary Material
Acknowledgements
We thank all farmers for their participation in the study. We are grateful to Alexandra Collaud and Alexandra Rossano (Institute of Veterinary Bacteriology, Vetsuisse Faculty, University of Bern) for bacteriological analyses and MIC determination.
Contributor Information
Jens Becker, Clinic for Ruminants, Vetsuisse Faculty, University of Bern, Bremgartenstrasse 109a, 3012 Bern, Switzerland.
Vincent Perreten, Institute of Veterinary Bacteriology, Vetsuisse Faculty, University of Bern, Länggassstrasse 122, 3012 Bern, Switzerland.
Gertraud Schüpbach-Regula, Veterinary Public Health Institute, Vetsuisse Faculty, University of Bern, Schwarzenburgstrasse 161, 3097 Liebefeld, Switzerland.
Dimitri Stucki, Clinic for Ruminants, Vetsuisse Faculty, University of Bern, Bremgartenstrasse 109a, 3012 Bern, Switzerland.
Adrian Steiner, Clinic for Ruminants, Vetsuisse Faculty, University of Bern, Bremgartenstrasse 109a, 3012 Bern, Switzerland.
Mireille Meylan, Clinic for Ruminants, Vetsuisse Faculty, University of Bern, Bremgartenstrasse 109a, 3012 Bern, Switzerland.
Funding
The study was financed by the National Research Program ‘Antimicrobial Resistance’ (NRP 72) of the Swiss National Science Foundation (Grant no. 407240_167083), IP-SUISSE and the Migros Genossenschaftsbund, as well as the Swiss Federal Office of Agriculture.
Transparency declarations
None of the people, institutions or organizations mentioned above was involved in the study design or the interpretation of the results.
Supplementary data
Tables S1 to S3 are available as Supplementary data at JAC Online.
References
- 1. WHO . Antimicrobial Resistance Global Report on Surveillance. 2014. https://apps.who.int/iris/rest/bitstreams/515657/retrieve.
- 2. Lava M, Schüpbach-Regula G, Steiner Aet al. Antimicrobial drug use and risk factors associated with treatment incidence and mortality in Swiss veal calves reared under improved welfare conditions. Prev Vet Med 2016; 126: 121–30. [DOI] [PubMed] [Google Scholar]
- 3. Becker J, Schüpbach-Regula G, Steiner Aet al. Effects of the novel concept ‘outdoor veal calf’ on antimicrobial use, mortality and weight gain in Switzerland. Prev Vet Med 2020; 176: 1–38. [DOI] [PubMed] [Google Scholar]
- 4. ARCH-Vet . Rapport sur les ventes d’antibiotiques et l’antibiorésistance en médecine vétérinaire en Suisse. 2018. https://www.blv.admin.ch/blv/fr/home/tiere/tierarzneimittel/antibiotika/vertrieb.html.
- 5. Schönecker L, Schnyder P, Overesch Get al. Associations between antimicrobial treatment modalities and antimicrobial susceptibility in Pasteurellaceae and E. coli isolated from veal calves under field conditions. Vet Microbiol 2019; 236: 108363. [DOI] [PubMed] [Google Scholar]
- 6. Di Labio E, Regula G, Steiner Aet al. Antimicrobial resistance in bacteria from Swiss veal calves at slaughter. Zoonoses Public Health 2007; 54: 344–52. [DOI] [PubMed] [Google Scholar]
- 7. Bosman AB, Wagenaar JA, Stegeman JAet al. Antimicrobial resistance in commensal Escherichia coli in veal calves is associated with antimicrobial drug use. Epidemiol Infect 2014; 142: 1893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gay E, Bour M, Cazeau Get al. Antimicrobial usages and antimicrobial resistance in commensal Escherichia coli from veal calves in France: evolution during the fattening process. Front Microbiol 2019; 10: 792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ceccarelli D, Hesp A, Van Der Goot Jet al. Antimicrobial resistance prevalence in commensal Escherichia coli from broilers, fattening turkeys, fattening pigs and veal calves in European countries and association with antimicrobial usage at country level. J Med Microbiol 2020; 69: 537–47. [DOI] [PubMed] [Google Scholar]
- 10. Yang D, Van Gompel L, Luiken RECet al. Association of antimicrobial usage with faecal abundance of aph(3′)-III, ermB, sul2 and tetW resistance genes in veal calves in three European countries. Int J Antimicrob Agents 2020; 56: 106131. [DOI] [PubMed] [Google Scholar]
- 11. Catry B, Dewulf J, Maes Det al. Effect of antimicrobial consumption and production type on antibacterial resistance in the bovine respiratory and digestive tract. PLoS One 2016; 11: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moser L, Becker J, Schüpbach-Regula Get al. Welfare assessment in calves fattened according to the “outdoor veal calf” concept and in conventional veal fattening operations in Switzerland. Animals 2020; 10: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Becker J, Steiner A, Meylan Met al. Vergleichende Wirtschaftlichkeitsanalyse des Kälbermastsystems «Freiluftkalb» und der konventionellen IP-SUISSE-Labelmast [Comparative economic analysis of the new concept for veal calf fattening «outdoor veal calf» and the conventional IP-SUISSE label]. Schweiz Arch Tierheilkd 2021; 163: 203–17. [DOI] [PubMed] [Google Scholar]
- 14. IP-SUISSE . Guidelines for Animal Husbandry. 2019: 13–5. https://www.ipsuisse.ch/richtlinien-tierhaltung/.
- 15. Catry B, Haesebrouck F, De Vliegher Set al. Variability in acquired resistance of pasteurella and mannheimia isolates from the nasopharynx of calves, with particular reference to different herd types. Microb Drug Resist 2005; 11: 387–94. [DOI] [PubMed] [Google Scholar]
- 16. EUCAST . Breakpoint tables for interpretation of MICs and zone diameters. Version 12 2022. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_12.0_Breakpoint_Tables.pdf.
- 17. CLSI . Performance Standards for Antimicrobial Susceptibility Testing—Thirty-First Edition: M100. 2021. [Google Scholar]
- 18. EMA . Revised ESVAC reflection paper on collecting data on consumption of antimicrobial agents per animal species, on technical units of measurement and indicators for reporting consumption of antimicrobial agents in animals. EMA/286416/2012-Rev1 2013; 44: 1–29. https://www.ema.europa.eu/en/documents/scientific-guideline/revised-european-surveillance-veterinary-antimicrobial-consumption-esvac-reflection-paper-collecting_en.pdf. [Google Scholar]
- 19. EMA . Defined daily doses for animals (DDDvet) and defined course doses for animals (DCDvet): European Surveillance of Veterinary Antimicrobial Consumption (ESVAC). Eur Surveill Vet Antimicrob Consum 2016; 44: 13–8. http://www.ema.europa.eu/docs/en_GB/document_library/Other/2016/04/WC500205410.pdf. [Google Scholar]
- 20. Kasabova S, Hartmann M, Werner Net al. Used daily dose vs. defined daily dose-contrasting two different methods to measure antibiotic consumption at the farm level. Front Vet Sci 2019; 6: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grave K, Greko C, Nilsson Let al. The usage of veterinary antibacterial drugs for mastitis in cattle in Norway and Sweden during 1990-1997. Prev Vet Med 1999; 42: 45–55. [DOI] [PubMed] [Google Scholar]
- 22. Jensen VF, Jacobsen E, Bager F. Veterinary antimicrobial-usage statistics based on standardized measures of dosage. Prev Vet Med 2004; 64: 201–15. [DOI] [PubMed] [Google Scholar]
- 23. Langford FM, Weary DM, Fisher L. Antibiotic resistance in gut bacteria from dairy calves: A dose response to the level of antibiotics fed in milk. J Dairy Sci 2003; 86: 3963–6. [DOI] [PubMed] [Google Scholar]
- 24. Berge ACB, Epperson WB, Pritchard RH. Assessing the effect of a single dose florfenicol treatment in feedlot cattle on the antimicrobial resistance patterns in faecal Escherichia coli. Vet Res 2005; 36: 723–34. [DOI] [PubMed] [Google Scholar]
- 25. Checkley SL, Campbell JR, Chirino-Trejo Met al. Antimicrobial resistance in generic fecal Escherichia coli obtained from beef cattle on arrival at the feedlot and prior to slaughter, and associations with volume of total individual cattle antimicrobial treatments in one western Canadian feedlot. Can J Vet Res 2008; 72: 101–8. [PMC free article] [PubMed] [Google Scholar]
- 26. Cassini A, Högberg LD, Plachouras Det al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis 2019; 19: 56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Berge ACB, Moore DA, Sischo WM. Field trial evaluating the influence of prophylactic and therapeutic antimicrobial administration on antimicrobial resistance of fecal Escherichia coli in dairy calves. Appl Environ Microbiol 2006; 72: 3872–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jarrige N, Cazeau G, Bosquet Get al. Effects of antimicrobial exposure on the antimicrobial resistance of Escherichia coli in the digestive flora of dairy calves. Prev Vet Med 2020; 185: 105177. [DOI] [PubMed] [Google Scholar]
- 29. Lava M, Pardon B, Schüpbach-Regula Get al. Effect of calf purchase and other herd-level risk factors on mortality, unwanted early slaughter, and use of antimicrobial group treatments in Swiss veal calf operations. Prev Vet Med 2016; 126: 81–8. [DOI] [PubMed] [Google Scholar]
- 30. Becker J, Perreten V, Steiner Aet al. Antimicrobial susceptibility in E. coli and Pasteurellaceae at the beginning and at the end of the fattening process in veal calves: comparing ‘outdoor veal calf’ and conventional operations. Vet Microbiol 2022; 269: 109419. [DOI] [PubMed] [Google Scholar]
- 31. Saini V, McClure JT, Scholl DTet al. Herd-level relationship between antimicrobial use and presence or absence of antimicrobial resistance in Gram-negative bovine mastitis pathogens on Canadian dairy farms. J Dairy Sci 2013; 96: 4965–76. [DOI] [PubMed] [Google Scholar]
- 32. Lai CC, Wang CY, Chu CCet al. Correlation between antibiotic consumption and resistance of Gram-negative bacteria causing healthcare-associated infections at a university hospital in Taiwan from 2000 to 2009. J Antimicrob Chemother 2011; 66: 1374–82. [DOI] [PubMed] [Google Scholar]
- 33. Sali V, Nykäsenoja S, Heikinheimo Aet al. Antimicrobial use and susceptibility of indicator Escherichia coli in finnish integrated pork production. Front Microbiol 2021; 12: 3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hordijk J, Mevius DJ, Kant Aet al. Within-farm dynamics of ESBL/AmpC-producing Escherichia coli in veal calves: A longitudinal approach. J Antimicrob Chemother 2013; 68: 2468–76. [DOI] [PubMed] [Google Scholar]
- 35. Berge ACB, Atwill ER, Sischo WM. Animal and farm influences on the dynamics of antibiotic resistance in faecal Escherichia coli in young dairy calves. Prev Vet Med 2005; 69: 25–38. [DOI] [PubMed] [Google Scholar]
- 36. Schnyder P, Schönecker L, Schüpbach-Regula Get al. Effects of management practices, animal transport and barn climate on animal health and antimicrobial use in Swiss veal calf operations. Prev Vet Med 2019; 167: 146–57. [DOI] [PubMed] [Google Scholar]
- 37. Pardon B, Catry B, Dewulf Jet al. Prospective study on quantitative and qualitative antimicrobial and anti-inflammatory drug use in white veal calves. J Antimicrob Chemother 2012; 67: 1027–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.