Abstract
The endophytic diazotroph Azoarcus sp. strain BH72 is capable of infecting rice roots and of expressing the nitrogenase (nif) genes there. In order to study the genetic background for nitrogen fixation in strain BH72, the structural genes of nitrogenase (nifHDK) were cloned and sequenced. The sequence analysis revealed an unusual gene organization: downstream of nifHDK, a ferredoxin gene (fdxN; 59% amino acid sequence identity to R. capsulatus FdxN) and open reading frames showing 52 and 36% amino acid sequence identity to nifY of Pseudomonas stutzeri A15 and ORF1 of Azotobacter vinelandii were located. Northern blot analysis, reverse transcriptase PCR and primer extension analysis revealed that these six genes are located on one transcript transcribed from a ς54-type promoter. Shorter transcripts sequentially missing genes of the 3′ part of the full-length mRNA were more abundantly detected. Mutational analyses suggested that FdxN is an important but not the essential electron donor for dinitrogenase reductase. An in-frame deletion of fdxN resulted in reduced growth rates (59% ± 9%) and nitrogenase activities (81%) in nitrogen-fixing pure cultures in comparison to the wild type. Nitrogenase activity was fully complemented in an fdxN mutant which carried a nifH promoter-driven fdxN gene in trans. Also, in coculture with the ascomycete Acremonium alternatum, where strain BH72 develops intracytoplasmic membrane stacks, the nitrogenase activity in the fdxN deletion mutant was decreased to 56% of the wild-type level. Surprisingly, the fdxN deletion also had an effect on the rapid “switch-off” of nitrogenase activity in response to ammonium. Wild-type strain BH72 and the deletion mutant complemented with fdxN in trans showed a rapid reversible inactivation of acetylene reduction, while the deletion mutant did not cease to reduce acetylene. In concordance with the hypothesis that changes in the redox state of NifH or electron flux towards nitrogenase may be involved in the mechanism of physiological nitrogenase switch-off, our results suggest that the ferredoxin may be a component involved in this process.
In addition to the root surface, inner tissues of roots may also be colonized by bacteria. Endophytic diazotrophic bacteria invade roots and shoots of grasses without causing symptoms of plant disease (23, 46). Establishment in inner tissues of agriculturally important crops such as sugar cane or rice has been shown for several gram-negative bacteria, such as Herbaspirillum seropedicae (23, 24), Acetobacter diazotrophicus (25), and Azoarcus spp. (19).
Azoarcus sp. strain BH72, which was isolated from the endorhizosphere of Kallar grass in Punjab, Pakistan (45), is also capable of infecting roots of rice seedlings in the laboratory (19). Reporter gene studies have demonstrated that nitrogenase (nif) genes of Azoarcus spp. can be expressed endophytically in the aerenchyma of these seedlings, suggesting that the interior of rice roots provides a microenvironment suitable for nitrogen fixation (6). Expression of nifHDK genes (6) as well as nitrogenase activity (16) requires low concentrations of oxygen and ammonium (below 0.5 mM); however, anaerobic conditions do not permit nitrogen fixation in this strictly respiratory bacterium.
Azoarcus sp. strain BH72 is unusual in that it can shift into a state of “hyperinduction” under certain growth conditions that include extremely low oxygen concentrations (30 nM). Moreover, in contrast to other Proteobacteria, this strain harbors three instead of two copies of PII-like proteins, the central signal transmitters of nitrogen metabolism (37). Hyperinduction of strain BH72 is characterized by increased activity and efficiency of nitrogen fixation (18), appearance of intracellular membrane stacks (diazosomes), and association of the iron protein of nitrogenase with diazosome membranes (20). Diazosome formation can be induced reproducibly in the laboratory by cocultivating Azoarcus sp. strain BH72 with the ascomycete Acremonium alternatum, which was isolated from the root interior of Kallar grass as well (17). The cells attach to the fungal mycelium, and the fungal respiration may provide sufficiently microaerobic niches for diazosome formation. The association of nitrogenase with these membranes suggests that they are involved in efficient nitrogen fixation, possibly by providing a more efficient electron flux to nitrogenase (20).
The electrons required to reduce N2 are carried to nitrogenase by either flavodoxins or ferredoxins. Little is known about the generation of reductant for N2 fixation in nonphototrophic bacteria. While the NifJF pathway via a pyruvate:flavodoxin oxidoreductase was characterized for Klebsiella pneumoniae (41), the generation of low-redox-potential electron carriers for nitrogenase reduction in many heterotrophic diazotrophs remains unclear. Nevertheless, the immediate molecule donating electrons to nitrogenase reductase had been identified for many diazotrophs. While in gram-positive bacteria (10) and in cyanobacteria (53) [2Fe-2S] ferredoxins have been shown to supply electrons to nitrogenase, the group of Proteobacteria favors more or less nif-specific 2[4Fe-4S] ferredoxins (30, 52). Some of the genes encoding these ferredoxins have been found to be localized in an operon with nif genes (other than nitrogenase genes) and are therefore regulated in a nif-dependent manner (26, 33).
In order to analyze nitrogen fixation in Azoarcus sp. strain BH72 genetically, nitrogenase structural genes were cloned and sequenced in the present study, revealing strong homologies with known nif genes of other Proteobacteria. In contrast to most other bacteria, strain BH72 was found to cotranscribe a ferredoxin gene with the structural nifHDK genes. Mutational analysis revealed that the ferredoxin is not obligatory for nitrogen fixation. However, it is essential for the rapid “switch-off” of nitrogenase activity in response to ammonium addition and thus a newly identified component involved in this process.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used in this study are listed in Table 1. Azoarcus strain BH72 originated from roots of Kallar grass, Leptochloa fusca (L.) Kunth (45).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| Azoarcus sp. strain BH72 | Wild type | 45 |
| BHΔfdxN | fdxN in-frame deletion in BH72 | This study |
| BHΔfdxN(pfdxN) | BHΔfdxN complemented with pfdxN | This study |
| E. coli DH5αF′ | F′ endA1 hsdR17(rK− mK+) supE44 thi-1 recA1 gyrA relA1 φ80lacZΔM15 Δ(lacZYA-argF)U169 | 13 |
| Plasmids | ||
| pUC19/18 | Apr | 59 |
| pBSKII/pBKSII | Apr | Stratagene |
| pLAFR3 | Tetr, cosmid vector | 56 |
| pEN9 | Apr, KpnI fragment from GenBank clone pBGVN9 carrying nif gene cluster in pUC19 | This study |
| pEN94 | Apr, subclone from pEN9 | This study |
| pEN9ΔF | Apr, fdxN deletion on pEN9 | This study |
| pfdxN | Tetr, fdxN gene fused to the nifH promoter region on pLAFR3 | This study |
Culture media and growth conditions.
If not stated otherwise, Azoarcus sp. strain BH72 was grown at 37°C in VM medium supplemented with ethanol (47, 48). For nitrogen fixation in pure culture, the cells were grown microaerobically in N-free SM medium (44) in a closed batch culture (32) or, for mixed-culture growth experiments, in a 2-liter fermentor (Biostat B; Braun Biotech, Melsungen, Germany) equipped with a regulated oxygen supply set at 0.1% O2 in N2, and stirred at 600 rpm. The cells were precultured overnight in SM medium supplemented with 0.01% yeast extract and 0.05% ammonium chloride, washed three times in N-free SM medium, and inoculated at an optical density at 578 nm (OD578) of 0.02 in the fermentor. Cells were harvested at the late exponential growth phase (OD578 of 1.5) after six generations. Cocultures with the ascomycete Acremonium alternatum were carried out as described previously (20), with wild-type and mutant cells added to the cultures in the same amounts. The flasks were sealed with gas-tight rubber stoppers and incubated until the oxygen in the headspace had decreased from initially atmospheric concentrations to approximately 2%. This occurred 5 to 8 days after inoculation.
Analysis of bacterial growth and nitrogen fixation.
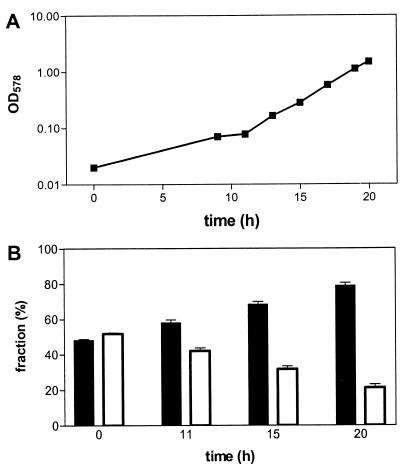
To analyze the growth of the ΔfdxN mutant strain compared to that of the wild type under exactly the same growth conditions, the strains were coinoculated in the fermentor in equal amounts. Growth parameters were as described above. Culture samples of the mixed culture were taken at different time points during exponential growth, diluted in saline (0.9% NaCl), and plated on VM agar plates to give approximately 200 colonies per plate. These colonies were restreaked on Hybond N membranes in six replicates of 30 randomly picked colonies in the first experiment. Thus, 180 colonies total from each time point were used for colony hybridization, using a digoxigenin-labeled fdxN probe (fragment amplified with primers TE29 and TE30) to differentiate mutant colonies from the wild type and thereby determine the ratio of both cell types (standard deviations are given for the six replicates at 11 and 15 h in Fig. 4). In two more independent fermentor experiments, only the first and the last time points were evaluated in this way (standard deviations are given in Fig. 4 for all three independent experiments). Growth rates for individual strains were estimated from calculating cell densities of the mutant and the wild type according to the percentage of distribution obtained by colony hybridization at different time points.
FIG. 4.
Growth comparison of the wild type and the fdxN deletion mutant in mixed culture. The wild type and the fdxN deletion mutant were grown in an oxygen-controlled bioreactor on N2 in mixed culture. (A) Growth measured as turbidity (OD578). (B) Relative amount of each of the two strains at different time points during exponential growth of the mixed culture, determined by colony hybridization. Black bar, wild type; white bar, fdxN deletion strain. Values are means with standard deviations. For details of the calculations, see Materials and Methods.
To analyze the mutant phenotype in cocultures with the ascomycete, independent cultures of wild-type and mutant strains were grown. Nitrogen-fixing capacity was measured using the acetylene reduction assay (32) 5 to 8 days after inoculation as described below, and the amount of ethylene formed per vial was determined by gas chromatography. The oxygen concentration in the headspace was also determined by gas chromatography (32). Cultures had been inoculated with equal amounts of bacterial cells (8 × 108 per ml [5]), which do not grow significantly in coculture but adhere to the fungal mycelium (approximately 20 mg per culture), reduce acetylene, and form dumbbell-shaped, diazosome-containing cells which appear to be arrested in cell division (reference 20 and unpublished results). Acetylene reduction of cultures was estimated in three independent experiments with three to five replicates. To demonstrate an equal yield of bacterial protein from both wild-type and mutant cultures in coculture with the ascomycete, in which fungal mycelium and bacteria cannot efficiently be separated from each other, equal volumes of cultures were harvested and roughly disrupted in a kitchen blender (Braun) for 30 s. The bacteria and fungal debris were sedimented by centrifugation at 20,000 × g for 2 min and resuspended in sample treatment buffer to give 140 mg (fresh weight) per ml. After addition of 1% sodium dodecyl sulfate, the cells were incubated at 95°C for 5 min and centrifuged at 20,000 × g for 5 min to pellet DNA and cell debris. The clear protein-containing supernatant was used for sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis, using antibodies against the iron protein of nitrogenase (kindly provided by R. Ludden, University of Wisconsin, Madison) to differentiate bacterial from fungal protein.
For analysis of switch-off of nitrogenase activity, nitrogen-fixing cells were grown in closed batch culture overnight as described above and then transferred to fresh N-free SM medium (5 to 15 ml of culture to give 30 ml total) in sealed Erlenmeyer flasks adjusted to 1% O2 and 1% acetylene in the headspace. Acetylene reduction and optical density were monitored before and after addition of NH4Cl (2 mM final concentration).
Statistical evaluations were carried out using the GraphPad Instat software, applying the Student t test or the Tukey-Kramer multiple comparison test.
Techniques for DNA and RNA manipulation.
General techniques for DNA analysis were carried out according to standard protocols (3, 47). Homologous DNA gene probes for Southern and Northern blot analysis were digoxigenin labeled in a PCR using a digoxigenin labeling and detection kit (Boehringer, Mannheim, Germany). Primer sequences are given in Table 2. For Northern blot analysis, RNA was isolated from exponentially growing bacterial cells by the hot phenol method (1), and Northern blot analysis was carried out according to standard protocols (3, 47). Reverse transcriptase PCR (RT-PCR) was carried out with 1.5 μg of RNA using Ready-To-Go RT-PCR beads (Amersham Pharmacia Biotech) according to manufacturer's instructions, using primers nifKfw and Fdxrev (Table 2) for the RT reaction at 42°C for 20 min and inactivation at 95°C for 5 min, followed by the PCR with 35 cycles of denaturation at 95°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min.
TABLE 2.
Primers used
| Application | Primer and sequence (position)a |
|---|---|
| nifK probe | —b |
| ORF1 probe | TE27: 5′CCTGCGCTTCACCTATTGTT3′ (6240–6259) |
| TE28: 5′AGCGAAATCCAACTCCGCCA3′ (110 nt downstream of ORF1) | |
| fdxN probe | TE29: 5′ACCAAGTCGATCACCGACAA3′ (5200–5219) |
| TE30: 5′CTACAAAAAACAGGGAGCC3′ (5384–5366) | |
| Test for fdxN deletion | TE14: 5′CCGCCAGTGGTGGGCTTTCG3′ (4911–4930) |
| TE19: 5′GACGACCTCGACACCGGGCA3′ (5499–5518) | |
| Amplification of nifHDK promoter region | fdxpro2(PstI): 5′AACTGCAGAGTCGAACAGCCAGCCGCGCA3′ (34–54) |
| fdxprorev(BamHI): 5′AAGGATCCAAGAAAAAAACTACCGGGTGTCCTGGG3′ (600–574) | |
| fdxN gene | fdxvor(BamHI): 5′CGGGATCCGGAGAAGGAATCATGGCTCTG3′ (5125–5145) |
| TE36(EcoRI): 5′ACGAGCCTCATGGGAATTC3′ (5421–5403) | |
| RT-PCR | nifKfw: 5′CGCCTTCGCGGCACCCGT3′ (4799–4816) |
| Fdxrev: 5′CAGTCACCACAGGACGTGCAT3′ (5183–5163) | |
| Primer extension | TH25rCy: 5′TGCCGATACCGCCCTTGCC (668–650) |
Primer extension was carried out with a CY5-labeled primer (TH25rCy) (Table 2) according to standard protocols (3) with 20 μg of RNA isolated from nitrogen-fixing cells, using an automated sequencer (ALFexpress; Amersham Pharmacia Biotech) for product analysis.
DNA sequencing and computational analysis.
DNA sequencing was carried out as described by Sanger et al. (51). Primers were 35S labeled and sequencing was carried out using the DNA sequencing kit Sequenase, version 2.0 (Amersham, Braunschweig, Germany). Products were separated on an 8% acrylamide gel (GENE-PAGE; Amresco, Solon, Ohio) and detected by autoradiography. Alternatively, CY5-labeled primers were used for sequencing with an automated sequencer as described by Hurek et al. (15).
Construction of fdxN mutants of Azoarcus sp. strain BH72.
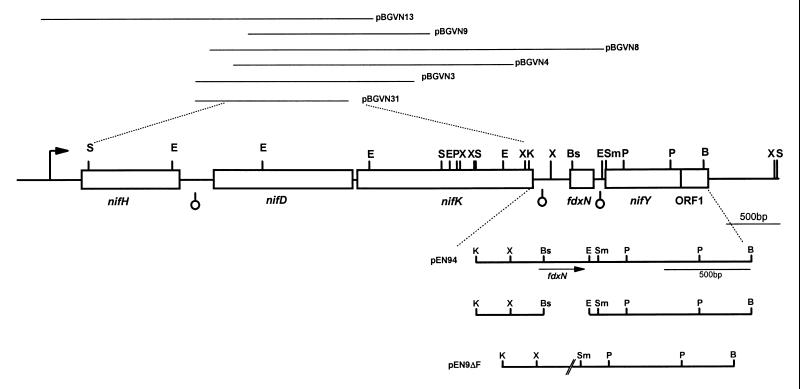
The construction of an fdxN in-frame deletion is depicted in Fig. 1. Plasmid pEN94, which carries the genomic region of the fdxN gene, was digested with BsmI and EcoRI. The sticky ends of the restriction fragments were blunted using Klenow large fragment, which removed the 3′ overhang of the BsmI site and filled the 5′ overhang at the EcoRI site. The blunt ends were religated, yielding a stop codon that interrupted the translation of the fdxN gene after the seventh amino acid. The mutation was confirmed by sequence analysis and reintroduced into pEN9 by cloning the KpnI/BamHI insert of pEN94ΔfdxN into pEN9 digested with the same restriction endonucleases, yielding pEN9ΔF. The mutation was introduced into the Azoarcus sp. strain BH72 chromosome by allelic exchange mutagenesis. Specifically, the plasmid pEN9ΔF, which does not replicate in Azoarcus spp., was integrated into the chromosome by a single homologous recombination event after electroporation. Recombinants carrying the vector-borne Apr gene were used for a second recombination event: colonies were replica plated on medium with and without antibiotic and screened for cells that had lost the vector-encoded resistance. Six in 5,000 colonies tested which had lost the vector were tested for the fdxN deletion by PCR with primers TE14 and TE18. These primers amplified a 609-bp fragment in the wild type, while the mutant showed a 373-bp product. Two of six double recombinants had exchanged the wild-type for the mutagenized gene (strain BHΔfdxN).
FIG. 1.
Gene organization of the nifHDK locus. The restriction map of the nifHDK operon in Azoarcus sp. strain BH72 shows nif gene-containing clones from a genomic library above and the construction of the fdxN deletion (pEN9ΔF) below the map. The bold arrow indicates the promoter region, and the light arrow shows the position of fdxN in plasmid pEN94. Sequences with inverted repeats for stem-loop formation are indicated by hairpins below the map. S, SalI; E, EcoRI; P, PstI; X, XhoI; K, KpnI; Bs, BsmI; Sm, SmaI; B, BamHI.
To complement the mutation, the fdxN gene was genetically fused to the original nifH promoter and provided in trans. The coding sequence and the promoter region were both amplified by PCR using Pfu polymerase (Stratagene) and the primers (Table 2) fdxpro2(PstI) and fdxprorev(BamHI) at 64°C with 1.5 mM MgCl2 for the promoter region or fdxvor(BamHI) and TE36(EcoRI) at 53°C with 1.5 mM MgCl2 for the coding sequence. The two amplified fragments were restriction digested with PstI/BamHI or EcoRI/BamHI, respectively, and cloned into PstI/EcoRI-digested vector pUC19, the two PCR fragments being fused at the BamHI site. A clone of the correct sequence was subcloned into the broad-host-range plasmid pLAFR3 (56) after EcoRI/PstI digestion, yielding plasmid pfdxN, which was conjugated into strain BH72 by triparental mating. Since the fragments provided in trans were very short (400 to 600 bp), a double recombination event of fdx into the chromosome was unlikely.
Nucleotide sequence accession number.
The sequences of the nifHDK operon were submitted to GenBank (accession no. AF200742).
RESULTS
Characterization of the nifHDK-fdxN region of Azoarcus sp. strain BH72.
The nitrogenase genes of Azoarcus sp. strain BH72 were obtained by screening a genomic library in pUC19 (Sau3A1-digested DNA cloned into the BamHI site [47]) with a heterologous nifH gene probe of Azorhizobium caulinodans. The resulting plasmids carrying the nifHDK region and results of the sequence analysis of 6.5 kb are shown in Fig. 1. Southern hybridization assays at low stringency with either a homologous nifH or nifK probe indicated that these nif genes are present in a single copy on the chromosome of Azoarcus sp. strain BH72 (data not shown). Nitrogenase structural gene products showed the highest homology to the FeMo nitrogenase from Azotobacter spp., with 88% identity to NifH of Azotobacter chroococcum (27) and 82% identity to NifD and 77% identity to NifK of Azotobacter vinelandii (21). Downstream of the nif genes, a sequence coding for a 2[4Fe-4S] ferredoxin was detected which was most closely related to the ferredoxin FdxN from Rhodobacter capsulatus, with 59% amino acid identity (11, 29). Downstream of this fdxN gene, a nifY homologue (52 and 42% amino acid identity to NifY of Pseudomonas stutzeri A15 and A. vinelandii, respectively) and an open reading frame (ORF1) with weak amino acid identity (36%) to an open reading frame (named ORF1) from the A. vinelandii nif region (21) were found.
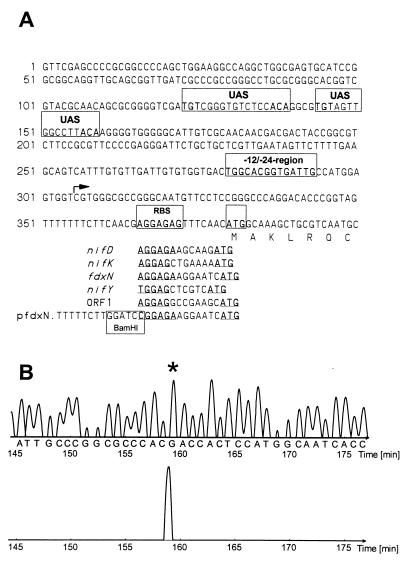
The upstream untranslated region of the nifHDK region harbored sequence homologies to the consensus of ς54-dependent promoters (Fig. 2A). Sequences for putative NifA binding sites (or putative upstream activating sequences) were detected approximately 120 and 140 bp upstream of the ς54 promoter. Putative ribosome binding sites (RBS) in Azoarcus sp. strain BH72 contained a minimal consensus of four or five bases (aGGAG) at a 6- or 7-base distance from the possible start codon (Fig. 2A), which is ATG. The transcriptional start site was verified by primer extension studies (Fig. 2B).
FIG. 2.
Promoter region of nifHDK. (A) Sequence of the genomic region upstream of nifH and of the fusion of the promoter region of nifH fused to fdxN in plasmid pfdxN. The arrow indicates the putative transcription start site, and the unlabeled box indicates the possible start codon. Regulatory sequences are boxed and labeled: UAS, putative upstream activating sequence; −12/−24 region, putative ς54-dependent promoter region; RBS, putative Shine-Dalgarno sequence of nifH. The putative RBS of the other genes of the nifHDK region are given at the bottom, as well as the sequence of the fusion region in plasmid pfdxN. (B) Primer extension analysis localizing the transcriptional start at minute 158.6, corresponding to nucleotides 541/542 (159 and 158.2 min). Top, sequencing reaction; bottom, primer extension.
Analysis of the nifHDK mRNA.
Nitrogenase genes in Azoarcus sp. strain BH72 are clustered in a region of approximately 6.3 kb (Fig. 1) covering six open reading frames. Since putative promoter sequences were detected only upstream of the nifH gene, these genes are likely to be part of the same operon.
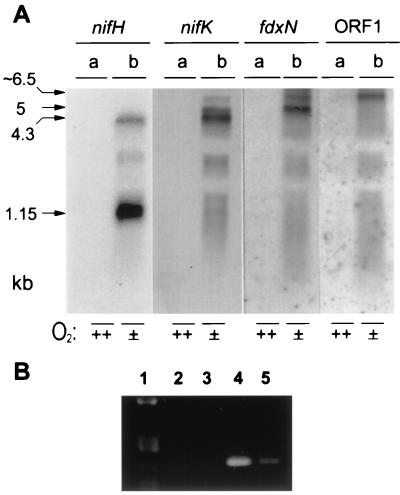
In order to analyze whether the six genes formed one transcriptional unit, Northern blot analysis of RNA extracted from Azoarcus sp. strain BH72 was carried out with gene probes targeted to different genes of the nifHDK region (Fig. 3). All four probes hybridized only to RNA extracted from N2-fixing cells, where they detected several apparently overlapping transcripts. A fragment of approximately 6 kb which is the same size as the entire nifH-orf1 region was detected with an orf1 probe; however, the signal was weak, requiring long exposure times. An RNA fragment of the same length hybridized with the fdxN probe; however, with this probe a stronger hybridization signal appeared at approximately 5 kb, corresponding in size to the nifH-fdxN region (Fig. 3). A nifK probe hybridized with three fragments of 6.5, 5, and 4.3 kb, the latter corresponding in size to the nifH-nifK region, which was most intensely stained. Lowest exposure times were required for the nifH probe, which hybridized most strongly with a 1.15-kb band (corresponding to nifH alone) and less intensely with the 4.3-kb fragment (Fig. 3). This mRNA hybridization pattern indicated that all genes are localized on one large transcript, which occurs, however, at low abundance. Shorter transcripts sequentially missing genes of the 3′ part of the region were more abundant, which might be due to multiple transcriptional termination sites or sequential degradation of the original transcript from its 3′ end in defined steps.
FIG. 3.
Analysis of the nifHDK mRNA. (A) Northern blot analysis with RNA from Azoarcus sp. strain BH72 cells grown aerobically (lanes a) in VM medium containing combined nitrogen and microaerobically (lanes b) on N2. Hybridization was carried out with probes directed against nifH, nifK, fdxN, and ORF1 as indicated above the lanes. Blank areas in lanes are of the sizes of rRNA. (B) RT-PCR using RNA of N2-fixing cells of strain BH72 and primers annealing to nifK and fdxN, spanning 384 bp. Products were separated on a 1.5% agarose gel. Lane 1, size marker (lambda DNA digested with PstI); lane 2, negative control (no RNA added); lane 3, RT inactivated by incubation at 95°C for 5 min prior to addition of 1.5 μg of RNA; lane 4, 1.5 μg of RNA added without heat inactivation; lane 5, 60 ng of chromosomal DNA of strain BH72 added to the RT-PCR mixture.
To prove that the nifHDK operon was transcriptionally linked with downstream genes, a PCR involving an RT step was carried out with RNA of the nitrogen-fixing strain BH72. The first primer for the RT reaction and the PCR was designed to anneal to the 5′ end of the ferredoxin gene, while the second primer was targeted to nifK. An RT-PCR product of the expected size was detected only in the presence of RNA and active RT (Fig. 3B, lane 4) and not after heat inactivation of RT (control for DNA contamination) (lane 3) or without addition of RNA (lane 2).
Role of the ferredoxin FdxN in nitrogen fixation.
In order to investigate the role of the nif operon-encoded ferredoxin in nitrogen fixation, we constructed an in-frame deletion mutant of fdxN in Azoarcus sp. strain BH72. The mutant (BHΔfdxN) was still able to fix nitrogen in semisolid (0.2% agar) N-free medium. Whether nitrogen fixation reached wild-type levels was elucidated by quantitation of growth rates under nitrogen-fixing conditions.
To subject the wild type and the ΔfdxN mutant of Azoarcus to exactly the same growth conditions, the strains were cultivated in a mixed culture on N2 in an oxygen-controlled bioreactor at initially identical cell numbers. The culture was grown to an OD578 of ∼1.5, corresponding to six generations of bacterial growth (Fig. 4A). In three independent experiments, cultures were tested for the distribution of the different genotypes at the first and the last time points by colony hybridization with a probe directed against the fdxN gene of strain BH72; in one experiment this was tested at several time points (Fig. 4B). While the proportions of wild-type and mutant strains were approximately 50% in the beginning of the experiment, the relative amount of the ΔfdxN mutant decreased during the course of the experiment (Fig. 4B). After 20 h or six generations, the wild type dominated the culture by a ratio of 4:1. Calculations of individual growth rates from these data revealed that the mutant BHΔfdxN had only 59% ± 9% of the growth rate of the wild type.
To investigate whether this growth deficiency was due to a decreased electron flux to nitrogenase or to other cellular processes, quantitative measurements of nitrogenase activity were carried out. To verify that the deficiency was due solely to the lack of FdxN and not to the destabilization of the nifHDK transcript, we complemented the fdxN gene in trans under the control of the nifH promoter of strain BH72, which was cloned into the mobilizable broad-host-range plasmid pLAFR3 to create pfdxN. Wild-type and mutant cells were precultured on N2 in separate closed batch cultures and then transferred to fresh medium in microaerobic flasks (1% O2) at 37°C (two experiments with three replicates each); after 3 h of incubation, acetylene (5%) was added, and the ethylene formation was quantified by gas chromatography after 1 h of incubation. While the wild type reduced 29.4 ± 2.4 μmol of acetylene/h/mg of protein, the fdxN deletion mutant reached significantly lower values (P < 1%) of 23.9 ± 1.5 μmol of acetylene/h/mg of protein. This corresponded to approximately 81% of the wild-type nitrogenase activity rate. The complemented strain BHΔfdxN(pfdxN) restored the rates to 32.3 ± 1.7 μmol/h/mg of protein (110%, not significantly different from the wild-type value). Thus, a decrease in nitrogenase activity was observed which could be fully complemented by adding the fdxN gene in trans, suggesting that nitrogenase activity and thus also growth on N2 were specifically affected by an impaired electron flow due to the lack of fdxN in strain BH72.
Because the diazotrophic growth was not completely abolished by the deletion of fdxN, we speculated that other electron donors, either other ferredoxins or flavodoxins, may be present in Azoarcus sp. strain BH72. In search of other, related ferredoxin genes chromosomal DNA was hybridized with a fdxN gene probe at low stringency. Specific hybridization signals were not observed at a hybridization temperature of 40°C with subsequent washing in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (data not shown). Thus, highly related ferredoxins were not detectable under these conditions. Whether flavodoxins may be alternatively used remains to be tested in the future.
Role of the ferredoxin FdxN for N2 fixation of diazosome-containing cells.
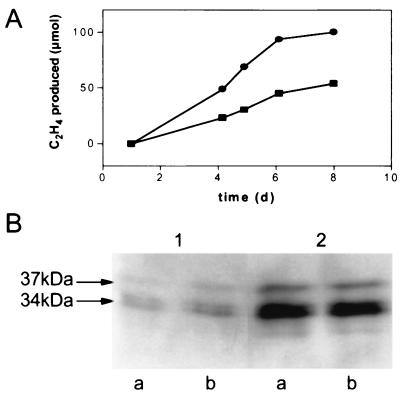
Coculture of Azoarcus sp. strain BH72 with the ascomycete Acremonium alternatum 2003 leads to formation of diazosomes, which occur in strain BH72 in a state of augmented activity and efficiency of nitrogen fixation (18, 20). Therefore, the effect of the fdxN mutation on nitrogen fixation was also analyzed in cocultures. The total ethylene formed per flask, measured when oxygen in the headspace had decreased to 2% at days 5 to 7, was determined in three independent experiments with five flasks each of strain BH72 and BHΔfdxN. Acetylene reduction of the mutant (6.9 ± 2.0 μmol of ethylene formed per flask) was significantly (P < 0.0001) different from that of the wild type (12.2 ± 2.6 μmol), corresponding to 56.6% of the wild-type fixation rate. A decreased acetylene reduction activity of the mutant was observed throughout the incubation period (Fig. 5A), when ethylene accumulates while the oxygen concentration decreases from 21 to 2% due to fungal and bacterial respiration (20, 32). Western blot analyses showed that comparable amounts of nitrogenase Fe protein were present in both cultures at the end of the experiment (Fig. 5B), indicating that the difference was caused not by decreased bacterial growth but by decreased nitrogenase activity. In both mutant and wild-type cells, diazosomes were detected (data not shown). Accordingly, an iron protein of nitrogenase of higher apparent molecular weight was observed (Fig. 5B), which is a covalently modified protein formed in diazosome-containing cells (20).
FIG. 5.
Nitrogen fixation of the wild type and the fdxN deletion mutant in coculture. (A) Nitrogen fixation was measured as the amount of acetylene reduction per flask of Acremonium alternatum 2003 cocultured with Azoarcus sp. strain BH72 (circles) or with the mutant BHΔfdxN (squares). (B) Western blot analysis of cocultures with BH72 (lanes a) or BHΔfdxN (lanes b). Total protein extract of the coculture was diluted 2-fold (lanes 1) and 10-fold (lanes 2) and used for Western blot analysis with antibodies directed against the Fe protein of nitrogenase (34 and 37 kDa).
Effect of FdxN on rapid switch-off of nitrogenase in response to ammonium addition.
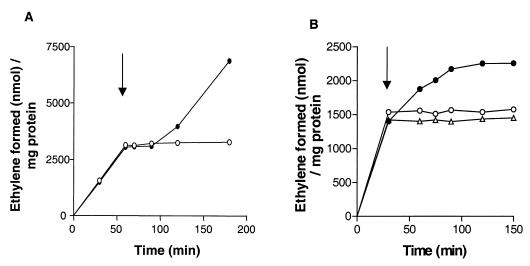
Addition of 2 mM NH4Cl to a nitrogen-fixing culture of Azoarcus sp. strain BH72 led to fast and complete (100%) inhibition of acetylene reduction, while cultures in N-free medium continued reducing acetylene. This nitrogenase switch-off was reversible, because nitrogenase activity was recovered within 30 to 50 min when only 0.2 mM ammonium was added (Fig. 6A), which was rapidly consumed to values below the detection limit (approximately 1 μM) by the bacteria within this time (data not shown). To assess whether the ferredoxin FdxN is involved in the process of nitrogenase inhibition, the deletion mutant BHΔfdxN with and without complementation of fdxN in trans was tested in switch-off experiments using 2 mM NH4Cl. The complemented fdxN mutant showed almost wild-type-level inhibition of nitrogenase activity, while the fdxN mutant continued reducing acetylene (Fig. 6B). A rapid, complete switch-off was thus not observed in the fdxN mutant, but a slow retardation of nitrogenase activity which might be due to the repression of transcription of nifHDK genes by ammonium was observed.
FIG. 6.
Effects of ammonium addition on nitrogenase activity (acetylene reduction) of N2-fixing cultures. (A) Reversible, fast, and complete inhibition (switch-off) of nitrogenase activity in Azoarcus sp. strain BH72 upon addition of 0.2 (closed circles) or 2 mM (open circles) NH4Cl (final concentration). (B) Influence of ammonium addition (2 mM) on nitrogenase activity of BH72 (wild type; open circles), BHΔfdxN(pfdxN) (complemented mutant; triangles), and BHΔfdxN (deletion mutant; closed circles). Results are from one representative of three independent experiments where similar kinetics were observed.
DISCUSSION
Structural genes for the nitrogenase enzyme complex are often cotranscribed in one operon in bacteria. The structural nitrogenase genes nifHDK in Azoarcus sp. strain BH72 occur in a single copy on a large transcript that includes three more putative open reading frames, as shown by Northern hybridization and RT-PCR. While in A. vinelandii and K. pneumoniae the nifHDK genes are followed by nifT, a gene of unknown and nonessential function for nitrogen fixation (21, 55), the downstream gene in strain BH72 showed high sequence identities to bacterial 2[4Fe-4S] ferredoxins. Downstream of the fdxN gene, the operon structure resembles the situation in A. vinelandii with a nifY homologue and an open reading frame with weak homology to ORF1 (21). NifY is known to be involved in the maturation of nitrogenase (14) or may have a role in sensing and signaling the activity status of nitrogenase with respect to regulating nifHDK mRNA stability in K. pneumoniae (54), while a mutation in ORF1 had no obvious phenotype in A. vinelandii (21).
The nifHDK operon in Azoarcus sp. strain BH72 is transcribed only under N-limiting and microaerobic conditions (6). This is a common feature of free-living nitrogen-fixing bacteria, mediated by ς54-dependent promoters (39). Also in this Azoarcus strain, the transcriptional start of the nifHDK mRNA corresponded in distance to −12/−24 promoter regions typical for ς54-dependent promoters. Initiation of transcription from a ς54-bound RNA polymerase needs to be facilitated by additional transcriptional activators (e.g., NifA or NtrC). Putative binding sequences for NifA were detected 120 and 140 bp upstream of the promoter region, suggesting a NifA-mediated regulation of nitrogen fixation in Azoarcus sp. strain BH72, as is commonly found in diazotrophic Proteobacteria (39).
In Northern blot analysis of the nifHDK fdxN nifY ORF1 operon of strain BH72, several different transcripts were observed, the full-length transcript appearing to be least abundant. As observed for Azospirillum brasilense (4), the nifH transcript appeared to be most abundant. Multiple transcripts of nifHDK mRNA were also observed for A. vinelandii (22) and R. capsulatus (57). Inverted repeat sequences potentially capable of forming stable stem-loop structures were detected in the intergenic regions of the latter two bacteria and also in strain BH72 between nifHD, nifK fdxN, and fdxN nifY. They might lead to differential termination of the transcript, or, as speculated for R. capsulatus (57), they may be a target for intramolecular processing of the nifHDK mRNA. Subsequent degradation of the full-length transcript from the 3′ end, giving stable intermediates that resisted RNA degradation, would be an alternative explanation for the transcript pattern observed in strain BH72. Formation of stem-loop structures may assist to protect RNA from 3′-end degradation (2). Differential stability of mRNA as a form of regulation was also demonstrated for other bacterial gene clusters, such as the malEFG operon in Escherichia coli (40) and the puf operon in R. capsulatus (34).
The close transcriptional linkage of the ferredoxin gene fdxN with the structural nif genes may imply a role for electron transport to nitrogenase in Azoarcus sp. strain BH72. 2[4Fe-4S] ferredoxins as electron carriers with a strong negative redox potential of −400 mV are known to play various roles in cellular electron transport. Ferredoxins and flavodoxins are proposed to be electron donors for nitrogenase in bacteria. In some organisms, such as A. vinelandii (26), R. capsulatus (52), or Sinorhizobium meliloti (33), nif-specific ferredoxins that are encoded in nif regions other than nifHDK have been identified. The S. meliloti ferredoxin was shown to be essential for symbiotic N2 fixation with legumes. Localization of a ferredoxin gene in an operon of structural nitrogenase genes, which we describe here for an Azoarcus sp., has been reported only for A. vinelandii, where a ferredoxin-like gene is localized downstream of vnfH, which encodes the iron protein of vanadium nitrogenase (50).
Mutational and genetic complementation experiments in Azoarcus sp. strain BH72 showed that FdxN plays an important but not essential role in nitrogen fixation. The in-frame deletion of the fdxN gene reduced nitrogenase activity and diazotrophic growth in pure culture as well as nitrogen fixation of diazosome-containing cells in coculture with the ascomycete Acremonium alternatum 2003 to a comparable degree (81, 59, and 56% of the wild-type rate, respectively). This defect is most likely due to a less efficient electron transport to dinitrogenase reductase in the absence of FdxN and not to destabilization of the nitrogenase gene mRNA, since nitrogenase activity could be fully restored by complementation of fdxN in trans. A similar nonessential role of a ferredoxin as an electron donor to nitrogenase was found, e.g., in Anabaena sp. (38), while in S. meliloti (33) and R. capsulatus (30) one ferredoxin was essential for nitrogen fixation. As no other ferredoxin gene with high sequence identity could be detected in Azoarcus sp., it is not clear whether the residual electron transport to nitrogenase is due to alternative ferredoxins or flavodoxins. As for Azoarcus sp., for most heterotrophic bacteria it is not yet known how ferredoxins are reduced. For R. capsulatus, the set of membrane-bound and Fe-S cluster-containing proteins of the rnf operon has been discussed as a candidate for electron donation to ferredoxins involved in N2 fixation (28). The proteins show homology to NADH:ubiquinone oxidoreductase from Vibrio alginolyticus. (35). Analogous operons are also present in the fully sequenced genomes of E. coli and Haemophilus influenzae and suggest a general occurrence in bacteria (28).
That a ferredoxin can be essential for the physiological nitrogenase inactivation as detected in the Azoarcus sp. is a novel observation. Certain bacteria fixing N2 react to a supply of ammonium rapidly by inactivation of nitrogenase activity (43, 49). The so-called nitrogenase switch-off by ammonium depends on two different mechanisms. In some diazotrophs, such as Rhodospirillum rubrum (43), R. capsulatus (31), and Azospirillum brasilense (9), the iron protein of nitrogenase (NifH) is subject to posttranslational modification, a reversible mono-ADP-ribosylation at a specific arginine residue. Additionally, a physiological switch-off mechanism which does not involve this covalent modification of nitrogenase exists in some bacteria (42). The mechanism is still unknown. Here we report that such a rapid reversible physiological switch-off mechanism also occurs in Azoarcus sp. strain BH72, similar to those in R. capsulatus and Azospirillum brasilense in terms of speed and extent of nitrogenase inhibition. Nitrogenase activity was almost completely abolished within a few minutes upon addition of ammonium. In contrast, nitrogenase switch-off in Rhodospirillum rubrum occurs more slowly and is incomplete (60). In Rhodospirillum rubrum, ammonium-induced switch-off was shown to be absolutely dependent on ADP ribosylation of NifH (36, 61), whereas in R. capsulatus and Azospirillum brasilense, modification of NifH is not absolutely required, indicating a second mechanism of regulation (7, 8, 42, 58, 61). Surprisingly, the rapid complete switch-off in response to ammonium was abolished in the fdxN deletion mutant of Azoarcus sp. strain BH72; only a slow decrease of nitrogenase activity was still observed. This suggested that the ferredoxin may be part of the mechanism of the process of rapid switch-off. This would be in concordance with the hypothesis that changes in the redox state of NifH or electron flux towards nitrogenase may be factors involved (12, 42), but no sensor or signal transduction proteins have been identified up to now. The essential role of FdxN for nitrogenase switch-off also suggests that it is the major electron donor for nitrogenase in wild-type BH72: putative alternative electron donors are apparently not able to compensate for the switch-off function in the deletion mutant, although they still allow approximately 50% of the nitrogen fixation activity. Therefore, they are unlikely to operate in the wild type but might be more abundant in the deletion mutant, as speculated for R. capsulatus (30).
ACKNOWLEDGMENTS
We are grateful to P. W. Ludden for the kind gift of antibodies to dinitrogenase reductase of Rhodospirillum rubrum, to Thomas Hurek (Max-Planck-Institut für marine Mikrobiologie, Bremen, Germany) for electron-microscopic inspection of cells for diazosomes, and to Jan Gielen in Marc Van Montagu's Laboratorium Genetika, Ghent, Belgium, for help with sequencing parts of the nifHDK operon.
We thank the Deutsche Forschungsgemeinschaft, who supported this work (Re756/5-1/2).
REFERENCES
- 1.Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 2.Alifano P, Bruni C B, Carlomango M S. Control of mRNA processing and decay in procaryotes. Genetica. 1994;94:157–172. doi: 10.1007/BF01443430. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 4.De Zamaroczy M, Delorme F, Elmerich C. Regulation of transcription and promoter mapping of the structural genes for nitrogenase nifHDK of Azospirillum brasilense Sp7. Mol Gen Genet. 1989;220:88–94. doi: 10.1007/BF00260861. [DOI] [PubMed] [Google Scholar]
- 5.Dörr J, Hurek T, Reinhold-Hurek B. Type IV pili are involved in plant-microbe and fungus-microbe interactions. Mol Microbiol. 1998;30:7–17. doi: 10.1046/j.1365-2958.1998.01010.x. [DOI] [PubMed] [Google Scholar]
- 6.Egener T, Hurek T, Reinhold-Hurek B. Endophytic expression of nif genes of Azoarcus sp. strain BH72 in rice roots. Mol Plant-Microb Interact. 1999;12:813–819. doi: 10.1094/MPMI.1998.11.1.71. [DOI] [PubMed] [Google Scholar]
- 7.Fedorov A S, Troshina O U, Laurinavichene T V, Glazer V M, Babykin M M, Zinchenko V V, Yakunin A F, Tsygankov A A. Regulatory effect of ammonium on the nitrogenase activity of Rhodobacter sphaeroides and Rhodobacter capsulatus is not mediated by ADP-ribosylation of the Fe-protein of nitrogenase. Microbiology. 1998;67:610–615. [Google Scholar]
- 8.Förster B, Maner K, Fassbinder F, Oelze J. Reversible inactivation of nitrogenase in Rhodobacter capsulatus strain W107I deleted in the draTG gene region. FEMS Microbiol Lett. 1999;170:167–171. [Google Scholar]
- 9.Fu H, Hartmann A, Lowery R G, Fitzmaurice W P, Roberts G P, Burris R H. Posttranslational regulatory system for nitrogenase activity in Azospirillum spp. J Bacteriol. 1989;171:4679–4685. doi: 10.1128/jb.171.9.4679-4685.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golinelli M P, Gagnon J, Meyer J. Specific interaction of the [2Fe-2S] ferredoxin from Clostridium pasteurianum with the nitrogenase MoFe protein. Biochemistry. 1997;36:11797–11803. doi: 10.1021/bi970528p. [DOI] [PubMed] [Google Scholar]
- 11.Grabau C, Schatt E, Jouanneau Y, Vignais P M. A new [2Fe-2S] ferredoxin from Rhodobacter capsulatus. J Biol Chem. 1991;266:3294–3299. [PubMed] [Google Scholar]
- 12.Halbleib C M, Zhang Y, Roberts G P, Ludden P W. Effects of perturbations of the nitrogenase electron transfer chain on reversible ADP-ribosylation of nitrogenase Fe protein in Klebsiella pneumoniae strains bearing the Rhodospirillum rubrum dra operon. J Bacteriol. 2000;182:3681–3687. doi: 10.1128/jb.182.13.3681-3687.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 14.Homer M J, Paustian T D, Shah V K, Roberts G P. The nifY product of Klebsiella pneumoniae is associated with apodinitrogenase and dissociates upon activation with the iron-molybdenum cofactor. J Bacteriol. 1993;175:4907–4910. doi: 10.1128/jb.175.15.4907-4910.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurek T, Egener T, Reinhold-Hurek B. Divergence in nitrogenases of Azoarcus spp., Proteobacteria of the β subclass. J Bacteriol. 1997;179:4172–4178. doi: 10.1128/jb.179.13.4172-4178.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hurek T, Reinhold B, Fendrik I, Niemann E G. Root-zone-specific oxygen tolerance of Azospirillum spp. and diazotrophic rods closely associated with Kallar grass. Appl Environ Microbiol. 1987;53:163–169. doi: 10.1128/aem.53.1.163-169.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hurek T, Reinhold-Hurek B. Interactions of Azoarcus sp. with rhizosphere fungi. In: Varma A, Hock B, editors. Mycorrhiza. 2nd ed. Berlin, Germany: Springer Verlag; 1998. pp. 595–614. [Google Scholar]
- 18.Hurek T, Reinhold-Hurek B, Turner G L, Bergersen F J. Augmented rates of respiration and efficient nitrogen fixation at nanomolar concentrations of dissolved O2 in hyperinduced Azoarcus sp. strain BH72. J Bacteriol. 1994;176:4726–4733. doi: 10.1128/jb.176.15.4726-4733.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurek T, Reinhold-Hurek B, Van Montagu M, Kellenberger E. Root colonization and systemic spreading of Azoarcus sp. strain BH72 in grasses. J Bacteriol. 1994;176:1913–1923. doi: 10.1128/jb.176.7.1913-1923.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurek T, Van Montagu M, Kellenberger E, Reinhold-Hurek B. Induction of complex intracytoplasmic membranes related to nitrogen fixation in Azoarcus sp. BH72. Mol Microbiol. 1995;18:225–236. doi: 10.1111/j.1365-2958.1995.mmi_18020225.x. [DOI] [PubMed] [Google Scholar]
- 21.Jacobson M R, Brigle K E, Bennett L T, Setterquist R A, Wilson M S, Cash V L, Beynon J, Newton W E, Dean D R. Physical and genetic map of the major nif gene cluster from A. vinelandii. J Bacteriol. 1989;171:1017–1027. doi: 10.1128/jb.171.2.1017-1027.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobson M R, Premakumar R, Bishop P E. Transcriptional regulation of nitrogen fixation by molybdenum in Azotobacter vinelandii. J Bacteriol. 1986;167:480–486. doi: 10.1128/jb.167.2.480-486.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.James E K, Olivares F L. Infection and colonization of sugar cane and other graminaceous plants by endophytic diazotrophs. Crit Rev Plant Sci. 1998;17:77–119. [Google Scholar]
- 24.James E K, Olivares F L, Baldani J I, Döbereiner J. Herbaspirillum, an endophytic diazotroph colonizing vascular tissue in leaves of Sorghum bicolor L. Moench. J Exp Bot. 1997;48:785–797. [Google Scholar]
- 25.James E K, Reis V M, Olivares F L, Baldandi J I, Döbereiner J. Infection of sugar cane by the nitrogen-fixing bacterium Acetobacter diazotrophicus. J Exp Bot. 1994;45:757–766. [Google Scholar]
- 26.Joerger R D, Bishop P E. Nucleotide sequence and genetic analysis of the nifB-nifQ region from Azotobacter vinelandii. J Bacteriol. 1988;170:1475–1487. doi: 10.1128/jb.170.4.1475-1487.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones R, Woodley P, Robson R. Cloning and organization of some genes for nitrogen fixation from Azotobacter chroococcum and their expression in Klebsiella pneumoniae. Mol Gen Genet. 1984;197:318–327. doi: 10.1007/BF00330980. [DOI] [PubMed] [Google Scholar]
- 28.Jouanneau Y, Jeong H S, Hugo N, Meyer C, Willison J C. Overexpression in Escherichia coli of the rnf genes from Rhodobacter capsulatus: characterization of two membrane-bound iron-sulfur proteins. Eur J Biochem. 1998;251:54–64. doi: 10.1046/j.1432-1327.1998.2510054.x. [DOI] [PubMed] [Google Scholar]
- 29.Jouanneau Y, Meyer C, Gaillard J, Forest E, Gagnon J. Purification and characterization of a novel dimeric ferredoxin (FdIII) from Rhodobacter capsulatus. J Biol Chem. 1993;268:10636–10644. [PubMed] [Google Scholar]
- 30.Jouanneau Y, Meyer C, Naud I, Klipp W. Characterization of an fdxN mutant of Rhodobacter capsulatus indicates that ferredoxin I serves as electron donor to nitrogenase. Biochim Biophys Acta. 1995;1232:33–42. doi: 10.1016/0005-2728(95)00106-x. [DOI] [PubMed] [Google Scholar]
- 31.Jouanneau Y, Roby C, Meyer C M, Vignais P M. ADP-ribosylation of dinitrogenase reductase in Rhodobacter capsulatus. Biochemistry. 1989;28:6524–6530. [Google Scholar]
- 32.Karg T, Reinhold-Hurek B. Global changes in protein composition of N2-fixing Azoarcus sp. strain BH72 upon diazosome formation. J Bacteriol. 1996;178:5748–5754. doi: 10.1128/jb.178.19.5748-5754.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klipp W, Reilander H, Schülter A, Krey R, Pühler A. The Rhizobium meliloti fdxn gene encoding a ferredoxin-like protein is necessary for nitrogen fixation and is cotranscribed with nifA and nifB. Mol Gen Genet. 1989;216:293–302. doi: 10.1007/BF00334368. [DOI] [PubMed] [Google Scholar]
- 34.Klug G. The role of mRNA degradation in the regulated expression of bacterial photosynthesis genes. Mol Microbiol. 1993;9:1–7. doi: 10.1111/j.1365-2958.1993.tb01663.x. [DOI] [PubMed] [Google Scholar]
- 35.Kumagai H, Fujiwara T, Matsubara H, Saeki K. Membrane localization, topology, and mutual stabilization of the rnfABC gene products in Rhodobacter capsulatus and implications for a new family of energy-coupling NADH oxidoreductases. Biochemistry. 1997;36:5509–5521. doi: 10.1021/bi970014q. [DOI] [PubMed] [Google Scholar]
- 36.Liang J, Nielsen G M, Lies D P, Burris R H, Roberts G P, Ludden P W. Mutations in the draT and draG genes of Rhodospirillum rubrum result in loss of regulation of nitrogenase by reversible ADP-ribosylation. J Bacteriol. 1991;173:6903–6909. doi: 10.1128/jb.173.21.6903-6909.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin D, Hurek T, Reinhold-Hurek B. Occurrence of three PII-like signal transmitter proteins in the diazotroph Azoarcus sp. BH72. Mol Microbiol. 2000;38:276–288. doi: 10.1046/j.1365-2958.2000.02095.x. [DOI] [PubMed] [Google Scholar]
- 38.Masepohl B, Schölisch K, Görlitz K, Kutzki C, Böhme H. The heterocyst-specific fdxH gene product of the cyanobacterium Anabaena sp. PCC 7120 is important but not essential for nitrogen fixation. Mol Gen Genet. 1997;253:770–776. doi: 10.1007/s004380050383. [DOI] [PubMed] [Google Scholar]
- 39.Merrick M. Regulation of nitrogen fixation genes in free-living and symbiotic bacteria. In: Stacey G, Burris R, Evans H, editors. Biological nitrogen fixation. New York, N.Y: Chapman & Hall; 1992. pp. 835–876. [Google Scholar]
- 40.Newbury S F, Smith N H, Higgins C F. Differential mRNA stability controls relative gene expression within a polycistronic operon. Cell. 1987;51:1131–1143. doi: 10.1016/0092-8674(87)90599-x. [DOI] [PubMed] [Google Scholar]
- 41.Nieva-Gomez D, Roberts G P, Klevickis S, Brill W. Electron transport to nitrogenase in Klebsiella pneumoniae. Proc Natl Acad Sci USA. 1980;77:2555–2558. doi: 10.1073/pnas.77.5.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pierrard J, Ludden P W, Roberts G P. Posttranslational regulation of nitrogenase in Rhodobacter capsulatus: existence of two independent regulatory effects of ammonium. J Bacteriol. 1993;175:1358–1366. doi: 10.1128/jb.175.5.1358-1366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pope M R, Murell S A, Ludden P W. Covalent modification of the iron protein of nitrogenase from Rhodospirillum rubrum by adenine diphosphoribosylation of a specific arginine residue. Proc Natl Acad Sci USA. 1985;82:3173–3177. doi: 10.1073/pnas.82.10.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reinhold B, Hurek T, Fendrik I. Strain-specific chemotaxis of Azospirillum spp. J Bacteriol. 1985;162:190–195. doi: 10.1128/jb.162.1.190-195.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reinhold B, Hurek T, Niemann E-G, Fendrik I. Close association of Azospirillum and diazotrophic rods with different root zones of Kallar grass. Appl Environ Microbiol. 1986;52:520–526. doi: 10.1128/aem.52.3.520-526.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reinhold-Hurek B, Hurek T. Life in grasses: diazotrophic endophytes. Trends Microbiol. 1998;6:139–144. doi: 10.1016/s0966-842x(98)01229-3. [DOI] [PubMed] [Google Scholar]
- 47.Reinhold-Hurek B, Hurek T, Claeyssens M, Van M M. Cloning, expression in Escherichia coli, and characterization of cellulolytic enzymes of Azoarcus sp., a root-invading diazotroph. J Bacteriol. 1993;175:7056–7065. doi: 10.1128/jb.175.21.7056-7065.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reinhold-Hurek B, Hurek T, Gillis M, Hoste B, Vancanneyt M, Kersters K, De Ley J. Azoarcus gen. nov., nitrogen-fixing proteobacteria associated with roots of Kallar grass (Leptochloa fusca (L.) Kunth), and description of two species, Azoarcus indigens sp. nov. and Azoarcus communis sp. nov. Int J Syst Bacteriol. 1993;43:574–584. [Google Scholar]
- 49.Roberts G P, Ludden P W. Nitrogen fixation by photosynthetic bacteria. In: Stacey G, Burris R H, Evans H J, editors. Biological nitrogen fixation. New York, N.Y: Chapman & Hall; 1992. pp. 135–165. [Google Scholar]
- 50.Robson R, Woodley P, Jones R. Second gene nifH coding for a nitrogenase iron protein in Azotobacter chroococcum is adjacent to a gene coding for a ferredoxin-like protein. EMBO J. 1986;5:1159–1164. doi: 10.1002/j.1460-2075.1986.tb04341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schatt E, Jouanneau Y, Vignais P M. Molecular cloning and sequence analysis of the structural gene of ferredoxin I from the photosynthetic bacterium Rhodobacter capsulatus. J Bacteriol. 1989;171:6218–6226. doi: 10.1128/jb.171.11.6218-6226.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schrautemeier B, Cassing A, Boehme H. Characterization of the genome region encoding an FdxH-type ferredoxin and a new 2(4F3–4S) ferredoxin from the nonheterocystous, nitrogen-fixing cyanobacterium Plectonema boryanum PCC 73110. J Bacteriol. 1994;176:1037–1046. doi: 10.1128/jb.176.4.1037-1046.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simon H M, Gosink M M, Roberts G P. Importance of cis determinants and nitrogenase activity in regulated stability of the Klebsiella pneumoniae nitrogenase structural gene mRNA. J Bacteriol. 1999;181:3751–3760. doi: 10.1128/jb.181.12.3751-3760.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simon H M, Homer M J, Roberts G P. Perturbation of nifT expression in Klebsiella pneumoniae has limited effect on nitrogen fixation. J Bacteriol. 1996;178:2975–2977. doi: 10.1128/jb.178.10.2975-2977.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willison J C, Pierrard J, Huebner P. Sequence and transcript analysis of the nitrogenase structural gene operon (nifHDK) of Rhodobacter capsulatus: evidence for intramolecular processing of nifHDK mRNA. Gene. 1993;133:39–46. doi: 10.1016/0378-1119(93)90222-o. [DOI] [PubMed] [Google Scholar]
- 58.Yakunin A F, Hallenbeck P C. Short-term regulation of nitrogenase activity by NH4+ in Rhodobacter capsulatus: multiple in vivo nitrogenase responses to NH4+ addition. J Bacteriol. 1998;180:6392–6395. doi: 10.1128/jb.180.23.6392-6395.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y, Burris R H, Ludden P W, Roberts G P. Comparison studies of dinitrogenase reductase ADP-ribosyl transferase/dinitrogenase reductase activating glycohydrolase regulatory systems in Rhodospirillum rubrum and Azospirillum brasilense. J Bacteriol. 1995;177:2354–2359. doi: 10.1128/jb.177.9.2354-2359.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y, Burris R H, Ludden P W, Roberts G P. Presence of a second mechanism for the posttranslational regulation of nitrogenase activity in Azospirillum brasilense in response to ammonium. J Bacteriol. 1996;178:2948–2953. doi: 10.1128/jb.178.10.2948-2953.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]