Abstract
Renal fibrosis is an unavoidable end result of all forms of progressive chronic kidney diseases (CKD). Discovery of efficacious drugs against renal fibrosis is in crucial need. In a preliminary study we found that a derivative of artemisinin, dihydroartemisinin (DHA), exerted strong renoprotection, and reversed renal fibrosis in adenine-induced CKD mouse model. In this study we investigated the anti-fibrotic mechanisms of DHA, particularly its specific target in renal cells. Renal fibrosis was induced in mice by unilateral ureteral obstruction (UUO) or oral administration of adenine (80 mg · kg−1), the mice received DHA (30 mg · kg−1 · d−1, i.g.) for 14 or 21 days, respectively. We showed that DHA administration markedly attenuated the inflammation and fibrotic responses in the kidneys and significantly improved the renal function in both the renal fibrosis mouse models. In adenine-treated mice, DHA was more effective than 5-azacytidine against renal fibrosis. The anti-fibrotic effects of DHA were also observed in TGF-β1-treated HK-2 cells. In order to determine the target protein of DHA, we conducted pull-down technology coupled with shotgun proteomics using a small-molecule probe based on the structure of DHA (biotin-DHA). As a results, DNA methyltransferase 1 (DNMT1) was identified as the anti-fibrotic target of DHA in 3 different types of renal cell lines (HK-2, HEK293 and 3T3). We demonstrated that DHA directly bound to Asn 1529 and Thr 1528 of DNMT1 with a Kd value of 8.18 μM. In primary mouse renal tubular cells, we showed that DHA (10 μM) promoted DNMT1 degradation via the ubiquitin–proteasome pathway. DHA-reduced DNMT1 expression effectively reversed Klotho promoter hypermethylation, which led to the reversal of Klotho protein loss in the kidney of UUO mice. This subsequently resulted in inhibition of the Wnt/β-catenin and TGF-β/Smad signaling pathways and consequently conferred renoprotection in the animals. Knockdown of Klotho abolished the renoprotective effect of DHA in UUO mice. Our study reveals a novel pharmacological activity for DHA, i.e., renoprotection. DHA exhibits this effect by targeting DNMT1 to reverse Klotho repression. This study provides an evidence for the possible clinical application of DHA in the treatment of renal fibrosis.
Keywords: renal fibrosis, dihydroartemisinin, DNMT1, Klotho, 5-azacytidine, affinity-based proteome profiling
Introduction
Chronic kidney disease (CKD) has emerged as a global health concern. It accounted for the loss of more than 7.1 million lives in 2010, which is projected to increase to 9.1 million by 2030 [1]. The number of patients with CKD and end-stage kidney disease (ESKD) increases annually due to the growing incidence of type 2 diabetes mellitus, hypertension, obesity, and aging [2]. Approximately 11% of stage 3 CKD cases are estimated to progress to ESKD, eventually requiring dialysis or kidney transplantation. The treatment outcomes of CKD patients remain poor due to limited available treatment options [3]. Renal fibrosis is comprised of a series of chronic and progressive pathological events that affect the kidneys regardless of cause [3]. Renal fibrosis which inevitably accelerates CKD progression is epitomized by fibroblast and myofibroblast proliferation. Myofibroblasts in turn are distinguished by the production of alpha-smooth muscle actin (α-SMA) fibers, collagen and extracellular matrix (ECM) proteins. Incessant ECM production leads to a decreased glomerular filtration rate and renal injury [4]. In the past 20 years, no new drug has been approved to specifically prevent CKD [5]. Thus, the discovery of efficacious drugs in this regard is crucial.
Artemisinin is a well-established antimalarial drug isolated from Artemisia annua L [6, 7]. However, its derivative, dihydroartemisinin (DHA) is usually preferred due to its enhanced water solubility and few side effects [8–10]. Aside from its known antimalarial activity, DHA is reported to possess a plethora of biological activities such as anti-fibrotic, anti-inflammatory and antitumor activities [11–14]. The results of preliminary in-house studies have shown that DHA exhibited strong renoprotection and reversed renal fibrosis in an adenine-induced CKD mouse model. However, the mechanism(s) that underlie this effect, particularly the specific target site of DHA remain unclear and therefore require elucidation. The identification of probable cellular targets of bioactive molecules is crucial in drug discovery [15], since it aids in the elucidation of underlying mechanisms and the prediction of possible side effects [16]. Affinity-based proteome profiling (AfBP) has recently been used for target protein–drug interactions [17, 18]. Through AfBP, Liao et al. showed that sappanone A (a small-molecule probe) possesses pronounced anti-neuroinflammatory effects by targeting cysteine residue 140 (Cys140) in the Bateman domain of inosine 5′-monophosphate dehydrogenase type II (IMPDH2) using an affinity-based probe [19]. Chen et al. found withangulatin A (WA) to be a novel inhibitor of Homo sapiens phosphoglycerate dehydrogenase (PHGDH) via proteomics and phenotypic profiling [20]. In this work, a small-molecule probe based on the structure of DHA (biotin-DHA) and AfBP was used to investigate the target protein of DHA.
To our knowledge, our study is the first to demonstrate that DHA markedly reversed renal fibrosis by directly binding to Asn 1529 and Thr 1528 of DNA methyltransferase 1 (DNMT1), resulting in DNMT1 degradation via the ubiquitin–proteasome pathway. Downregulation of DNMT1 expression effectively reversed Klotho promoter hypermethylation, consequently reversing Klotho protein loss and protecting the kidneys against injury. Knockdown of Klotho abolished the renoprotective effect of DHA. Our study therefore reveals renoprotection by DHA, a previously unknown biological property of this established drug. DHA exerted this effect by targeting DNMT1 to reverse Klotho repression. This study therefore provides a proof-of-concept of the possible clinical application of DHA to treat renal fibrosis.
Materials and methods
Human renal tissues
Renal tissues from patients with category III immunoglobulin A nephropathy and patients with Henoch Schonlein purpura nephritis were provided by the Affiliated Hospital of Nanjing University of Chinese Medicine.
Animal models
All animals were used for designated studies after a week of acclimatization to laboratory conditions. Male ICR mice (weight, 18–20 g) were freely given water and regular mice chow and their body weights taken every week. To establish the adenine mouse model, male ICR mice were orally administered 80 mg/kg per day of adenine (73-24-5; J&K) for 21 days. Dihydroartemisinin and 5-azacytidine (J&K) were administered by oral gavage for 21 days starting on day 22. The experimental protocol comprised of five groups: (i) the control group (mice that received oral saline daily, n = 8), (ii) DHA group (mice that received DHA at a dose of 30 mg/kg daily by oral gavage from day 22 to 42, n = 8), (iii) adenine group (mice that received oral gavage of 80 mg/kg adenine every day from day 1 to 21, n = 8), (iv) 5-Aza + adenine group (mice that received 80 mg/kg adenine every day from day 1 to 21 and 2 mg/kg 5-aza daily by oral gavage from day 22 to 42, n = 8), and (v) DHA + adenine group (mice that received 80 mg/kg adenine every day from day 1 to 21 and 30 mg/kg DHA daily by oral gavage from day 22 to 42, n = 8).
Renal fibrosis mouse models were established using the Unilateral Ureteral Obstruction (UUO) protocol. The experimental mice, were then randomly grouped as follows: (i) the sham operated-group (mice that received a normal diet, n = 10); (ii) dihydroartemisinin group (mice that received 30 mg/kg dihydroartemisinin daily by oral administration); and (iii) renal fibrotic group [mice that underwent UUO (14 days)]. After the completion of the experiment, the mice were anaesthetized with isoflurane, and blood was collected from ventriculus dexter and their kidneys were harvested and stored at −80 °C until further analysis. A minimum of 6 mice was used for each group and a minimum of three independent cell experiments were performed. No animal was excluded from the experiments except for technical reasons.
Serotype 9 AAV vectors (AAV9) carrying shRNA against Klotho were obtained from Hanheng Biotechnology (Shanghai, China). Randomly selected mice were subjected to intra-renal pelvic injection of either AAV9 carrying shRNA against Klotho or control vector at 50 μL (2 × 1012 v g/mL) per mouse. After 3 weeks, UUO model was carried out.
Blood parameter measurement, histology, and Western blot analysis
Blood analysis was performed on an Olympus AU640 automatic analyser. The method for histological analysis was the same as previously reported [21]. Western blot protocol used was same as previously described [22]. The following antibodies were used: anti-collagen I (14695-1-AP; Proteintech), anti-collagen III (22734-1-AP; Proteintech), anti-DNMT1 (ab92314; Abcam), anti-KL (28100-1-AP; Proteintech), anti-ubiquitin (10201-1-AP; Proteintech), anti-fibronectin (ab2413; Abcam), anti-PAI-1 (13801-1-AP; Proteintech), anti-α-SMA (ab124964; Abcam), anti-GAPDH (HRP-60004; Proteintech), anti-Smad2 (#5339; CST), anti-Smad3 (#9523; CST), anti-phospho-smad2 (#18338; CST), anti-phospho-smad2 (#9520; CST), anti-β-catenin (#8480; CST), anti-active β-catenin (#19874T), anti-Dvl2 (#3224; CST), anti-Dvl3 (#3218; CST), and HRP-conjugated goat anti-rabbit IgG (#7074s, CST). Semi-quantitation of the proteins was determined using ImageJ software (version 1.48), with a normalization of their band densities to that of GAPDH.
Co-immunoprecipitation
Protein lysates were collected in RIPA buffer (P0013D, Beyotime) supplemented with protease inhibitor (P1005, Beyotime) and phosphatase inhibitor A (P1082, Beyotime) cocktails. For immunoprecipitation, the cell lysates were sonicated and centrifuged for 10 min at 14,000 × g, and the supernatants were normalized by measuring the total protein concentration. Lysates were incubated with a monoclonal mouse antibody against DNMT1 (1:20; ab92314; Abcam, Cambridge, MA) overnight at 4 °C before pull-down with protein A/G magnetic beads (sc-2003; Santa Cruz). The magnetic beads were washed three times with lysis buffer before elution in PBS buffer. All samples were resolved by SDS-PAGE and transferred to nitrocellulose membranes. The membranes were immunoblotted using the following antibodies: DNMT1 (1:1000; ab92314; Abcam) and ubiquitin (1:1000; 10201-2-AP; Proteintech). The membranes were then incubated with the appropriate horseradish peroxide-conjugated secondary antibody and visualized with enhanced chemiluminescence reagent (GE Healthcare). For all protein bands, densitometry was performed with the ImageJ Software (version 1.48), and the density of the protein bands was normalized to the density of the GAPDH band.
Immunohistochemical staining
Sections (5 μm) of kidney tissues were obtained and deparaffinized in xylene, hydrated in graded ethanol solutions, and rinsed with tap water and distilled water. Then, endogenous peroxide activity was blocked by incubation in 0.3% hydrogen peroxide in methanol for 30 min. For antigen retrieval, the kidney tissue sections were incubated with 10 μmol/L citrate buffer solution (pH: 6.0) and boiled for 10–15 min. Subsequently, the sections were blocked with 10% normal goat serum for 1 h at room temperature and then incubated overnight at 4 °C with rabbit Megalin (19700-1-AP), DNMT1 (ab88453, Abcam) or KL (28100-1-AP, Proteintech). After washing with PBS, the secondary antibody was added, and the sections were incubated at 37 °C for 1 h. Finally, the kidney tissue sections were exposed to diaminobenzidine peroxidase substrate for 5 min and counterstained with haematoxylin and eosin. Fluorescence images of the sections were obtained using a Leica DMi8 fluorescence microscope (Leica, Germany).
Cell culture and treatment
Epithelial cells from the proximal convoluted tubules of the human renal cortex (HK-2 cells, Stem-Cell Bank, Chinese Academy of Sciences) were cultured in DMEM/F-12 (C11330500BT; Gibco) supplemented with 10% FBS (04-001-1ACS, BI) and 1% penicillin/streptomycin (15140122, ThermoFisher). Mouse embryonic fibroblast (3T3 cells, Stem-Cell Bank, Chinese Academy of Sciences) and Human embryonic kidney cells (HEK-293 cells, Stem-Cell Bank, Chinese Academy of Sciences) were cultured in DMEM (01-052-1ACS; BI) supplemented with 10% FBS (04-001-1ACS, BI) and 1% penicillin/streptomycin (15140122, ThermoFisher). Prior to treatment, the cells were plated at a density of 8 × 105 cells/well in 6-well plates overnight. The cells were then treated with the following reagents for 24 h unless otherwise indicated: TGF-β1 (10 ng/mL; R&D Systems, Minneapolis, MN), the protein synthesis inhibitor cycloheximide (20 µg/kg; Glpbio), dihydroartemisinin (Yuanyeshengwu), and the proteosomal inhibitor MG-132 (0.5 µM; Glpbio). All inhibitors except for MG-132 were added 30 min prior to the addition of TGF-β1. MG-132 was added 1 h prior to the addition of TGF-β1.
Isolation of tubular epithelial cells
The isolation of mouse renal tubular epithelial cells involved simply taking out the 1–3 days old mice, and after aseptically separating the kidney tissues, the capsule and connective tissues were removed. After washing 3 times with PBS, the shredded kidney tissues were sequentially passed through 80 mesh and 200 mesh screens in PBS. The cells were centrifuged at 1500 rpm for 5 min. Afterwards, the collected tubular tissue was digested with 1 mg/mL type IV collagenase (V900893-100; Sigma) at 37 °C for 25 min. They were then passed through a 100-mesh screen after the digestion is terminated. The cells were centrifuged at 1600 rpm for 5 min. Finally, the tubular cells were resuspended in DMEM/F-12 (36254; Stemcell) supplemented with 10% FBS (10099141 C; Gibco). The medium was changed after the cells were cultured in cell culture dishes for 48 h.
Sulforhodamine B colorimetric assay
A total of 8000 HK-2 cells were seeded in 96-well plates. On the first day after inoculation, HK-2 cells were cultured with DHA for 24 h, and then the drug was removed. The cells were fixed with 50% acetic acid at room temperature for 1 h, washed with water 3 times, and stained with 0.4% (w/v) sulforhodamine B (SRB) solution for 20 min at room temperature. After staining, the HK-2 cells were washed with washing buffer (3 times) and dried in an oven. The protein binding dye was dissolved in 10 mM Tris alkali, and the absorbance was measured at 492 nm using an enzyme-labeled instrument (Sunrise, TECAN; Mènnedorf, Switzerland). The data were then graphically presented as a histogram using GraphPad Prism (8.0)
mRNA isolation and qRT-PCR
Total mRNA was extracted using a High Pure RNA Isolation Kit (RNAiso Plus, Takara Bio, Japan) according to the manufacturer’s instructions. Total RNA was reverse transcribed with HiScript II Q RT SuperMix for qPCR according to the instructions of the manufacturer (Vazyme, Nanjing, China). Quantitative real-time PCR (qRT-PCR) was carried out on the Step One system (A&B) using AceQ qPCR SYBR Green Master Mix (Vazyme). The mRNA levels of genes were calculated by normalization to the level of GAPDH.
The synthesis and characterization for biotin-DHA
The synthesis and characterization of biotin-DHA are shown in Supplementary Figs. 1–5.
Preparation of pull-down samples
The cells were lysed with RIPA buffer and divided into three parts. The cells were incubated overnight at 4 °C with biotin-DHA or 0.2% dimethyl sulfoxide as a control, and then streptavidin agarose (20357, Thermo Scientific) was added to the cell lysates, and incubated at room temperature for 1 h. Centrifugation was performed at low temperature, the immobilized beads were washed with RIPA buffer 4 times, 2× SDS-PAGE buffer was added, the samples were boiled, and then the pull-down samples were placed into a refrigerator at −20 °C.
Identification of DHA target proteins
Identification of DHA target proteins was based on pull-down technology coupled with shotgun proteomics analysis. Details of the methods are presented in the SI Methods.
Target validation by Western blotting
Pull-down samples from labeled lysates along with pull-down samples from DMSO-treated and unlabeled lysates (negative controls) were separated on 7% SDS-PAGE gels. After SDS-PAGE, the proteins were transferred to a PVDF membrane and subsequently blocked with 2.5% (w/v) BSA. the membranes were incubated for 1 h at room temperature with DNMT1 antibodies. After three washes with PBST, the blots were further incubated with the appropriate secondary antibody for 1 h at room temperature. After incubation, the blots were washed three times with PBST and exposed with a luminescence imaging system.
Molecular docking and modeling
The crystal structure of human DNMT1 was downloaded from a protein data bank (PDB id: 3pta). Then, DHA was docked into the protein by using the AutoDock program [23]. The genetic algorithm was applied for conformational analysis. To assess the conformational space of DHA as completely as possible, we performed 100 individual genetic algorithm runs to generate 100 docked conformations. The size of the docking box was properly set to enclose the possible binding pocket. The protein structure was fixed during molecular docking. The docked protein-ligand complex structure was further relaxed through molecular dynamics (MD) simulation by using the Amber program [24]. The most populated structure during the 500 ns MD simulation was obtained via cluster analysis.
Methylation-specific PCR (MSP)
Mouse genomic DNA was extracted from the kidney and modified. Prediction of a CpG island in the Klotho promoter and design of a primer for methylation-specific PCR were performed with MethPrimer software. Mouse genomic sequences near the Klotho transcription starting site (−700 to +950 bp) were analysed. The relative locations of CpG islands and methylation-specific PCR primers are plotted in Fig. 4c. Mouse genomic DNA was extracted from the kidney and modified by bisulfate (59824, EpiTect Fast DNA Bisulfite Kit, QIAGEN) treatment. Methylation-specific PCR primers were designed to amplify the -352/-176 locus, and PCR was performed with the specific methylated and unmethylated primers ME068-S1-MF, ME068-S1-MR, ME068-S1-UF, and ME068-S1-UR, which are listed in Supplementary Table 1. Genomic DNA not treated with bisulfate was used as the input control. The PCR products were analysed on a 2% agarose gel and visualized under ultraviolet light.
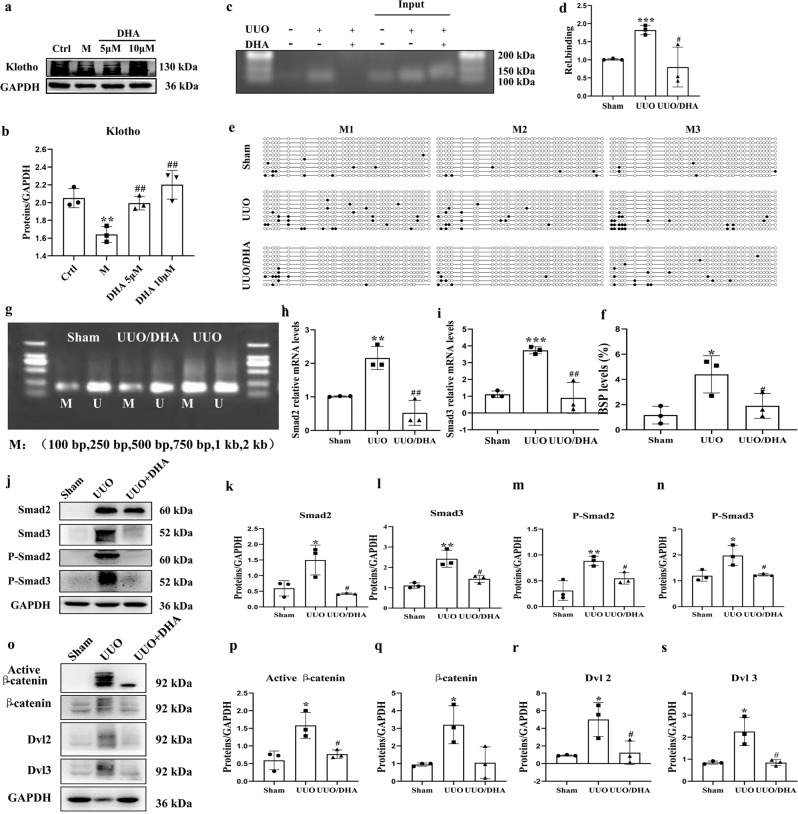
Fig. 4. DHA reverses Klotho promoter hypermethylation and inhibiting the TGF-β/Smad and Wnt/β-catenin signaling pathways in UUO mice.
a, b Expression of Klotho in primary mouse renal tubular cells (PRTCs) treated with TGF-β (10 ng/mL) in the presence or absence of DHA for 48 h as assayed by Western blotting. c, d ChIP assay. The renal tissues as indicated were immunoprecipitated with an antibody for DNMT1, and then the genomic DNA (input) and the antibody-bound DNA fragments were PCR-amplified with primers covering the DNMT1 site on the Klotho promoter. PCR products were analyzed on agarose gels and analyzed by ImageJ software. e BSP analysis of sham, UUO, and DHA-treated UUO mouse kidneys (2 weeks). Each box represents the indicated mouse kidney (M); each row of circles in the boxes represents the Klotho CpG island; each dot represents a single CpG site. Open circles indicate unmethylated (Unmethy) CpGs; filled circles indicate methylated CpGs. Each row represents a single sequenced clone (ten clones for each mouse are presented). f Quantification analysis of (e). g MSP analysis of kidney Klotho promoter methylation (Methy) from sham, UUO, and DHA-treated UUO mice (2 weeks, 3 randomly selected samples in each group). h, i Renal expression of Smad2 and Smad3 mRNA in different groups as assessed by real-time PCR. j Kidney expressions of Smad2, phosphorhospho-Smad2 Smad3, phosphorhospho-Smad3 from sham, UUO and DHA-treated UUO mice as assayed by Western blotting (three samples in each group). k–n Quantification analysis of (j). o Kidney expression of active β-catenin, β-catenin, DVL2, and DVL3 from sham, UUO and DHA-treated UUO mice as assayed by Western blot (three samples in each group). p–s Quantification analysis of (j). The results are the means ± SD of at least three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 (compared with the sham group or control group); #P < 0.05; ##P < 0.01 (compared with the UUO group or M group).
Bisulfite-sequencing PCR (BSP)
Purified cellular DNA was treated with bisulfite using the Methylation-Lightning Kit (59824, EpiTect Fast DNA Bisulfite Kit, QIAGEN) according to the manufacturer’s protocol. To amplify the Kl promoter fragments, touchdown PCR was performed using AceTaq DNA Polymerase (P401, AceTaq DNA Polymerase, Vazyme). This round of PCR consisted of the following cycling conditions: 95 °C for 10 min and 40 cycles of 30 s at 94 °C, 30 s at 55 °C, and 30 s at 70 °C. The final elongation was performed at 72 °C for 1 min. The sequences of the PCR primers are listed in Supplementary Table 1. The PCR products were purified using the Agarose gel DNA Recovery Kit (DP219, TianGen), cloned into the Pucm-T Vector (D2006, Beyotime) and transformed into DH5α competent cells (CB101, TianGen). The plasmid DNA was then purified with a DNA StarPrep Miniprep Kit (DP105, TianGen) and sequenced (Sangon Biotech).
Chromatin immunoprecipitation
ChIP assay was performed with renal tissues. The immunoprecipitation was performed with ChIP quality antibody to DNMT1. The starting (input) and immunoprecipitated DNAs were analyzed by PCR and quantitative real-time PCR (qRT-PCR) using primer set NbF: 5′-GAATGGGAGAAAGGGGAAA-3′ and NbR: 5′-CTTCCTGGGACTGGTGTTC-3′, which cover a putative DNMT1 binding motif. Regular PCR products were separated on 1.0% agarose gels and analysis of PCR product densitometry was performed with Image J Software. The qRT-PCR was performed by a StepOne Plus Real- Time PCR System (Applied Biosystems, USA).
DNMT1 mutant assay
DNMT1 plasmids containing the mutation (Thr1528 → Ala1528 and Asn1529 → Ala1529) and wild type (WT) were constructed by Geneppl technology (Nanjing, China). HK-2 cells were seeded onto 6-well plates and transfected with mutation plasmid and control plasmid for 48 h at 70% confluence using JetPRIMER Transfection reagent (22Y0302M2, Polyplus Transfection, France) in jetPRIME buffer (Polyplus Transfection) for 48 h. Then, the transfected cells were collected and the total lysates were prepared.
Statistical analysis
Multigroup comparisons were performed using one‑way ANOVA or Kruskal–Wallis test. Student’s t test or Mann–Whitney–Wilcoxon test was used for comparisons between two groups. A P value of <0.05 was considered significant. At least three independent experiments were performed. Analyses were performed with SPSS version 16.0 (IBM) for Windows (Microsoft Corp.) and Prism Software (GraphPad Software). Image fluorescence analysis was performed using HCImage Hamamatsu’s image acquisition and analytical software (Hamamatsu Corporation).
Results
The renoprotective effects of DHA
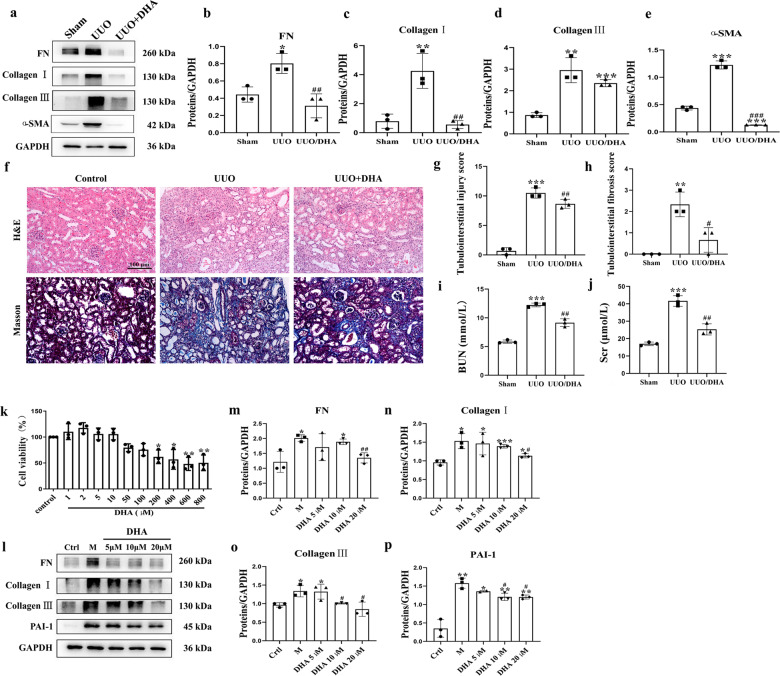
Mice that received DHA (30 mg · kg−1 p.o.) daily for 21 days starting on the 1st day before the UUO procedure exhibited marked reductions in the levels of various pro‑fibrotic markers, such as pro-fibrotic proteins collagen I, collagen III, fibronectin and alpha-smooth muscle actin (α-SMA) (Fig. 1a–e). Hematoxylin and eosin (H&E) and Masson staining exhibited attenuations in dilations, atrophy of the tubes and widening of the interstitial space with severe inflammatory cell infiltration by DHA in the UUO mice (Fig. 1f–h). Treatment with DHA significantly reduced the levels of BUN and Scr, suggesting an improved renal function in the UUO mice (Fig. 1i, j).
Fig. 1. DHA exhibited good renoprotective effects in vivo and in vitro (n = 8 for UUO mice).
a Expressions of fibronectin, collagen I, collagen III, and α-SMA in HK2 cells treated with TGF-β (10 ng/mL) in presence or absence of DHA for 48 h as assayed by Western blotting. b–e Quantification analysis of (a). f Representative photomicrographs showing the H&E staining and Masson’s trichrome staining of the left kidneys from sham, UUO, and DHA-treated UUO mice (H&E and Masson’s staining; scale bar, 100 μm). g, h Bar graph depicts renal injury scores based on H&E staining. The bar graph depicts renal interstitial fibrosis scores based on Masson’s trichrome staining. i, j Biochemical parameters, including blood urea nitrogen (BUN) and serum creatinine (Scr) of each mouse. k In vitro cell viability test of HK-2 cells treated with DHA for 24 h. l Kidney expression of fibronectin, collagen I, collagen III, and PAI-1 from sham, UUO and DHA-treated UUO mice as assayed by Western blotting (three samples in each group). m–p Quantification analyses of (l). The results are the means ± SD of at least three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 (compared with sham group or control group); #P < 0.05; ##P < 0.01; ###P < 0.001 (compared with the UUO group or M group).
We then examined the possible protective effects of DHA on fibrosis in the transformation of growth factor beta 1 (TGF-β1)-induced HK-2 cells. First, we assessed the cytotoxicity of DHA in normal HK-2 cells. DHA (5, 10, and 20 μM) did not influence the viability of normal HK-2 cells, while DHA (200 μM) significantly inhibited the viability of HK-2 cells (Fig. 1k). Therefore, DHA was used at 5, 10, and 20 μM in subsequent experiments. Treatment with DHA significantly inhibited pro-fibrotic mediators including collagen I, collagen III, fibronectin and plasminogen activator inhibitor (PAI-1) (Fig. 1l–p). Holistically, these findings indicated that DHA exerts strong anti-fibrotic effects in vitro and in vivo.
DNMT1 is targeted by DHA
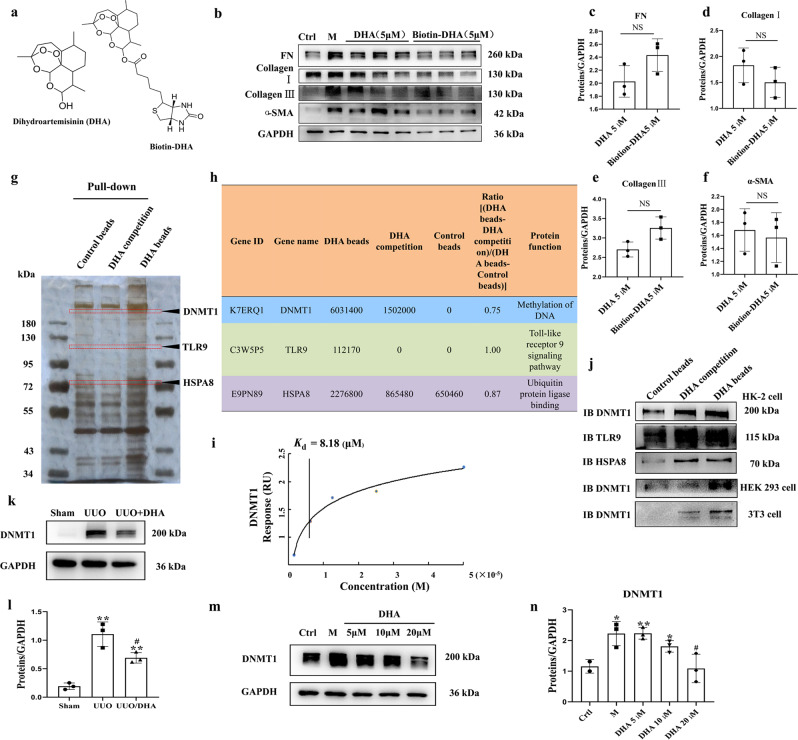
To examine the potential target of DHA, affinity purification was conducted using prepared probes (chemical) (Fig. 2a). A biotin-tagged DHA probe (biotin-DHA) was used to pull-down the cellular target of DHA. Biotin-DHA probe retained anti-fibrotic ability, indicating the lack of influence of chemical modification on the bioactivity of DHA (Fig. 2b–f). Using a pull-down assay together with shotgun proteomics, we tried to identify the target protein. As shown in Fig. 2g, several obvious protein bands could be observed in the samples of the group treated with DHA beads. However, a weaker protein band was observed in the group treated with excess DHA. We scraped these bands off and performed proteomics analysis. The significant decrease in the levels of proteins in the presence of an excess amount of DHA for competition are shown in Supplementary Table 2. We selected three proteins that might be associated with kidney diseases [25–27], namely, DNMT1, Toll-Like Receptor 9 (TLR9) and HSPA8 (Fig. 2h), and used Western blot analysis to verify the presence of these proteins with specific anti-DNMT1, anti-TLR9 and anti-HSPA8 antibodies, respectively (Fig. 2j). The results confirmed DNMT1 as the target of DHA and excluded the other proteins as alternative targets. Mammals generally have three active DNMT isoforms, DNMT1, DNMT3a, and DNMT3b, which have distinct functions. DNMT3a and DNMT3b were not found in the protein list obtained by the proteomics analysis. We therefore examined the interaction of DHA with DNMT3a and DNMT3b using Western blot analysis. The results indicated that DNMT3a and DNMT3b were not the target proteins of DHA (Supplementary Fig. 6b). In addition, we confirmed that DNMT1 was the target of DHA in HEK293 and 3T3 cell lysates (Fig. 2j). To verify that DHA interacted with DNMT1, surface plasmon resonance (SPR) analysis was performed. The target affinity [Kd (equilibrium dissociation constant) value] of DHA for DNMT1 was 8.18 μM (Fig. 2i).
Fig. 2. DNMT1 was identified as the target of DHA.
a The chemical structures of DHA and the biotin-DHA analogs. b Expression of fibronectin, collagen I, collagen III and α-SMA in HK2 cells treated with TGF-β (10 ng/mL) in the presence or absence of DHA and Biotin-DHA for 48 h as assayed by Western blotting. c–f Quantification analysis of (b). g DHA target proteins were identified using pull-down technology coupled with shotgun proteomics. The HK-2 lysates were incubated with DHA beads or control beads, and then the proteins bound to the beads were resolved by SDS/PAGE, followed by silver staining. h The results of silver staining as analyzed by proteomics. i SPR analysis of DHA binding to DNMT1. j HK-2, HEK293, and 3T3 cell lysates were incubated with DHA beads or control beads, and the proteins bound to the beads were resolved by SDS/PAGE, followed by Western blotting. k Protein expression of DNMT1 from sham, UUO and DHA-treated UUO mice as assayed by Western blotting (three samples in each group). l Quantification analysis of (k). m Protein expressions of DNMT1 in HK2 cells treated with TGF-β (10 ng/mL) in presence or absence of DHA for 48 h as assayed by Western blotting. n Quantification analysis of (m). The results are the means ± SD of at least three independent experiments. *P < 0.05; **P < 0.01 (compared with sham group or control group); #P < 0.05 (compared with the UUO group or M group).
In the kidneys of UUO mice, the expression of DNMT1 was markedly increased (Fig. 2k, l). However, the increase in DNMT1 expression was markedly inhibited in DHA-treated UUO mice. We next measured the level of DNMT1 by Western blot analysis in TGF-β1-induced HK-2 cells treated with or without DHA. The results showed that the expression of DNMT1 was significantly increased in TGF-β1-induced HK-2 cells (Fig. 2m, n), but decreased under the treatment with DHA.
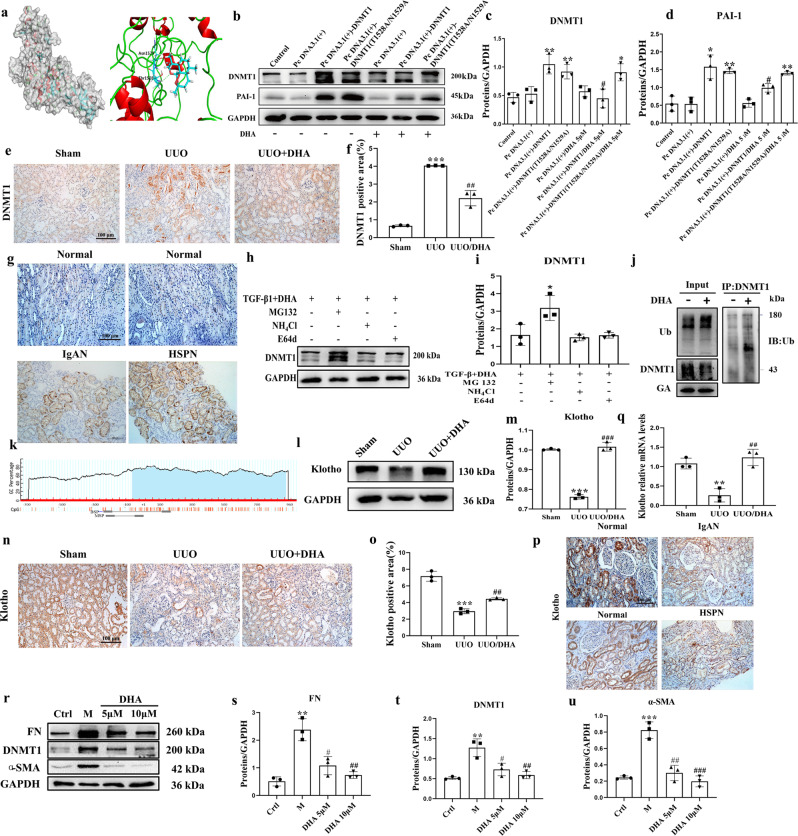
Asparagine 1529 and threonine 1528 are key sites for the anti-fibrotic effect of DHA
To explore the binding between DHA and DNMT1, in silico molecular docking and molecular dynamics (MD) simulations were performed. The binding modes of DHA to DNMT1 were determined through the MD simulation. As shown in Fig. 3a, DHA formed two hydrogen bonds with DNMT1 by interacting with Asn1529 and Thr1528. To evaluate whether Asn1529 and Thr1528 are key sites for the anti-fibrotic effect of DHA, we examine the DNMT1 level using Western blot analysis in HK-2 cells transfected with a DNMT1 WT plasmid or a mutant DNMT1 (T1528A/N1529A) plasmid. The results indicated that the cells transfected with the mutant plasmid diminished the effect of DHA on DNMT1 and its anti-fibrosis effects (PAI-1) (Fig. 3b, c, d), providing direct evidence that Asn1529 and Thr1528 are key sites for the binding of DHA to DNMT1.
Fig. 3. DHA reduces DNMT1 and enhances Klotho levels.
a Molecular dynamics model of DHA binding to DNMT1. b After transfection of the DNMT1 overexpression plasmid into HK-2 cells for 48 h, the DNMT1 and PAI-1 proteins were detected by Western blotting. c, d Quantification analysis of (b). e, f Immunohistochemical micrographs of DNMT1 in the kidney tissues of mice from each group (scale bar, 100 μm). g Immunohistochemical micrographs of DNMT1 in human kidney tissues. h Representative Western blot analyses of DNMT1 expression are shown in primary mouse renal tubular cells (PRTCs) that were treated with TGF-β (10 ng/mL) and DHA (10 μM) in combination with MG132 (0.5 µM), E64d (10 µg/mL), or ammonium chloride (NH4Cl; 1 mM) for 24 h. i Quantification analysis of 3 g. j DNMT1 and ubiquitin were analyzed by immunoprecipitation with or without DHA. k Schematic diagram of the mouse Klotho promoter. The CpG island is shown in blue. The relative locations of methylation-specific polymerase chain reaction (MSP) and bisulfite-sequencing polymerase chain reaction (BSP) primers are indicated. l, m Protein expressions of Klotho from sham, UUO, and DHA-treated UUO mice as assayed by Western blotting (three samples in each group). n, o Immunohistochemical photomicrographs of Klotho in kidney tissues of mice from each group. p Immunohistochemical photomicrographs of Klotho in human kidney tissues. q Renal expression of Klotho mRNA in different groups was assessed by real-time PCR. r Expression of fibronectin, DNMT1, and α-SMA in primary mouse renal tubular cells (PRTCs) treated with TGF-β (10 ng/mL) in the presence or absence of DHA for 48 h are assayed by Western blotting. s–u Quantification analysis of 3q. The results are the means ± SD of at least three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 (compared with sham group or control group); #P < 0.05; ##P < 0.01; ###P < 0.001 (compared with the UUO group or M group).
DHA reduces DNMT1 levels via the ubiquitin–proteasome pathway
DHA significantly reduced the increased expression of DNMT1. Immunohistochemical staining showed an increase in DNMT1 in the UUO group, while DHA reduced the level of DNMT1 (Fig. 3e, f). Consistent with the results in the UUO mice, immunohistochemical staining demonstrated that the level of DNMT1 in the patients with IgA nephropathy and Henoch Schonlein purpura nephritis (HSPN) was also markedly increased (Fig. 3g).
Since there was a direct interaction between DHA and DNMT1 and the DNMT1 levels were markedly decreased under DHA treatment, we surmised that there DNMT1 was post-transcriptionally regulated following DHA treatment. To assess if this was the underlying mechanism, we treated primary mouse renal tubular cells (PRTCs) challenged with TGF-β1 + DHA in combination with the proteasome inhibitor MG132 or the lysosome inhibitor NH4Cl or E64d. MG132, but not NH4Cl or E64d, blocked the decrease in DNMT1 protein expression in response to TGF-β1 + DHA treatment (Fig. 3h, i). These results indicated that DMNT1 was degraded via the ubiquitin–proteasome pathway. To assess the effect of DHA on DNMT1 ubiquitination and protein stability, lysates from TGF-β1-induced PRTCs treated with or without DHA were immunoprecipitated with DNMT1 and immunoblotted for ubiquitin. DHA-treated cells exhibited more DNMT1 ubiquitination than control cells (Fig. 3j), suggesting that DHA promoted DNMT1 ubiquitination.
DHA reverses Klotho levels and Klotho promoter hypermethylation in the kidneys of UUO mice
Human and mouse Klotho promoters contain typical CpG islands (Fig. 3k). To investigate the possible changes in DNA methylation in Klotho alterations, we first measured Klotho levels in UUO mice. The kidneys of UUO mice exhibited a reduction in the Klotho level, and treatment with DHA almost completely reversed this reduction (Fig. 3l, m). Immunohistochemical staining also indicated the decreased level of Klotho in the UUO group, while treatment with DHA increased the level of Klotho (Fig. 3n, o). Consistent with the results in UUO mice, immunohistochemical staining showed that the Klotho levels of the IgA nephropathic and HSPN patients were also markedly decreased (Fig. 3p). We also measured Klotho mRNA levels using quantitative polymerase chain reaction (qPCR). The results indicated that kidneys of UUO mice showed downregulated Klotho mRNA expression while DHA almost toally reversed this effect (Fig. 3q). We then evaluated the protective effects of DHA on fibrosis in TGF-β1-induced primary mouse renal tubular cells (PRTCs). Treatment with DHA (10 μM) inhibited the increase in the pro-fibrotic mediators, including fibronectin, DNMT1 and alpha-smooth muscle actin (α-SMA) (Fig. 3r–u). We also measured Klotho levels in the TGF-β1-induced PRTCs. The Klotho expression was markedly decreased in TGF-β1-induced PRTCs, and treatment with DHA almost completely reversed this reduction (Fig. 4a, b). Taken together, our findings indicated that DHA can restore reduced Klotho level in vitro and in vivo.
Next, we assessed the methylation-specific polymerase chain reaction (PCR) in a CpG-rich area (−352/−176). The results indicated that the kidneys of UUO mice displayed increased methylation, whereas treatment with DHA significantly decreased the methylation level (Fig. 4g). Additionally, we performed more methylation analysis by bisulfite-specific PCR (Fig. 4e, f). We also performed chromatin immunoprecipitation analysis to assess if DNMT1 binds to the central Klotho promoter region. The results revealed clearly that DNMT1 was bound to the −338 to −223 promoter region of Klotho. Chromatin immunoprecipitation by the DNMT1 antibodies in the UUO mice was greater than that in DHA-treated mice (Fig. 4c, d). The UUO mice demonstrated a marked methylation increase and treatment with DHA significantly reduced the increase in methylation (Fig. 4e, f), suggesting that the ability of DHA to reverse Klotho loss was probably the result of its demethylation effects.
DHA attenuates renal fibrosis by inhibiting the TGF-β/Smad and Wnt/β-catenin signaling pathways
It has been reported that low renal Klotho expression enhances TGF-β1 activity and causes renal fibrosis [28]. In addition, Klotho is a known antagonist of endogenous Wnt/β-catenin signaling [29]. Thus, we evaluated the effect of DHA on the TGF-β/Smad and Wnt/β-catenin signaling pathways in UUO mice treated with DHA. UUO led to marked increases in the expression of Dvl2, Dvl3, β-catenin and active β-catenin, an indication of Wnt/β-catenin signaling activation (Fig. 4o–s). DHA significantly attenuated the UUO-induced increase in expression of Dvl2, Dvl3, β-catenin and active β-catenin. Furthermore, UUO significantly increased expression of Smad2, Smad3, P-Smad2 and P-Smad3, an indication of TGF-β/Smad signaling activation (Fig. 4j–n). DHA significantly reduced expression of these proteins. In addition, the mRNA levels of Smad2 and Smad3 exhibited similar patterns as their protein levels (Fig. 4h, i). These results indicated that DHA can inhibit the activations of these signaling pathways in vivo.
DHA reverses established renal fibrosis in an adenine model
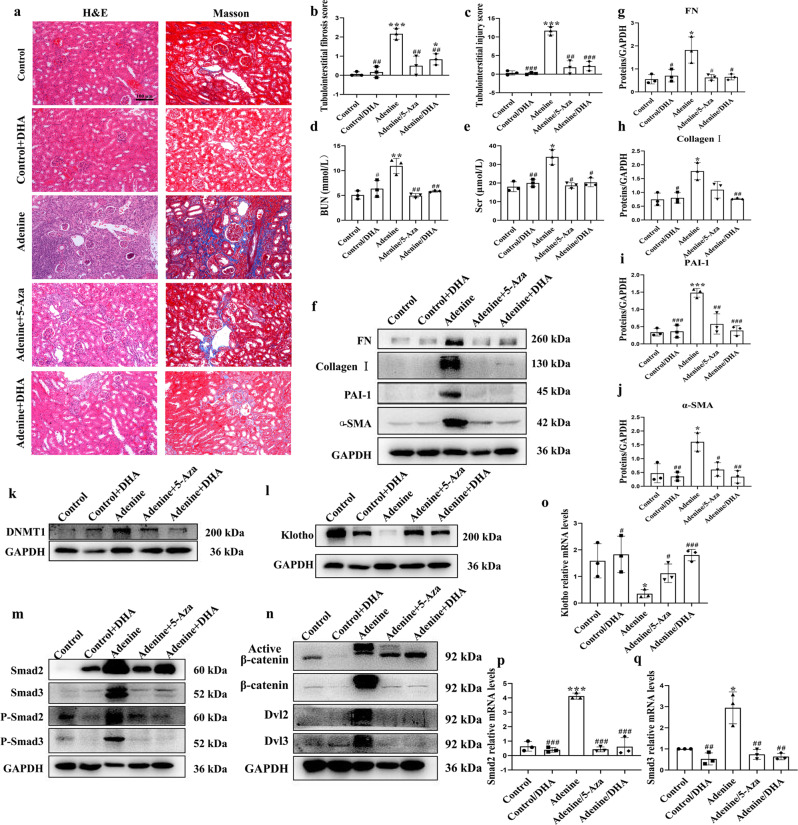
Mice that received DHA (30 mg· kg−1 p.o.) every day for 21 days starting on Day 21 post‑adenine (80 mg · kg−1, p.o.)-induced renal injury (Supplementary Fig. 6a) showed marked decreases in the levels of pro‑fibrotic markers, such as the pro-fibrotic proteins collagen I, PAI-1, fibronectin and α-SMA (Fig. 5f–j). H&E and Masson staining showed that the dilations, atrophy of the tubes, and widening of the interstitial space by inflammatory cells infiltration were attenuated by DHA in the adenine-treated mice (Fig. 5a–c). Treatment with DHA markedly prevented the increased BUN and serum creatinine levels, suggesting that DHA improved renal function in adenine-induced CKD mice (Fig. 5d, e). These results together indicated that DHA exerted strong anti-fibrotic effects and that the anti-fibrotic effects of DHA were stronger than those of 5-aza in adenine-treated mice.
Fig. 5. DHA reversed adenine-induced renal fibrosis in mice (n = 10).
a Representative photomicrographs of the H&E staining and Masson’s trichrome staining of left kidneys from control, DHA, adenine, and 5-Aza or DHA-treated adenine mice (H&E and Masson’s staining; scale bar, 100 μm). b, c Bar graph depicts renal injury scores based on H&E staining. The bar graph depicts renal interstitial fibrosis scores based on Masson’s trichrome staining. d, e Biochemical parameters, including blood urea nitrogen (BUN) and serum creatinine (Scr) of each mouse. f–n Kidney expression of fibronectin, collagen I, PAI-1, α-SMA, DNMT1, Klotho, Smad2, Smad3, phospho-Smad2, phospho-Smad3, active β-catenin, β-catenin, DVL2 and DVL3 from control, DHA, adenine-, and 5-Aza- or DHA-treated adenine mice as assayed by Western blotting (three samples in each group). o–q Renal expression of Klotho, Smad2 and Smad3 mRNA in different groups were assessed by real-time PCR. The results are the means ± SD of at least three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 (compared with the control group); #P < 0.05; ##P < 0.01; ###P < 0.001 (compared with the adenine group).
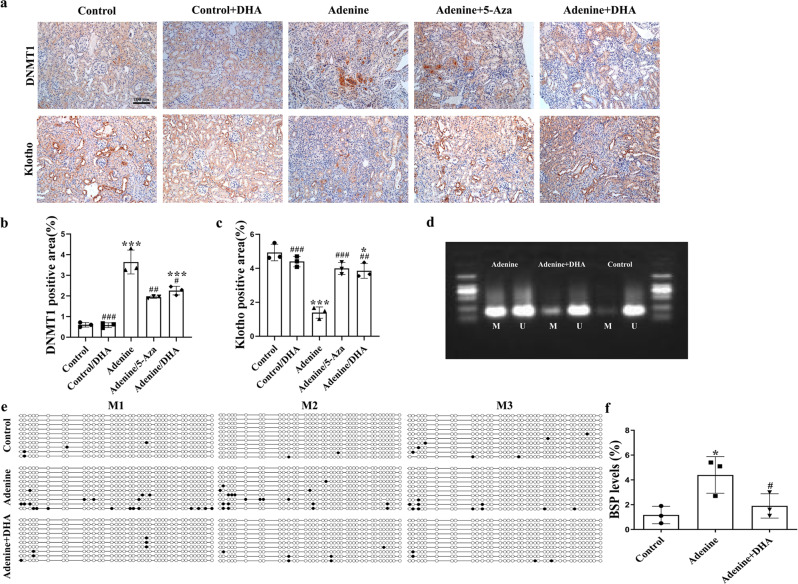
In the adenine model, the DNMT1 level significantly increased and the Klotho level significantly decreased, whereas DHA treatment significantly decreased the DNMT1 level and increased the Klotho level (Fig. 5k, l). The results of the immunohistochemical staining were similar to the aforementioned data (Fig. 6a–c). In addition, the kidneys of mice from this model exhibited reduced Klotho mRNA expression, and DHA treatment led to an almost complete restoration to normalcy (Fig. 5o). Moreover, DHA inhibited the TGF-β/Smad and Wnt/β-catenin signaling pathways in the adenine-treated mice (Fig. 5m, n). Additionally, the mRNA levels of Smad2 and Smad3 exhibited similar patterns as their protein levels (Fig. 5p, q). The results of the methylation studies (Fig. 6d–f) indicated an increased level of same in the adenine-treated mice. However, these observations were reversed after treatment with DHA.
Fig. 6. DHA reverses Klotho promoter hypermethylation in adenine-induced mice.
a Immunohistochemical photomicrographs of DNMT1 and Klotho in kidney tissues of mice in each group (scale bar, 100 μm). b, c Quantification analysis of (a). d MSP analysis of kidney Klotho promoter methylation (Methy) from control, adenine-, and DHA-treated adenine mice (6 weeks, three randomly selected samples in each group). e BSP analysis of kidneys from control, adenine- and DHA-treated adenine mice kidneys (6 weeks). Each box represents the indicated mouse kidney (M); each row of circles in the boxes represents the Klotho CpG island; each dot represents a single CpG site. Open circles indicate unmethylated (Unmethy) CpGs; filled circles indicate methylated CpGs. Each row represents a single sequenced clone (ten clones for each mouse are presented). f Quantification analysis of (e). The results are the means ± SD of at least three independent experiments. *P < 0.05; ***P < 0.001 (compared with the sham group); #P < 0.05; ##P < 0.01; ###P < 0.001 (compared with the adenine group).
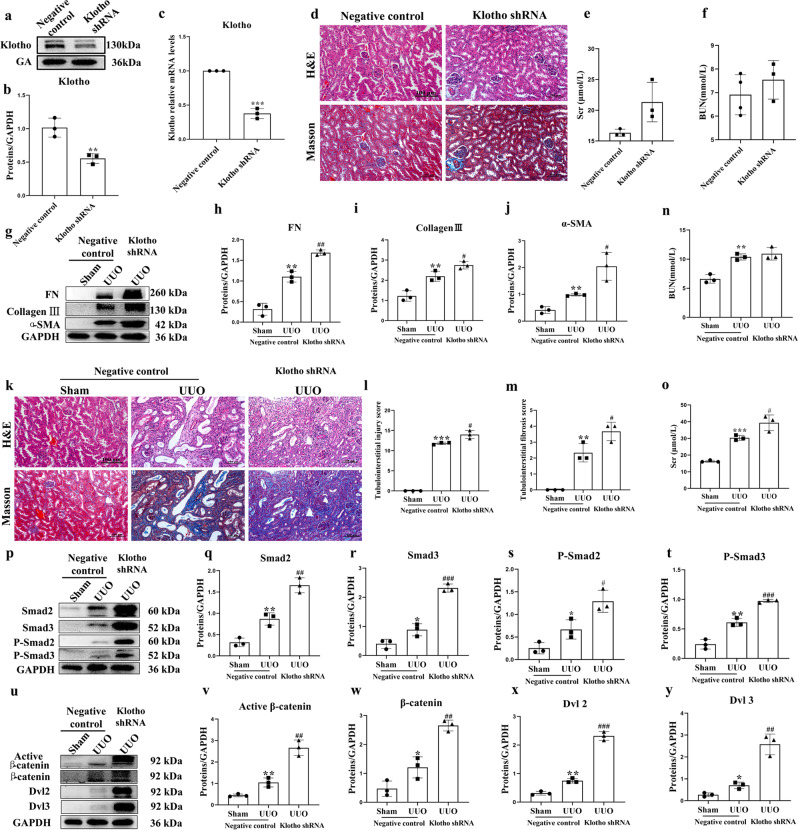
Knockdown of Klotho accelerates UUO-induced renal fibrosis
To determine the physiological function of Klotho and its role in renal fibrosis, we knocked down Klotho expression in mouse kidneys by renal pelvis injection of adeno-associated virus (AAV) carrying shRNA against Klotho. Klotho expression in knockdown mice was visibly reduced relative to control mice (Fig. 7a, b). Likewise, the mRNA level of Klotho was significantly reduced (Fig. 7c). These results confirmed the knockdown of Klotho in mice. Knockdown of Klotho had nearly no impacts on renal function, histopathology and pro-fibrotic proteins in the mice (Fig. 7d–f). UUO resulted in significantly increased pro‑fibrotic markers, including pro-fibrotic proteins collagen III, fibronectin and α-SMA (Fig. 7g–j), while knockdown of Klotho further enhanced the expression of these pro‑fibrotic proteins. H&E and Masson staining showed that knockdown of Klotho further aggravated dilation and atrophy of the tubes and widening of the interstitial space by inflammatory cells infiltration in UUO mice (Fig. 7k–m). Knockdown of Klotho further increased BUN and Scr levels (Fig. 7n, o). Moreover, UUO induced the TGF-β/Smad and Wnt/β-catenin signaling pathways. Knockdown of Klotho strongly induced the activation of these pathways (Fig. 7p–y). These indicated that expression of Klotho in the kidney negatively correlates with renal fibrosis, and that knockdown of Klotho expression results in the acceleration of renal fibrosis.
Fig. 7. Knockdown of Klotho significantly aggravated renal fibrosis in UUO mice (n = 6–8).
a, b Kidney expression of Klotho from mice injected with AAV9 carrying shRNA against Klotho or control vector as assayed by Western blotting (three samples in each group). c Renal expression of Klotho mRNA in different groups as assessed by real-time PCR. d Representative photomicrographs of the H&E staining and Masson’s trichrome staining from the left kidneys of negative control and Klotho shRNA mice (H&E and Masson’s staining; scale bar, 100 μm). e, f Biochemical parameters, including blood urea nitrogen (BUN) and serum creatinine (Scr), of each mouse. g Kidney expression of fibronectin, collagen III and α-SMA from sham, UUO and Klotho shRNA-treated UUO mice as assayed by Western blotting (three samples in each group). h–j Quantification analysis of (g). k Representative photomicrographs of the H&E staining and Masson’s trichrome staining of the left kidneys from Sham, UUO and Klotho shRNA-treated UUO mice (H&E and Masson’s staining; scale bar, 100 μm). l, m Quantification analysis of (k). n, o Biochemical parameters, including blood urea nitrogen (BUN) and serum creatinine (Scr), of each mouse. p–y Kidney expression levels of Smad2, Smad3, phospho-Smad2, phospho-Smad3, active β-catenin, β-catenin, DVL2 and DVL3 from sham, UUO and Klotho shRNA-treated UUO mice were assayed by Western blotting (three samples in each group). Results are means ± SD of at least three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 (compared with the sham group); #P < 0.05; ##P < 0.01; ###P < 0.001 (compared with the UUO group).
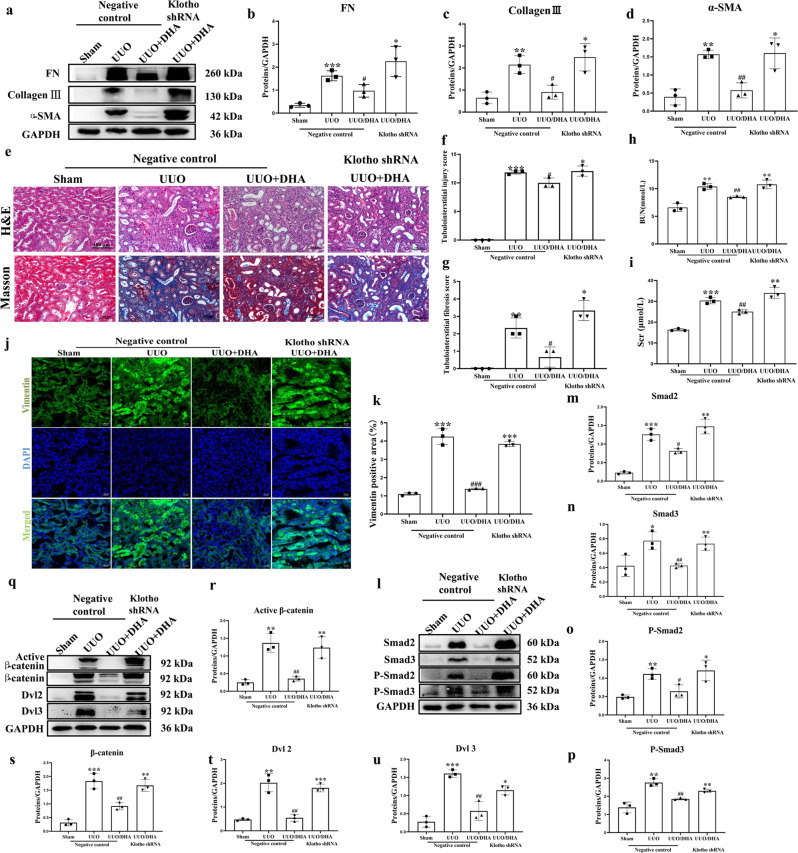
Knockdown of Klotho abolishes the renal protective effect of DHA in UUO mice
DHA markedly decreased pro-fibrotic proteins in UUO mice. However, knockdown of Klotho in UUO mice resulted in increased expressions of pro‑fibrotic markers, including pro-fibrotic proteins collagen III, fibronectin and α-SMA (Fig. 8a–d) and thus eliminated the protection conferred by DHA. Likewise, DHA attenuated the tubulointerstitial injury, but was reversed by knockdown of Klotho (Fig. 8e–g). Treatment with DHA significantly reduced the increased BUN and Scr levels. However, this effect was reversed by Klotho knockdown (Fig. 8h, i). The results of the immunofluorescence studies exhibited similar patterns (Fig. 8j, k). In addition, DHA inhibited the activation of the TGF-β/Smad and Wnt/β-catenin signaling pathways, while knockdown of Klotho abolished the inhibitory effect of DHA on these pathways (Fig. 8l–u).
Fig. 8. Klotho is essential for the antirenal fibrotic effect of DHA in UUO mice (n = 6–8).
a The fibrosis-related proteins (fibronectin, collagen III and α-SMA) were measured in different groups of mice by Western blotting (three samples in each group). b–d Quantification analysis of (a). e–g Representative H&E staining and Masson’s trichrome staining of kidneys from different groups of mice (H&E and Masson’s staining; scale bar, 100 μm). h, i Biochemical parameters, including blood urea nitrogen (BUN) and serum creatinine (Scr) of each mouse. j Vimentin staining for the detection of actin filaments in different groups of mice (Scale bar represents 20 µm). k Quantification analysis of (j). l–u Kidney expressions of Smad2, Smad3, phospho-Smad2, phospho-Smad3, active β-catenin, β-catenin, DVL2 and DVL3 from different groups of mice as assayed by Western blotting (three samples in each group). The results are the means ± SD of at least three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 (compared with the sham group); #P < 0.05; ##P < 0.01; ###P < 0.001 (compared with the UUO group).
Discussion
It is estimated that almost 700 million people are diagnosed with CKD worldwide [30]. Treatment of CKD remains challenging due to the unavailability of approved efficacious drugs. In spite of the progress made in respect of our understanding of the pathogenesis of CKD pathogenesis and the attention devoted to developing new classes of drugs, the known classes of medication that slows down kidney function decline remain the angiotensin-converting–enzyme (ACE) inhibitors and angiotensin-receptor blockers (ARBs). However, much of these data have been obtained from type 2 diabetes patients [31]. DHA, a derivative of artemisinin, is a safe and efficacious antimalarial drug. In recent times, DHA has gained much traction for its relatively diverse pharmacological activities [32–34]. Herein, we give scientific proof for the role of DHA as a novel and effective CKD suppressor that aids in resolving kidney dysfunction and renal fibrosis in a coordinated manner. To our knowledge, this is the first study to identify the molecular targets that underlie the renoprotective effect of DHA. We provide insights into the mechanisms of the anti-fibrotic effect of DHA, which include DHA-induced DNMT1 degradation, which leads to the deactivation of the TGF-β/Smad and Wnt/β-catenin signaling pathways via reduction in Klotho hypermethylation (Fig. 9).
Fig. 9.
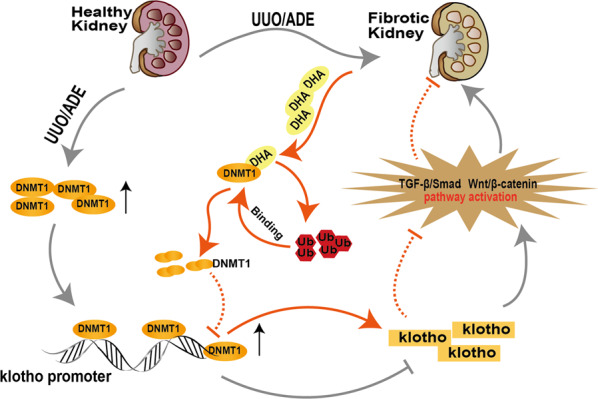
Schematic illustration of the antirenal fibrotic effect of DHA.
Druggable target identification plays a prominent role in the identification of therapeutic drugs [19, 35, 36]. To determine the direct target for DHA in cells, a small-molecule probe based on the structure of DHA identified DNMT1 as the key antirenal fibrotic target in three different types of renal cells, including HK-2, HEK-293 and 3T3 cells. Molecular dynamics (MD) simulations indicated that the DHA formed two hydrogen bonds with DNMT1 by interacting with Asn1529 and Thr1528, suggesting these might be the key sites for the binding of DHA to DNMT1. We then confirmed the inhibitory effect of DHA on DNMT1 was diminished in cells transfected with DNMT1/Asn1529A/Thr1528A mutant, providing direct evidence that Asn1529 and Thr1528 are key sites for the anti-fibrotic effect of DHA.
DNA methylation is a crucial epigenetic mechanism comprised of the covalent addition of a methyl group to the 5-carbon position of cytosine in the CpG dinucleotide sequences by DNA methyltransferases (DNMTs) [37]. The DNMT family includes DNMT1, DNMT3a, and DNMT3b [38]. DNMT1, which is the most abundant form, is responsible for maintenance of DNA methylation, while DNMT3a and DNMT3b are responsible for de novo methylation [39, 40]. Notably, it is known that epigenetic modifications, especially DNA methylation, are critical contributors to both acute kidney disease and CKD progression [41–43]. Zhang et al. [43] reported that 5-Aza, a DNA methylation inhibitor, mitigates the urinary excretion of albumin, glomerular mesangial expansion, and podocyte injury and improves kidney function [40]. In this work, we found marked increase in DNMT1 in patients with IgA nephropathy. Our results also demonstrated that DHA treatment effectively alleviated the disruption of serum biochemistry and attenuated renal fibrosis in vitro and in vivo. Moreover, we showed that DHA directly targeted the Asn1529 and Thr1528 residues in DNMT1 to induce the degradation of DMNT1 via the ubiquitin–proteasome pathway and that the decrease in DNMT1 levels induced by DHA relied on neither a decrease in DNMT1 gene transcription nor protein synthesis. The reduction in DNMT1 expression led to the decrease in promoter hypermethylation of CpG-rich genes, and resulted in the increased translation of these genes. Thus, we speculate that DHA restored the loss of certain key proteins and conferred protection on the kidney.
Klotho is an anti-aging protein that is predominantly expressed in the kidney [44]. It is a protein with a single transmembrane domain. Its extracellular domain consists of two tandem domains (KL1 and KL2), which can be cleaved by ADAM proteases and released [45, 46]. Therefore, Klotho exists in two forms (transmembrane and secretory forms) in vivo. Secreted Klotho can act as an endocrine factor to exert effects against renal fibrosis [47]. When Klotho expression is inhibited, renal fibrosis is aggravated [48]. The Klotho promoter contains abundant CpG islands, and its expression in renal cells and silencing in other tissues are mainly influenced by epigenetic modifications [49]. Atypical methylation of Klotho is linked to aging-related diseases and different types of cancer [50–52]. Current studies have suggested an association of the hypermethylation of the Klotho promoter with CKD pathogenesis [53, 54]. In this study, the protein expression level of Klotho was notably downregulated in the UUO model and adenine-induced model, and its mRNA expression was also significantly downregulated, which indicated that the transcription of Klotho was inhibited. Thus, the decreased Klotho protein expression might result from Klotho promoter hypermethylation. Therefore, we predicted the CpG island in the Klotho promoter and assessed the methylation of different groups in the two models. Our findings showed an increased methylation level of Klotho in the model, but this was decreased after DHA administration. On the basis of these findings and the results of chromatin immunoprecipitation, it can be concluded that DNMT1 binds to the Klotho promoter region and that DHA can affect Klotho expression through DNMT1. In addition, secreted Klotho antagonizes Wnt/β-Catenin signaling by physical interaction and functional isolation through its kl1 domain. Klotho directly binds to type II TGF-β receptor and interferes with the binding of TGF-β to same, leading to inhibition of TGF-β signaling [29, 55]. Therefore, we measured the amounts of proteins linked to the Wnt/β-catenin and TGF-β/Smad signaling pathways by Western blot analysis and qPCR and found that Klotho inhibited the activation of the two signaling pathways. In addition, knockdown of Klotho in UUO mice abolished the protective effect mediated by DHA, suggesting that Klotho was the key protein for the antirenal fibrotic effect of DHA. These data together indicate that DHA can reverse the loss of Klotho by targeting DNMT1 and downregulate its expression in UUO and adenine-treated mice. Upregulation of Klotho expression conferred protection on the kidneys via inhibition of the Wnt/β-catenin and TGF-β/Smad signaling pathways.
In conclusion, the results of our study revealed the Asn1529 and Thr1528 residues as the key regulatory sites for DHA-mediated DNMT1 degradation in the setting of renal fibrosis. Downregulation of DNMT1 expression effectively reversed Klotho promoter hypermethylation, consequently restoring Klotho protein and ameliorating renal fibrosis. Thus, this study provides proof-of-concept that, regulation of the DNMT1-Klotho signaling axis by DHA or other pharmacological agents that deactivate fibrotic pathways may be crucial for the treatment of renal fibrosis.
Supplementary information
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (Grants 81703691, 81803694), and the Fundamental Research Funds for the Central Universities (Grant No. 2632020ZD07). We are grateful to Dr. Raphael N. Alolga for his aid in the English polish. We are also grateful to Dr. Jin-feng Liu for his aid in molecular dynamics simulations.
Author contributions
Conceptualization: WZ, ZHZ; Investigation and data curation: MMC, HLL, ZLS, WHW, HJ, and LXW; Resources: XFA, CC, and KS; Funding acquisition: ZHZ, WZ, and NHT; Supervision: ZHZ, NHT, and WZ; Writing-original draft: WZ, ZHZ, and NHT; Writing-review and editing: WZ, NDV, ZHZ, and NHT.
Data availability
The additional data collected during this study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Ethics approval
The studies involving human participants were reviewed and approved by the medical ethics committee of the Affiliated Hospital of Nanjing University of Chinese Medicine and followed the tenets of the Declaration of Helsinki (2019NL-109-02). The studies involving animal experiments were reviewed and approved by the institutional ethics committee for animal experimentation of China Pharmaceutical University and the application approval number was 202002001.
Informed consent
Written consents from participants were also acquired.
Footnotes
These authors contributed equally: Wei Zhou, Min-min Chen, Hui-ling Liu
Contributor Information
Ning-hua Tan, Email: nhtan@cpu.edu.cn.
Zhi-hao Zhang, Email: zzh-198518@163.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41401-022-00898-3.
References
- 1.Moosa MR, Norris KC. Sustainable social development: tackling poverty to achieve kidney health equity. Nat Rev Nephrol. 2021;17:3–4. doi: 10.1038/s41581-020-00342-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ortiz A, Fernandez-Fernandez B. Humble kidneys predict mighty heart troubles. Lancet Diabetes Endocrinol. 2015;3:489–91. doi: 10.1016/S2213-8587(15)00092-3. [DOI] [PubMed] [Google Scholar]
- 3.Humphreys BD. Mechanisms of renal fibrosis. Annu Rev Physiol. 2018;80:309–26. doi: 10.1146/annurev-physiol-022516-034227. [DOI] [PubMed] [Google Scholar]
- 4.Humphreys BD. Mechanisms of renal fibrosis. Annu Rev Physiol. 2017;7:10–7. doi: 10.1146/annurev-physiol-022516-034227. [DOI] [PubMed] [Google Scholar]
- 5.Turner JM, Bauer C, Abramowitz MK, Melamed ML, Hostetter TH. Treatment of chronic kidney disease. Kidney Int. 2012;81:351–62. doi: 10.1038/ki.2011.380. [DOI] [PubMed] [Google Scholar]
- 6.Klayman LD. Qinghaosu (artemisinin): an antimalarial drug from China. Pediatr Infect Dis J. 1985;4:709. doi: 10.1097/00006454-198511000-00046. [DOI] [PubMed] [Google Scholar]
- 7.Kong LY, Tan RX. Artemisinin, a miracle of traditional Chinese medicine. Nat Prod Rep. 2015;32:1617–21. doi: 10.1039/C5NP00133A. [DOI] [PubMed] [Google Scholar]
- 8.Tu YY. The development of new antimalarial drugs: qinghaosu and dihydro-qinghaosu. Chin Med J. 1999;112:976–7. [PubMed] [Google Scholar]
- 9.Tu YY. The discovery of artemisinin (qinghaosu) and gifts from Chinese medicine. Nat Med. 2011;17:1217–20. doi: 10.1038/nm.2471. [DOI] [PubMed] [Google Scholar]
- 10.Smithuis F, Kyaw MK, Phe O, Aye KZ, Htet L, Barends M, et al. Efficacy and effectiveness of dihydroartemisinin-piperaquine versus artesunate-mefloquine in falciparum malaria: an open-label randomised comparison. Lancet. 2006;367:2075–85. doi: 10.1016/S0140-6736(06)68931-9. [DOI] [PubMed] [Google Scholar]
- 11.Wang JX, Tang W, Zhou R, Wan J, Shi LP, Zhang Y, et al. The new water-soluble artemisinin derivative SM905 ameliorates collagen-induced arthritis by suppression of inflammatory and Th17 responses. Br J Pharmacol. 2008;153:1303–10. doi: 10.1038/bjp.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin R, Zhang Z, Chen L, Zhou Y, Zou P, Feng C, et al. Dihydroartemisinin (DHA) induces ferroptosis and causes cell cycle arrest in head and neck carcinoma cells. Cancer Lett. 2016;381:165–75. doi: 10.1016/j.canlet.2016.07.033. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Z, Yao Z, Zhao S, Shao J, Chen A, Zhang F, et al. Interaction between autophagy and senescence is required for dihydroartemisinin to alleviate liver fibrosis. Cell Death Dis. 2017;8:e2886. doi: 10.1038/cddis.2017.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang DX, Qiu J, Zhou HH, Yu Y, Zhou DL, Xu Y, et al. Dihydroartemisinin alleviates oxidative stress in bleomycin-induced pulmonary fibrosis. Life Sci. 2018;205:176–83. doi: 10.1016/j.lfs.2018.05.022. [DOI] [PubMed] [Google Scholar]
- 15.Ziegler S, Pries V, Hedberg C, Waldmann H. Target identification for small bioactive molecules: finding the needle in the haystack. Angew Chem Int Ed Engl. 2013;52:2744–92. doi: 10.1002/anie.201208749. [DOI] [PubMed] [Google Scholar]
- 16.Schenone M, Dančík V, Wagner BK, Clemons PA. Target identification and mechanism of action in chemical biology and drug discovery. Nat Chem Biol. 2013;9:232–40. doi: 10.1038/nchembio.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, Hobson S, et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat Biotechnol. 2007;25:1035–44. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- 18.Parker CG, Galmozzi A, Wang Y, Correia BE, Sasaki K, Joslyn CM, et al. Ligand and target discovery by fragment-based screening in human cells. Cell. 2017;168:527–41.e29. doi: 10.1016/j.cell.2016.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liao LX, Song XM, Wang LC, Lv HN, Chen JF, Liu D, et al. Highly selective inhibition of IMPDH2 provides the basis of antineuroinflammation therapy. Proc Natl Acad Sci USA. 2017;114:E5986–94. doi: 10.1073/pnas.1706778114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen C, Zhu T, Liu X, Zhu D, Zhang Y, Wu S, et al. Identification of a novel PHGDH covalent inhibitor by chemical proteomics and phenotypic profiling. Acta Pharm Sin B. 2022;12:246–61. [DOI] [PMC free article] [PubMed]
- 21.Zhang ZH, Vaziri ND, Wei F, Cheng XL, Bai X, Zhao YY. An integrated lipidomics and metabolomics reveal nephroprotective effect and biochemical mechanism of Rheum officinale in chronic renal failure. Sci Rep. 2016;6:22151. doi: 10.1038/srep22151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang ZH, He JQ, Zhao YY, Chen HC, Tan NH. Asiatic acid prevents renal fibrosis in UUO rats via promoting the production of 15d-PGJ2, an endogenous ligand of PPAR-γ. Acta Pharmacol Sin. 2020;41:373–82. doi: 10.1038/s41401-019-0319-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19:1639–62. doi: 10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B. [DOI] [Google Scholar]
- 24.Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, et al. The AMBER biomolecular simulation programs. J Comput Chem. 2005;26:1668–88. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L, Zhang Q, Liu S, Chen Y, Li R, Lin T, et al. DNA methyltransferase 1 may be a therapy target for attenuating diabetic nephropathy and podocyte injury. Kidney Int. 2017;92:140–53. doi: 10.1016/j.kint.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki H, Suzuki Y, Narita I, Aizawa M, Kihara M, Yamanaka T, et al. Toll-like receptor 9 affects severity of IgA nephropathy. J Am Soc Nephrol. 2008;19:2384–95. doi: 10.1681/ASN.2007121311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pai MY, Lomenick B, Hwang H, Schiestl R, McBride W, Loo JA, et al. Drug affinity responsive target stability (DARTS) for small-molecule target identification. Methods Mol Biol. 2015;1263:287–98. doi: 10.1007/978-1-4939-2269-7_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugiura H, Yoshida T, Shiohira S, Kohei J, Mitobe M, Kurosu H, et al. Reduced Klotho expression level in kidney aggravates renal interstitial fibrosis. Am J Physiol Ren Physiol. 2012;302:F1252–64. doi: 10.1152/ajprenal.00294.2011. [DOI] [PubMed] [Google Scholar]
- 29.Zhou L, Li Y, Zhou D, Tan RJ, Liu Y. Loss of Klotho contributes to kidney injury by derepression of Wnt/β-catenin signaling. J Am Soc Nephrol. 2013;24:771–85. doi: 10.1681/ASN.2012080865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Webster AC, Nagler EV, Morton RL, Masson P. Chronic kidney disease. Lancet. 2017;389:1238–52. doi: 10.1016/S0140-6736(16)32064-5. [DOI] [PubMed] [Google Scholar]
- 31.Heerspink HJL, Stefánsson BV, Correa-Rotter R, Chertow GM, Greene T, Hou FF, et al. Dapagliflozin in patients with chronic kidney disease. N Engl J Med. 2020;383:1436–46. doi: 10.1056/NEJMoa2024816. [DOI] [PubMed] [Google Scholar]
- 32.Qin G, Zhao C, Zhang L, Liu H, Quan Y, Chai L, et al. Dihydroartemisinin induces apoptosis preferentially via a Bim-mediated intrinsic pathway in hepatocarcinoma cells. Apoptosis. 2015;20:1072–86. doi: 10.1007/s10495-015-1132-2. [DOI] [PubMed] [Google Scholar]
- 33.Zhang XG, Li GX, Zhao SS, Xu FL, Wang YH, Wang W. A review of dihydroartemisinin as another gift from traditional Chinese medicine not only for malaria control but also for schistosomiasis control. Parasitol Res. 2014;113:1769–73. doi: 10.1007/s00436-014-3822-z. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Z, Guo M, Zhao S, Shao J, Zheng S. ROS-JNK1/2-dependent activation of autophagy is required for the induction of anti-inflammatory effect of dihydroartemisinin in liver fibrosis. Free Radic Biol Med. 2016;101:272–83. doi: 10.1016/j.freeradbiomed.2016.10.498. [DOI] [PubMed] [Google Scholar]
- 35.Fetz V, Prochnow H, Brönstrup M, Sasse F. Target identification by image analysis. Nat Prod Rep. 2016;33:655–67. doi: 10.1039/C5NP00113G. [DOI] [PubMed] [Google Scholar]
- 36.Wang J, Gao L, Lee YM, Kalesh KA, Ong YS, Lim J, et al. Target identification of natural and traditional medicines with quantitative chemical proteomics approaches. Pharmacol Ther. 2016;162:10–22. doi: 10.1016/j.pharmthera.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 37.Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mau T, Yung R. Potential of epigenetic therapies in non-cancerous conditions. Front Genet. 2014;5:438. doi: 10.3389/fgene.2014.00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin B, Robertson KD. DNA methyltransferases, DNA damage repair, and cancer. Adv Exp Med Biol. 2013;12:620–36.. doi: 10.1007/978-1-4419-9967-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem. 2002;3:274–93. doi: 10.1002/1439-7633(20020402)3:4<274::AID-CBIC274>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 41.Smyth LJ, McKay GJ, Maxwell AP, McKnight AJ. DNA hypermethylation and DNA hypomethylation is present at different loci in chronic kidney disease. Epigenetics. 2013;9:366–76. doi: 10.4161/epi.27161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodríguez-Romo R, Berman N, Gómez A, Bobadilla NA. Epigenetic regulation in the acute kidney injury (AKI) to chronic kidney disease transition (CKD) Nephrology. 2015;20:736–43. doi: 10.1111/nep.12521. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Q, Liu L, Lin W, Yin S, Duan A, Liu Z, et al. Rhein reverses Klotho repression via promoter demethylation and protects against kidney and bone injuries in mice with chronic kidney disease. Kidney Int. 2017;91:144–56. doi: 10.1016/j.kint.2016.07.040. [DOI] [PubMed] [Google Scholar]
- 44.Clinton SM, Glover ME, Maltare A, Laszczyk AM, Mehi SJ, Simmons RK, et al. Expression of klotho mRNA and protein in rat brain parenchyma from early postnatal development into adulthood. Brain Res. 2013;1527:1–14. doi: 10.1016/j.brainres.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Loon EP, Pulskens WP, van der Hagen EA, Lavrijsen M, Vervloet MG, van Goor H, et al. Shedding of klotho by ADAMs in the kidney. Am J Physiol Ren Physiol. 2015;309:F359–68. doi: 10.1152/ajprenal.00240.2014. [DOI] [PubMed] [Google Scholar]
- 46.Lindberg K, Amin R, Moe OW, Hu MC, Erben RG, Östman Wernerson A, et al. The kidney is the principal organ mediating klotho effects. J Am Soc Nephrol. 2014;25:2169–75. doi: 10.1681/ASN.2013111209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mencke R, Olauson H, Hillebrands JL. Effects of Klotho on fibrosis and cancer: a renal focus on mechanisms and therapeutic strategies. Adv Drug Deliv Rev. 2017;121:85–100. doi: 10.1016/j.addr.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 48.Sugiura H, Yoshida T, Shiohira S, Kohei J, Mitobe M, Kurosu H, et al. Reduced Klotho expression level in kidney aggravates renal interstitial fibrosis. Am J Physiol Ren Physiol. 2012;302:F1252–64. doi: 10.1152/ajprenal.00294.2011. [DOI] [PubMed] [Google Scholar]
- 49.Smyth LJ, Duffy S, Maxwell AP, McKnight AJ. Genetic and epigenetic factors influencing chronic kidney disease. Am J Physiol Ren Physiol. 2014;307:F757–76. doi: 10.1152/ajprenal.00306.2014. [DOI] [PubMed] [Google Scholar]
- 50.Jin SL, Zhang Y, Chen ZH, Qian DW, Qine YJ, Yongjie Q, et al. Epigenetic changes of the Klotho gene in age-related cataracts. Eur Rev Med Pharmacol. 2015;19:2544–53. [PubMed] [Google Scholar]
- 51.Xie B, Zhou J, Yuan L, Ren F, Liu DC, Li Q, et al. Epigenetic silencing of Klotho expression correlates with poor prognosis of human hepatocellular carcinoma. Hum Pathol. 2013;44:795–801. doi: 10.1016/j.humpath.2012.07.023. [DOI] [PubMed] [Google Scholar]
- 52.Rubinek T, Shulman M, Israeli S, Bose S, Avraham A, Zundelevich A, et al. Epigenetic silencing of the tumor suppressor klotho in human breast cancer. Breast Cancer Res Treat. 2011;133:649–57. doi: 10.1007/s10549-011-1824-4. [DOI] [PubMed] [Google Scholar]
- 53.Young GH, Wu VC. KLOTHO methylatio is linked to uremic toxins and chronic kidney disease. Kidney Int. 2012;81:611–2. doi: 10.1038/ki.2011.461. [DOI] [PubMed] [Google Scholar]
- 54.Chen J, Zhang X, Zhang H, Lin J, Zhang C, Wu Q, et al. Elevated Klotho promoter methylation is associated with severity of chronic kidney disease. PLoS ONE. 2013;8:e79856. doi: 10.1371/journal.pone.0079856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Azuma M, Koyama D, Kikuchi J, Yoshizawa H, Thasinas D, Shiizaki K, et al. Promoter methylation confers kidney-specific expression of the Klotho gene. FASEB J. 2012;26:4264–74. doi: 10.1096/fj.12-211631. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The additional data collected during this study are available from the corresponding author upon reasonable request.