Abstract
INTRODUCTION:
As the number of biomarkers used to study Alzheimer’s disease (AD) continues to increase, it is important to understand the utility of any given biomarker, as well as what additional information a biomarker provides when compared to others.
METHODS:
We used hierarchical clustering to group 19 cross-sectional biomarkers in autosomal dominant AD. Feature selection identified biomarkers that were the strongest predictors of mutation status and estimated years from symptom onset (EYO). Biomarkers identified included clinical assessments, neuroimaging, cerebrospinal fluid amyloid, and tau, and emerging biomarkers of neuronal integrity and inflammation.
RESULTS:
Three primary clusters were identified: neurodegeneration, amyloid/tau, and emerging biomarkers. Feature selection identified amyloid and tau measures as the primary predictors of mutation status and EYO. Emerging biomarkers of neuronal integrity and inflammation were relatively weak predictors.
DISCUSSION:
These results provide novel insight into our understanding of the relationships among biomarkers and the staging of biomarkers based on disease progression.
Keywords: Autosomal dominant Alzheimer’s disease, biomarkers, machine learning
1 |. INTRODUCTION
Alzheimer’s disease (AD) is defined pathologically by the accumulation of amyloid beta (Aβ) plaques, neurofibrillary tangles (NFTs), neuroinflammation, and neuronal/synaptic loss that leads to brain atrophy and decreased glucose metabolism.1 These changes manifest clinically with cognitive decline and functional impairment. Autosomal dominant Alzheimer’s disease (ADAD) accounts for less than 1% of all AD cases and is caused by pathogenic mutations in the amyloid precursor protein (APP), presenilin 1 (PSEN1), or presenilin 2 (PSEN2) genes that lead to early increases in Aβ deposition in the brain.2,3 The mean age at onset of cognitive impairment in ADAD mutation carriers (MCs) is earlier than in sporadic AD and remains consistent within a family, allowing for calculation of the estimated number of years from symptom onset (EYO).4,5
Multiple neuroimaging methods have been used to evaluate in vivo changes in the brain due to ADAD. [11C]Pittsburgh Compound-B (PiB) has high affinity for Aβ plaques, with distributions similar to those seen at autopsy.6 In ADAD, PiB PET has identified Aβ deposition occurring more than 20 years prior to estimated age at symptom onset in MCs.5,7–9 [18F]Fluorodeoxyglucose (FDG) uptake reflects glucose metabolism and has shown the ability to discriminate symptomatic MCs from non-carriers (NCs).6,8,9 Decreases in glucose metabolism occur ≈5 to 10 years before symptom onset in MCs.9,10 Finally, structural magnetic resonance imaging (MRI) provides a method to evaluate volumetric and cortical thickness changes associated with disease progression.11,12 ADAD is characterized by progressive atrophy that affects temporal and subcortical regions ≈5 years from the estimated age at symptom onset.9
Apart from neuroimaging, changes in cerebrospinal fluid (CSF) Aβ and tau biomarkers have been identified. CSF markers of Aβ, including Aβ42 and CSF Aβ42/40 ratio, decrease during the early stages of the disease and correlate with amyloid deposition in the brain.13,14 Tau is an abundant microtubule-associated protein that is regulated by phosphorylation. Tau that is in a hyperphosphorylated state can form aggregates that lead to NFTs. Levels of CSF total tau are elevated in MCs ≈10 years from the estimated age at symptom onset.15 More recently, mass spectrometry studies showed that phosphorylated/unphosphorylated ratios of tau isoforms are associated with specific stages of the disease.16
Additional CSF and plasma biomarkers, generally referred to as “emerging” biomarkers, have recently been studied in AD. Neurogranin is a postsynaptic protein expressed in dendritic spines, and it correlates with elevated levels of tau.17 Synaptosomal-associated protein 25 (SNAP-25) is involved in the fusion of synaptic vesicles to the pre-synaptic membrane. Increases in CSF neurogranin and SNAP-25 are thought to reflect synaptic damage.18 Visinin-like protein 1 (VILIP-1) is a calcium sensor protein and is also a marker of neuronal injury.18 Chitinase-3-like protein 1 (YKL-40) is a secreted glycoprotein expressed primarily by astrocytes and has been associated with neuroinflammation.19,20 Finally, neurofilament light chain (NfL) is found within axonal cytoskeleton and is elevated in symptomatic AD.21,22
Although the number of biomarkers used to study AD continues to increase, the utility of a given measure in providing additional information when compared to other biomarkers remains unclear. Data driven methods capable of identifying which biomarkers are most reflective of AD progression and staging would be beneficial to clinical trials and patient care. Machine learning (ML) is a method for generating models that learn from existing data rather than being constrained by a priori rules.23 ML methods do not rely on heuristics, are robust to feature interactions, and are sensitive to complex association patterns. The ability to identify MCs and accurately determine an individual’s position relative to symptomatic impairment provides a unique opportunity for ML to evaluate similarities among biomarkers and identify which biomarkers are most reflective of mutation status and EYO.
The goal of this study was to identify natural groupings of biomarkers based on disease progression, as well as identify what biomarkers are most reflective of disease status and EYO within a cross-sectional cohort of MCs (n = 130) and NCs (n = 79). We used hierarchical clustering to group biomarkers based on similarity, and decision tree–based feature selection to identify biomarkers that were the strongest predictors of mutation status and EYO. This data-driven approach provides insight into both the behavior and utility of existing biomarkers for ADAD and could be important for clinical trials evaluating therapies. In addition, these results could provide a better understanding of other forms of AD.
2 |. METHODS
2.1 |. Participants
MCs (n = 130) with pathogenic mutations in PSEN1, PSEN2, or APP and healthy, mutation-negative sibling NCs (n = 79) were recruited from sites participating in the Dominantly Inherited Alzheimer Network (DIAN). Participants from the 14th data freeze with genetic, clinical, and neuroimaging data, who passed quality control procedures and had a complete set of biomarkers were included. A single time point from each participant was used for analysis. The Washington University in Saint Louis (WUSTL) Institutional Review Board provided supervisory review and human subject approval. Participants provided written, informed consent or assent with proxy consent. All study procedures were approved by the WUSTL Human Research Protection Office and the institutional review boards of the participating sites.
2.2 |. Clinical classification
The clinical dementia rating (CDR) Dementia Staging Instrument, which includes its sum of boxes (CDR-SB), and the Folstein Mini-Mental State Examination (MMSE) were performed at each clinical assessment.24,25 Depression was measured using the Geriatric Depression Scale (GDS).26 A participant’s EYO was calculated based on the participant’s current age relative to the family mutation–specific expected age at onset of dementia symptoms.4 Parental age at first progressive cognitive decline was used if the mutation-specific age of onset was unknown. EYO was calculated identically for both MCs and NCs. In the context of NCs, increasing EYO essentially serves as a proxy measure for increased aging. Mutation status was determined using polymerase chain reaction (PCR) amplification followed by Sanger sequencing.5
2.3 |. MRI acquisition and processing
MRI was performed using the Alzheimer’s Disease Neuroimaging Initiative (ADNI) protocol.27 Sites used a 3T scanner that passed quality control assessments. The ADNI Imaging Core screened images for compliance. T1 weighted images at 1.1 × 1.1 × 1.2 mm voxel resolution were acquired for participants. FreeSurfer 5.328,29 was used to perform volumetric segmentation and cortical surface reconstruction, and to define cortical and subcortical regions of interest (ROIs). A regression approach was used to correct subcortical volumes for intracranial volumes. FreeSurfer-defined ROIs were used for regional processing of positron emission tomography (PET) data. Cortical signature (CorSig) was calculated for subsequent analyses.30
2.4 |. PET acquisition and processing
Amyloid PET was performed using a bolus injection of 8 to 18 mCi PiB. Data from the 40 to70 minute post-injection timeframe were converted to regional standardized uptake value ratios (SUVRs) relative to the cerebellar gray matter.31 PET glucose metabolism imaging was performed with a single bolus injection of 5 mCi FDG. A 30-minute dynamic acquisition beginning 30 minutes post-injection was acquired. The last 20 minutes of each FDG scan were converted to SUVRs using the cerebellar gray matter as a reference region. All PET data were partial volume corrected and aligned to the T1 image.32,33 Scanner-specific filters were applied to achieve a common resolution (8 mm).34 A summary measure representing the arithmetic mean of SUVRs from the precuneus, superior and rostral middle frontal, lateral and medial orbitofrontal, and superior and middle temporal was used for PET analyses.31
2.5 |. CSF acquisition and processing
CSF was collected using methods previously described.13 CSF (10 to 20 mL) was collected during a fasting state via standard lumbar puncture (LP) at 08:00 hours under gravity flow. Samples were collected, flash frozen on dry ice, and shipped to the DIAN Biomarker Core at WUSTL, at which point they were thawed on wet ice, aliquoted (0.5 mL), flash frozen, and stored at −80°C. Concentrations of CSF Aβ40, Aβ42, total tau, and total p-tau (181) were measured by chemiluminescent enzyme immunoassay using an automated platform (LUMIPULSE G1200, Fujirebio) according to the manufacturer’s specifications. CSF Neurogranin, SNAP-25, and VILIP-1 were measured with microparticle-based immunoassays using Single Molecule Counting technology.17,35,36 CSF YKL-40 (Quidel) was measured via commercial enzyme-linked immunosorbent assays (ELISAs) according to manufacturer’s recommendations. CSF p-tau extracts were analyzed by nanoLC-MS/HRMS using Parallel Reaction Monitoring using higher energy collisional dissociation (HCD) fragmentation. NanoLC-MS/MS experiments were performed using a nanoAcquity UPLC system (Waters) coupled to a Fusion Tribrid mass spectrometer (Thermo Scientific). CSF tau phosphorylation levels were calculated using ratios between MS/HRMS transitions of endogenous unphosphorylated peptides and 15N labeled peptides from protein internal standard. Ratios of phosphorylation on T181, S202, T205, and T217 were measured using the ratio of the MS/HRMS transitions from phosphorylated peptides and corresponding unphosphorylated peptides (pT217/T217, pT205/T205, and pT181/T181).16
2.6 |. Blood acquisition and processing
Serum NfL was acquired and processed using methods described previously.22 In short, blood was collected in the morning under fasting conditions by venipuncture using red top plain Vacutainer tubes (Becton, Dickinson and Company). Tubes were centrifuged at 2000 × g at room temperature for 15 minutes after clotting. Serum was taken into a single transfer tube (SARSTEDT AG & Co.) and frozen on dry ice. Measurements were performed using a single-molecule array assay using the capture monoclonal antibody 47:3 and the biotinylated detection antibody 2:1 (UmanDiagnostics AB).37 The samples were measured in duplicate on a Simoa HD-1 platform (Quanterix) using a two-step neat assay. Serum samples were measured at 1:4 dilution (Tris-buffered saline, 0.1% Tween 20, 1% non-fat milk powder, HeteroBlock [300 ug ml−1; Omega Biologicals]). The amount of time between biomarker collections (imaging, CSF, plasma) per visit was 1 day ± 12.4 days (median, interquartile range [IQR]).
2.7 |. Machine learning and statistical analyses
Analyses were performed in MATLAB R2021a. All measures were standardized to zero mean and unit variance. Predictive features of mutation status and EYO were ranked according to importance using decision tree-based feature selection.38 This method utilizes a curvature test that identifies the strongest predictors by minimizing the P-value of chi-square tests of independence between each predictor and the response, as well as each pair of predictors and response. When evaluating the strongest features of mutation status, classes were weighted such that each class accounted for 50% of the accuracy. All decision trees were validated with 10-fold cross-validation. Clustering was performed with agglomerative hierarchical clustering. We chose hierarchical clustering because (1) ease of interpretability of results, (2) the hierarchical nature of the algorithm does not require a predetermined number of clusters, and (3) the results can be viewed at different scales based on the needs of a given study. Agglomerative hierarchical clustering is a bottom-up approach, where each observation starts in its own cluster, and clusters are merged as they move up the cluster tree. The distance metric utilized was the absolute value of the biomarker correlations. The linkage criterion, which determines the distance between sets as a function of the pairwise correlations between biomarkers, used Weighted Pair Group Method with Arithmetic Mean.39 (Supplemental material provides further details on the cluster analysis.)
3 |. RESULTS
3.1 |. Demographics
Detailed demographics are presented in Table 1. NC and MC participants were similar with regards to age, sex, race, and education.
Table 1.
Demographics
| Mutation carriers (MC) | Mutation-negative non-carriers (NC) | p-values | |
|---|---|---|---|
| N | 130 | 79 | |
| Age (years) ± SD | 39.9 ± 10.7 | 41.7 ± 11.3 | .24 |
| Sex (% Male) | 45% | 39% | .39 |
| Race (% Caucasian) | 92% | 92% | .98 |
| Education (years) ± SD | 14.6 ± 3.1 | 14.8 ± 2.3 | .62 |
| EYO (years) ± SD | −6.6 ± 9.8 | −5.5 ± 11.5 | .48 |
Abbreviations: SD, standard deviation; EYO, estimated years until symptom onset.
3.2 |. Clustering
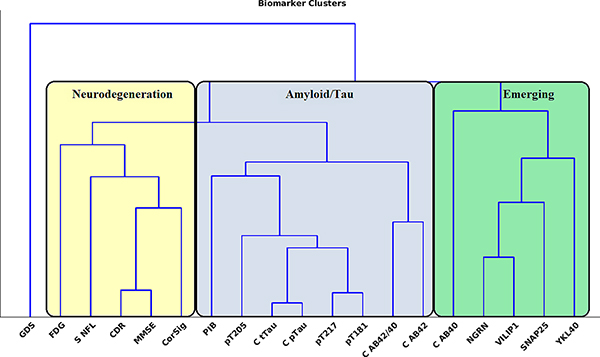
The clustering results can be seen in Figure 1, and the similarity matrix used for clustering in Figure S1. At the highest level, the data were grouped into three categories: neurodegeneration, Aβ/tau, and emerging biomarkers of neuronal integrity and inflammation. The Aβ/tau cluster consisted of all CSF tau-related variables, PiB, and CSF Aβ42 and Aβ42/40 ratio. Within the amyloid/tau cluster, PiB showed greater similarity with CSF tau variables compared to Aβ variables. The neurodegeneration cluster consisted of clinical (CDR-SB), neuropsychological (MMSE), and measures of neuronal dysfunction/damage (FDG, cortical signature, and serum NfL). Finally, except for CSF Aβ40, the emerging biomarkers of neuronal integrity and inflammation clustered together and consisted of CSF neurogranin, SNAP-25, VILIP-1, and YKL-40. GDS was the least similar to any other biomarker. Supplemental material and Figure S6 provide further details on cluster-validation results.
FIGURE 1.
At the highest level, biomarkers grouped by neurodegeneration (yellow), amyloid and tau (blue), and emerging biomarkers of neuronal integrity and inflammation (green). The amyloid/tau cluster encompassed amyloid PET (PiB), CSF Aβ40, and Aβ42/40, and all CSF tau measures, including tau mass spectrometry ratios (pT217, pT181, pT205) and Lumipulse total tau and total pTau (C tTau, C pTau). The neurodegeneration cluster included measures of metabolism (FDG), atrophy (CorSig), serum NfL, and cognitive assessment measures. The final cluster consisted of the emerging biomarkers of neuronal integrity and inflammation as well as CSF Aβ40. Of note, depression (GDS) was separate
3.3 |. Mutation status feature selection
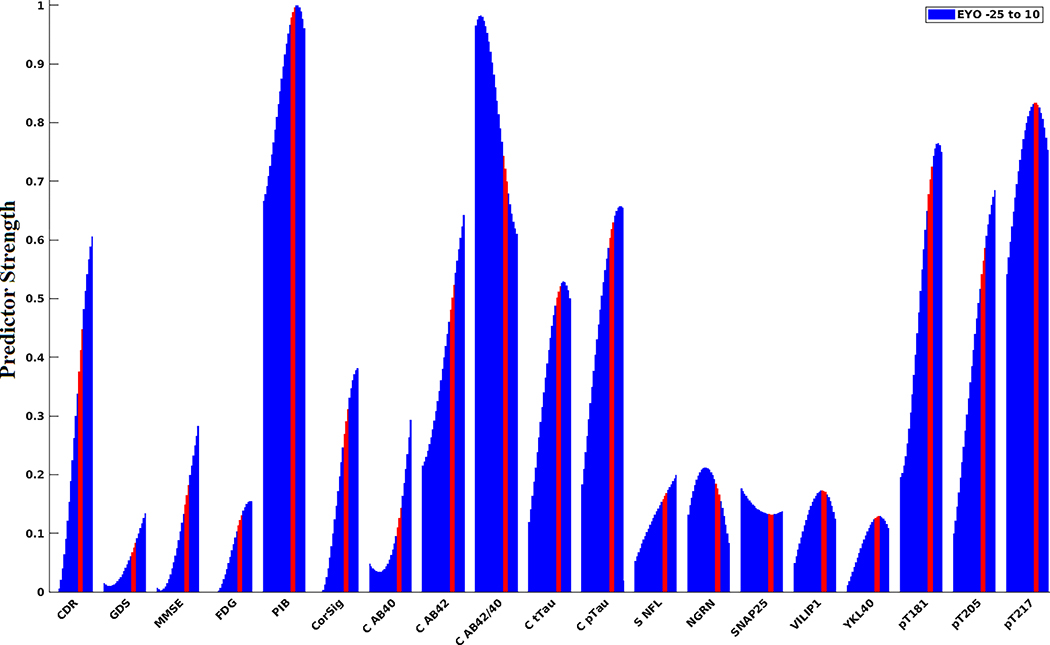
The decision tree was able to classify mutation status with an average accuracy of 92.4%. Figure 2 shows the strongest predictors of mutation status by EYO calculated over a 15-year sliding window. The strongest predictors were markers of Aβ and mass spectrometry tau ratios (CSF Aβ42/40, PiB, and CSF pTau 217, pT181).CSFAβ42/40 and PiB showed an inverse trajectory, with Aβ42/40 being the strongest predictor during the early phase of the disease (EYO <−15) and decreasing in predictive strength later in the disease process (EYO >0), whereas PiB showed the opposite pattern and was a stronger predictor at a later EYO. The CDR-SB was the strongest clinical predictor of progression but not until EYO ≥0. The emerging biomarkers of neuronal integrity and inflammation, FDG, and cortical signature were not identified as strong predictors.
FIGURE 2.
The strongest predictors of mutation status with regard to estimated years to symptom onset (EYO). Features were calculated for a sliding 15-year window that incremented 1 year at a time that encompassed EYOs between −25 and +10. Red line indicates 15-year window that was centered at EYO of 0 (±1 year). The strongest predictors of mutation status were amyloid markers (CSF Aβ42/40, PiB, and pTau 217 ratio). CSF Aβ42/40 and PiB showed an inverse trajectory. CSF Aβ42/40 was the strongest predictor early in the disease process followed by a relative decrease later in the disease course. In contrast, PiB was a slightly weaker predictor earlier in the disease process but gradually increased in predictive strength later in the disease course. Results coincide with the predictive strengths of each of the measures when not using a sliding window and all EYOs were combined into a single analysis (see Figure S2)
3.4 |. EYO feature selection
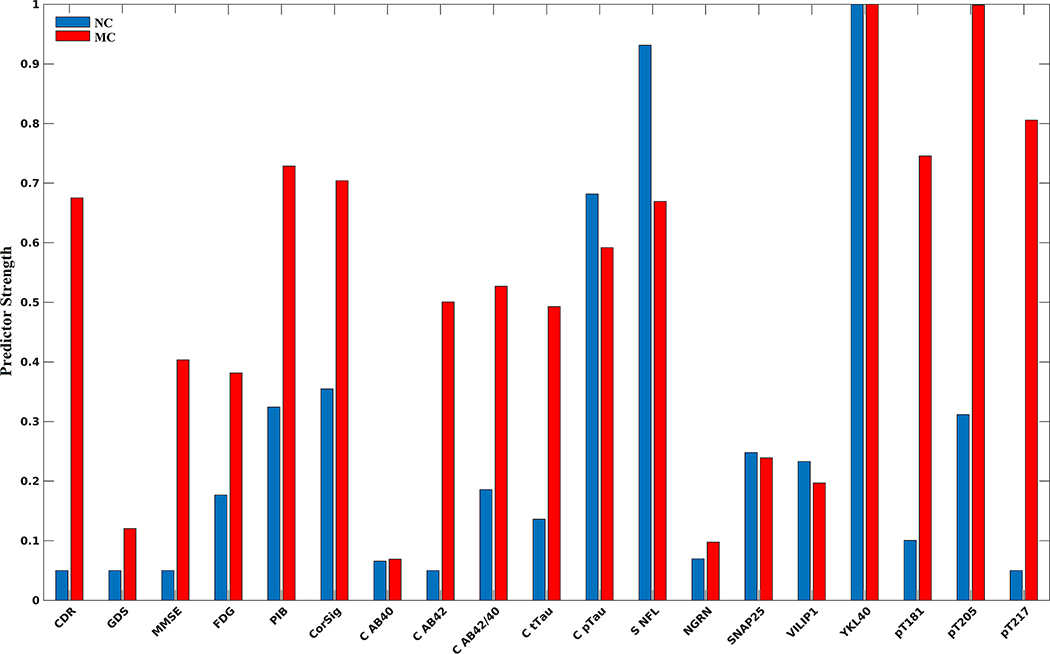
The utility of each biomarker for predicting EYO is shown in Figure 3 for MC (red) and NC (blue). The decision tree was able to predict EYO with a mean squared error of 6.4 years. Feature selection identified phosphorylated tau measures (pT205, pT217, pT181 ratios) as the strongest predictors of EYO. Of interest, YKL-40, ptau181 measured with immunoassay, and serum NfL were strong predictors for both MCs and NCs. Within NC, YKL-40, serum NfL, and ptau181 by immunoassay were the strongest predictors, indicating a potential aging component regardless of disease pathology (Figure 4 and Figure S4). When accounting for age, the strongest predictors of EYO were CSF pT217, pT205, and pT181 ratios by mass spectrometry, as well as CDR-SB.
FIGURE 3.
Strongest predictors of EYO for mutation carrier (MC) and non-carrier (NC) participants. Overall, the strongest predictors for MC participants were phosphorylated tau mass spectrometry ratios (pT205, pT217, pT181). YKL-40 and serum neurofilament light (NfL) were strong predictors for both MC and NC participants, suggesting a strong aging component regardless of disease pathology (see Figures 4 and S4 bottom right for trajectories)
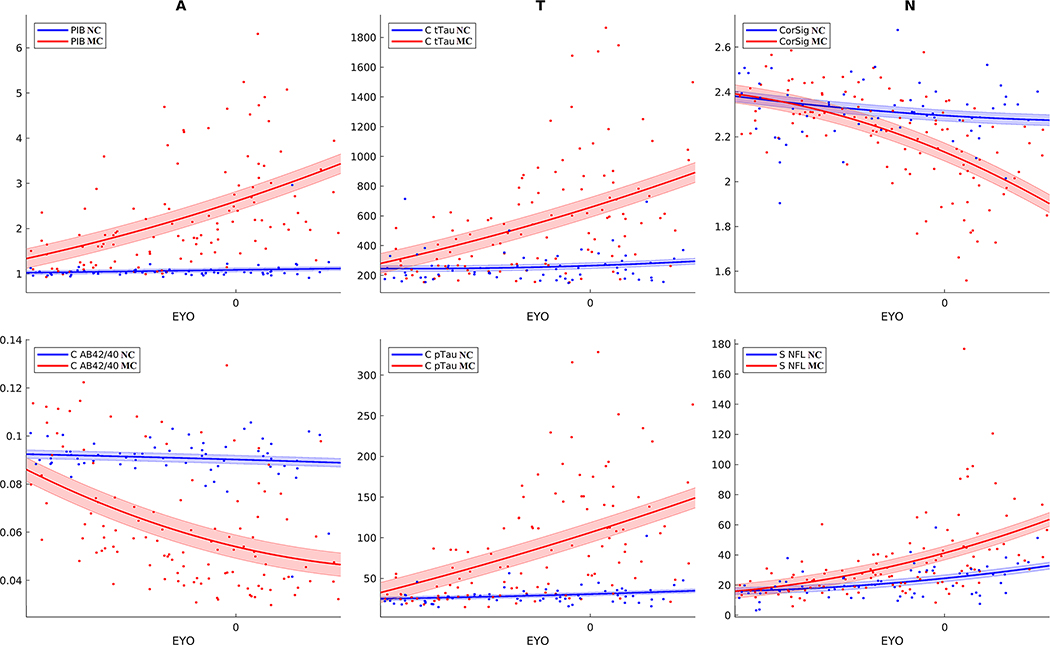
FIGURE 4.
Trajectories for biomarkers of amyloid, tau, and neurodegeneration (AT(N)) for NC (blue) and MC (red) from −20 to +10 EYO. Trajectories for amyloid biomarkers (PiB, CSF Aβ42/40), tau (Lumipulse CSF total tau and CSF total pTau [181]), and neurodegeneration (cortical signature and serum NfL) were fit using a two-degree polynomial with shaded regions representing standard error. Differences between MC and NC were observed early for amyloid and tau (−20 to −15 EYO). Differences were also observed between MC and NC for neurodegeneration markers but were closer to EYO (−10 to −5 EYO). Of note, changes in cortical signature and serum NfL were seen with aging regardless of mutation status
4 |. DISCUSSION
We used hierarchical clustering to identify groupings of biomarkers reflective of ADAD pathology. Neurodegeneration, amyloid/tau, and emerging biomarkers of neuronal integrity and inflammation were the three primary clusters. Decision tree–based feature selection recognized measures of Aβ and tau as the primary predictors of mutation status throughout disease progression, whereas neurodegenerative and clinical measures were moderate predictors in the later stages of the disease (EYO ≥0). The emerging biomarkers of neuronal integrity and inflammation were not identified as strong predictors. When evaluating time from symptom onset, CSF phosphorylated tau measures and clinical metrics were the strongest predictors for MCs. Finally, serum NfL and CSF YKL-40 were strong predictors of aging regardless of mutation status.
Clustering revealed three primary groupings for biomarkers. The amyloid/tau cluster encompassed all CSF and PET measures except for CSF Aβ40. Because Aβ40 did not start to deviate for MCs compared to NCs until −10 to −5 EYO (Figure S5), it was likely grouped into a different cluster because other amyloid and tau measures showed differences at earlier stages of the disease. The CSF Aβ42 and CSF Aβ42/40 ratio grouped together, whiles phosphorylated tau measures also grouped together. This indicates that although specific phosphorylation sites may be more correlated with measures of amyloid or tau, the overall progression is similar for each of the tau phosphorylation measures. PiB’s position in the dendrogram indicated it was more like CSF tau than Aβ. PiB also showed a higher correlation (absolute value) with tau measures (Aβ42 = .49, Aβ42/40 = .54, pT181 = .54, pT205 = .67, pT217 = .66, all P < .001). This is likely due to the plateau that occurs in Aβ42/40 around EYO = 0, which does not occur in PiB and CSF tau species (Figures 4 and 5).
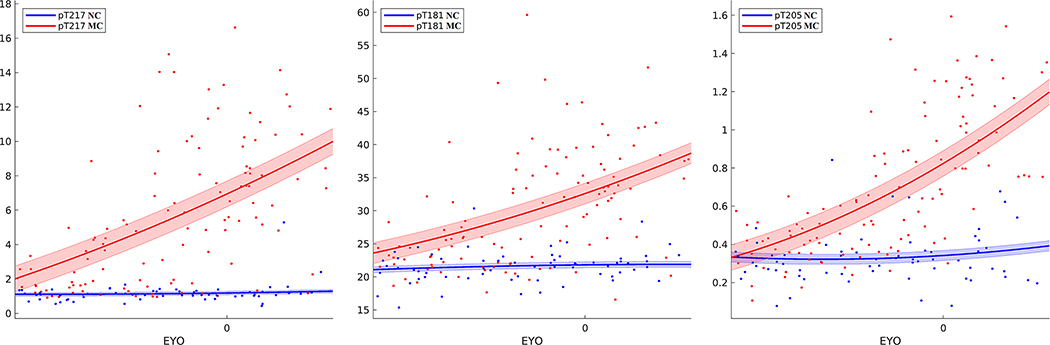
FIGURE 5.
Trajectories for CSF mass spectrometry phosphorylated tau ratios pT217, pT181, and pT205 for NC (blue) and MC (red) from −20 to +10 EYO. Trajectories were fit using a two-degree polynomial with shaded regions representing standard error. For both CSF pT217 and CSF pT181 changes were seen very early (20 EYO), whereas changes in CSF pT205 occurred slightly later
The neurodegeneration cluster consisted of clinical and psychological measures (CDR-SB and MMSE), neuroimaging measures of neurodegeneration (CortSig and FDG), and serum NfL. Consistent with the amyloid-tau-neurodegeneration (AT(N)) framework,40 differences in neurodegenerative measures between MCs and NCs deviated at roughly −5 EYO (Figure 4 and Figure S5). Serum NfL showed the earliest changes and was highly correlated with CDR-SB and MMSE (.67 and .62, P < .001). All biomarkers in the neurodegeneration cluster were highly correlated (>.5) with CSF pT205, which is consistent with previous findings.16
The last cluster consisted primarily of the emerging biomarkers of neuronal integrity and inflammation. The trajectories for these biomarkers can be seen in Figure S4. Among these measures, SNAP-25 and neurogranin, both presumed to be measures of synaptic damage, showed the greatest consistent ability to distinguish between MCs and NCs (Figure 2). SNAP-25 showed a slight elevation in MCs that remained relatively consistent regardless of EYO. VILIP, SNAP-25, and YKL-40 all showed increases with age regardless of mutation status. This suggest many of these biomarkers are more likely measures of aging and not specific to AD.
Both Figure 2 and Figure S2 illustrate that CSF Aβ42/40 and PiB were the strongest predictors of mutation status. This could be explained by a dramatic increase in amyloid deposition within MC brains ≈15 years before EYO; alternatively, it could be explained by more MCs beginning to accumulate amyloid plaques during this phase of the disease. CSF Aβ42/40 was the strongest predictor earlier in the disease process but lost predictive strength in later stages. This result agrees with a previous study5 that has shown that changes in CSF Aβ42 occur early in the disease and begin to plateau as they approach EYO = 0. In contrast, PiB became a stronger predictor with time. This is likely due to the trajectory exhibited by amyloid accumulation in the brain, where early in the disease the rate of accumulation is moderate, but around EYO =−15 deposition begins to drastically increase.41 Overall, these results are consistent with other studies, which have shown that CSF amyloid markers are the earliest to diverge in MCs compared to NCs, followed by PiB.5 The third strongest predictor was CSF pT217. Like PiB, CSF pT217 was a moderate predictor early in the disease and became stronger with time, surpassing CSF Aβ42/40 as the second-strongest predictor of mutation status. Although not as strong as CSF pT217, both CSF pT181 and pT205 were good predictors of mutation status, which has been observed.42 Because measures of amyloid are believed to be the earliest to change in AD, it has been hypothesized that amyloid pathology may lead to an increase in the phosphorylation of tau.43 It is, therefore, not surprising that these biomarkers were the strongest predictors of mutation status. Finally, neurodegenerative measures and clinical/cognitive metrics, known to change only in the later stages of the disease, gained predictive strength at EYO ≥0 (Figure S5). The weakest predictors of mutation status were GDS, MMSE, FDG, CSF Aβ40, serum NFL, and the emerging biomarkers of neuronal integrity and inflammation. Except for GDS and CSF Aβ40, these measures generally group within the neurodegeneration category of the AT(N) framework.
Figure S3 shows the trained decision tree and optimal cutoffs used to classify participants by mutation status. PiB, CSF Aβ42/40, pT217 ratio, and pT181 ratio gave the optimal results. At the top level, the optimal cutoff for PiB was 1.23. This corresponds to an EYO of −22.2 based on the fitted line shown in Figure 4. If the PiB value was less than this cutoff, CSF Aβ42/40 was then evaluated. Of interest, a participant was classified as mutation positive if the CSF Aβ42/40 was greater than .11 (EYO ≤−28). This corresponds to early EYOs where PiB is relatively low (EYO ≤−22) and CSF Aβ42/40 is elevated compared to NCs (see Figure 4). Finally, if CSF Aβ42/40 and PiB did not meet their cutoffs (EYOs between −28 and −22), a pT217 ratio greater than 1.24 (EYO −24) and a pT181 ratio greater than 23.7 (EYO −20, Figure 5) resulted in a participant being classified as mutation positive.
The relative strength of a biomarker’s ability to predict EYO in MC and NC participants is shown in Figure 3. Phosphorylated tau ratios (pT205, pT217, and pT181; Figure 5), CDR-SB, cortical signature, CSF pTau, serum NFL, and CSF YKL-40 were the strongest predictors of EYO in MC participants. In NC participants, CSF pTau, serum NFL, and YKL-40 were the strongest predictors of EYO. This equates to changes that occurs with normal aging regardless of disease status. Figure 4 and Figure S4 show the trajectories for these biomarkers. For each of these biomarkers there was a consistent increase with age in NCs. When compensating for the aging component, the strongest predictors of EYO were pT217, pT205, CDR-SB, and pT181.
These findings have clinical importance for people with ADAD. As the number of biomarkers used to study AD pathology continues to increase, it is important to understand which biomarkers are most reflective of disease pathology. This study is the first to use ML-based methods to directly identify how biomarkers cluster, as well as identify the strongest combination of biomarkers that predict mutation status and EYO within each cluster. We have demonstrated that the optimal set of biomarkers can be reduced to a small set of Aβ and tau measures, and also show that numerous biomarkers that have gained popularity offer little to no additional information about ADAD pathology when compared to others. This is significant when considering that these biomarkers may be used in clinical trials to evaluate the effectiveness of therapies. Furthermore, using our trained decision tree, we can identify biomarker cutoffs, as well as map the cutoffs to specific disease stages, and therefore identify what biomarker is best to use at a specific point in time. This demonstrates the utility of ML in understanding the complex disease progression associated with ADAD that goes beyond group-level statistics. Finally, although identifying mutation status in ADAD can easily be achieved with blood test and EYO calculated based on familial information, the fact that we are able to use this information to identify the optimal set of biomarkers reflective of disease state and pathology is of great importance. Specifically, we believe that these results are directly applicable to other forms of AD, such as sporadic AD, where this information is not available.
The main limitation of our study is the use of a cross-sectional research design. Research using prospective designs is needed to further delineate the staging and progression of various biomarkers in relation to EYO. Furthermore, additional variables, such as the specific mutation type should be considered if sufficient numbers are present in future analysis. Alternative forms of feature selection and/or clustering should also be considered to evaluate the relationship among biomarkers and ensure concordance of results. Because many Aβ and tau intervention trials involve participants with ADAD, similar research should be conducted in other genetic forms of AD (eg, Down syndrome) and late-onset AD to ensure correspondence of the biomarker groupings and behavior. Finally, with the increasing adoption of blood-based testing, it is important to evaluate the similarities and differences between blood and CSF biomarkers.
4.1 |. Conclusion
Understanding the similarities and differences among AD biomarkers, and which biomarkers best represent disease processes, is vital for maximizing the efficiency of clinical trials and patient care. In this study, we have identified three primary cluster of AD biomarkers (neurodegeneration, amyloid/tau, and emerging). Feature selection identified measures of amyloid and tau as the strongest predictors of mutation status, and measures of tau and atrophy as the strongest predictors of EYO. Our analyses also suggest that a model for diagnosing ADAD (classifying mutation status) independent of disease stage requires only four AD biomarkers (PiB PET, CSF Aβ42/40, pT217, and pT181 ratios).
Supplementary Material
RESEARCH IN CONTEXT.
Systematic review: The authors reviewed the literature using traditional (eg, PubMed) sources and meeting abstracts and presentations. Relevant citations are included where appropriate.
Interpretation: Our findings suggest that biomarkers of autosomal dominant Alzheimer’s disease (ADAD) can be grouped into three primary clusters: neurodegeneration, amyloid/tau, and emerging biomarkers of neuronal integrity and inflammation. Furthermore, the primary predictors of ADAD status consist of amyloid and tau measures, whereas the emerging biomarkers of neuronal integrity and inflammation are relatively weak predictors.
Future directions: Future work will focus on (1) performing similar analysis in other forms of Alzheimer’s disease (eg, Down syndrome and sporadic Alzheimer’s disease) to ensure correspondence of the biomarker groupings and behavior, and (2) performing the analysis on longitudinal data.
ACKNOWLEDGEMENTS
We would like to acknowledge the participants and their families, without whom these studies would not be possible. In addition, we thank all the participating researchers and coordinators (https://dian.wustl.edu/our-research/observational-study/dian-observational-study-sites/) who support the studies. DIAN Clinical-Trials.gov identifier: NCT00869817. This research was funded by the National Institutes of Health (NIH) (grant numbers K01AG053474, K23AG046363, R01AG052550, UFAG 032438, UL1TR000448, P30NS098577, R01EB009352, P50AG05131, U01AG042791, U01AG042791-S1 [FNIH and Accelerating Medicines Partnership], R1AG046179]; the German Center for Neurodegenerative Diseases (DZNE); the National Institute for Health Research (NIHR) Queen Square Dementia Biomedical Research Centre; and the Medical Research Council Dementias Platform United Kingdom (UK) (grant numbers MR/L023784/1, MR/009076/1), Alzheimer’s Association International Research Grant Program #AARFD-20-681815, NSF DMS 156243, DIAN-J by AMED, and an anonymous organization. Furthermore, we acknowledge the support of Fred Simmons and Olga Mohan, the Barnes-Jewish Hospital Foundation, the Charles F. and Joanne Knight Alzheimer Research Initiative, the Hope Center for Neurological Disorders, the Mallinckrodt Institute of Radiology, the Paula and Rodger O. Riney fund, and the Daniel J. Brennan fund.
Funding information
National Institutes of Health, Grant/Award Numbers: K01AG053474, K23AG046363, R01AG052550, UFAG 032438, UL1TR000448, P30NS098577, R01EB009352, P50AG05131, U01AG042791, U01AG042791-S1; German Center for Neurodegenerative Diseases; National Institute for Health Research; Medical Research Council Dementias Platform UK, Grant/Award Numbers: MR/L023784/1, MR/009076/1; Alzheimer’s Association International Research Grant Program, Grant/Award Numbers: #AARFD-20-681815, NSF DMS 156243
CONFLICT OF INTEREST
The authors declare no competing interest. Anne Fagan has received research funding from the National Institute on Aging of the National Institutes of Health, Biogen, Centene, Fujirebio, and Roche Diagnostics. She is a member of the scientific advisory boards for Roche Diagnostics and Genentech and consults for Diadem, DiamiR, and Siemens Healthcare Diagnostics Inc. Carlos Cruchaga receives research support from Biogen, EISAI, Alector, and Parabon. The funders of the study had no role in the collection, analysis, or interpretation of data; in the writing of the report; or in the decision to submit the paper for publication. Dr. Cruchaga is also a member of the advisory board of ADx Healthcare, Halia Therapeutics, and Vivid Genomics. Jasmeer P. Chhatwal served on the medical advisory board for Otsuka Pharmaceuticals. Johannes Levin reports speaker’s fees from Bayer Vital, speaker’s fees from Willi Gross Foundation, consulting fees from Axon Neuroscience, consulting fees from Ionis Pharmaceuticals, author fees from Thieme medical publishers and W. Kohlhammer GmbH medical publishers, compensation for work as part-time CMO from MODAG GmbH, and nonfinancial support from AbbVie outside the submitted work. John Morris is funded by NIH grants numbers P50AG005681, P01AG003991, P01AG026276, and UF1AG032438. Dr. Jack serves on an independent data monitoring board for Roche and has served as a speaker for Eisai, but he receives no personal compensation from any commercial entity. He receives research support from NIH and the Alexander Family Alzheimer Disease Research Professorship of the Mayo Clinic. Eric McDade is involved in a clinical trial on AV-1451 sponsored by Avid and serves on a data safety monitoring committee for Eli-Lilly and Alector and is on the Scientific Advisory Board for Alzamend; and receives research support from Eli-Lilly and Hoffman-La Roche. Dr. McDade is a co-inventor of the “Methods of diagnosing AD with phosphorylation changes” technology licensed by Washington University to C2N Diagnostics. Washington University also holds 5% equity in C2N. Through these relationships, Washington University and Dr. McDade are entitled to receive royalties from the license agreement with C2N. Dr. David Holtzman, Chair of Neurology at Washington University Medical School, is a co-founder of C2N and serves on C2N’s advisory board. Randall Bateman is on the scientific advisory board of C2N Diagnostics and reports research support from AbbVie, Biogen, Eisai, Eli Lilly, Co/Avid Radiopharmaceuticals, Roche, Janssen, and United Neuroscience. Dr. Weiner receives support for NIH grants: 5U19AG024904-14; 1R01AG053798-01A1; R01 MH098062; U24 AG057437-01; 1U2CA060426-01; 1R01AG058676-01A1; and 1RF1AG059009-01, DOD: W81XWH-15-2-0070; 0W81XWH-12-2-0012; W81XWH-14-1-0462; W81XWH-13-1-0259, PCORI: PPRN-1501-26817, California Dept. of Public Health: 16-10054, U. Michigan: 18-PAF01312, Siemens: 444951-54249, Biogen: 174552, Hillblom Foundation: 2015-A-011-NET, Alzheimer’s Association: BHR-16-459161; The State of California: 18-109929. He also receives support from Johnson & Johnson, Kevin and Connie Shanahan, GE, VUmc, Australian Catholic University (HBI-BHR), The Stroke Foundation, and the Veterans Administration. He has served on Advisory Boards for Eli Lilly, Cerecin/Accera, Roche, Alzheon, Inc., Merck Sharp & Dohme Corp., Nestle/Nestec, PCORI/PPRN, Dolby Family Ventures, National Institute on Aging (NIA), Brain Health Registry and ADNI. He serves on the Editorial Boards for Alzheimer’s & Dementia, TMRI and MRI. He has provided consulting and/or acted as a speaker/lecturer to Cerecin/Accera, Inc., BioClinica, Nestle/Nestec, Roche, Genentech, NIH, The Buck Institute for Research on Aging, FUJIFILM-Toyama Chemical (Japan), Garfield Weston, Baird Equity Capital, University of Southern California (USC), Cytox, and Japanese Organization for Medical Device Development, Inc. (JOMDD) and T3D Therapeutics. He holds stock options with Alzheon, Inc., Alzeca, and Anven.
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of the article at the publisher’s website.
REFERENCES
- 1.Serrano-Pozo A, Frosch MP, Masliah E, Hyman BT. Neuropathological alterations in Alzheimer disease. Cold Spring Harb Perspect Med 2011;1:6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bateman RJ, Aisen PS, De Strooper B, et al. Autosomal-dominant Alzheimer’s disease: A review and proposal for the prevention of Alzheimer’s disease. Alzheimers Res Ther 2011;3:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schindler SE, Fagan AM. Autosomal dominant Alzheimer disease: A unique resource to study CSF biomarker changes in preclinical AD. Front Neurol 2015;6:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryman DC, Acosta-Baena N, Aisen PS, et al. Symptom onset in autosomal dominant Alzheimer disease: a systematic review and meta-analysis. Neurology 2014;83:253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bateman RJ, Xiong C, Benzinger TLS, et al. Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N Engl J Med 2012;367:795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marcus C, Mena E, Subramaniam RM. Brain PET in the diagnosis of Alzheimer’s disease. Clin Nucl Med 2014;39:413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yau WYW, Tudorascu DL, McDade EM, et al. Longitudinal assessment of neuroimaging and clinical markers in autosomal dominant Alzheimer’s disease: a prospective cohort study. Lancet Neurol 2015;14:804–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDade E, Wang G, Gordon BA, et al. Longitudinal cognitive and biomarker changes in dominantly inherited Alzheimer disease. Neurology 2018;91:1295–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordon BA, Blazey TM, Su Y, et al. Spatial patterns of neuroimaging biomarker change in individuals from families with autosomal dominant Alzheimer’s disease: a longitudinal study. Lancet Neurol 2018;17:241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan L, Liu CY, Wong KP, et al. Regional association of pCASL-MRI with FDG-PET and PiB-PET in people at risk for autosomal dominant Alzheimer’s disease. Neuroimage Clin 2018;17:751–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson KA, Fox NC, Sperling RA, Klunk WE. Brain imaging in Alzheimer disease. Cold Spring Harb Perspect Med 2012;2:6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowe VJ, Lundt E, Knopman D, et al. Comparison of [18F]Flutemetamol and [11C]Pittsburgh Compound-B in cognitively normal young, cognitively normal elderly, and Alzheimer’s disease dementia individuals. Neuroimage Clin 2017;16:295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fagan AM, Xiong C, Jasielec MS, et al. Longitudinal change in CSF biomarkers in autosomal-dominant Alzheimer’s disease. Sci Transl Med 2014;6:226ra30–226ra30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tarasoff-Conway JM, Carare RO, Osorio RS, et al. Clearance systems in the brain—implications for Alzheimer disease. Nat Rev Neurol 2015;11:457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Llibre-Guerra JJ, Li Y, Schindler SE, et al. Association of longitudinal changes in cerebrospinal fluid total tau and phosphorylated tau 181 and brain atrophy with disease progression in patients with Alzheimer disease. JAMA Netw Open 2019;2:e1917126–e1917126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barthélemy NR, Li Y, Joseph-Mathurin N, et al. A soluble phosphorylated tau signature links tau, amyloid and the evolution of stages of dominantly inherited Alzheimer’s disease. Nat Med 2020;26(3):398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kester MI, Teunissen CE, Crimmins DL, et al. Neurogranin as a cerebrospinal fluid biomarker for synaptic loss in symptomatic Alzheimer disease. JAMA Neurol 2015;72(11):1275–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schindler SE, Li Y, Todd KW, et al. Emerging cerebrospinal fluid biomarkers in autosomal dominant Alzheimer’s disease. Alzheimers Dement 2019;15(5):655–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prakash M, Bodas M, Prakash D, et al. Diverse pathological implications of YKL-40: answers may lie in ‘outside-in’signaling. Cell Signalling 2013;25:1567–1573. [DOI] [PubMed] [Google Scholar]
- 20.Craig-Schapiro R, Perrin RJ, Roe CM, et al. YKL-40: a novel prognostic fluid biomarker for preclinical Alzheimer’s disease. Biol Psychiatry 2010;68:903–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bridel C, Van Wieringen WN, Zetterberg H, et al. Diagnostic value of cerebrospinal fluid neurofilament light protein in neurology: a systematic review and meta-analysis. JAMA Neurol 2019;76(9):1035–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Preische O, Schultz SA, Apel A, et al. Serum neurofilament dynamics predicts neurodegeneration and clinical progression in presymptomatic Alzheimer’s disease. Nat Med 2019;25(2):277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med 2019;380:1347–1358 [DOI] [PubMed] [Google Scholar]
- 24.Morris JC. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology 2012;41:1588–1592. [DOI] [PubMed] [Google Scholar]
- 25.Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res 1975;12(3):189–198. [DOI] [PubMed] [Google Scholar]
- 26.Yesavage JA. Geriatric Depression Scale. Psychopharmacol Bull 1988;24(4):709–711. 10.1007/978-3-319-6989-22_736-1. [DOI] [PubMed] [Google Scholar]
- 27.Jack CR, Bernstein MA, Borowski BJ, et al. Update on the magnetic resonance imaging core of the Alzheimer’s Disease Neuroimaging Initiative. Alzheimers Dement 2010;6:212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischl B FreeSurfer. Neuroimage 2012;62:774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Nat Acad Sci USA 2000;97:11050–11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dincer A, Gordon BA, Hari-Raj A, et al. Comparing cortical signatures of atrophy between late-onset and autosomal dominant Alzheimer disease. Neuroimage Clin 2020;28:102491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su Y, D’Angelo GM, Vlassenko AG, et al. Quantitative analysis of PiB-PET with FreeSurfer ROIs. PLoS One 2013;8:73377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su Y, Blazey TM, Snyder AZ, et al. Partial volume correction in quantitative amyloid imaging. Neuroimage 2015;107:55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rousset OG, Ma Y, Evans AC. Correction for partial volume effects in PET: principle and validation. J Nucl Med. 1998;39:904–911. [PubMed] [Google Scholar]
- 34.Joshi A, Koeppe RA, Fessler JA. Reducing between scanner differences in multi-center PET studies. Neuroimage 2009;46:154–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sutphen CL, McCue L, Herries EM, et al. Longitudinal decreases in multiple cerebrospinal fluid biomarkers of neuronal injury in symptomatic late onset Alzheimer’s disease. Alzheimers Dement 2018;14(7):869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crimmins DL, Herries EM, Ohlendorf MF, et al. Double monoclonal immunoassay for quantifying human visinin-like protein-1 in CSF: to the editor. Clin Chem 2017;63(2):603–604. [DOI] [PubMed] [Google Scholar]
- 37.Disanto G, Barro C, Benkert P, et al. Serum Neurofilament light: a biomarker of neuronal damage in multiple sclerosis. Ann Neurol 2017;1(6):857–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loh WY. Regression trees with unbiased variable selection and interaction detection. Stat Sin 2002;12:361–386. [Google Scholar]
- 39.Sokal RR. A statistical method for evaluating systematic relationships. Univ Kans Sci Bull 1958;38:1409–1438. [Google Scholar]
- 40.Jack CR, Bennett DA, Blennow K, et al. A/T/N: an unbiased descriptive classification scheme for Alzheimer disease biomarkers. Neurology 2016;87(5):539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luckett PH, McCullough A, Gordon BA, et al. Modeling autosomal dominant Alzheimer’s disease with machine learning. Alzheimers Dement 2021;17(6):1005–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barthélemy NR, Bateman RJ, Hirtz C, et al. Cerebrospinal fluid phospho-tau T217 outperforms T181 as a biomarker for the differential diagnosis of Alzheimer’s disease and PET amyloidpositive patient identification. Alzheimers Res Ther 2020;12:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janelidze S, Berron D, Smith R, et al. Associations of plasma phospho-Tau217 levels with tau positron emission tomography in early Alzheimer disease. JAMA Neurol 2021;78:149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.