Abstract
The opportunistic fungal pathogen Candida albicans can switch spontaneously and reversibly between different cell forms, a capacity that may enhance adaptation to different host niches and evasion of host defense mechanisms. Phenotypic switching has been studied intensively for the white-opaque switching system of strain WO-1. To facilitate the molecular analysis of phenotypic switching, we have constructed homozygous ura3 mutants from strain WO-1 by targeted gene deletion. The two URA3 alleles were sequentially inactivated using the MPAR-flipping strategy, which is based on the selection of integrative transformants carrying a mycophenolic acid (MPA) resistance marker that is subsequently deleted again by site-specific, FLP-mediated recombination. To investigate a possible cell type-independent switching in the expression of individual phase-specific genes, two different reporter genes that allowed the analysis of gene expression at the single-cell level were integrated into the genome, using URA3 as a selection marker. Fluorescence microscopic analysis of cells in which a GFP reporter gene was placed under the control of phase-specific promoters demonstrated that the opaque-phase-specific SAP1 gene was detectably expressed only in opaque cells and that the white-phase-specific WH11 gene was detectably expressed only in white cells. When MPAR was used as a reporter gene, it conferred an MPA-resistant phenotype on opaque but not white cells in strains expressing it from the SAP1 promoter, which was monitored at the level of single cells by a significantly enlarged size of the corresponding colonies on MPA-containing indicator plates. Similarly, white but not opaque cells became MPA resistant when MPAR was placed under the control of the WH11 promoter. The analysis of these reporter strains showed that cell type-independent phase variation in the expression of the SAP1 and WH11 genes did not occur at a detectable frequency. The expression of these phase-specific genes of C. albicans in vitro, therefore, is tightly linked to the cell type.
Pathogenic microorganisms must be able to adapt to changing environmental conditions during the course of an infection to ensure survival and growth in different host niches. This adaptation is achieved by the regulated expression of appropriate sets of genes whose products are needed by the pathogen at a particular stage of an infection, in response to corresponding environmental signals (23). In addition, many prokaryotic and eukaryotic pathogens have the capacity to generate variants with altered properties in a reversible and often random fashion, a phenomenon known as phase or antigenic variation (10). Such variability not only provides an opportunity to produce progeny that are better adapted to a special host niche, but it also enables evasion of the host's immune response to surface-exposed or secreted antigens produced by the initially infecting organisms. Certain traits can be both advantageous and disadvantageous for the microorganism, depending on the host niche or infection stage. The capacity to alter their expression state independently of environmental signals may enhance pathogens' ability to cause disease (6, 12, 21, 31, 45).
Antigenic variation can be achieved by different molecular mechanisms, including inversion of promoter elements or structural genes by site-specific recombination (1, 20), differential methylation of regulatory sequences (7), insertion and deletion of single nucleotides, oligonucleotide repeats, or mobile genetic elements into either promoter or coding regions (12, 44, 46, 50), or movement of silent genes into expression loci by homologous recombination (11, 13, 22). These specific mechanisms ensure that variants are generated at a much higher frequency than would be possible by random mutation alone but still result in a semistable phenotype to allow the expansion of new variants that eventually take over the population, especially after passing through infection bottlenecks (29).
The opportunistic fungal pathogen Candida albicans displays a remarkable capacity to alter its cellular characteristics. Apart from a transition between growth in yeast and hyphal forms, generally referred to as dimorphism, C. albicans can reversibly switch between different cell types that are distinguishable by the appearance of the resulting colonies on agar plates (35). In contrast to the above-mentioned bacterial and protozoal examples of antigenic variation, where usually a single trait is affected, phenotypic switching in C. albicans alters many different cellular properties, including morphology, adhesion, antigenicity, and pathogenicity, in a programmed, coordinated fashion. Most C. albicans strains are capable of phenotypic switching, but the process has been most intensely studied using the white-opaque switching of strain WO-1 as a model system. This strain predominantly switches between the typical budding yeast form, which produces hemispherical, white colonies, and an elongated cell type, which forms flat, opaque colonies (34). A growing list of phase-specifically expressed genes has been discovered, including the white-phase-specific genes WH11 and EFG1 and the opaque-phase-specific genes SAP1 (PEP1), SAP3, OP4, and CDR3 (2, 25, 26, 37, 38, 47). The transition between the white and opaque phases involves the coordinated activation of opaque-phase-specific genes and deactivation of white-phase-specific genes and vice versa, which has been postulated to be governed by a master regulatory switch, the nature of which is still unknown. Switching among a set of preselected cell types may allow a more efficient adaptation to new host niches than randomly altering individual cellular properties (36).
The specific role of many phase-specific genes has not yet been elucidated, but some of them may have a function in the maintenance of the distinct cellular phenotype. The WH11 gene encodes a protein with homology to the heat shock protein Hsp12 and is localized in the cytoplasm of white cells (33). The SAP genes encode secreted aspartic proteinases that have been shown to contribute to the virulence of WO-1 and other C. albicans strains (14, 16). However, Sap antigens are also recognized by the host's immune system (19). Sap antigens have been detected on the surfaces of C. albicans organisms (4, 5, 32) and may thus allow the host to recognize and destroy the invading cells. On the other hand, forced expression of the white-phase-specific gene WH11 in opaque cells resulted in enhanced virulence in a mouse model of systemic candidiasis (17), whereas forced expression of the opaque-phase-specific gene SAP1 in white cells increased the virulence of these cells in a model of cutaneous infection (16). It is therefore conceivable that phase variation of single genes might also be advantageous under conditions that otherwise favor the maintenance of a specific cell type and could thus occur independently from switching between the white and opaque phases by a different molecular mechanism. Such an additional level of variability would further enhance the flexibility of an infecting C. albicans strain.
To date, the expression of phase-specific genes has been analyzed only at the population level, either by Northern hybridization or by using a luciferase reporter enzyme (25, 26, 38, 40). A rare switching in individual cells from the expressed to a nonexpressed state of an otherwise phase-specific gene would therefore not have been detected. Similarly, activation of such a gene in the “wrong” cell type also would probably not be detected, since this could be explained by the presence of a minor proportion of the other cell type in populations of white or opaque cells.
Recently, the GFP gene, encoding the green fluorescent protein (GFP) from Aequorea victoria, has been adapted to the noncanonical codon usage of C. albicans, allowing researchers to monitor the expression of target genes at the level of single cells (8, 27). In addition, we have developed a method for the genetic manipulation of C. albicans wild-type strains (48). Sequential gene disruption can now also be performed in uridine-prototrophic strains like WO-1 by using a marker conferring resistance against mycophenolic acid (MPA) for the selection of transformants and its subsequent deletion by FLP-mediated, site-specific recombination (MPAR flipping). In the present study, we have used these new molecular tools to analyze the expression of phase-specific genes in strain WO-1 at the single-cell level.
MATERIALS AND METHODS
Strains and growth media.
The C. albicans strains used in this study are listed in Table 1. Strains were subcultured separately in the white and opaque phases at room temperature on agar plates containing Lee's medium, pH 6.8 (3), and 5 μg of phloxine B ml−1, which selectively stains opaque colonies pink (35). For routine growth of the strains, YPD liquid medium (10 g of yeast extract, 20 g of peptone, and 20 g of glucose per liter) was used. Cells were grown overnight in YCB-BSA (23.4 g of yeast carbon base, 4 g of bovine serum albumin per liter [pH 4.0]) to induce the SAP2 promoter for excision of the MPAR flipper from MPA-resistant transformants. Screening for MPA-sensitive derivatives was performed after growth on minimal agar plates (6.7 g of yeast nitrogen base without amino acids [Bio 101, Vista, Calif.], 20 g of glucose, 0.77 g of complete supplement medium [Bio 101], and 15 g of agar per liter) containing 1 μg of MPA ml−1 (48). Uridine (100 μg ml−1) was added to the media to support growth of ura3 mutant strains.
TABLE 1.
C. albicans strains used in this study
| Strain | Parent | Genotypea | Reference |
|---|---|---|---|
| WO-1 | Wild-type strain | 34 | |
| WUM1A | WO-1 | ura3-1Δ::MPAR-FLIP/URA3-2 | This study |
| WUM1B | WO-1 | URA3-1/lura3-2Δ::MPAR-FLIP | This study |
| WUM2A | WUM1A | ura3-1Δ::FRT/URA3-2 | This study |
| WUM2B | WUM1B | URA3-1/ura3-2Δ::FRT | This study |
| WUM3A | WUM2A | ura3-1Δ::FRT/ ura3-2Δ::MPAR-FLIP | This study |
| WUM3B | WUM2B | ura3-1Δ::MPAR-FLIP/ura3-2Δ::FRT | This study |
| WUM5A | WUM3A | ura3-1Δ::FRT/ura3-2Δ::FRT | This study |
| WUM5B | WUM3B | ura3-1Δ::FRT/ura3-2Δ::FRT | This study |
| WUM4A | WUM2A | ura3-1Δ::FRT/ura3-2::MPAR-FLIP | This study |
| WUM6A | WUM4A | ura3-1Δ::FRT/ura3-2::FRT | This study |
| WUGS1A | WUM5A | sap1-1::PSAP1-GFP/SAP1-2 | This study |
| WUGS1B | WUM5A | SAP1-1/sap1-2::PSAP1-GFP | This study |
| WUMS1A | WUM5A | sap1-1::PSAP1-MPAR/SAP1-2 | This study |
| WUMS1B | WUM5A | SAP1-1/sap1-2::PSAP1-MPAR | This study |
| WUGW11A and-B | WUM5A | WH11/wh11::PWH11-GFP | This study |
| WUMW11A and-B | WUM5A | WH11/wh11::PWH11-MPAR | This study |
Apart from the indicated features the strains are identical to their parents. MPAR-FLIP denotes the MPAR flipper cassette.
Plasmid constructions.
For inactivation of the URA3 gene, two different plasmid constructions were made. First, a DNA fragment comprising the URA3 coding region plus 420 bp of the upstream and 126 bp of the downstream region that was originally cloned as a SalI-PstI fragment into pBluescript (27) was removed by SalI-BamHI digestion and cloned into pUC18 to yield pURA7. An internal fragment from positions 22 to 228 of the URA3 coding region was subsequently deleted by inverse PCR with the primers URA9 (5′-GCTCTCTCACCGCGGTCTTAGTGTTGAC-3′) and URA8 (5′-CCTATGAATCCACTCGAGAACCATTATTAG-3′), thereby introducing SacII and XhoI sites (underlined) into which a SacII-XhoI fragment containing the MPAR flipper from pSFI1 (48) was inserted instead of the deleted URA3 sequences, resulting in pSFIU2 (Fig. 1A). To delete the whole URA3 gene, larger flanking regions were obtained by screening a C. albicans fosmid library (kindly provided by Stewart Scherer, Acacia Biosciences, Richmond, Calif.) using the insert from pURA7 as a probe. A 4.7-kb XhoI-SacI fragment containing the URA3 gene from a positive clone (16H6) was ligated into pBluescript. Sequence analysis of the resulting plasmid, pURA300, allowed us to design primers for the amplification of URA3-flanking regions. The upstream region from positions −2462 to −394 was obtained by PCR with the primers URA15 (5′-TTTGTGCATGCTGTATTTCCAAAACG-3′) and URA16 (5′-TGTTTCCGCGGATACCATCCAAATCAATTCC-3′) containing introduced SphI and SacII sites (underlined), respectively. The downstream region ranging from positions 114 to 1102 behind the stop codon was amplified with the primers URA17 (5′-CTATTTACAATCTCGAGGTGGTCCTTC-3′) and URA18 (5′-CCATTAATTGCGAGCTCTGCTACTGGA-3′) containing introduced XhoI and SacI sites (underlined), respectively. The SacII-XhoI MPAR flipper fragment from pSFI1 was then ligated together with the SphI-SacII URA3 upstream and the XhoI-SacI URA3 downstream fragments into the SphI/SacI-digested pUC18, resulting in pSFIU4 (Fig. 1A). The SphI-SacI URA3 deletion/disruption fragments from pSFIU4 and pSFIU2 were then used to replace the wild-type URA3 alleles in C. albicans WO-1.
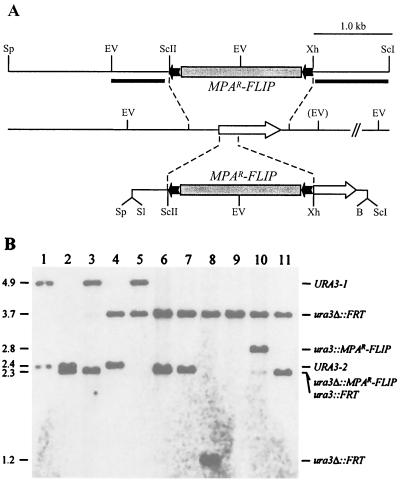
FIG. 1.
Inactivation of the URA3 gene by MPAR flipping. (A) Structure of the URA3 locus in strain WO-1 and allelic replacements using the inserts from pSFIU4 (upper part) or pSFIU2 (lower part). Open arrow, URA3 coding region; solid lines, URA3 upstream and downstream sequences. Only relevant restriction sites are shown: B, BamHI; EV, EcoRV; ScI, SacI; ScII, SacII; Sl, SalI; Sp, SphI; Xh, XhoI. The EcoRV site shown in parentheses is absent from the genomic URA3-1 allele and also from the cloned downstream URA3 fragment in pSFIU4. The 5.6-kb MPAR flipper, details of which have been presented elsewhere (48), is not drawn to scale. Solid bars, DNA fragments used as probes for verification of the correct allelic replacements by Southern hybridization. (B) Southern hybridization of EcoRV-digested genomic DNA of the ura3 mutants using the 5′URA3 fragment as probe. Molecular sizes (in kilobases) are on the left. Lanes: 1, WO-1 (URA3/URA3); 2, WUM1A (ura3-1Δ::MPAR-FLIP/URA3-2); 3, WUM1B (URA3-1/ura3-2Δ::MPAR-FLIP); 4, WUM2A (ura3-1Δ::FRT/URA3-2); 5, WUM2B (URA3-1/ura3-2Δ::FRT); 6, WUM3A (ura3-1Δ::FRT/ura3-2Δ::MPAR-FLIP); 7, WUM3B (ura3-1Δ::MPAR-FLIP/ura3-2Δ::FRT); 8, WUM5A (ura3-1Δ::FRT/ura3-2Δ::FRT); 9, WUM5B (ura3-1Δ::FRT/ura3-2Δ::FRT); 10, WUM4A (ura3-1Δ::FRT/ura3-2::MPAR-FLIP); 11, WUM6A (ura3-1Δ::FRT/ura3-2::FRT).
To construct reporter gene fusions, the promoter regions of the SAP1 and WH11 genes were PCR amplified with the primer pair SAP1P1 (5′-GGTTACGGAAAATCTAGAAGATGGCCC-3′) and SAP1P2 (5′-TGTGTGTCGACTTAGAAATGGAAGAGTGA-3′) and the primer pair WHS5 (5′-CTTGTTTCATCTTCTAGACCCATAGC-3′) and WHS6 (5′-GACATGTCGACTTGTTCTGCTTGTTGTTTTG-3′), respectively, using genomic DNA from strain WO-1 as the template. In addition, sequences from the SAP1 coding and WH11 downstream regions were obtained by PCR with the primer pair SAP1C (5′-GTTATGCTGCAGACATCACTATTGG-3′) and SAP1D (5′-GACCGTTAGCGGAGCTCAACGGAGC-3′) and the primer pair WHS7 (5′-AAACAATGCATGAGTAACCTCAATTGAGTTG-3′) and WHS8 (5′-GTACACCTAACCCGAGCTCACAAGACCTTTG-3′), respectively. The primers were derived from the published C. albicans genome sequence (http://www-sequence.stanford.edu/group/candida); the introduced XbaI, SalI, PstI, NsiI, and SacI sites are underlined. The XbaI-SalI SAP1 and WH11 promoter fragments and the PstI-SacI 3′ SAP1 and NsiI-SacI 3′ WH11 fragments were then substituted for the corresponding XbaI-SalI PSAP2 and PstI-SacI 3′ SAP2 fragments in plasmid pGFP41 (27) from which the PstI site in the polylinker had been removed. These substitutions generated pGFP61 and pGFP68 containing the PSAP1-GFP and PWH11-GFP reporter gene fusions, respectively (Fig. 2A and 3A). To construct fusions with the MPAR marker, a promoterless MPAR gene was first obtained by PCR amplification with the primers IMH1 (5′-AAAACGTCGACAATGGTGTTTGAAACTTCAAAAGC-3′) and IMH2 (5′-GTAGACCGCGGAGGAATGGGCATTATTGTAG-3′) and, after digestion at the introduced SalI and SacII sites (underlined) and blunting of the overhanging ends of the SacII site, used to replace the GFP gene in pGFP61 and pGFP68, thereby generating pS1IU2 and pWH11IU2, which contain the PSAP1-MPAR and PWH11-MPAR reporter gene fusions, respectively (Fig. 2A and 3A). The XbaI-SacI fragments from pGFP61, pGFP68, pS1IU1, and pWH11IU1 were then used to insert the reporter gene fusions into one of the SAP1 or WH11 alleles of ura3-negative derivatives of strain WO-1.
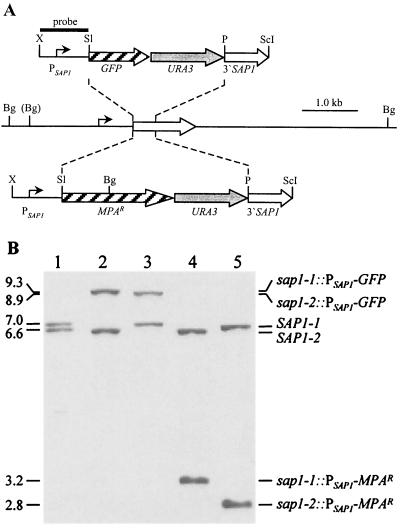
FIG. 2.
Integration of the PSAP1-GFP and PSAP1-MPAR reporter gene fusions into the SAP1 alleles of strain WUM5A. (A) Genomic structure of the SAP1 locus in strain WO-1 and its ura3 derivatives and structure of the inserted reporter gene fusions from plasmids pGFP61 (upper part) and pS1IU2 (lower part). Open arrow, SAP1 coding region; solid lines, SAP1 upstream and downstream regions. Only relevant restriction sites are shown: Bg, BglII; P, PstI; ScI, SacI; Sl, SalI; X, XbaI. The BglII site shown in parentheses is absent from the genomic SAP1-1 allele. Solid bar, DNA fragment used as a probe for verification of the correct allelic replacements by Southern hybridization. (B) Southern hybridization of BglII-digested genomic DNA of the parent strain WUM5A and derivatives carrying the PSAP1-GFP and PSAP1-MPAR reporter gene fusions inserted in either of the SAP1 alleles. Molecular sizes (in kilobases) are on the left. Lanes: 1, WUM5A (SAP1-1/SAP1-2); 2, WUGS1A (sap1-1::PSAP1-GFP/SAP1-2); 3, WUGS1B (SAP1-1/sap1-2::PSAP1-GFP); 4, WUMS1A (sap1-1::PSAP1-MPAR/SAP1-2); 5, WUMS1B (SAP1-1/sap1-2::PSAP1-MPAR).
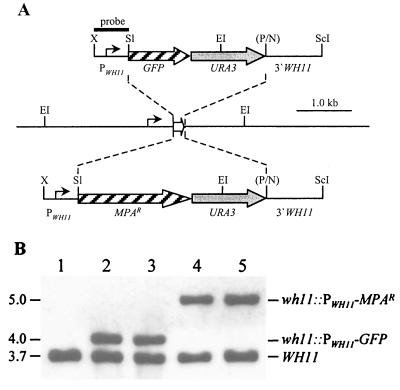
FIG. 3.
Integration of PWH11-GFP and PWH11-MPAR reporter gene fusions into the WH11 locus of strain WUM5A. (A) Genomic structure of the WH11 locus in strain WO-1 and its ura3 derivatives, and structure of the inserted reporter gene fusions from plasmids pGFP68 (upper part) and pWH11IU2 (lower part). Open arrow, WH11 coding region; solid lines, WH11 upstream and downstream regions. Only relevant restriction sites are shown: EI, EcoRI; N, NsiI; P, PstI; ScI, SacI; Sl, SalI; X, XbaI. The PstI and NsiI sites shown in parentheses were destroyed during the cloning procedure. Solid bar, DNA fragment used as a probe for verification of the correct allelic replacements by Southern hybridization. (B) Southern hybridization of EcoRI-digested genomic DNA of the parent strain WUM5A and derivatives carrying the PWH11-GFP and PWH11-MPAR reporter gene fusions inserted in one of the WH11 alleles. Molecular sizes (in kilobases) are on the left. Lanes: 1, WUM5A (WH11/WH11); 2, WUGW11A (WH11/wh11::PWH11-GFP); 3, WUGW11B (WH11/wh11::PWH11-GFP); 4, WUMW11A (WH11/wh11::PWH11-MPAR); 5, WUMW11B (WH11/wh11::PWH11-MPAR).
C. albicans transformation.
C. albicans strains were transformed by electroporation (15) with gel-purified linear DNA fragments from the plasmids described above. MPA-resistant transformants were selected on minimal agar plates containing 10 μg of MPA ml−1. Single colonies were picked after 5 to 7 days of growth at 30°C and restreaked on the same medium. Uridine-prototrophic transformants of ura3 mutants were selected on minimal agar plates without uridine. After confirmation of the correct allelic replacements, white and opaque colonies of the strains were identified by plating on agar plates containing Lee's medium and 5 μg of phloxine B ml−1 and then maintained separately on the same plates at room temperature.
Isolation of chromosomal DNA and Southern hybridization.
Genomic DNA from C. albicans strains was isolated as described by Millon et al. (24). DNA (10 μg) was digested with appropriate restriction enzymes, separated on a 1% (wt/vol) agarose gel, and, after ethidium bromide staining, transferred by vacuum blotting onto a nylon membrane and fixed by UV cross-linking. Southern hybridization with enhanced chemiluminescence (ECL)-labeled probes was performed with the ECL labeling and detection kit from Amersham (Braunschweig, Germany) according to the instructions of the manufacturer.
Fluorescence microscopy.
White and opaque cells of the strains were grown separately or as mixed populations overnight at 25°C in liquid Lee's medium, pH 6.8, and aliquots of the cultures were spotted on microscope slides. Fluorescence was detected using a Zeiss Axiolab microscope equipped for epifluorescence microscopy with a 50-W mercury high-pressure bulb and with the Zeiss fluorescein-specific filter set at 09.
Screening for MPA-resistant and MPA-sensitive white and opaque colonies.
The MPA resistance conferred by the expression of the PSAP1-MPAR and PWH11-MPAR reporter gene fusions was initially assessed by suspending cells from white or opaque colonies of strain WO-1 and the strains carrying these fusions in water and plating appropriate dilutions on agar plates (7.5-cm diameter) containing Lee's medium supplemented with 5 μg of phloxine B ml−1 and 7.5 μg of MPA ml−1 at a density of about 50 cells per plate. The plates were incubated at 25°C for 5 days or at room temperature for 7 days, after which the colony size was scored. To screen large numbers of cells of the reporter strains, suspended cells from different colonies were treated in the same way but using larger plates (14-cm diameter) and a density of approximately 300 cells per plate. All colonies whose diameters differed by at least 20% from the average were isolated, and the cells were reexamined for MPAR expression in the same way.
RESULTS
Construction of ura3 mutants of strain WO-1 by targeted gene deletion.
To facilitate the genetic manipulation of strain WO-1, we constructed a ura3 mutant of this strain by targeted gene deletion using the MPAR-flipping strategy (48). A fragment comprising about 0.4 kb of upstream and 0.1 kb of downstream sequences in addition to the coding region of the URA3 gene is sufficient for selection of prototrophic transformants of C. albicans ura3 mutants. In plasmid pSFIU4 (Fig. 1A) almost all URA3 sequences present in the selection marker were replaced by the MPAR flipper cassette. Strain WO-1 was transformed with the insert from this plasmid, and MPA-resistant transformants were analyzed by Southern hybridization. The two URA3 alleles in WO-1 can be distinguished by an EcoRV restriction site polymorphism downstream of the coding region (Fig. 1A) and are located on 4.9- and 2.4-kb EcoRV fragments (Fig. 1B, lane 1). Two independent transformants in which either URA3-1 (WUM1A) (Fig. 1B, lane 2) or URA3-2 (WUM1B) (lane 3) had been replaced by the deletion cassette were selected, and the MPAR flipper was subsequently excised by induced, FLP-mediated recombination as described previously (48). All 10 tested MPA-sensitive derivatives of the two parent strains exhibited the hybridization pattern expected after specific deletion of the MPAR flipper cassette. The presence of a new 3.7-kb EcoRV fragment in both WUM2A (Fig. 1B, lane 4) and WUM2B (lane 5) demonstrates that, during integration into the URA3-2 allele, the polymorphic EcoRV site had been eliminated by the homologous cloned URA3 downstream sequence that did not contain this site. The insert from pSFIU4 was then used again to delete the remaining URA3 wild-type allele in strains WUM2A and WUM2B. MPA-resistant transformants were selected and screened for uridine auxotrophy. Southern hybridization analysis demonstrated the correct replacement of the second URA3 allele in the resulting strains, WUM3A (Fig. 1B, lane 6) and WUM3B (lane 7). Excision of the MPAR flipper from these strains resulted in strains WUM5A (Fig. 1B, lane 8) and WUM5B (lane 9). The presence of a 1.2-kb hybridizing EcoRV fragment in strain WUM5A demonstrates that, in this case, the polymorphic EcoRV site had been maintained after integration of the cassette into the URA3-2 allele.
In addition to the desired intrachromosomal recombination between FRT sites flanking the MPAR flipper, deletion of the cassette after the second round of excision could also have occurred by mitotic interchromosomal recombination between the centromere-proximal FRT site of the MPAR flipper and the FRT site present on the homologous chromosome. Such an event is not distinguishable from intrachromosomal recombination when the same replacement construct is used for deletion of both alleles of a target gene. Although interchromosomal recombination was never observed in previous experiments in which we used the flipper strategy for sequential disruption of different genes in various C. albicans strains (28, 48), the possibility remained that this might occur more frequently at the URA3 locus and, in that case, would lead to mutants that had become homozygous for all chromosomal regions located centromere distal to the site of crossover. To test for this possibility, we generated a second disruption construct in which only a part of the URA3 coding region was replaced by the MPAR flipper, such that different flanking regions were present on both sides of the cassette compared with the first deletion cassette (Fig. 1A). The insert from plasmid pSFIU2 was then used for a new transformation of the heterozygous strain WUM2A. A resulting transformant, strain WUM4A (Fig. 1B, lane 10), in which the remaining URA3 wild-type allele had been correctly replaced by the disruption cassette was used to excise the MPAR flipper after induction of the FLP gene. All seven tested MPA-sensitive derivatives exhibited the 2.3-kb fragment expected after specific excision of the MPAR flipper, demonstrating that interchromosomal recombination between FRT sites located on homologous chromosomes did not occur at an appreciable frequency also at the URA3 locus. One of these derivatives in which the two URA3 alleles had been inactivated with different deletion/disruption constructs (strain WUM6A) (Fig. 1B, lane 11) was kept for further analysis. For clarity, the different inactivated URA3 alleles were designated ura3Δ::FRT (obtained with pSFIU4) and ura3::FRT (obtained with pSFIU2). All integration and excision events were also confirmed with a URA3 downstream fragment (Fig. 1A) as an additional probe for Southern analysis of the strains, yielding the expected hybridization patterns (data not shown).
Phenotypic analysis of the ura3 mutants.
The three independently constructed ura3 mutants of strain WO-1, strains WUM5A, WUM5B, and WUM6A, were then tested for possible differences in their growth characteristics. All three ura3 mutants exhibited identical growth in uridine-supplemented YPD or minimal medium. In addition, reintroduction of the URA3 gene into strain WUM5A (see below) restored growth in media without uridine to the same level as that of the parent strain WO-1.
All three ura3 mutants displayed the white-opaque switching typical of strain WO-1, as monitored at the colony level on agar plates containing Lee's medium supplemented with phloxine B and by the morphology of individual cells observed under the microscope (see also below).
Integration specificity in WO-1 ura3 mutants using the URA3 gene as a selection marker.
The homozygous ura3 mutants WUM5A and WUM6A were both derived from the heterozygous mutant WUM2A but differ from each other in that virtually all URA3 sequences contained in the URA3 selection marker were removed from the genome of WUM5A whereas a considerable portion of these sequences was still present in one of the disrupted URA3 alleles of WUM6A. The presence of these sequences might interfere with specific integration at other genomic loci when the URA3 marker is used for selection of transformants. To test for the specificity of integration, the two strains were transformed with a DNA fragment containing a fusion of the SAP1 promoter and a C. albicans-adapted GFP gene. The cassette contained the URA3 selection marker and was flanked on both sides by SAP1 sequences, such that specific integration into one of the SAP1 alleles should occur by a double-crossover event, resulting in allelic replacement (Fig. 2A).
The two SAP1 alleles in strain WO-1 and its ura3 derivatives can be distinguished by a BglII restriction site polymorphism in the SAP1 upstream region (Fig. 2A) and are located on 7.0- and 6.6-kb BglII fragments (Fig. 2B, lane 1). Of seven prototrophic transformants of strain WUM5A, three had integrated the PSAP1-GFP fusion into the SAP1-1 allele, as exemplified by strain WUGS1A (Fig. 2B, lane 2), and two had integrated the reporter gene fusion in the SAP1-2 allele, as in strain WUGS1B (lane 3). The two other transformants had undergone additional recombination events, but the disrupted URA3 loci remained unchanged in all seven transformants (not shown). Of 12 tested prototrophic transformants of strain WUM6A, four contained the fusion in the SAP1-1 allele and six had it in the SAP1-2 allele. In the two remaining transformants, the URA3 allele disrupted in the second mutagenesis round was reconstituted. These results demonstrate that the presence of sequences homologous to the URA3 selection marker can indeed promote integration of the marker into its original locus, but overall, the specificity of targeted integration at a heterologous site was high and comparable between the two strains.
Analysis of phase-specific gene expression at the single-cell level.
In white and opaque cells of strain WO-1, the expression of certain genes is coupled to the cell type, the WH11 gene being white phase specific and other genes, like SAP1, being detectably expressed only in opaque cell cultures. However, phase variation of single genes might be advantageous also under conditions that otherwise favor the maintenance of a specific cell type and could thus occur independently from switching between the white and opaque phases. If such a gene-specific phase variation happened at relatively low frequency, as observed in other pathogenic microorganisms, it might not be detected with the methods used up to now to analyze phenotypic switching in C. albicans. GFP can be used as a reporter of gene expression at the level of single cells. We therefore analyzed the expression of the opaque-phase-specific SAP1 gene in single cells using strains carrying a transcriptional fusion of the SAP1 promoter with the GFP gene integrated into either of the two SAP1 alleles (see above). In addition, the GFP gene was also placed under the control of the white-phase-specific gene WH11 and introduced into the genome of strain WUM5A (Fig. 3A). From six uridine-prototrophic transformants, five had correctly integrated the reporter gene fusion into one of the WH11 alleles, as demonstrated by the appearance of an expected 4.0-kb EcoRI fragment in addition to the 3.7-kb fragment representing the wild-type WH11 gene after Southern hybridization with a WH11-specific probe. Two independent transformants (strains WUGW11A and WUGW11B) (Fig. 2B, lanes 2 and 3) were selected for further study.
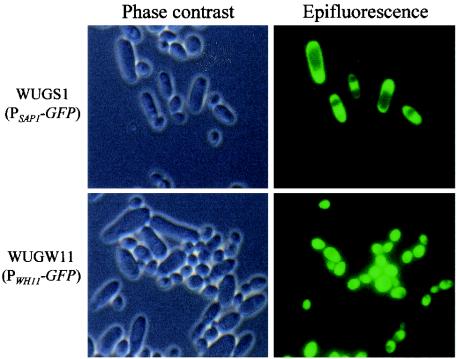
Analysis of the reporter strains by epifluorescence microscopy demonstrated that GFP was a useful reporter for qualitative analysis of phase-specific gene expression at the single-cell level. White and opaque cells of strain WO-1 can be distinguished by their morphologies, white cells being round to oval (length <1.5 times the width) and opaque cells being significantly elongated (length >2 times the width). Opaque but not white cells of strains WUGS1A and WUGS1B expressing GFP under the control of the SAP1 promoter fluoresced, and vice versa: fluorescence was observed in white but not opaque cells of strains WUGW11A and WUGW11B expressing GFP under the control of the WH11 promoter (Fig. 4). Activation of the WH11 gene in an opaque cell should therefore be detectable as a fluorescent elongated cell in opaque cell populations of strains WUGW11A and -B, whereas activation of the SAP1 gene in a white cell would be detected as a fluorescent round cell in white cell populations of strains WUGS1A and -B. Upon screening such populations of the reporter strains by epifluorescence microscopy (>104 cells of each strain), we could not detect expression of SAP1 or WH11 in the wrong cell type. It should be noted that the elongate morphology of opaque cells may sometimes not be distinct when the cells are viewed from one of the poles. However, the few fluorescent cells detected in opaque cell populations of strains WUGWH11A and -B had the appearance of white cells, with none of them being elongated, and very likely represented cells that had undergone switching. Similarly, all fluorescent cells detected in white cell populations of strains WUMS1A and -B matched the opaque phenotype. In no case could a fluorescent cell unambiguously be allocated to the wrong phase. These results demonstrated, at the single-cell level, that expression of the phase-specific SAP1 and WH11 genes is linked to the cell type.
FIG. 4.
Expression of PSAP1-GFP and PWH11-GFP reporter gene fusions in white and opaque cells of strain WO-1. For better illustration, cells from white and opaque colonies of the strains were mixed in this particular experiment. Shown are phase-contrast and the corresponding fluorescence micrographs of white and opaque cells. Identical results were obtained with strains WUGS1A and WUGS1B and with strains WUGW11A and WUGW11B.
Phase variation of SAP1 and WH11 expression does not occur independently of white-opaque switching in vitro.
The results obtained with the strains carrying the GFP reporter gene fusions showed that a switch from the “OFF” to the “ON” state of the phase-specific SAP1 and WH11 genes does not occur in the wrong cell type, since single fluorescent cells of the corresponding phase would have been easily detected in the background of a nonfluorescent cell population. This indicates that activation of phase-specific genes is under the tight control of the postulated master switching regulator. Nevertheless, a switch from the ON to the OFF state for individual phase-specific genes might occur even in the normally appropriate cell type by an independent mechanism. We also searched for white cells that do not detectably express WH11 and for opaque cells that do not detectably express SAP1 by looking at several hundred individual cells of white cell populations of strains WUGW11A and -B and of opaque cell populations of strains WUGS1A and -B first by phase-contrast and then by epifluorescence microscopy. In this way we were unable to detect such an ON-OFF switch. However, at an expected low frequency, a corresponding nonfluorescent cell might be missed in a population of fluorescent cells. For this reason, we devised a second reporter system in which a drug resistance gene was placed under the control of phase-specific promoters. Since the MPAR marker confers resistance against MPA upon cells in which it is expressed, a promoterless MPAR gene was fused to the SAP1 and WH11 promoters and integrated into the corresponding loci in strain WUM5A with the help of the URA3 selection marker (Fig. 2A and 3A). Two transformants carrying the PSAP1-MPAR fusion inserted in either of the two SAP1 alleles (strains WUMS1A and WUMS1B) (Fig. 2B, lanes 4 and 5) and two transformants in which the PWH11-MPAR fusion was integrated in one of the WH11 alleles (strains WUMW11A and WUMW11B) (Fig. 3B, lanes 4 and 5) were selected for analysis.
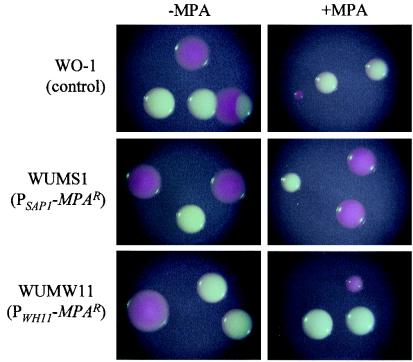
The strains carrying these reporter gene fusions should express an MPA-resistant phenotype only in the appropriate cell type in which the promoter is activated, i.e., only opaque cells of the strains with the PSAP1-MPAR fusion should exhibit enhanced MPA resistance and only white cells of the strains with the PWH11-MPAR fusion should become MPA resistant. Figure 5 shows that growth of the parent strain WO-1 was sensitive to the presence of 7.5 μg of MPA ml−1 in the medium. Opaque cells displayed a higher sensitivity than white cells, since they formed smaller colonies than white cells, although in the absence of MPA opaque colonies are larger than white colonies due to their flat morphology. The expression of the MPAR gene resulted in the expected phenotype. The strains carrying the MPAR gene under the control of the SAP1 promoter exhibited enhanced MPA resistance in the opaque phase, whereas the sensitivity of white cells remained unchanged. In contrast, the strains carrying the MPAR gene under the control of the WH11 promoter showed the reverse phenotype, although, for unknown reasons, opaque cells exhibited slightly enhanced growth on plates with or without MPA compared with the parent strain WO-1. Nevertheless, these results demonstrate that the reporter system worked sufficiently well in both white and opaque cells to allow screening for colonies that did not express the MPAR gene.
FIG. 5.
White and opaque colonies of strain WO-1 and its derivatives carrying PSAP1-MPAR and PWH11-MPAR reporter gene fusions after 7 days of growth at room temperature on agar plates containing Lee's medium and 5 μg of phloxine B ml−1 without or with 7.5 μg of MPA ml−1. For better illustration, a mixture of white and opaque cells of the strains was spread on the plates in this particular experiment. Identical results were obtained with strains WUMS1A and WUMS1B and with strains WUMW11A and WUMW11B.
We then used this reporter system to screen for rare cells that might have shut off SAP1 expression in the opaque phase in which the gene is normally expressed. If such a cell type-independent phase variation of SAP1 expression did exist, we expected that it would result in a semistable phenotype, similar to the white-opaque switching itself, and be detectable by the appearance of MPA-sensitive opaque colonies from which SAP1-expressing, MPA-resistant revertants should arise again with a similar low frequency. Altogether, 12,000 colonies of strains WUMS1A and WUMS1B were screened. Small colonies of various sizes arose with a frequency of about 1% (80 colonies). However, none of these small colonies had switched off expression of the SAP1 gene, since upon rescreening they gave rise to large, MPA-resistant colonies in the same manner as the parent strains WUMS1A and WUMS1B. The small size of these colonies was, therefore, not caused by repression of the PSAP1-MPAR fusion but due to stochastic, MPA-independent slower growth of a minor proportion of the cell population, at a frequency similar to that observed in previous experiments in which MPA-containing plates were used to screen for the presence or absence of the MPAR marker (43). Among 35,000 screened white colonies of strains WUMS1A and WUMS1B, no MPA-resistant white clones were observed, confirming the result obtained with the GFP reporter strains that the SAP1 gene was not activated in the wrong cell type.
When the same experiment was performed with strains WUMW11A and WUMW11B, we found that cell type-independent phase variation of WH11 expression also did not occur at a detectable frequency. All 18,000 white colonies screened retained the MPA-resistant phenotype, and none of the 13,000 opaque colonies tested exhibited elevated MPA resistance. From these results we conclude that, at least under the in vitro conditions tested, the expression of the phase-specific SAP1 and WH11 genes in strain WO-1 is tightly linked to the cell type.
DISCUSSION
The white-opaque switching of strain WO-1 has served for many years as a model system to study phenotypic switching in C. albicans and has led to the discovery of phase-specific genes, expression or down-regulation of which might provide selective advantages in specific host niches encountered by the fungus at particular stages of an infection (36). An ade2 mutant generated by chemical mutagenesis of strain WO-1 has been used in the past as a host for genetic analyses of phenotypic switching, for example, to introduce luciferase reporter gene fusions and to allow misexpression of phase-specific genes (16, 17, 18, 39, 40, 41). However, until recently no ura3 mutant of strain WO-1 was available that could be used for genetic manipulations requiring a recyclable marker, like sequential gene disruption. While our work was in progress, the generation of a ura3-auxotrophic derivative from the WO-1 ade2 mutant was reported, and this, for the first time, enabled the construction of a specific mutant (efg1/efg1) in this strain background by targeted gene deletion using the URA3 blaster strategy (42). The ura3 mutants described in our present study have the advantage that they were generated directly from strain WO-1 by two independent rounds of specific allelic replacement of both URA3 copies and, therefore, have not suffered from unspecific mutagenesis procedures or selection of mitotic recombinants. Although the MPAR marker in principle can be used for all genetic manipulations in any C. albicans wild-type strain, the availability of a specifically generated ura3 mutant is desirable for model strains like WO-1. First, using URA3 as a selection marker, prototrophic transformants can be recovered after only 2 days of selection on uridine-deficient medium, whereas the primary isolation of MPA-resistant transformants usually requires about 1 week, with additional time needed for clone purification. Therefore, the generation of mutants by sequential gene disruptions is significantly accelerated when the URA3 gene can be used as selection marker. Second, many already available URA3-based genetic constructions (e.g., gene disruption cassettes) that have been used for the molecular analysis of the C. albicans model strain CAI4 can probably be used directly also in the ura3 mutants of WO-1. Finally, the availability of a second selection marker like URA3 allows the alternative use of MPAR as a reporter gene, as demonstrated in the present study for the analysis of phase-specific gene expression at the level of individual colonies to detect switch events in the cells from which they were derived.
We have used two different reporter systems, GFP and MPAR, that allowed the analysis of phase-specific gene expression at the single-cell level, with the aim of detecting a possible cell type-independent switching in the expression status of individual genes. Our results demonstrate that the expression of the phase-specific WH11 and SAP1 genes is tightly coupled to the white or opaque cell morphology, since all white cells but no opaque cells examined expressed the WH11 gene, and all opaque cells but no white cells expressed the SAP1 gene. A significant activation of a phase-specific gene in the wrong cell type should have been easily detected with both reporter systems, since forced misexpression of the WH11 and SAP1 genes does not affect the cell morphology (16, 17). It should be noted that both reporter systems are useful only for qualitative rather than quantitative analyses, and we probably would have been unable to detect a low level of gene expression. However, a true morphology-independent phase variation in the expression of a gene would be expected to result in expression levels in the wrong cell type that are comparable to those in the correct phase and, vice versa, in down-regulation of the gene in the normally appropriate cell type to the level of the wrong phase. According to the fluorescence signals obtained with the GFP gene and the difference in MPA resistance between cells expressing and not expressing the MPAR gene, such a phase variation should have been detected with our reporter systems. Therefore, we conclude that cell type-independent switching of individual phase-specific genes does not occur at an appreciable frequency (<10−4) under the in vitro conditions tested, at least not at a frequency comparable to that of the white-opaque switching itself (34). However, this does not exclude such a possibility during infection. In fact, extreme differences in the frequency of antigenic variation in vitro and in vivo have been observed in other pathogens (49). Therefore, we intend to analyze switching of phase-specific genes in different animal models of candidiasis. The MPAR-based reporter system will be especially useful for this purpose, since clones of possible variants would be detected after simple plating of recovered cells and then be directly accessible for an analysis of the underlying molecular changes.
Phenotypic switching in C. albicans has been proposed to be under the control of a master regulatory switch, since many different cellular properties are altered in a programmed, coordinated fashion (36). However, the nature of this master regulator is unknown, as is the pathway leading to activation and deactivation of phase-specific genes. Inactivation of the SIR2 gene results in deregulated phenotypic switching in the C. albicans strain CAI4 (30), but switching in this strain is by far less well characterized than the white-opaque switching of strain WO-1. Such specific mutations can now also be introduced into strain WO-1 to assess their effect on white-opaque switching. The reporter gene fusions described in this study will be useful for addressing the question of whether in such regulatory mutants the expression of phase-specific genes is still linked to cell morphology or decoupled from the cell type. This approach should allow the dissection of the signal transduction pathway from the master regulatory switch to the different genes whose expression is controlled by phenotypic switching.
ACKNOWLEDGMENTS
This study was supported by the Bundesministerium für Bildung und Forschung (BMBF grant O1 K1 8906-0).
We thank Klaus Schröppel for his advice concerning growth of strain WO-1.
REFERENCES
- 1.Abraham J M, Freitag C S, Clements J R, Eisenstein B I. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:5724–5727. doi: 10.1073/pnas.82.17.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balan I, Alarco A-M, Raymond M. The Candida albicans CDR3 gene codes for an opaque-phase ABC transporter. J Bacteriol. 1997;179:7210–7218. doi: 10.1128/jb.179.23.7210-7218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedell G W, Soll D R. Effects of low concentrations of zinc on the growth and dimorphism of Candida albicans: evidence for zinc-resistant and -sensitive pathways for mycelium formation. Infect Immun. 1979;26:348–354. doi: 10.1128/iai.26.1.348-354.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borg M, Rüchel R. Expression of extracellular acid proteinase by proteolytic Candida species during experimental infection of oral mucosa. Infect Immun. 1988;56:626–631. doi: 10.1128/iai.56.3.626-631.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borg-von-Zepelin M, Beggah S, Boggian K, Sanglard D, Monod M. The expression of the secreted aspartyl proteinases Sap4 to Sap6 from Candida albicans in murine macrophages. Mol Microbiol. 1998;28:543–554. doi: 10.1046/j.1365-2958.1998.00815.x. [DOI] [PubMed] [Google Scholar]
- 6.Borst P, Rudenko G. Antigenic variation in African trypanosomes. Science. 1994;264:1872–1873. doi: 10.1126/science.7516579. [DOI] [PubMed] [Google Scholar]
- 7.Braaten B A, Nou X, Kaltenbach L S, Low D A. Methylation patterns in pap regulatory DNA control pyelonephritis-associated pili phase variation in E. coli. Cell. 1994;76:577–588. doi: 10.1016/0092-8674(94)90120-1. [DOI] [PubMed] [Google Scholar]
- 8.Cormack B P, Bertram G, Egerton M, Gow N A R, Falkow S, Brown A J P. Yeast-enhanced green fluorescent protein (yGFP): a reporter of gene expression in Candida albicans. Microbiology. 1997;143:303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- 9.De Backer M D, Magee P T, Pla J. Recent developments in molecular genetics of Candida albicans. Annu Rev Microbiol. 2000;54:463–498. doi: 10.1146/annurev.micro.54.1.463. [DOI] [PubMed] [Google Scholar]
- 10.Deitsch K W, Moxon E R, Wellems T E. Shared themes of antigenic variation and virulence in bacterial, protozoal, and fungal infections. Microbiol Mol Biol Rev. 1997;61:281–293. doi: 10.1128/mmbr.61.3.281-293.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haas R, Meyer T F. The repertoire of silent pilus genes in Neisseria gonorrhoeae: evidence for gene conversion. Cell. 1986;44:107–115. doi: 10.1016/0092-8674(86)90489-7. [DOI] [PubMed] [Google Scholar]
- 12.Hammerschmidt S, Hilse R, van Putten J P, Gerardy-Schahn R, Unkmeir A, Frosch M. Modulation of cell surface sialic acid expression in Neisseria gonorrhoeae via a transposable genetic element. EMBO J. 1996;15:192–198. [PMC free article] [PubMed] [Google Scholar]
- 13.Hoeijmakers J H J, Frasch A C C, Bernards A, Borst P, Cross G A M. Novel expression-linked copies of the genes for variant surface antigens in trypanosomes. Nature. 1980;284:78–80. doi: 10.1038/284078a0. [DOI] [PubMed] [Google Scholar]
- 14.Hube B, Sanglard D, Odds F, Hess D, Monod M, Schäfer W, Brown A J P, Gow N A R. Disruption of each of the secreted aspartyl proteinase genes SAP1, SAP2, and SAP3 of Candida albicans attenuates virulence. Infect Immun. 1997;65:3529–3538. doi: 10.1128/iai.65.9.3529-3538.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Köhler G A, White T C, Agabian N. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J Bacteriol. 1997;179:2331–2338. doi: 10.1128/jb.179.7.2331-2338.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kvaal C, Lachke S A, Srikantha T, Daniels K, McCoy J, Soll D R. Misexpression of the opaque-phase-specific gene PEP1 (SAP1) in the white phase of Candida albicans confers increased virulence in a mouse model of cutaneous infection. Infect Immun. 1999;67:6652–6662. doi: 10.1128/iai.67.12.6652-6662.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kvaal C, Srikantha T, Soll D R. Misexpression of the white-phase-specific gene WH11 in the opaque phase of Candida albicans affects switching and virulence. Infect Immun. 1997;65:4468–4475. doi: 10.1128/iai.65.11.4468-4475.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lockhart S R, Nguyen M, Srikantha T, Soll D R. A MADS box protein consensus binding site is necessary and sufficient for activation of the opaque-phase-specific gene OP4 of Candida albicans. J Bacteriol. 1998;180:6607–6616. doi: 10.1128/jb.180.24.6607-6616.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macdonald F, Odds F C. Inducible proteinase of Candida albicans in diagnostic serology and the pathogenesis of systemic candidosis. J Med Microbiol. 1980;13:423–435. doi: 10.1099/00222615-13-3-423. [DOI] [PubMed] [Google Scholar]
- 20.Marrs C F, Ruehl W W, Schoolnik G K, Falkow S. Pilin gene phase variation of Moraxella bovis is caused by an inversion of the pilin genes. J Bacteriol. 1988;170:3032–3039. doi: 10.1128/jb.170.7.3032-3039.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez J J, Mulvey M A, Schilling J D, Pinkner J S, Hultgren S J. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J. 2000;19:2803–2812. doi: 10.1093/emboj/19.12.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meier J T, Simon M I, Barbour A G. Antigenic variation is associated with DNA rearrangements in a relapsing fever Borrelia. Cell. 1985;41:403–409. doi: 10.1016/s0092-8674(85)80013-1. [DOI] [PubMed] [Google Scholar]
- 23.Mekalanos J J. Environmental signals controlling expression of virulence genes in bacteria. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Millon L, Manteaux A, Reboux G, Drobacheff C, Monod M, Barale T, Michel-Briand Y. Fluconazole-resistant recurrent oral candidiasis in human immunodeficiency virus-positive patients: persistence of Candida albicans strains with the same genotype. J Clin Microbiol. 1994;32:1115–1118. doi: 10.1128/jcm.32.4.1115-1118.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrow B, Srikantha T, Soll D R. Transcription of the gene for a pepsinogen, PEP1, is regulated by the white-opaque switching in Candida albicans. Mol Cell Biol. 1992;12:2997–3005. doi: 10.1128/mcb.12.7.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morrow B, Srikantha T, Anderson J, Soll D R. Coordinate regulation of two opaque-phase-specific genes during white-opaque switching in Candida albicans. Infect Immun. 1993;61:1823–1828. doi: 10.1128/iai.61.5.1823-1828.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morschhäuser J, Michel S, Hacker J. Expression of a chromosomally integrated, single-copy GFP gene in Candida albicans, and its use as a reporter of gene regulation. Mol Gen Genet. 1998;257:412–420. doi: 10.1007/s004380050665. [DOI] [PubMed] [Google Scholar]
- 28.Morschhäuser J, Michel S, Staib P. Sequential gene disruption in Candida albicans by FLP-mediated site-specific recombination. Mol Microbiol. 1999;32:547–556. doi: 10.1046/j.1365-2958.1999.01393.x. [DOI] [PubMed] [Google Scholar]
- 29.Moxon E R, Rainey P B, Novak M A, Lenski R E. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr Biol. 1994;4:24–33. doi: 10.1016/s0960-9822(00)00005-1. [DOI] [PubMed] [Google Scholar]
- 30.Pérez-Martín J, Uría J A, Johnson A D. Phenotypic switching in Candida albicans is controlled by a SIR2 gene. EMBO J. 1999;18:2580–2592. doi: 10.1093/emboj/18.9.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts D J, Craig A G, Berendt A R, Pinches R, Nash G, Marsh K, Newbold C I. Rapid switching to multiple antigenic and adhesive phenotypes in malaria. Nature. 1992;357:689–692. doi: 10.1038/357689a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaller M, Korting H C, Schäfer W, Bastert J, Chen W, Hube B. Secreted aspartic proteinase (Sap) activity contributes to tissue damage in a model of human oral candidosis. Mol Microbiol. 1999;34:169–180. doi: 10.1046/j.1365-2958.1999.01590.x. [DOI] [PubMed] [Google Scholar]
- 33.Schröppel K, Srikantha T, Wessels D, DeCock M, Lockhart S R, Soll D R. Cytoplasmic localization of the white phase-specific WH11 gene product of Candida albicans. Microbiology. 1996;142:2245–2254. doi: 10.1099/13500872-142-8-2245. [DOI] [PubMed] [Google Scholar]
- 34.Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll D R. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bacteriol. 1987;169:189–197. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soll D R, Morrow B, Srikantha T. High-frequency phenotypic switching in Candida albicans. Trends Genet. 1993;9:61–65. doi: 10.1016/0168-9525(93)90189-O. [DOI] [PubMed] [Google Scholar]
- 36.Soll D R. Gene regulation during high-frequency switching in Candida albicans. Microbiology. 1997;143:279–288. doi: 10.1099/00221287-143-2-279. [DOI] [PubMed] [Google Scholar]
- 37.Sonneborn A, Tebarth B, Ernst J F. Control of white-opaque phenotypic switching in Candida albicans by the Efg1p morphogenetic regulator. Infect Immun. 1999;67:4655–4660. doi: 10.1128/iai.67.9.4655-4660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Srikantha T, Soll D R. A white-specific gene in the white-opaque switching system in Candida albicans. Gene. 1993;131:53–60. doi: 10.1016/0378-1119(93)90668-s. [DOI] [PubMed] [Google Scholar]
- 39.Srikantha T, Morrow B, Schröppel K, Soll D R. The frequency of integrative transformation at phase-specific genes of Candida albicans correlates with their transcriptional state. Mol Gen Genet. 1995;246:342–352. doi: 10.1007/BF00288607. [DOI] [PubMed] [Google Scholar]
- 40.Srikantha T, Klapach A, Lorenz W W, Tsai L K, Laughlin L A, Gorman J A, Soll D R. The sea pansy Renilla reniformis luciferase serves as a sensitive bioluminescent reporter for differential gene expression in Candida albicans. J Bacteriol. 1996;178:121–129. doi: 10.1128/jb.178.1.121-129.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srikantha T, Tsai L K, Soll D R. The WH11 gene of Candida albicans is regulated in two distinct developmental programs through the same transcriptional activation sequences. J Bacteriol. 1997;179:3837–3844. doi: 10.1128/jb.179.12.3837-3844.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Srikantha T, Tsai L K, Daniels K, Soll D R. EFG1 null mutants of Candida albicans switch but cannot express the complete phenotype of white-phase budding cells. J Bacteriol. 2000;182:1580–1591. doi: 10.1128/jb.182.6.1580-1591.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Staib P, Kretschmar M, Nichterlein T, Köhler G, Michel S, Hof H, Hacker J, Morschhäuser J. Host-induced, stage-specific virulence gene activation in Candida albicans during infection. Mol Microbiol. 1999;32:533–546. doi: 10.1046/j.1365-2958.1999.01367.x. [DOI] [PubMed] [Google Scholar]
- 44.Stern A, Brown M, Nickel P, Meyer T F. Opacity genes in Neisseria gonorrhoeae: control of phase and antigenic variation. Cell. 1986;47:61–71. doi: 10.1016/0092-8674(86)90366-1. [DOI] [PubMed] [Google Scholar]
- 45.Stoenner H G, Dodd T, Larsen C. Antigenic variation of Borrelia hermsii. J Exp Med. 1982;156:1297–1311. doi: 10.1084/jem.156.5.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiser J N, Love J M, Moxon E R. The molecular mechanism of phase-variation of Haemophilus influenzae lipopolysaccharide. Cell. 1989;59:657–665. doi: 10.1016/0092-8674(89)90011-1. [DOI] [PubMed] [Google Scholar]
- 47.White T C, Miyasaki S H, Agabian N. Three distinct secreted aspartyl proteinases in Candida albicans. J Bacteriol. 1993;175:6126–6133. doi: 10.1128/jb.175.19.6126-6133.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wirsching S, Michel S, Morschhäuser J. Targeted gene disruption in Candida albicans wild-type strains: the role of the MDR1 gene in fluconazole resistance of clinical Candida albicans isolates. Mol Microbiol. 2000;36:856–865. doi: 10.1046/j.1365-2958.2000.01899.x. [DOI] [PubMed] [Google Scholar]
- 49.Zhang J R, Norris S J. Kinetics and in vivo induction of genetic variation of vlsE in Borrelia burgdorferi. Infect Immun. 1998;66:3689–3697. doi: 10.1128/iai.66.8.3689-3697.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ziebuhr W, Krimmer V, Rachid S, Lößner I, Götz F, Hacker J. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol Microbiol. 1999;32:345–356. doi: 10.1046/j.1365-2958.1999.01353.x. [DOI] [PubMed] [Google Scholar]