Abstract
The presence of SARS-CoV-2 in untreated sewage has been confirmed in many countries but its incidence and infection risk in contaminated waters is poorly understood. The River Thames in the UK receives untreated sewage from 57 Combined Sewer Overflows (CSOs), with many discharging dozens of times per year. This study investigated if such discharges provide a pathway for environmental transmission of SARS-CoV-2. Samples of wastewater, surface water, and sediment collected close to six CSOs on the River Thames were assayed over eight months for SARS-CoV-2 RNA and infectious virus. Bivalves were also sampled as an indicator species of viral bioaccumulation. Sediment and water samples from the Danube and Sava rivers in Serbia, where raw sewage is also discharged in high volumes, were assayed as a positive control. No evidence of SARS-CoV-2 RNA or infectious virus was found in UK samples, in contrast to RNA positive samples from Serbia. Furthermore, this study shows that infectious SARS-CoV-2 inoculum is stable in Thames water and sediment for <3 days, while SARS-CoV-2 RNA is detectable for at least seven days. This indicates that dilution of wastewater likely limits environmental transmission, and that detection of viral RNA alone is not an indication of pathogen spillover.
Keywords: SARS-CoV-2, London, Combined sewer overflows, Faecal pollution, Untreated wastewater
Graphical abstract
1. Introduction
Early detection of the severe acute respiratory syndrome coronavirus SARS-CoV-2 is essential to contain community outbreaks of COVID-19 (Dhama et al., 2020). While the primary route of viral transmission between humans is via exposure to respiratory fluids carrying infectious SARS-CoV-2 (Zhang et al., 2020; Zhou et al., 2021), evidence of faecal-oral transmission has raised concerns regarding possible environmental transmission to humans and wildlife through spillover from sewage (Kitajima et al., 2020; Thakur et al., 2021; Tran et al., 2021). Numerous studies have recorded SARS-CoV-2 in feces at up to 107 genome copies/mL (reviewed by Jones et al. (2020)) and infectious SARS-CoV-2 has been isolated from feces and urine (Sun et al., 2020; Xiao et al., 2020). Faulty sewerage systems have previously been linked to an earlier SARS-CoV-1 (another highly pathogenic human coronavirus) outbreak (Peiris et al., 2003), and the presence and infectious potential of other coronaviruses in water and sewage ranges from days to weeks (Casanova et al., 2009). Together, these studies indicate transmission of SARS-CoV-2 via sewage is a potential concern for SARS-CoV-2 outbreaks (Sharif et al., 2021). Numerous reviews have discussed the possibility of SARS-CoV-2 transmission to humans from exposure to raw sewage or waters receiving untreated or inadequately treated wastewater, however these have been based on limited empirical evidence (Ahmed et al., 2021) and the ability of enveloped viruses to remain infectious in wastewater is still debated (Wurtzer et al., 2021).
Like other coronaviruses, SARS-CoV-2 RNA has been detected in wastewater in multiple countries (reviewed by Kitajima et al. (2020)), and genome concentrations have correlated positively with the number of human cases within the catchment (Fitzgerald et al., 2021). Wastewater has also been used as a proxy for tracking circulating SARS-CoV-2 variants (Amman et al., 2022), indicating that wastewater-based epidemiology is an efficient way to monitor SARS-CoV-2 dynamics in human populations at large scales. SARS-CoV-2 RNA has also been found in rivers, due to inadequate wastewater treatment or sewage spillover prior to treatment (Guerrero-Latorre et al., 2020; Kolarević et al., 2021; Rimoldi et al., 2020), indicating a potential route for transmission of SARS-CoV-2 to humans and wildlife, particularly in urban areas. While one study has unsuccessfully attempted to isolate infectious SARS-CoV-2 from river water (Rimoldi et al., 2020), there is a paucity of data on infectious SARS-CoV-2 in river water in regions with high disease prevalence.
The River Thames is the UK's second longest river, with a catchment covering over 16,000 km2 (Richardson and Soloviev, 2021). Its Greater London area houses about 14 million people (Whitehead et al., 2013) – one fifth of the entire UK population – with many more visiting the area daily, and it provides about 2/3 of London's water supplies (Greater London Authority, 2011). The Thames supports many species of wildlife and is also used for recreation, which brings humans, potential hosts and animal vectors of disease into close contact. It also acts as the outlet for 57 Combined Sewer Overflows (CSOs; (Munro et al., 2019)), which release both raw and processed sewage directly into the river. These overflows are designed to reduce the risk of sewage flooding homes and businesses and, although they operate throughout the year, they are used particularly during periods of heavy rain in the winter (Richardson and Soloviev, 2021), when SARS-CoV-2 is also at its seasonal peak in the human population (Nichols et al., 2021). Although recent improvements to the Thames sewerage network have reduced sewage discharges from around 40 million tonnes in 2011 to 18 million tonnes per year (Richardson and Soloviev, 2021), individual sewage works are still discharging 3.5 billion L of untreated sewage a year, with occasions during the initial pandemic in 2020 of >1 billion L being released in one day (House of Commons Environmental Audit Committee, 2022).
CSO discharges can increase human health risks, with polluted waters providing transmission routes for enteric pathogens, either through direct exposure (e.g. through swimming, angling or boating) or through the consumption of contaminated foods (e.g. riverine fishes and shellfish) (Potasman et al., 2002). Although the urban Thames itself is below the water quality standards required to merit formal bathing water status, it is still used by many people for this purpose, as are many of the ponds and lakes within the catchment area, including the popular Hampstead Heath Bathing Ponds, which saw over 120,000 visitors over nine weeks in summer 2020 (Hampstead Heath Annual Report 2020/21 www.cityoflondon.gov.uk, 2021). The latter were forced to close in September 2020 after a sewage surcharge, and again in October 2020 after high levels of Enterococci were found in the water (Hampstead Heath Annual Report 2020/21 www.cityoflondon.gov.uk, 2021). Neither these ponds nor the River Thames have thus far been tested for the presence of SARS-CoV-2. Along with measuring bacteria in water samples to assess water quality (e.g. Escherichia coli and intestinal enterococci; (DEFRA, 2021)), filter-feeding bivalves are often used as indicators of water quality as they concentrate micro-organisms, including viruses, in their tissues (Fiorito et al., 2019) as well as posing a potential risk to human health if ingested (EFSA, 2019). Porcine epidemic diarrhoea virus, used as a surrogate for SARS-CoV-2, and heat inactivated SARS-CoV-2 have recently been shown to contaminate bivalves in laboratory trials, highlighting the importance of testing these filter-feeders for SARS-CoV-2 in the wild, as biosensors and possible transmission routes to other wildlife and humans (Desdouits et al., 2021).
This study investigated whether both SARS-CoV-2 RNA and/or infectious particles can be detected from running and standing surface waters and sediments in the Thames catchment, including the river itself and the Hampstead Heath bathing ponds. Bivalve samples were also collected adjacent to major CSOs, and the presence of the human gut bacteriophage crAssphage was used as evidence of sewage contamination (Kongprajug et al., 2019). These surveys were compared to the Sava and Danube rivers in Serbia, which receive large quantities of raw sewage from the Serbian capital of Belgrade (1,700,000 inhabitants): only 13 % of collected municipal wastewaters are processed before release (Ministry of Environmental Protection, 2019). The goals of this study were (i) to test a novel methodology for concentrating and detecting the RNA and infectious particles of enveloped RNA viral pathogens from high volume water samples and (ii) to evaluate whether the London waterways studied here are viable conduits for SARS-CoV-2 transmission.
2. Materials and methods
2.1. Sampling sites and sample collection
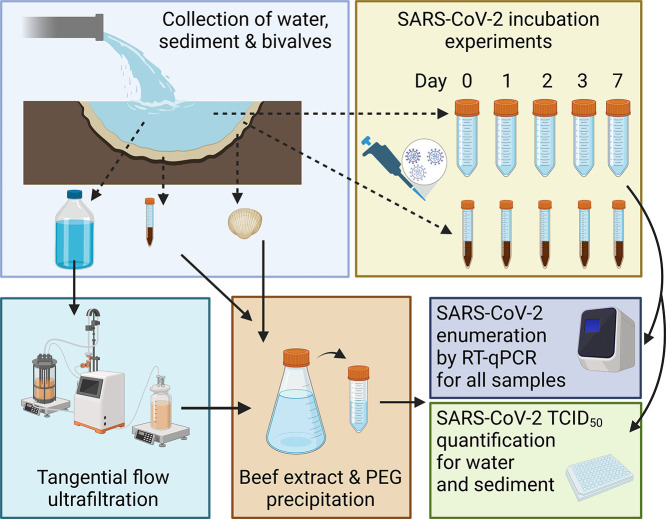
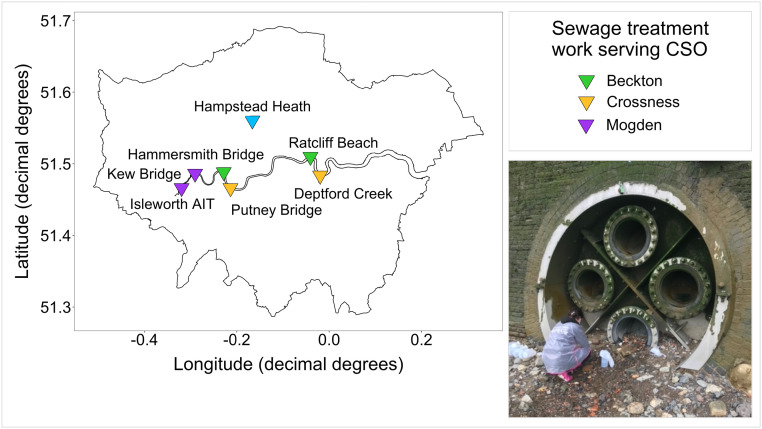
In the River Thames basin 38 water and 140 sediment samples were collected between the 14th of January and the 25th of August 2021, and tested for the presence of SARS-CoV-2 RNA. From March 17th 2021 any samples collected were also tested for infectious SARS-CoV-2 (n = 152). Sampling sites on the Thames were chosen based on case load estimates, accessibility and discharge rate (size of sewage works). The former were calculated using openly available data obtained from the UK government COVID-19 website (https://coronavirus.data.gov.uk/details/) classified by lower tier local authority level. Sampling sites covered 19 miles of the river, with two sites downstream of CSOs servicing each of London's three largest sewage treatment works (Fig. 1 ): Beckton (Hammersmith Bridge and Ratcliff Beach, Limehouse), Crossness (Putney Bridge and Deptford Creek) and Mogden (Isleworth AIT and Kew Bridge), with the former being one of the largest in Europe (Richardson and Soloviev, 2021). Wherever possible sampling was carried out during or within 24 h of a sewage discharge event from the CSOs, as determined by rower notification email alerts from Thames Water. Samples from Hampstead Heath were collected during the summer peak of case numbers in the Thames basin on June 21st and July 23rd 2021, from each of the three (Female, Male and Mixed) swimming ponds.
Fig. 1.
Map of London, showing the River Thames and the seven main sampling sites, coloured by the sewage treatment works that the CSO services. The image shows an example of the sampling effort: collecting samples from Ratcliff Beach CSO at low tide.
From each site on the Thames, at low tide, 10 L of wastewater were collected directly from discharging CSOs when possible. Where CSOs were inaccessible (Isleworth AIT and Kew Bridge), 10 L of surface water was collected immediately downstream of the CSO. Three samples of 250 g surface sediment (top 2 cm) were collected 1 m from the shore, at 0 m, 5 m and 10 m downstream of each CSO. Forty Asian Clam (Corbicula fluminea) samples of varying size were also collected from Isleworth AIT and Kew Bridge and 6 water and 12 sediment samples were collected from Hampstead Heath at the point of access to the three bathing ponds. Samples were processed on the day of collection.
In the metropolitan area of Belgrade, Serbia, an additional 17 samples (11 sediment and 6 surface water samples) were collected from the Sava and Danube rivers, between February 28th and April 21st 2021. These sites receive high volumes of unprocessed wastewater and were already found to contain SARS-CoV-2 RNA at concentrations of 5.97 × 103 to 1.32 × 104 copies/L in December 2020 (Kolarević et al., 2021). Samples were collected from six different sites across five weeks, with two samples collected the same week from two different sites (see supplementary Table 1). Samples were collected using the methodology described above for the Thames, from sites downstream of CSOs, except that only 1 L of water was collected due to transport restrictions. Samples from Serbia were processed within 4 days of collection (see supplementary materials for details of Serbian sites).
For quality assurance all samples were collected in sterilised containers, footwear was sterilised between sites and timepoints and full PPE was worn at all times to avoid cross-contamination of samples. Samples were transported in the dark to avoid UV degradation. Sampling dates for all sites can be seen in Fig. 2 , alongside caseload data for each region.
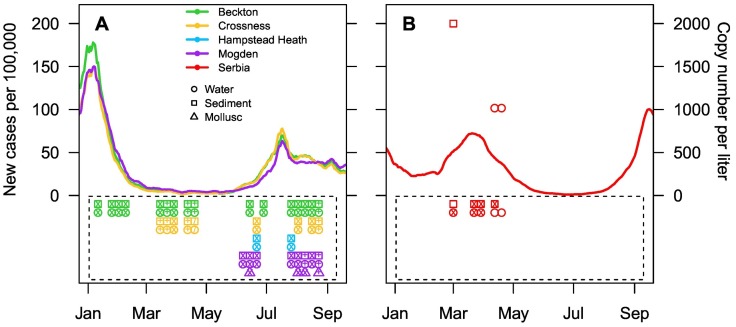
Fig. 2.
New cases of SARS-CoV-2 per 100,000 people in 2021 throughout the sampling period, in London, UK, by sewerage local authority: Beckton (green), Crossness (yellow) and Mogden (purple) (A) and in Serbia (red) (B), as data was not available for Belgrade alone. Copy number of the N1 gene per litre of water for sampling times in London (A) and in Belgrade (B) show positive RT-qPCR results for Serbian samples only (B). Samples contained in boxes were below the limit of detection. The symbols within the plotting characters represent the number of overlapping samples per character (clear = no overlap, two samples = x, three samples = +, and four or more samples = /). Overlapping samples could be samples collected from two sites in the same sewerage local authority (e.g. from Hammersmith Bridge and Ratcliff Beach for Beckton), or multiple collections in the same two week block.
2.2. Culture of SARS-CoV-2 and murine hepatitis virus controls
SARS-CoV-2 (SARS-CoV-2/England/IC19/2020; (McKay et al., 2020)) was provided by Professor Barclay's lab of Imperial College, London. Vero E6 cells were used for the propagation of SARS-CoV-2 using methods described previously (Case et al., 2020), with an amendment of Dulbecco's Modified Eagle Medium (DMEM) high glucose with 10 % Fetal Bovine Serum (FBS) for cell culture and viral propagation. Vero E6 cells were obtained from Sigma-Aldrich (St Louis, Missouri, USA). Murine hepatitis virus (MHV; strain: MHV-A59) and NCTC clone 1469 derivative cell line were obtained from the American Type Culture Collection (ATCC, Manassas, Virginia, USA). NCTC 1469 cells were used for propagation and infectivity assays of MHV with the manufacturers' protocol using DMEM high glucose with 10 % horse serum. All viral stocks were stored at −80 °C. Stock viruses were quantified using the TCID50 method as described previously (Harcourt et al., 2020; Hover et al., 2016; Lei et al., 2021), and via quantitative, reverse transcriptase, polymerase chain reaction (RT-qPCR) as described below.
2.3. Concentration of water and validation for viral detection
To maximise the probability of detection of SARS-CoV-2 from dilute CSO and river samples, 10 L of water samples from the Thames Basin, and 1 L of water samples from the Danube and Sava rivers, were concentrated using a tangential flow ultrafiltration (TFUF) - PEG precipitation technique for enteric viruses (Farkas et al., 2018), with one modification: TFUF concentrate (40 mL) was eluted and precipitated using PEG 8000, instead of PEG 6000. Detailed protocols are provided in the Supplementary Material. As this method has only been validated with RNA detection for non-enveloped viruses, for which >10 % of viral RNA was recovered across all experiments (Farkas et al., 2018), to validate it for SARS-CoV-2 infectivity, 10 L of Milli-Q water was spiked with MHV to a final concentration of 1.5 × 104 gc/L, and run through the TFUF. MHV belongs to the same genus as SARS-CoV-2, is structurally and morphologically similar (Gorbalenya et al., 2020), and has been shown to have similar decay rates in wastewater (Ahmed et al., 2020c), but it does not have the rigorous biosafety requirements necessary for working with infectious SARS-CoV-2. Ultraclean water was used for this validation to determine the specific impact of the TFUF-PEG concentration method on viral infectivity, rather than the effect of the specific sample type being tested. Two millilitres of sample was collected at the end of the process, filtered (0.22 μM) and 1.5 mL of sample was used to carry out a TCID50 infectivity assay, as above. TCID50/mL was converted to the expected concentration, and compared to values recovered in spiked water. Replicate runs (n = 3) showed the concentration of MHV after PEG precipitation to range from a maintenance of the original concentration in spiked water to 160× more concentrated than spiked water (see supplementary Table 2). While this is a large range in recovery efficiency (0.2–80 %, as calculated in (Ahmed et al., 2020a)), this method still amplifies viral signal in environmental samples, enhancing the detection of infectious SARS-CoV-2 above what has been attempted in previous studies (Rimoldi et al., 2020).
2.4. Preparation of sediment and bivalve samples for viral detection
Sediment samples were processed using beef extract elution (as in (Farkas et al., 2017)). Five grams of sediment were added to 15 mL of 3 % beef extract, 2 M sodium nitrate, pH 5.5. Solid matter was removed by centrifugation at 3000 rpm for 10 min and a PEG precipitation was carried out as above (Section 2.3; following (Farkas et al., 2018)).
The digestive tissue of bivalves was extracted following Desdouits et al. (2021). In brief, up to 2 g of homogenised digestive tissue was incubated in 1 mL 0.2 mg/mL proteinase K solution for 60 min at 37 °C followed by 15 min at 60 °C. The liquid phase was separated by centrifugation at 3000 rpm for 10 min. Supernatant was added to an equal volume of 2× DNA/RNA Shield and stored at −20 °C.
2.5. Detection of SARS-CoV-2 RNA and crAssphage DNA
Total DNA/RNA was extracted from water, sediment, and from bivalve and infectivity assay eluent (see Section 2.6), using an OT-2 Liquid Handling Robot (Opentron, Long Island City, New York, USA) and Quick-DNA/RNA Viral MagBead kits (Zymo Research, Irvine, CA, USA), following the manufacturer's protocol for DNA/RNA extraction from liquids. DNA/RNA was eluted in a final volume of 60 μL and stored at −20 °C. SARS-CoV-2 RNA was quantified by targeting the N1 and E genes (primer and probe sets shown in Table 1 ). Detailed protocols for RT-qPCR assays are provided in the Supplementary Information. In brief, RT-qPCR assays were carried out on a LightCycler 480 system (Roche Life Sciences, Basel, Switzerland). Standard curves were derived from commercial plasmid controls 2019-nCoV N Positive Control and 2019-nCoV E positive controls (Integrated DNA Technologies, Coralville, Iowa, USA) (Freire-Paspuel et al., 2021) and the limit of detection for SARS-CoV-2 was determined using a curve-fitting method (as in (Klymus et al., 2020)). All reactions were considered positive if the cycle threshold was below 40 cycles (as in (Randazzo et al., 2020)). SARS-CoV-2 viral loads were quantified as GC by plotting the Ct value to an external standard curve built with a tenfold serial dilution of plasmid control.
Table 1.
Primer and probe sequences for detection and quantification of SARS-CoV-2 (E and N1 genes), Murine hepatitis virus (MHV), and CrAssphage by RT-qPCR.
| Target | Name | Sequence (5′-3′) |
|---|---|---|
| E gene | E Sarbeco F | ACAGGTACGTTAATAGTTAATAGCGT |
| E Sarbeco R | ATATTGCAGCAGTACGCACACA | |
| E Sarbeco P | [6FAM]-ACACTAGCCATCCTTACTGCGCTTCG-[BHQ1] | |
| N1 gene | N1 Forward | GACCCCAAAATCAGCGAAT |
| N1 Reverse | TCTGGTTACTGCCAGTTGAATCTG | |
| N1 Probe | [6FAM]-ACCCCGCATTACGTTTGGTGGACC[BHQ1] | |
| MHV | Forward | GGAACTTCTCGTTGGGCATTATACT |
| Reverse | ACCACAAGATTATCATTTTCACAACATA | |
| Probe | [Cyanine5]-ACATGCTACGGCTCGTGTAACCGAACTGT [BHQ3] | |
| CrAssphage | CPQ56 Forward | CAGAAGTACAAACTCCTAAAAAACGTAGAG |
| CPQ65 Reverse | GATGACCAATAAACAAGCCATTAGC | |
| CPQ65 Probe | [6FAM]-AATAACGATTTACGTGATGTAAC[BHQ1] |
CrAssphage DNA was quantified in a selection of concentrated water samples from each sampling site on the Thames to confirm that the sites chosen contained human waste. CrAssphage detection in wastewater has been highlighted as a way to improve interpretations of wastewater surveillance data and detection of sewage in river water (Kongprajug et al., 2019), particularly as SARS-CoV-2: crAssphage DNA ratios have been found to be significantly associated with the number of positive tests per 10,000 individuals (Wilder et al., 2021). Reaction mixture concentrations and reaction conditions were as previously described for SARS-CoV-2 with the substitution of CrAssphage specific primers and probe (Table 1) and adjustment of annealing phase to 56 °C for 60 s.
To assess PCR inhibition from environmental samples, an internal amplification control was used in place of water in the qPCR mix as in Staley et al. (2012). The internal control was an MHV plasmid designed with primer sites complementary to the RT-qPCR primers (Table 1). All water and bivalve samples, and one sediment sample per site and timepoint were tested for inhibition. Five hundred target copies of MHV were added per reaction and all samples were run alongside three no template controls (Ahmed et al., 2020a), using the RT-qPCR assay described previously (Besselsen et al., 2002); see Table 1 for primers and probe sets. The expected C T value for amplification of the control in uninhibited samples was determined as the mean for all blanks, since they did not contain inhibitory compounds. Reactions were deemed inhibited if the C T value of the RNA sample was greater than three standard deviations of the average MHV C T (Staley et al., 2012).
2.6. SARS-CoV-2 infectivity assay
To assess the presence of infectious SARS-CoV-2 in environmental samples (n = 166), concentrated, PEG precipitated, water and sediment samples were sequentially filtered through 0.45 μM and 0.22 μM filters and a limited dilution was performed (as described by Harcourt et al. (2020)), with serum-free DMEM containing 8× penicillin, streptomycin and amphotericin B. Cells were diluted to 200,000 cells/mL and 100 μL of cell suspension was added to each well. If CPE were present cells were scraped and 100 μL of media and cells were added to 300 μL of DNA/RNA shield for RNA extraction and RT-qPCR analysis (as above). Samples that were negative for SARS-CoV-2 RNA via RT-qPCR were used as controls to ensure that the sample itself did not inhibit Vero E6 cell growth.
2.7. Evaluating how long SARS-CoV-2 remains infectious in river water
To assess the impact of environmental samples on SARS-CoV-2 viability, SARS-CoV-2 was spiked into water and sediment samples collected from the Hammersmith CSO on the River Thames, at three timepoints in September 2021. This CSO had the most regular discharge record of raw sewage from Thames Water over the sampling period. Detailed protocols are provided in the Supplementary Material. In brief, pasteurised and unpasteurised experiments separated the effect of abiotic and biotic factors on virus recovery, respectively. Four thousand TCID50 of SARS-CoV-2 was added to 1 mL aliquots of pasteurised and unpasteurised samples, and incubated at room temperature for 1, 2, 3 and 7 days. After incubation, samples were vortexed, filtered through a 0.22 μM filter and serially diluted to achieve a starting dilution of 200 TCID50/well on Vero E6 cells (Perera et al., 2020). After 6 days incubation, TCID50/mL was determined as above, and 100 μL of sample extracted for RT-qPCR as above.
3. Results
3.1. SARS-CoV-2 RNA was detected in the Danube and Sava rivers but not in the Thames
Samples from three of the six sites from the Danube and Sava rivers in Belgrade were over the limit of detection (LoD; as quantified by Gerrity et al. (2021)) for the N1 gene, and samples from three sites were negative. Positive samples included one sediment sample (Site 1; from the Danube), and two water samples (Site 4 and 5; from the Sava) (Fig. 2). No samples were over the LoD of 100 gc/μL for the E gene. Copy numbers of the N1 gene were over the LoD (10 gc/μL) but under the limit of quantification (80 gc/μL). This indicates concentrations of SARS-CoV-2 RNA were over 1000 gc/L in river water and 2000 gc/g in sediment. No infectious SARS-CoV-2 was recovered from any of these samples. None of the 218 samples collected from the Thames Basin were positive for the N1 or E gene (Fig. 2), and no infectious SARS-CoV-2 was recovered from any of these samples. All sites tested positive for crAssphage, with the exception of Kew Bridge (see Fig. 1 for site locations), confirming sewage pollution.
Only one water sample (from Kew Bridge) and four sediment samples (three from Hammersmith and one from Putney) out of 126 samples tested were found to inhibit the qPCR assay using the internal MHV amplification control. Twelve samples were not tested due to lack of RNA. With regard to SARS-CoV-2 infectivity assays, out of 152 samples tested for SARS-CoV-2 infectivity, only 23 were found to inhibit Vero E6 cell growth and only when either not diluted or diluted 1:2 (further dilutions were not inhibitive). This included 10 sediment samples and 13 water samples. No SARS-CoV-2 RNA was detected in any of these samples, indicating that it was not responsible for the cell death observed in these assays.
3.2. SARS-CoV-2 persists less than one week in river water
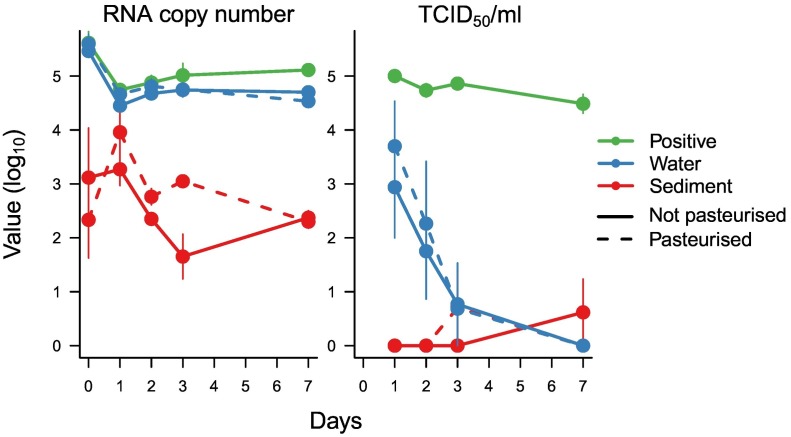
Because SARS-CoV-2 was not found in the River Thames, we evaluated how long it could remain infectious in such an environment if there was a contamination event (from any source). River water samples spiked with inoculum of infectious SARS-CoV-2 showed that while RNA was relatively stable over a week-long incubation, the ability to recover infectious virus from the samples (TCID50; Fig. 3 ) declined rapidly over the first three days with no viable virus present after one week. SARS-CoV-2 RNA recovered from sediment samples was lower than that found in water, but it too remained relatively stable over seven days (Fig. 3). In contrast, no viable SARS-CoV-2 was recovered from all replicates (n = 3) of the sediment incubations. Diluted, unspiked Hammersmith sediment and water samples had no detectable impact on cell growth. No significant differences were found between pasteurised (which removes the biological activity in the samples) and unpasteurised water or sediment samples for RNA or infectious virus recovery (two-way ANOVA).
Fig. 3.
Recovery of SARS-CoV-2 RNA and TCID50 of infectious SARS-CoV-2 after incubations in pasteurised and unpasteurised Hammersmith water and sediment over 7 days. For the positive control, SARS-CoV-2 was incubated in PBS for the same duration.
4. Discussion
Across 218 samples of CSO water, surface water, river sediment and bivalves collected from the Thames Basin no SARS-CoV-2 RNA was found, despite the study being carried out during two periods of the highest reported cases in London (e.g., 11,536 new cases reported on January 11th and 7641 on July 15th 2021) (https://coronavirus.data.gov.uk/details/cases), and in a region with one of the highest country-wide SARS-CoV-2 cases throughout the pandemic (https://coronavirus.data.gov.uk/details/cases). As crAssphage, was found at five out of the six sites on the Thames, this confirmed the presence of sewage (Kongprajug et al., 2019), and highlighted the importance of understanding SARS-CoV-2 transmission risk from CSOs discharges, given that SARS-CoV-2 can be detected in urine and faecal samples (Sun et al., 2020; Xiao et al., 2020). CrAssphage presence, evidence for the detection of SARS-CoV-2 RNA in samples from the Danube and Sava rivers in Serbia, with the same methodology, and the lack of site level qPCR inhibition indicates that if SARS-CoV-2 was present in the Thames, above its limit of detection, it would have been found. From Serbia, concentrations of the N1 gene from water samples collected from the Sava River in the declining phase of the fourth COVID-19 wave, were of the same order of magnitude as those found in December 2020 (5.97 × 103 to 1.32 × 104 copies/L (Kolarević et al., 2021)), confirming that the method presented here can concentrate SARS-CoV-2 RNA. Further, ten times more water was concentrated per sample from the Thames (10 L) than from Serbia (1 L). This all indicates that while London sewage works do discharge raw sewage into the Thames on a regular basis, the water intake from storms and surface water, and the tidal nature of the Thames, is sufficient to dilute SARS-CoV-2 RNA to below the limit of detection and reduce the threat of SARS-CoV-2 environmental spillover. It is noteworthy that the Sava and Danube rivers do not experience tides like the Thames, maybe partly explaining the differences found here regarding the presence of SARS-CoV-2 RNA in river systems.
Other studies have found SARS-CoV-2 RNA in river water in Ecuador and Serbia (Guerrero-Latorre et al., 2020; Kolarević et al., 2021), which is unsurprising given that there is far less sewage treatment in those countries. These studies focused on water sampling, in line with standard sampling of surface waters for environmental monitoring (e.g. for bathing water quality) (DEFRA, 2021). In contrast, the present study provides the first evidence for the accumulation of high concentrations of SARS-CoV-2 RNA (at least 2000 gc/g) in one sediment sample, where water samples from the same site were negative. This positive sample was collected downstream from the largest CSO on the Danube River (Pančevo Bridge), where there is a high dilution potential due to an average discharge of 5600 m3/s in this section of river (Kolarević et al., 2021), highlighting the limitations of point samples from surface water for environmental monitoring. Research suggests that high percentages of enveloped viruses (26 %) can adsorb to the solid fraction of wastewater (Ye et al., 2016), that sediments may protect viruses from inactivation (Hassard et al., 2016), and that sediments can provide a source of pathogens to the water column (Fluke et al., 2019). Studying the presence of viruses in sediments, rather than only in water, would provide greater insight into sites that are susceptible to accumulating and harbouring potential pathogens.
Most studies have focused on the presence of SARS-CoV-2 RNA in wastewater (reviewed by Kitajima et al. (2020)), and river water (Guerrero-Latorre et al., 2020; Kolarević et al., 2021), but very little is known about the potential infectivity of SARS-CoV-2 in waterways (Ahmed et al., 2020b; Naddeo and Liu, 2020). While previous studies have shown that SARS-CoV-2 infectivity is strongly reduced by 2 days in wastewater (Bivins et al., 2020; Wurtzer et al., 2021) and the degree of reduction likely depends on wastewater chemical and or microbial composition (Wurtzer et al., 2021), SARS-CoV-2 can persist in wastewater for 7 days (Bivins et al., 2020). This study evaluated whether the TFUF-PEG protocol that has been established to concentrate non-enveloped enteric viruses from river water (Farkas et al., 2018) can also be used to concentrate viruses such as SARS-CoV-2 and MHV for infectivity assays. This method had an average recovery efficiency of 28 % of MHV, but with great variation (0.2–80 %). Other currently available methods recover 1–25 % of infectious virus from more concentrated samples (e.g. sewage influent) (reviewed by Rusiñol et al. (2020)). Consequently, although the TFUF-PEG method is an improvement from studies that do not attempt concentration (e.g.(Rimoldi et al., 2020)), further development of viral concentration methods is critically needed for highly diluted samples, such as river water.
While clinical studies of faecal material from hospitalized patients have isolated infectious SARS-CoV-2 (Xiao et al., 2020), others have failed to do so (e.g. (Wölfel et al., 2020)). The findings presented here add to recent evidence that detected SARS-CoV-2 RNA from the natural environment does not occur as infectious viral particles, and thus do not represent a health hazard (Bivins et al., 2020; Rimoldi et al., 2020; Westhaus et al., 2021). Here, experiments with river water also provide evidence of relatively rapid degradation of SARS-CoV-2 infectivity within a few days, while RNA remains stable for longer. This is in agreement with experiments in wastewater, that show stable RNA concentrations (Ahmed et al., 2020c) but declining infectivity within 7 days (Bivins et al., 2020; Wurtzer et al., 2021). Infectivity data should therefore be embedded within risk assessments of pathogen spillover, because RNA surveys can be misleading, as suggested by Bivins et al. (2020). For SARS-CoV-2, these findings indicate that while some viral particles may remain infectious long enough to reach surface waters, they are unlikely to accumulate over time. However, further work is needed to confirm that SARS-CoV-2 does not survive waters and sediments during the colder winter months, when coronaviruses may remain infectious for longer (Casanova et al., 2009), as viruses are generally more stable at lower temperatures.
The COVID-19 pandemic has highlighted the dramatic consequences of novel outbreaks of viral pathogens. Public health organizations such as the Centre for Disease Control and Prevention, and the World Health Organisation, have therefore prioritized scientific research to enhance the ability to rapidly identify, track and contain novel human pathogens. Although the presence of SARS-CoV-2 RNA in raw sewage is abundant enough to be used to monitor levels of infection in human populations, this study indicates that sewage concentrations in the Thames is low enough to reduce the threat of environmental spillover. Reduced threat does not mean, however, that the Thames is safe. While no infectious SARS-CoV-2 was detected in the Thames, 23 samples did induce cell death, which is likely due to either chemical inhibition or the presence of other viruses that are capable of lysing Vero E6 cells. It is also still unclear how many SARS-CoV-2 particles are needed to cause an infection in humans, so although infectious SARS-CoV-2 was not detected, those particles might still be there, below the limit of detection. They may also be present in non-tidal areas, where river water is not diluted daily. What is needed is resilient and modernised sewerages to keep rivers uncontaminated. High concentrations of SARS-CoV-2 RNA in the Danube and Sava rivers in Serbia is concerning, especially as SARS-CoV-2 RNA has now been found in mollusc tissue (e.g. (Polo et al., 2021)). While this study focuses on SARS-CoV-2, rivers are conduits of disease transmission via sewage pollution and will remain a threat as long as water companies continue to release such extraordinary amounts of raw sewage into natural waterways.
CRediT authorship contribution statement
ER, VS, CC, TB, and GW conceptualised the project and acquired the funding. ER and VS administered the project. ER and VS supervised the research, which was mostly investigated by SJ and FH, with contributions from DH and JT. DH and TB created visualisations. SK and MK collected Serbian samples. ER wrote the initial draft manuscript with substantive and significant contributions from all authors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Wendy Barclay for providing SARS-CoV-2, and the UK Natural Environment Research Council for funding (NE/V010387/1). In Serbia, research activities were supported by Ministry of Education, Science and Technological Development of Republic of Serbia grant No. 451-03-9/2021-14/200007. The graphical abstract was created with BioRender.com.
Editor: Warish Ahmed
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2022.159161.
Appendix A. Supplementary material
Supplementary material containing additional methods and details of all samples collected.
Data availability
Data will be made available on request.
References
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R., Mueller J.F., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Science of The Total Environment. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Science of the Total Environment. 2020:728. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bibby K., D’Aoust P.M., Delatolla R., Gerba C.P., Haas C.N., Hamilton K.A., Hewitt J., Julian T.R., Kaya D., Monis P., Moulin L., Naughton C., Noble R.T., Shrestha A., Tiwari A., Simpson S.L., Wurtzer S., Bivins A. Differentiating between the possibility and probability of SARS-CoV-2 transmission associated with wastewater: empirical evidence is needed to substantiate risk. FEMS Microbes. 2021;2 doi: 10.1093/femsmc/xtab007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amman F., Markt R., Endler L., Hupfauf S., Agerer B., Schedl A., Richter L., Zechmeister M., Bicher M., Heiler G., Triska P., Thornton M., Penz T., Senekowitsch M., Laine J., Keszei Z., Klimek P., Nägele F., Mayr M., Bergthaler A.… Viral variant-resolved wastewater surveillance of SARS-CoV-2 at national scale. Nat. Biotechnol. 2022 doi: 10.1038/s41587-022-01387-y. [DOI] [PubMed] [Google Scholar]
- Besselsen D.G., Wagner A.M., Loganbill J.K. Detection of rodent coronaviruses by use of fluorogenic reverse transcriptase-polymerase chain reaction analysis. Comp. Med. 2002;52(2):111–116. [PubMed] [Google Scholar]
- Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M., Munster V.J., Bibby K. Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. Lett. 2020;7(12):937–942. doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova L., Rutala W.A., Weber D.J., Sobsey M.D. Survival of surrogate coronaviruses in water. Water Res. 2009;43(7):1893–1898. doi: 10.1016/j.watres.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Bailey A.L., Kim A.S., Chen R.E., Diamond M.S. Growth, detection, quantification, and inactivation of SARS-CoV-2. Virology. 2020;548:39–48. doi: 10.1016/j.virol.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEFRA . 2021. Outcome Indicator Framework for the 25 Year Environment Plan: 2021 Update. [Google Scholar]
- Desdouits M., Piquet J.C., Wacrenier C., le Mennec C., Parnaudeau S., Jousse S., Rocq S., Bigault L., Contrant M., Garry P., Chavanon F., Gabellec R., Lamort L., Lebrun L., le Gall P., Meteigner C., Schmitt A., Seugnet J.L., Serais O., le Guyader F.S.… Can shellfish be used to monitor SARS-CoV-2 in the coastal environment? Sci. Total Environ. 2021;778 doi: 10.1016/j.scitotenv.2021.146270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhama K., Khan S., Tiwari R., Sircar S., Bhat S., Malik Y.S., Singh K.P., Chaicumpa W., Bonilla-Aldana D.K., Rodriguez-Morales A.J. Coronavirus disease 2019–COVID-19. Clinical Microbiology Reviews. 2020;33(4) doi: 10.1128/CMR.00028-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA Analysis of the European baseline survey of norovirus in oysters. EFSA J. 2019;17(7) doi: 10.2903/j.efsa.2019.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas K., McDonald J.E., Malham S.K., Jones D.L. Two-step concentration of complex water samples for the detection of viruses. Methods Protoc. 2018;1(3):1–6. doi: 10.3390/mps1030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas K., Peters D.E., McDonald J.E., de Rougemont A., Malham S.K., Jones D.L. Evaluation of two triplex one-step qRT-PCR assays for the quantification of human enteric viruses in environmental samples. Food Environ. Virol. 2017;9(3):342–349. doi: 10.1007/s12560-017-9293-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorito F., Amoroso M.G., Lambiase S., Serpe F.P., Bruno T., Scaramuzzo A., Maglio P., Fusco G., Esposito M. A relationship between environmental pollutants and enteric viruses in mussels(Mytilus galloprovincialis) Environ. Res. 2019;169:156–162. doi: 10.1016/j.envres.2018.11.001. [DOI] [PubMed] [Google Scholar]
- Fitzgerald S.F., Rossi G., Low A.S., McAteer S.P., O’Keefe B., Findlay D., Cameron G.J., Pollard P., Singleton P.T.R., Ponton G., Singer A.C., Farkas K., Jones D., Graham D.W., Quintela-Baluja M., Tait-Burkard C., Gally D.L., Kao R., Corbishley A. Site specific relationships between COVID-19 cases and SARS-CoV-2 viral load in wastewater treatment plant influent. Environ. Sci. Technol. 2021;55(22):15276–15286. doi: 10.1021/acs.est.1c05029. [DOI] [PubMed] [Google Scholar]
- Fluke J., González-Pinzón R., Thomson B. Riverbed sediments control the spatiotemporal variability of E. Coli in a highly managed, Arid River. FrontiersWater. 2019;1 doi: 10.3389/frwa.2019.00004. [DOI] [Google Scholar]
- Freire-Paspuel B., Vega-Mariño P., Velez A., Cruz M., Perez F., Garcia-Bereguiain M.A. Analytical and clinical comparison of viasure (CerTest Biotec) and 2019-nCoV CDC (IDT) RT-qPCR kits for SARS-CoV2 diagnosis. Virology. 2021;553:154–156. doi: 10.1016/j.virol.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrity D., Papp K., Stoker M., Sims A., Frehner W. Early-pandemic wastewater surveillance of SARS-CoV-2 in southern Nevada: methodology, occurrence, and incidence/prevalence considerations. Water Res. X. 2021;10 doi: 10.1016/j.wroa.2020.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J. Nature Microbiology. 2020;5(4):536–544. doi: 10.1038/s41564-020-0695-z. Nature Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greater London Authority . 2011. Securing London’s Water Future. The Mayor’s Water Strategy. [Google Scholar]
- Guerrero-Latorre L., Ballesteros I., Villacrés-Granda I., Granda M.G., Freire-Paspuel B., Ríos-Touma B. SARS-CoV-2 in river water: implications in low sanitation countries. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampstead Heath Annual Report 2020/21 www.cityoflondon.gov.uk . 2021. Hampstead Heath Annual Report 2020/21. (Hampstead Heath Bathing Ponds and Lido Annual Report 2020/21). [Google Scholar]
- Harcourt J., Tamin A., Lu X., Kamili S., Sakthivel S.K., Murray J., Queen K., Tao Y., Paden C.R., Zhang J., Li Y., Uehara A., Wang H., Goldsmith C., Bullock H.A., Wang L., Whitaker B., Lynch B., Gautam R., Thornburg N.J.… Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus diseaseUnited States. Emerging Infectious Diseases. 2020;26(6):1266–1273. doi: 10.3201/EID2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassard F., Gwyther C.L., Farkas K., Andrews A., Jones V., Cox B., Brett H., Jones D.L., McDonald J.E., Malham S.K. Abundance and distribution of enteric bacteria and viruses in coastal and estuarine sediments-A review. Frontiers in Microbiology. 2016;7(NOV) doi: 10.3389/fmicb.2016.01692. Frontiers Media S.A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House of Commons Environmental Audit Committee . Fourth Report of Session 2021–22. 2022. Water quality in rivers. [Google Scholar]
- Hover S., King B., Hall B., Loundras E.A., Taqi H., Daly J., Dallas M., Peers C., Schnettler E., Mckimmie C., Kohl A., Barr J.N., Mankouri J. Modulation of potassium channels inhibits bunyavirus infection. J. Biol. Chem. 2016;291(7):3411–3422. doi: 10.1074/jbc.M115.692673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.L., Baluja M.Q., Graham D.W., Corbishley A., McDonald J.E., Malham S.K., Hillary L.S., Connor T.R., Gaze W.H., Moura I.B., Wilcox M.H., Farkas K. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci. Total Environ. 2020;749 doi: 10.1016/j.scitotenv.2020.141364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: State of the knowledge and research needs. Science of the Total Environment. 2020;739 doi: 10.1016/j.scitotenv.2020.139076. Elsevier B.V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymus K.E., Merkes C.M., Allison M.J., Goldberg C.S., Helbing C.C., Hunter M.E., Jackson C.A., Lance R.F., Mangan A.M., Monroe E.M., Piaggio A.J., Stokdyk J.P., Wilson C.C., Richter C.A. Reporting the limits of detection and quantification for environmental DNA assays. Environ. DNA. 2020;2(3):271–282. doi: 10.1002/edn3.29. [DOI] [Google Scholar]
- Kolarević S., Micsinai A., Szántó-Egész R., Lukács A., Kračun-Kolarević M., Lundy L., Kirschner A.K.T., Farnleitner A.H., Djukic A., Čolić J., Nenin T., Sunjog K., Paunović M. Detection of SARS-CoV-2 RNA in the Danube River in Serbia associated with the discharge of untreated wastewaters. Sci. Total Environ. 2021;783 doi: 10.1016/j.scitotenv.2021.146967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kongprajug A., Mongkolsuk S., Sirikanchana K. CrAssphage as a potential human sewage marker for microbial source tracking in Southeast Asia. Environ. Sci. Technol. Lett. 2019;6(3):156–164. doi: 10.1021/acs.estlett.9b00041. [DOI] [Google Scholar]
- Lei C., Yang J., Hu J., Sun X. On the calculation of TCID50 for quantitation of virus infectivity. Virologica Sinica. 2021;36(1):141–144. doi: 10.1007/s12250-020-00230-5. Science Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay P.F., Hu K., Blakney A.K., Samnuan K., Brown J.C., Penn R., Zhou J., Bouton C.R., Rogers P., Polra K., Lin P.J.C., Barbosa C., Tam Y.K., Barclay W.S., Shattock R.J. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat. Commun. 2020;11(1):3523. doi: 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ministry of Environmental Protection E.P.A. 2019. Environment in Serbia 2004–2019. [Google Scholar]
- Munro K., Martins C.P.B., Loewenthal M., Comber S., Cowan D.A., Pereira L., Barron L.P. Evaluation of combined sewer overflow impacts on short-term pharmaceutical and illicit drug occurrence in a heavily urbanised tidal river catchment (London, UK) Sci. Total Environ. 2019;657:1099–1111. doi: 10.1016/j.scitotenv.2018.12.108. [DOI] [PubMed] [Google Scholar]
- Naddeo V., Liu H. Editorial perspectives: 2019 novel coronavirus (SARS-CoV-2): what is its fate in urban water cycle and how can the water research community respond? Environmental Science: Water Research and Technology. 2020;6(5):1213–1216. doi: 10.1039/d0ew90015j. [DOI] [Google Scholar]
- Nichols G.L., Gillingham E.L., Macintyre H.L., Vardoulakis S., Hajat S., Sarran C.E., Amankwaah D., Phalkey R. Coronavirus seasonality, respiratory infections and weather. BMC Infect. Dis. 2021;21(1):1101. doi: 10.1186/s12879-021-06785-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S.M., Chu C.M., Cheng V.C.C., Chan K.S., Hung I.F.N., Poon L.L.M., Law K.I., Tang B.S.F., Hon T.Y.W., Chan C.S., Chan K.H., Ng J.S.C., Zheng B.J., Ng W.L., Lai R.W.M., Guan Y., Yuen K.Y. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361(9371):1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera Ranawaka A.P.M., Mok Chris K.P., Tsang Owen T.Y., Huibin Lv, Ko Ronald L.W., Wu Nicholas C., Meng Yuan, Shing Leung Wai, Chan Jacky M.C., Chik Thomas S.H., Choi Chris Y.C., Kathy Leung, Ho Chan Kin, Chan Karl C.K., Ka-Chi Li, Wu Joseph T., Wilson Ian A., Monto Arnold S., Poon Leo L.M., Malik Peiris. Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Euro Surveill. 2020;25(16) doi: 10.2807/1560-7917.ES.2020.25.16.2000421. March 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo D., Lois M., Fernández-Núñez M.T., Romalde J.L. Detection of SARS-CoV-2 RNA in bivalve mollusks and marine sediments. Sci. Total Environ. 2021;786 doi: 10.1016/j.scitotenv.2021.147534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potasman I., Paz A., Odeh M. Shellfish-Associated Infectious Outbreaks. CID; 2002. Infectious outbreaks associated with bivalve shellfish consumption: a worldwide perspective.http://apps.fao.org/ Science Press. [DOI] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson M., Soloviev M. The Thames: arresting ecosystem decline and building Back better. Sustainability. 2021;13(11):6045. doi: 10.3390/su13116045. [DOI] [Google Scholar]
- Rimoldi S.G., Stefani F., Gigantiello A., Polesello S., Comandatore F., Mileto D., Maresca M., Longobardi C., Mancon A., Romeri F., Pagani C., Cappelli F., Roscioli C., Moja L., Gismondo M.R., Salerno F. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020;744 doi: 10.1016/j.scitotenv.2020.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiñol M., Martínez-Puchol S., Forés E., Itarte M., Girones R., Bofill-Mas S. Vol. 17. Elsevier B.V; 2020. Concentration methods for the quantification of coronavirus and other potentially pandemic enveloped virus from wastewater; pp. 21–28. (Curr. Opin. Environ. Sci. Health). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif S., Ikram A., Khurshid A., Salman M., Mehmood N., Arshad Y., Ahmed J., Safdar R.M., Rehman L., Mujtaba G., Hussain J., Ali J., Angez M., Alam M.M., Akthar R., Malik M.W., Baig M.Z.I., Rana M.S., Usman M., Ali N. Detection of SARs-CoV-2 in wastewater using the existing environmental surveillance network: a potential supplementary system for monitoring COVID-19 transmission. PLoS ONE. 2021;16(6 June) doi: 10.1371/journal.pone.0249568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley C., Gordon K.V., Schoen M.E., Harwood V.J. Performance of two quantitative PCR methods for microbial source tracking of human sewage and implications for microbial risk assessment in recreational waters. Applied and Environmental Microbiology. 2012;78(20):7317–7326. doi: 10.1128/AEM.01430-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Zhu A., Li H., Zheng K., Zhuang Z., Chen Z., Shi Y., Zhang Z., Chen S., Liu X., Dai J., Li X., Huang S., Huang X., Luo L., Wen L., Zhuo J., Li Y., Wang Y., Li Y.… Isolation of infectious SARS-CoV-2 from urine of a COVID-19 patient. Emerg. Microbes Infect. 2020;9(1):991–993. doi: 10.1080/22221751.2020.1760144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur A.K., Sathyamurthy R., Velraj R., Lynch I., Saidur R., Pandey A.K., Sharshir S.W., Kabeel A.E., Hwang J.-Y., GaneshKumar P. Secondary transmission of SARS-CoV-2 through wastewater: concerns and tactics for treatment to effectively control the pandemic. J. Environ. Manag. 2021;290 doi: 10.1016/j.jenvman.2021.112668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H.N., Le G.T., Nguyen D.T., Juang R.-S., Rinklebe J., Bhatnagar A., Lima E.C., Iqbal H.M.N., Sarmah A.K., Chao H.-P. SARS-CoV-2 coronavirus in water and wastewater: a critical review about presence and concern. Environ. Res. 2021;193 doi: 10.1016/j.envres.2020.110265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhaus S., Weber F.A., Schiwy S., Linnemann V., Brinkmann M., Widera M., Greve C., Janke A., Hollert H., Wintgens T., Ciesek S. Detection of SARS-CoV-2 in raw and treated wastewater in Germany – suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead P.G., Crossman J., Balana B.B., Futter M.N., Comber S., Jin L., Skuras D., Wade A.J., Bowes M.J., Read D.S. A cost-effectiveness analysis of water security and water quality: impacts of climate and land-use change on the river Thames system. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2013;371(2002):20120413. doi: 10.1098/rsta.2012.0413. [DOI] [PubMed] [Google Scholar]
- Wilder M.L., Middleton F., Larsen D.A., Du Q., Fenty A., Zeng T., Insaf T., Kilaru P., Collins M., Kmush B., Green H.C. Co-quantification of crAssphage increases confidence in wastewater-based epidemiology for SARS-CoV-2 in low prevalence areas. Water Res. X. 2021;11 doi: 10.1016/j.wroa.2021.100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wurtzer S., Waldman P., Ferrier-Rembert A., Frenois-Veyrat G., Mouchel J.M., Boni M., Maday Y., Marechal V., Moulin L. Several forms of SARS-CoV-2 RNA can be detected in wastewaters: implication for wastewater-based epidemiology and risk assessment. Water Res. 2021;198 doi: 10.1016/j.watres.2021.117183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Sun J., Xu Y., Li F., Huang X., Li H., Zhao J., Huang J., Zhao J. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg. Infect. Dis. 2020;26(8):1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50(10):5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Zhang R., Li Y., Zhang A.L., Wang Y., Molina M.J. Identifying airborne transmission as the dominant route for the spread of COVID-19. Proc. Natl. Acad. Sci. U. S. A. 2020;117(26):14857–14863. doi: 10.1073/pnas.2009637117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Ayeh S.K., Chidambaram V., Karakousis P.C. Modes of transmission of SARS-CoV-2 and evidence for preventive behavioral interventions. BMC Infect. Dis. 2021;21(1):496. doi: 10.1186/s12879-021-06222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material containing additional methods and details of all samples collected.
Data Availability Statement
Data will be made available on request.