Original citation: J Clin Invest. 2022;132(7):e148667. https://doi.org/10.1172/JCI148667
Citation for this corrigendum: J Clin Invest. 2022;132(19):e165107. https://doi.org/10.1172/JCI165107
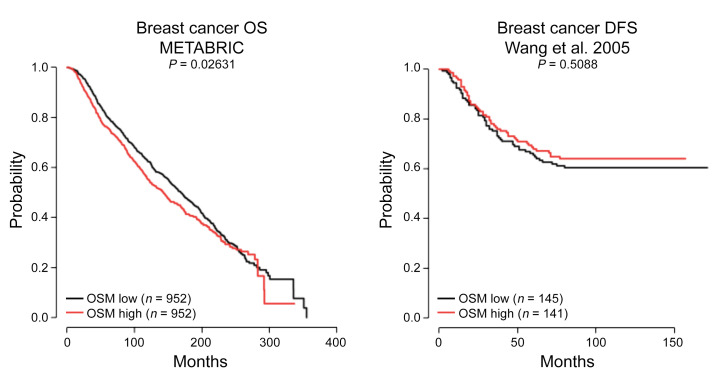
The authors recently became aware that, following the recent update of Cancertool (http://genomics.cicbiogune.es/CANCERTOOL/index.html), the two panels included in Supplemental Figure 3B needed to be modified. These panels included data downloaded from Cancertool reporting the prognostic potential of OSM in two independent breast cancer patient datasets, namely METABRIC (PMID 22522925) and Wang et al. (PMID 15721472). Based on the results from the updated Cancertool data, we cannot conclude that OSM is shown to exhibit prognostic potential in the Wang dataset. In addition, the analysis of the METABRIC dataset presented in Supplemental Figure 3B pertains to overall survival, but data were inadvertently reported as showing disease-free survival. The corrected panels are shown below. Corrected sentences referring to Supplemental Figure 3B data in Results and Methods are also shown below. The online supplemental file has been updated.
Results
We also observed that increased OSM mRNA levels associated with decreased overall survival in the METABRIC (19) breast cancer dataset (Supplemental Figure 3B). Of note, the prognostic analysis of OSM in the Wang (20) dataset did not reveal a significant association with disease-free survival (Supplemental Figure 3B).
Methods
Gene expression analyses of clinical datasets and bioinformatics analyses. Overall survival (OS) and disease-free survival (DFS) data for breast cancer patients based on OSM mRNA expression in the METABRIC (19) and Wang (20) datasets were analyzed and represented using the CANCERTOOL interface (59) in combination with cBioPortal (www.cbioportal.org).
The authors regret the errors.
Version 1. 09/28/2022
Electronic publication
Footnotes
See the related article at Stromal Oncostatin M cytokine promotes breast cancer progression by reprogramming the tumour microenvironment.