Abstract
Surface translocation has been described in a large variety of microorganisms, including some gram-negative enteric bacteria. Here, we describe the novel observation of the flagellum-independent migration of Vibrio cholerae and Escherichia coli on semisolid surfaces with remarkable speeds. Important aspects of this motility are the form of inoculation, the medium composition, and the use of agarose rather than agar. Mutations in several known regulatory or surface structure proteins, such as ToxR, ToxT, TCP, and PilA, did not affect migration, whereas a defect in lipopolysaccharide biosynthesis prevented translocation. We propose that the observed surface migration is an active process, since heat, protease, or chloramphenicol treatments of the cells have strong negative effects on this phenotype. Furthermore, several V. cholerae strains strongly expressing the hemagglutinin/protease but not their isogenic hap-negative mutants, lacked the ability of surface motility, and the treatment of migrating strains with culture supernatants from hap strains but not hap-null strains prevented surface translocation.
Bacterial translocation is arguably one of the most impressive features in the bacterial life. Perhaps the planktonic or free-swimming bacterial phase is primarily a mechanism of translocation from one surface to another, since in nature microbial activity is probably mostly associated with surfaces (4). Surface translocation enables the bacteria to establish symbiotic and pathogenic associations with plants and animals, and potential benefits of translocation include increased access to nutrients, avoidance of toxic substances, access to preferred colonization sites within hosts, and increased efficiency of transmission. The study of bacterial interactions with various surfaces is a newly emerging field and especially the remarkable ability of bacteria to form surface-associated structured and cooperative consortia, called biofilms, has received increasing attention in recent years (30).
Movement in aqueous environments by swimming or along surfaces by using different modes of translocation has been classified into several distinct forms. Six different types of translocation have been recognized by Hendrichsen (13) as (i) swarming, dependent on excessive development of flagella and partly on cell-to-cell interaction; (ii) swimming, dependent on flagella and fluid; (iii) gliding, dependent on intrinsic motive force and partly on cell-to-cell interactions; (iv) twitching, dependent on intrinsic motive force and, as we know now, type IV pili; (v) sliding, dependent on growth and reduced friction (i.e., spreading by expansion); and (vi) darting, dependent on growth of capsulated aggregates (i.e., spreading by ejection). All of these different modes of translocation require special surface structures or components, such as flagella, pili, surfactants, slime, or capsules. The flagellum-independent surface spreading of Serratia marcescens can be classified as sliding since it was dependent on growth and the production of the extracellular wetting agent serrawettin (21, 24). Surface translocation of Escherichia coli and some Vibrio spp. characterized as swarming have been reported and are associated with the differentiation of the bacteria into elongated hyperflagellated swarm cells (10). In fact, some Vibrio spp. produce special lateral flagella in addition to their single polar flagella that are required for swarming. Vibrio cholerae, the causative agent of cholera, is capable of swimming motility via a single polar flagellum but, to date, no surface translocation has been reported for this organism.
In the present report, we demonstrate flagellum-independent surface translocation on semisolid media by V. cholerae and E. coli that does not appear to fall into any of the known classifications of motility.
MATERIALS AND METHODS
Strains, media, and culture condition.
Table 1 summarizes the strains used in this study. Some strains were transformed with the plasmid pGFP-2 carrying a gene for green fluorescent protein (GFP) (kindly provided by S. Falkow). Standard motility plates were made with Luria-Bertani (LB) medium and 0.25% agar (Difco) and, in some cases, 0.3% agarose. Motility plates for surface translocation were made on the basis of a special mineral medium containing (in g/liter): K2HPO4 · 3H2O, 0.04; NaNO3, 1.5; MgSO4 · 7H2O, 0.076; CaCl2 · 2H2O, 0.036; Na2CO3, 0.02; yeast extract, 0.1; Bis-Tris Propane, 11.3; succinic acid, 5.9; Casamino Acids (Difco), 10; and NaCl, 8.76 (pH 7.0). The media were typically hardened with 0.3% low-melting-temperature agarose (SeaPlaque) and, in some cases, agar. All motility plates were kept in a humid chamber at 4°C. The bacteria were grown in standard K2HPO4 · 3H2O LB broth for 20 to 22 h in an orbital shaker at 250 rpm at 37°C. Then, 1 to 10 ml of bacterial suspension was centrifuged for 10 min at 5,100 × g at 20°C. The supernatants were carefully aspired, and the bacterial pellets were vortex mixed. In some cases, the cells were washed twice in different media or supernatants.
TABLE 1.
Bacterial strains
| Strain | Relevant phenotype(s) | Source or reference |
|---|---|---|
| V. cholerae | ||
| O395N1 | O1 serotype, classical biotype, parental | 23 |
| O395N1 ΔmotX | Nonmotile, paralyzed flagellum | 8 |
| O395N1 ΔfliG | Nonmotile, nonflagellate | 11 |
| O395N1 ΔcheA | Chemotaxis deficient | This study |
| SC512 | O395 rfbB::pSC95, LPS negative | S. L. Chiang (2) |
| TCP-2 | O395 tcpA::TnphoA, TCP negative | 14 |
| O395N1 ΔtoxR | ToxR negative | 12 |
| O395N1 ΔtoxT | ToxT negative | 12 |
| KFV9 | O395N1 ΔpilA, PilA negative | K. J. Fullner (6) |
| N16961 | O1 serotype, El Tor biotype, parental | 15 |
| N16961 Δhap | HAP negative | This study |
| Peru-2 | O1 serotype, El Tor biotype, parental | 28 |
| Peru-2 Δhap | HAP negative | This study |
| Bengal-2 | O139 serotype, parental | 3 |
| Bengal-2 Δhap | HAP negative | This study |
| CA401 | O1 serotype, classical biotype, parental | 7 |
| CA401 Δhap | HAP negative | This study |
| E. coli | ||
| RP437 | parental | J. S. Parkinson |
| RP6665 | RP437 ΔmotAB, paralyzed flagella | J. S. Parkinson |
| DFB225 | RP437 ΔfliG, nonflagellate | D. F. Blair (18) |
| RP9535 | RP437 ΔcheA, chemotaxis deficient | J. S. Parkinson (25) |
| TA17 | Parental | J. S. Parkinson |
Motility assays.
Next, 1.5 to 2.0 μl of a highly concentrated bacterial suspension was inoculated onto the surface of the motility plates by pipette. In some cases a toothpick was used for inoculation. The inoculated plates were incubated at 37°C for various times.
Microscopy.
Epifluorescens was viewed on an Olympus BX60 microscope attached to a charge-coupled device Hamamatsu C5810 camera. Electron microscopic (EM) analyses were performed by the St. Jude Children's Research Hospital Scientific Imaging Shared Resource Facility.
RESULTS
Surface migration of different V. cholerae and E. coli strains.
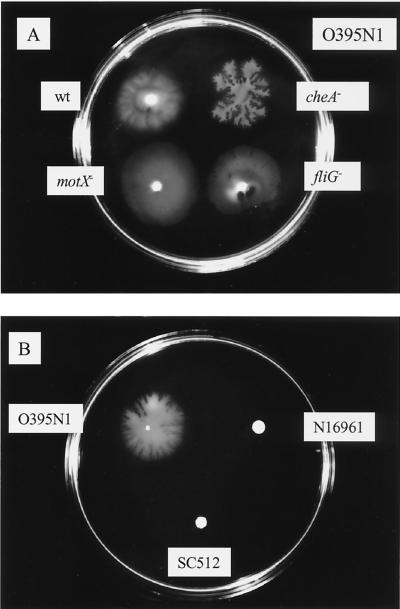
We observed that cells of the V. cholerae classical biotype strain O395N1 moved outward from an inoculum onto the semisolid surface of a mineral medium hardened with 0.3% agarose (Fig. 1A, see movie at www.stjuderesearch.org/ids/chase/index.html). Furthermore, cells of a chemotaxis-deficient mutant derivative as well as nonmotile mutant derivatives of this strain deleted in motX (paralyzed flagellum) or in fliG (nonflagellate) also moved on the surface of these plates (Fig. 1A, see movies at www.stjuderesearch.org/ids/chase/index.html). Mutation in various other genes, including toxR, toxT, tcpA, or pilA, did not significantly affect the ability of O395N1 to migrate on this surface (data not shown). In contrast, a mutant derivative strain of V. cholerae O395N1 carrying an insertion in the rfb gene cluster (SC512) was not able to translocate on this semisolid surface (Fig. 1B). However, most tested V. cholerae strains, including classical biotype strain CA401 (see Fig. 7), and several El Tor biotype strains, including N16961 (Fig. 1B) and Peru-2, as well as the O139 serotype strain Bengal-2 (see Fig. 7), did not show surface migration in most of the experiments.
FIG. 1.
Surface translocation of different V. cholerae strains. (A) Saturated bacterial suspensions of the parental (wt) strain O395N1, as well as a chemotaxis-deficient strain (cheA−) and two nonmotile mutant derivative strains with paralyzed flagella (motX−) or no flagella (fliG−) were inoculated onto the surface of semisolid plates. (B) Surface translocation of the classical biotype strain O395N1 was compared to that of its LPS-negative rfbB insertion derivative (SC512) and the El Tor biotype strain N16961. Plates were incubated at 37°C for 4 h.
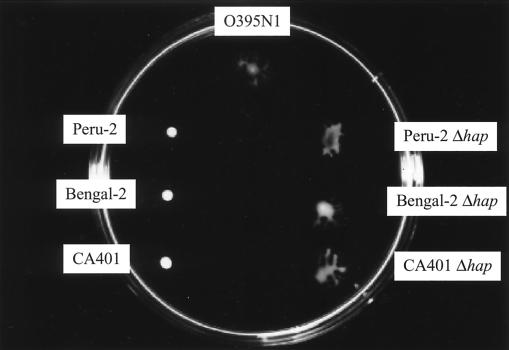
FIG. 7.
Surface translocation of several parental and hap-deleted V. cholerae strains. Strains O395N1, Peru-2, Peru-2 Δhap, Bengal-2, Bengal-2 Δhap, CA401, and CA401 Δhap are shown. The plate was incubated at 37°C for 2 h.
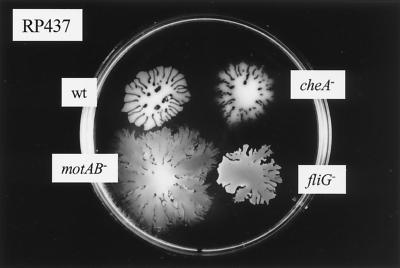
We also observed that cells of E. coli RP437 and its cheA, motAB, and fliG mutants (Fig. 2), as well as another E. coli strain, TA17 (data not shown), translocated on this semisolid surface.
FIG. 2.
Surface translocation of different E. coli strains. Saturated bacterial suspensions of the parental (wt) strain PR437, as well as a chemotaxis-deficient strain (cheA−) and two nonmotile mutant derivative strains with paralyzed flagella (motAB−) or no flagella (fliG−) were inoculated onto the surface of semisolid plates. Plates were incubated at 37°C for 4 h.
Macrocolony formation.
Significant speeds (Ca. 14 mm/h) are achieved by the bacteria during the first 1 to 3 h, and the diameters of the bacterial zones continued to increase rapidly during the first 6 h. The moving cells were able to form arrays of spots and stripes that arise sequentially, and in most cases the macrocolonies had a fractal structure (22) (Fig. 1 and 2). The average size of the macrocolonies was dependent on the strain, the substrate, the agarose concentration, and the time of incubation.
An investigation of the macrocolonies under the microscope showed that migrating spots are cell suspensions in surface liquid (see movies at www.stjuderesearch.org/ids/chase/index.html). It was easy to observe regular swimming of the parental bacteria inside wet spots and its branches, whereas the nonmotile strains did not show detectable motility of individual cells (see movies at www.stjuderesearch.org/ids/chase/index.html). Analysis of the bacterial cells recovered from the plate following surface migration by EM did not reveal any elongated or hyperflagellated cells for either V. cholerae or E. coli.
Conditions for surface migration.
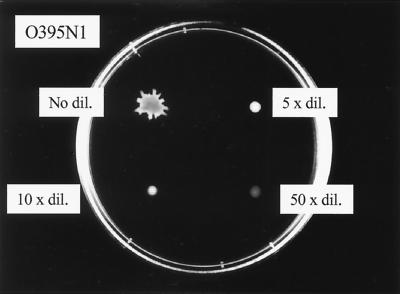
Unlike most investigators studying bacterial surface migration, we spot a dense cell suspension obtained after centrifugation of liquid cultures onto the surface of the semisolid plates rather than using a toothpick for inoculation. All available liquid is removed from the bacterial pellet, and the cells are “resuspended” by vortexing. In fact, we noted that dilution of the cells by either fresh LB broth (Fig. 3) or culture supernatants inhibited the rate of cell migration outward from the inoculation place. No surface migration of either V. cholerae or E. coli was observed after inoculation via a toothpick. Bacterial pellets of both V. cholerae and E. coli did not display any migration following heat treatment at 80°C for 15 min. Furthermore, the treatment of cells with pronase or proteinase K prevented any surface migration. Treatment of the bacterial cells with chloramphenicol during the last hour of growth, combined with the addition of chloramphenicol in the plates, resulted in the loss of surface migration of both V. cholerae and E. coli cells.
FIG. 3.
Surface translocation of V. cholerae strain O395N1 following dilution of the bacterial suspension in LB medium. The effects of no dilution, as well as 5-, 10-, and 50-fold dilutions, are shown. The plate was incubated at 37°C for 2 h.
An increase of agarose concentration in the plates of up to 0.7% significantly decreased the speed of surface migration roughly proportional to the amount of agarose. Using agar instead of agarose to harden the mineral media resulted in the loss of surface migration; similarly, semisolid plates made of LB medium solidified with agarose did not sustain surface migration. Different stocks of carbon, including glucose, glycerol, fumarate, and succinate were found to support surface migration of both V. cholerae and E. coli.
Mixing experiments.
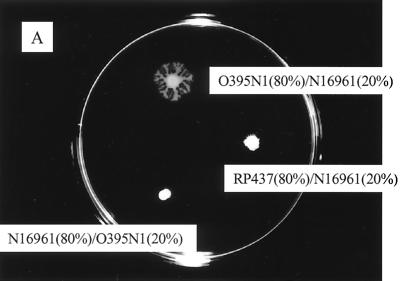
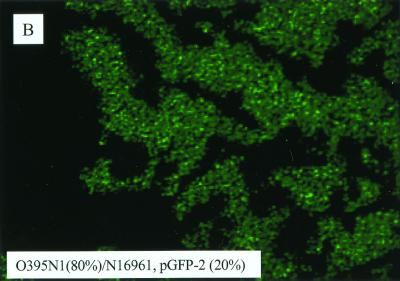
To better understand the lack of migration by most V. cholerae strains, we mixed 80% of V. cholerae O395N1 with 20% of V. cholerae N16961 carrying the gene encoding GFP and vice versa. When these composite pellets were inoculated onto the semisold surface of plates, the “green” cells of the nonmigrating N16961 strain were found to migrate together with the O395N1 cells (Fig. 4B), whereas 20% of O395N1 cells were not sufficient for the surface migration of the N16961 cells (Fig. 4A). The mixing of E. coli RP437 cells with V. cholerae N16961 cells in a 4:1 proportion showed virtually no migration (Fig. 4A).
FIG. 4.
Surface translocation of mixed bacterial suspensions. (A) A mixture of 80% V. cholerae O395N1 cells with 20% V. cholerae N16961 cells and vice versa was inoculated onto the semisolid surface of the plate. Similarly, 80% E. coli RP437 mixed with 20% V. cholerae N16961 cells were spotted. (B) Epifluorescent microscopy of the edge of a macrocolony derived from an inoculate consisting of 80% O395N1 mixed with 20% N16961 harboring a plasmid encoding GFP (pGFP-2).
Effects of a surfactant.
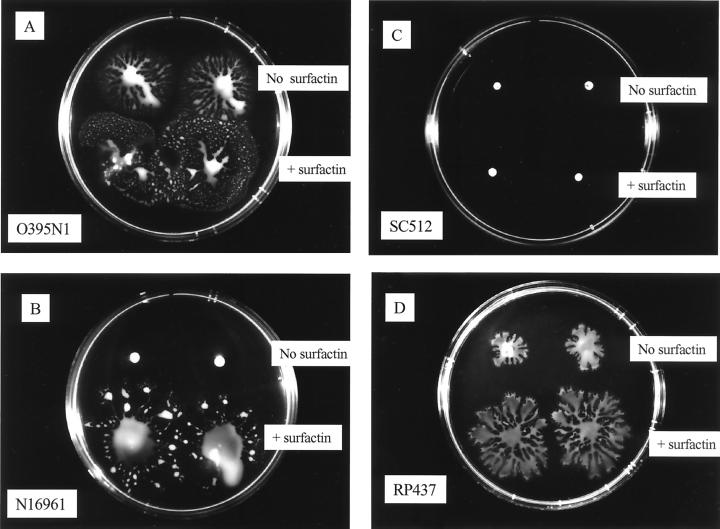
To start investigating the possible mechanism of this flagellum-independent surface migration, we tested for the production of a wetting agent by the cells that would lower the surface tension of the water and create a conditioning film for the bacteria to move on. However, we have been unable to detect the presence of surfactants in the V. cholerae or E. coli cultures by the drop collapsing test (14a). However, the addition of surfactin (Sigma), produced by B. subtilis, to the bacterial pellets stimulated surface migration of the E. coli strain and both the V. cholerae O395N1 and the N16961 strains (Fig. 5A, B, and D). In contrast, the derivative strain of O395N1 carrying an insertion in the rfbB gene did not show any surface translocation even in the presence of surfactin (Fig. 5C).
FIG. 5.
Surface translocation in the presence or absence of surfactin. A total of 5 μl of LB medium or 5 μg of surfactin per ml diluted in LB medium was were added to 100 μl of bacterial cell suspensions of V. cholerae O395N1 (A), N16961 (B), or SC512 (C) or E. coli RP437 (D). Then, 2 μl of the treated cell suspensions was inoculated onto the surface of the plates and incubated at 37°C for 4 h.
Effects of culture supernatants.
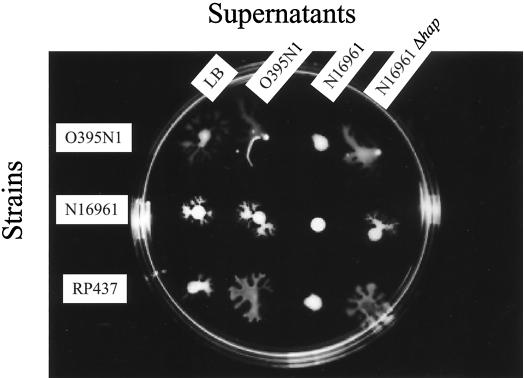
If the cells produce some extracellular compound necessary for this surface migration, we might be able to complement the nonmigrating strains by the addition of sterile culture supernatants from the migrating strain. However, washing the cell pellet of N16961 with culture supernatants from O395N1 did not greatly enhance the surface translocation of this strain compared to a washing step with LB medium (Fig. 6). However, it does not appear that the washing with LB medium or O395N1 supernatant leads to slightly improved movement of the N16961 cells outward from the inoculation site, compared to treatment with supernatants from N16961 which does not result in any migration (Fig. 6). Interestingly, washing with the supernatants of the nonmigrating strain N16961 inhibited the migration of O395N1 cells (Fig. 6), whereas heat treatment of this supernatant at 65°C for 10 min prevented this inhibitory property. Similarly, the N16961 supernatants inhibited the surface migration of E. coli RP437, whereas LB medium, O395N1 supernatants, or N16961 Δhap supernatants did not (Fig. 6). By using sizing spin columns, we demonstrated that this inhibitory compound is retained by 10-kDa and 30-kDa, but not by a 100-kDa, cutoff membrane.
FIG. 6.
Effects of washing on the surface translocation of V. cholerae O395N1 and N16961 and E. coli RP437. Treatments with LB medium or sterile supernatants from V. cholerae O395N1, N16961, or N16961 Δhap are shown. The plate was incubated at 37°C for 2 h.
Effects of HAP.
Since we had found that protease treatment prevented surface migration (see above), we postulated that the inhibitory factor observed in the culture supernatants of nonmigrating V. cholerae strains might be one of the several proteases produced by V. cholerae. Indeed, mutant derivatives of several nonmigrating V. cholerae strains deleted in the hap gene, encoding hemagglutinin/protease (HAP), showed surface migration comparable to the O395N1 strain (Fig. 7). Furthermore, treatments of V. cholerae O395N1 or E. coli RP437 with supernatants of the hap deletion strains, unlike the parental supernatants, did not inhibit their surface translocation (Fig. 6).
DISCUSSION
The ability to move on solid surfaces is widespread among bacteria. The swarming of Proteus spp., the gliding of Myxococcus spp., and the twitching of Pseudomonas spp. are well-known examples of bacterial surface translocation and, in all cases, the production of various special surface structures are required for motility. Several flagellated gram-negative species including Serratia spp., Vibrio alginolyticus, Vibrio parahaemolyticus, E. coli, Salmonella enterica serovar Typhimurium and, more recently, Yersinia and Pseudomonas spp. (5, 16, 26, 31), have been shown to display swarming behavior. For swarming, the bacteria form specialized elongated swarm cells, and the production of flagella is required for this type of surface migration. Flagellum-independent spreading has been reported for S. marcescens and requires growth and the production of the extracellular wetting agent serrawettin (21, 24) and thus can be classified as sliding. Here, we report the novel finding of flagellum-independent surface translocation of V. cholerae and E. coli on semisolid surfaces.
Unlike most investigators, we used a highly saturated bacterial pellet recovered following growth in liquid media rather than a toothpick to inoculate the surface of a medium plate. Thus, in our system the bacteria initially migrate outward from the inoculation point very rapidly without the requirement for growth. Indeed, the density of the bacterial pellet seems to be important for this phenomenon, as dilutions significantly reduce the ability of the cells to migrate. Following growth in LB medium, the cells are ready for this migration since spreading occurs almost immediately after inoculation. Another intriguing aspect of this surface migration is the remarkable speed of about 14 mm/h by which the bacteria can travel during the first few hours of incubation. Interestingly, regular swimming behavior, i.e., movement of the bacteria within the semisolid plates, is only observed after several hours in strains that do not show this surface migration or if the surface migration is artificially inhibited. The combination of a relatively low nutrient media solidified by agarose rather than agar is also important for this phenomenon, as has been observed in several other motility systems (17, 19, 20). We therefore propose that this type of surface translocation requires specific interactions between the surface of the bacteria and the surface of the medium plate.
A series of V. cholerae mutants defective in known regulatory proteins or surface structures were analyzed for this migrating behavior. No effects were observed in strains deficient in the ToxR, ToxT, TcpA, or PilA proteins. However, the loss of lipopolysaccharide (LPS) production in a strain with an insertion in the rfbB gene completely abolished surface migration. Similarly, LPS was found to be important for the swarming of Salmonella (29), and the surface-exposed glycopeptidolipids (GPL), a component of the mycobacterial cell wall, was required for the sliding of this nonflagellated microorganism (19). In addition to LPS, it appears that the production of an as-yet-unknown surface protein(s) is required for the surface migration of V. cholerae and E. coli, since chloramphenicol treatment, as well as heat or protease treatment, of the cells prevented migration. Most intriguingly, production of the V. cholerae HAP or treatment with culture supernatants from HAP-producing, but not hap-negative mutant strains inhibited migration. This strongly suggests the production of a proteinaceous factor involved in this process. In the nonflagellated cyanobacterium Synechococcus, a cell-surface-associated polypeptide, SwmA, has been shown to be required for motility (1). Analysis of the bacterial cells by EM did not show hyperflagellation or any kind of obvious surface structure, such as pili, that might be associated with this surface migration.
Although we were not able to demonstrate the production of a surfactant by the V. cholerae or E. coli strains by using the drop collapse test, the addition of surfactin, a surfactant produced by B. subtilis, further enhanced the surface translocation of the migrating V. cholerae and E. coli strains. Moreover, it enabled the nonmigrating strain N16961, but not the LPS-deficient mutant, to move outward from the inoculation site. Similarly, several different surfactants did not complement the nonsliding phenotype of GPL-negative Mycobacterium strains (27). In contrast, the addition of surfactin did rescue the swarm defect of an LPS-negative Salmonella strain (29). Although we do not yet understand the mechanism of this novel type of surface translocation, we propose the production of some surface tension-reducing compound by the bacteria that allows the cells to migrate. In addition to intact LPS, some unknown surface protein(s) appears to be required for this surface translocation. Clearly, further experiments, such as transposon mutagenesis and characterization of the protease-sensitive surface structure, are required to elucidate the mechanism underlying this novel phenotype.
V. cholerae, unlike some closely related Vibrio species, does not appear to differentiate into swarm cells and showed no evidence of lateral flagellum production or laf gene homologs in its genome. Its infection cycle includes colonization of the human gut as well as an environmental phase. This type of surface translocation might play a role in either or both of these phases of this pathogen's life cycle.
ACKNOWLEDGMENTS
We thank K. Gosink, T. Penfound, and R. Harshey for many helpful discussions and critical reading of the manuscript and S. Sarkisova for technical assistance. We are grateful to S. Parkinson, D. Blair, and K. Fullner for providing strains.
This study was supported in part by a NATO Linkage Grant (LST.CLG 975168), Cancer Center Support Grant (CA 21765), and ALSAC (American Lebanese Syrian Associated Charities).
REFERENCES
- 1.Brahamsha B. An abundant cell-surface polypeptide is required for swimming by the nonflagellated marine cyanobacterium Synechococcus. Proc Natl Acad Sci USA. 1996;93:6504–6509. doi: 10.1073/pnas.93.13.6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiang S L, Mekalanos J J. rfb mutations in Vibrio cholerae do not affect surface production of toxin-coregulated pili but still inhibit intestinal colonization. Infect Immun. 1999;67:976–980. doi: 10.1128/iai.67.2.976-980.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coster T S, Killeen K P, Waldor M K, Beattie D T, Spriggs D R, Kenner J R, Trofa A, Sadoff J C, Mekalanos J J, Taylor D N. Safety, immunogenicity, and efficacy of live attenuated Vibrio cholerae O139 vaccine prototype. Lancet. 1995;345:949–952. doi: 10.1016/s0140-6736(95)90698-3. [DOI] [PubMed] [Google Scholar]
- 4.Costerton J W, Cheng K-J, Geesey G G, Ladd T I, Nickel J C, Dasgupta M, Marrie T J. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 5.Fraser G M, Hughes C. Swarming motility. Curr Opin Microbiol. 1999;2:630–635. doi: 10.1016/s1369-5274(99)00033-8. [DOI] [PubMed] [Google Scholar]
- 6.Fullner K J, Mekalanos J J. Genetic characterization of a new type IV-A pilus gene cluster found in both classical and El Tor biotypes of Vibrio cholerae. Infect Immun. 1999;67:1393–1404. doi: 10.1128/iai.67.3.1393-1404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardner E W, Lyles S T, Lankford C E, Hagens S J. Vibrio cholerae infection in the embryonated egg. J Infect Dis. 1963;112:264–272. doi: 10.1093/infdis/112.3.264. [DOI] [PubMed] [Google Scholar]
- 8.Gosink K K, Häse C C. Requirements for conversion of the Na+-driven flagellar motor of Vibrio cholerae to the H+-driven motor of Escherichia coli. J Bacteriol. 2000;182:4234–4240. doi: 10.1128/jb.182.15.4234-4240.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harshey R M. Bees aren't the only ones: swarming in gram-negative bacteria. Mol Microbiol. 1994;13:389–394. doi: 10.1111/j.1365-2958.1994.tb00433.x. [DOI] [PubMed] [Google Scholar]
- 10.Harshey R M, Matsuyama T. Dimorphic transition in Escherichia coli and Salmonella typhimurium: surface-induced differentiation into hyperflagllate swarmer cells. Proc Natl Acad Sci USA. 1994;91:8631–8635. doi: 10.1073/pnas.91.18.8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Häse C C, Mekalanos J J. Effects of changes in membrane sodium flux on virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 1999;96:3183–3187. doi: 10.1073/pnas.96.6.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Häse C C, Mekalanos J J. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 1998;95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendrichsen J. Bacterial surface translocation: a survey and a classification. Bacteriol Rev. 1972;36:478–503. doi: 10.1128/br.36.4.478-503.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrington D A, Hall R H, Losonsky G, Mekalanos J J, Taylor R K, Levine M M. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J Exp Med. 1988;168:1487–1492. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Jain D K, Thompson D K C, Lee H, Trevors J T. A drop-collapsing test for screening surfactant producing microorganisms. J Microbiol Methods. 1991;13:271–279. [Google Scholar]
- 15.Kaper J B, Lockman H, Baldini M M, Levine M M. Recombinant nontoxinogenic Vibrio cholerae strains as attenuated cholera vaccine candidate. Nature. 1984;308:655–658. doi: 10.1038/308655a0. [DOI] [PubMed] [Google Scholar]
- 16.Kinscherf T G, Willis D K. Swarming by Pseudomonas syringae B728a requires gacS (lemA) and gacA but not the acyl-homoserine lactone biosynthestic gene ahlI. J Bacteriol. 1999;181:4133–4136. doi: 10.1128/jb.181.13.4133-4136.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohler T, Curty L K, Barja F, van Delden C, Pechere J C. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J Bacteriol. 2000;182:5990–5996. doi: 10.1128/jb.182.21.5990-5996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lloyd S A, Blair D F. Charged residues in the rotor protein FliG essential for torque generation in the flagellar motor of Escherichia coli. J Mol Biol. 1997;266:733–744. doi: 10.1006/jmbi.1996.0836. [DOI] [PubMed] [Google Scholar]
- 19.Martinez A, Torello S, Kolter R. Sliding motility in mycobacteria. J Bacteriol. 1999;181:7331–7338. doi: 10.1128/jb.181.23.7331-7338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuyama T, Bhasin A, Harshey R M. Mutational analysis of flagellum-independent surface spreading of Serratia marcescens 274 on a low-agar medium. J Bacteriol. 1995;177:987–991. doi: 10.1128/jb.177.4.987-991.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuyama T, Kaneda K, Nakagawa Y, Isa K, Hara-Hotta H, Yano I. A novel extracellular cyclic lipopeptide which promotes flagellum-dependent and-independent spreading growth of Serratia marcescens. J Bacteriol. 1992;174:1769–1776. doi: 10.1128/jb.174.6.1769-1776.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuyama T, Matsushita M. Fractal morphogenesis by a bacterial cell population. Crit Rev Microbiol. 1993;19:117–135. doi: 10.3109/10408419309113526. [DOI] [PubMed] [Google Scholar]
- 23.Mekalanos J J, Schwartz S J, Pearson G D N, Hardford N, Groyne F, de Wilde M. Cholera toxin genes: nucleotide sequences, deletion analysis and vaccine development. Nature. 1983;306:551–557. doi: 10.1038/306551a0. [DOI] [PubMed] [Google Scholar]
- 24.O'Rear J, Alberti L, Harshey R M. Mutations that impair swarming motility in Serratia marcescens 274 include but are not limited to those affecting chemotaxis or flagellar function. J Bacteriol. 1992;174:6125–6137. doi: 10.1128/jb.174.19.6125-6137.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parkinson J S, Houts S E. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis function. J Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rashid M H, Kornberg A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2000;97:4885–4890. doi: 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Recht J, Martinez A, Torello S, Kolter R. Genetic analysis of sliding motility in Mycobacterium smegmatis. J Bacteriol. 2000;182:4348–4351. doi: 10.1128/jb.182.15.4348-4351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor D N, Killeen K P, Hack D C, Kenner J R, Coster T S, Beattie D T, J. E, Hyman T, Trofa A, Sjogren M H, Friedlander A, Mekalanos J J, Sadoff J C. Development of a live, oral, attenuated vaccine against El Tor cholera. J Infect Dis. 1994;170:1518–1523. doi: 10.1093/infdis/170.6.1518. [DOI] [PubMed] [Google Scholar]
- 29.Toguchi A, Siano M, Burkart M, Harshey R M. Genetics of swarming motility in Salmonella enterica serovar Typhimurium: critical role for lipopolysaccharide. J Bacteriol. 2000;182:6308–6321. doi: 10.1128/jb.182.22.6308-6321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watnick P, Kolter R. Biofilm, city of microbes. J Bacteriol. 2000;182:2675–2679. doi: 10.1128/jb.182.10.2675-2679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Young G M, Smith M J, Minnich S A, Miller V L. The Yersinia enterocolitica motility master regulatory operon, flhDC, is required for flagellin production, swimming motility, and swarming motility. J Bacteriol. 1999;181:2823–2833. doi: 10.1128/jb.181.9.2823-2833.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]