Abstract
Cancer cells are known for their ability to adapt variable metabolic programs depending on the availability of specific nutrients. Our previous studies have shown that uptake of fatty acids alters cellular metabolic pathways in colon cancer cells to favor fatty acid oxidation. Here, we show that fatty acids activate Drp1 to promote metabolic plasticity in cancer cells. Uptake of fatty acids (FAs) induces mitochondrial fragmentation by promoting ERK-dependent phosphorylation of Drp1 at the S616 site. This increased phosphorylation of Drp1 enhances its dimerization and interaction with Mitochondrial Fission Factor (MFF) at the mitochondria. Consequently, knockdown of Drp1 or MFF attenuates fatty acid-induced mitochondrial fission. In addition, uptake of fatty acids triggers mitophagy via a Drp1- and p62-dependent mechanism to protect mitochondrial integrity. Moreover, results from metabolic profiling analysis reveal that silencing Drp1 disrupts cellular metabolism and blocks fatty acid-induced metabolic reprograming by inhibiting fatty acid utilization. Functionally, knockdown of Drp1 decreases Wnt/β-catenin signaling by preventing fatty acid oxidation-dependent acetylation of β-catenin. As a result, Drp1 depletion inhibits the formation of tumor organoids in vitro and xenograft tumor growth in vivo. Taken together, our study identifies Drp1 as a key mediator that connects mitochondrial dynamics with fatty acid metabolism and cancer cell signaling.
Subject terms: Metabolomics, Cancer metabolism
Introduction
A number of epidemiological studies indicate that obesity is a major risk factor for several types of cancer including colorectal cancer [1, 2]. Recent studies on determining the interaction between obesity and cancer have identified a role of adipocytes and FAs in modulating cancer cell metabolism [3–5]. We and others have shown that uptake of FAs allows cancer cells to survive energy stress conditions by upregulating mitochondrial fatty acid oxidation (FAO) [6–10]. This increased FAO stimulates the expression of genes associated with colon cancer stem cells by controlling Wnt/β-catenin signaling [7, 8]. However, the molecular mechanisms by which FAs communicate with mitochondria remain largely elusive.
Mitochondrial dynamics refers to a collection of mitochondrial movements, including continuous cycles of membrane fusion and fission, transporting along the cytoskeleton, and interactions with other organelles [11, 12]. The fission of mitochondria is controlled by Drp1 (dynamin-related protein 1, gene name DNM1L), a small GTPases in the dynamin family. A number of adapter proteins that localize on the outer membrane of mitochondria, including Fis1, MFF, MiD49, and MiD51, function as receptors to recruit Drp1 to mitochondria [12, 13]. Moreover, Drp1 is known to be phosphorylated at S616 by ERK or cyclin dependent kinases, which increases its ability to induce mitochondrial fission [14, 15]. In contrast, reversible phosphorylation of Drp1 at S637 by PKA and calcineurin inhibits mitochondrial fission by disrupting the interaction between Drp1 and MiD49/51 [16–18].
Recent studies have linked mitochondrial dynamics to a number of tumorigenesis processes [19]. It has been shown that Drp1 functions downstream of oncogenic RAS and BRAF signaling to promote cell transformation and tumor growth [14, 20, 21]. Interestingly, mitochondrial fission is also involved in regulating mitophagy, a mitochondrial quality control mechanism that removes damaged mitochondria using the autophagic machinery [22, 23]. Nevertheless, it is not entirely clear how changes in mitochondria dynamic and mitophagy allow cancer cells to adapt to a FA-enriched microenvironment.
In this study, we show that FA uptake triggers mitochondrial fission by inducing ERK-dependent phosphorylation and mitochondrial translocation of Drp1. Knockdown of Drp1 inhibits FA-mediated upregulation of Wnt/β-catenin signaling. Taken together, our findings provide new insights into the molecular mechanisms by which FAs stimulate Drp1 to promote tumorigenesis in colon cancer.
Materials and methods
Cell culture and reagents
Patient-derived colon cancer PT130 cells were established as previously described [7, 24]. HCT116 and SW480 cells were purchased from ATCC. All cell lines were cultured in DMEM (high glucose) supplemented with 10% fetal bovine serum (FBS, Sigma-Aldrich) and 1% penicillin-streptomycin. The cell lines were authenticated using short tandem repeat (STR) DNA profiling and tested negative for mycoplasma contamination (Genetica). The following reagents were obtained from commercial sources: oleic acid (OA-albumin complex), palmitic acid, trametinib, CHIR99021, GW501516, and SGC-CBP30 were from Sigma-Aldrich; MitoTracker Green FM, MitoTracker Deep Red FM, BODIPY 558/568-C12, BODIPY 493/503 lipid probe, N-2 and B-27 supplement were from Thermo Fisher Scientific. Bovine serum albumin (BSA)-conjugated palmitate was prepared as described previously [8]. For fatty acid treatment, cells were cultured in DMEM (low glucose, Sigma-Aldrich) supplemented with 10% lipoprotein-deficient bovine serum (Alfa Aesar, J65182AMG) and in the presence of either BSA or BSA-conjugated FAs (100 μM final concentration). The SignalSilence SQSTM1/p62 siRNA I and control siRNA were purchased from Cell Signaling. The siRNA targeting ERK1, ERK2, and MFF was obtained from Santa Cruz Biotechnology.
Plasmids and lentivirus
The GFP-tagged Drp1, Flag-tagged MFF, Myc-MiD49, Myc-MiD51 and Fis1 expression plasmids were obtained from Addgene (pEYFP-C1-Drp1, #45160; pcDNA3-Flag-MFF-iso2, #74389; pcDNA3.1(-)-MiD49-Myc/His, #44596; pcDNA3.1(-)-MiD51-Myc/His, #44598 and pcDNA3.1(+) mFis1, #44600). The Su9-mCherry-GFP [25] expression plasmid was kindly provided by Dr. Hiromi Sesaki (Johns Hopkins University). The Drp1-S616A mutant was generated using QuikChange mutagenesis. The WT and S616A mutant Drp1 were subcloned into p3XFlag-CMV vector to create Flag-tagged expression plasmids. Stable Drp1 knockdown cells and mouse tumor organoids were generated using lentivirus-based RNAi as previously described [8, 26]. The shRNA targeting sequences for human Drp1 (DNM1L) are as the following: 5′-GCTACTTTACTCCAACTTATT-3′ (B3) and 5′-CGGTGGTGCTAGAATTTGTTA-3′ (B4); and for mouse Dnm1l is: 5′-CGGTGGTGCTAGGATTTGTTA-3′ (C1) and 5′-GGCAATTGAGCTAGCGTATAT-3′ (C2). The shRNA targeting sequences for human ACLY are: 5′-CCTATGACTATGCCAAGACTA-3′ (G5) and 5′-GCCTAAGTACTCTTGCCAGTT-3′ (G7).
Immunoprecipitation (IP) and western blot analysis
Total protein lysates were prepared by incubating cells or tumor tissues in lysis buffer (50 mM Na2HPO4, pH 7.4, 1 mM sodium pyrophosphate, 20 mM NaF, 2 mM EDTA, 2 mM EGTA, 1% Triton X-100, 1 mM DTT, 200 mM benzamidine, 40 mg ml−1 leupeptin, 200 mM PMSF) for 5–10 min on ice and the detergent-insoluble debris was removed after centrifugation for 5 min at 16,000 g at 4 °C [27–30]. For immunoprecipitation, cell lysates were incubated with indicated antibodies and protein A/G agarose beads (Thermo Fisher). GFP-tagged Drp1 and Flag-tagged p62 were immunoprecipitated using the anti-GFP nanobody affinity gel (BioLegend) and the anti-FLAG M2 affinity gel (Sigma-Aldrich), respectively. The resulted beads were washed with lysis buffer and subjected for western blot analysis. The following antibodies were used: from Cell Signaling, Drp1 (#5391), phospho-Drp1 (S616, #4494), phospho-Drp1 (S637, #6319), ERK1/2 (#4695), phospho-ERK1/2 (#4370), LC3A/B (#12741), COX4 (#4850), ACLY (#13390), acetylated-lysine (Ac-Lys, #9441), active-β-catenin (#8814) and β-catenin (#8480); from BioLegend, MFF (clone N382/14) and Fis1 (clone Poly28613); from Abnova, p62 (SQSTM1, H00008878-M01); from Santa Cruz, ubiquitin (sc-8017); and from Sigma-Aldrich, β-actin (A1978) and Flag M2 (F3165). The uncropped image files for western blots can be found in the Supplementary Information.
Mitochondrial fractionation
The mitochondria and cytosol fractions were prepared as previously described with the following modifications [30]. Briefly, cells were trypsinized, washed twice with ice-cold PBS and collected into microcentrifuge tubes. Cell pellets were homogenized using a micro pestle and lysed in mitochondrial lysis buffer (250 mM sucrose, 20 mM HEPES, pH 7.4, 20 mM NaF, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 200 mM benzamidine, 40 mg ml−1 leupeptin and 200 mM PMSF). The homogenates were centrifuged at 1000 g for 10 min at 4 °C. The supernatant was collected and centrifuged again at 13,000 g for 20 min at 4 °C. The resulting supernatant was saved as the cytosol fraction. The pellet was washed twice with mitochondrial lysis buffer, resuspended in lysis buffer, and saved as the mitochondria fraction. The mitochondria and cytosol fractions were subjected to western blot analysis. COX4 and ERK were used as markers for mitochondrial and cytoplasmic fractions, respectively.
Seahorse extracellular flux analysis
The Seahorse XF96 Extracellular Flux Analyzer (Agilent) was used to measure the respiration activity of colon cancer cells as described previously [7, 8, 24, 30]. For FAO tests, cells were seeded in 96-well Seahorse plates and cultured in substrate-limited medium (DMEM containing 0.5 mM glucose, 1.0 mM glutamine, 0.5 mM carnitine, and 1% FBS) for overnight. Approximately 45 min prior to the beginning of measurements, cells were switched to FAO assay media (200 μM palmitate-BSA, 111 mM NaCl, 4.7 mM KCl, 2.0 mM MgSO4, 1.2 mM Na2HPO4, 2.5 mM glucose, 0.5 mM carnitine, and 5 mM HEPES). FCCP (3 μM), ETO (200 μM), and antimycin A (4 μM) were added at the indicated time. The number of cells per well was determined using Seahorse XF Imaging and Cell Counting software and all OCR measurements were normalized to cell numbers in each well. The relative levels of fatty acid-driven basal and maximal mitochondrial respiration were calculated using Seahorse Wave software (Agilent). In addition, mitochondrial stress tests were performed according to manufacturer’s protocol as described previously [24, 30].
Tumor organoid colony formation assay
Tumor organoids generated from Apc/Kras double mutant mice were cultured as previously described [7, 8]. To silence Drp1 expression, tumor organoids were dissociated to single cell suspension using Accumax (Sigma-Aldrich) and incubated with sh-Drp1 lentivirus in tumor organoid media (Advanced DMEM/F12 supplemented with 1×Glutamax, 1×HEPES, 1×N-2, 1×B-27, 1 mM NAcetyl-L-cysteine and 1% penicillin/streptomycin) for 1 h in a 37 °C incubator. Cells were subsequently embedded in 50% Matrigel in organoid media and puromycin (1 μg/ml) was used for the selection of Drp1 knockdown cells. To determine colony formation capacity, single cell suspensions of tumor organoids were generated using Accumax. Total of 2000 cells were embedded in 50% Matrigel in organoid media, and the number of tumor organoids formed after 6 days were counted under an inverted microscope.
Quantitative RT-PCR (RT-qPCR)
To determine the relative gene expression, total RNAs were extracted from cells or tumor tissues using the RNeasy Mini kit (Qiagen). The reverse transcription and quantitative PCR reactions were carried out as described previously [7, 8, 24]. Primers used in this study are shown in the supplemental materials. All values were normalized to the level of β-actin.
Immunofluorescence (IF) imaging
To examine fatty acid-induced changes in mitochondrial morphology, cells grown on glass coverslips were labeled with MitoTracker Green and BODIPY 558/568-C12 for 45 min. After washing out excess labeling reagents, cells were cultured in DMEM low glucose supplemented with 10% lipoprotein-deficient bovine serum. Trafficking of labeled FAs was monitored in live cells for up to 3 h using Nikon A1R+ confocal microscope. Alternatively, cells were fixed with 4% paraformaldehyde at indicted time points and analyzed using confocal microscopy. In addition, cells treated with unlabeled fatty acids were fixed and co-stained with antibodies against Drp1 and MFF or antibodies against COX4 and LAMP1 (Thermo, #14-1079-80). To examine mitophagy, cells transfected with Su9-mCherry-GFP were treated with FAs and subsequently fixed with paraformaldehyde to visualize mCherry and GFP signals. The mitophagy index was calculated based on the degree of GFP and mCherry colocalization as measured by Pearson coefficient. Decreased colocalization (i.e., a decrease in mitophagy index) indicates increased mitophagy in cells. Images were taken using Nikon A1+ confocal microscope and Pearson coefficient was determined using Nikon NIS-elements software. The characteristics of mitochondrial network was quantitatively determined using ImageJ with the Mitochondrial Network Analysis (MiNA) toolset [31]. Mitochondrional phenotypes were quantified in 20 cells randomly selected from images taken from at least different ten fields for each group. Although the MiNA analysis allows for quantitative comparisons among different treatment groups, one potential limitation is that results obtained from MiNA only reflect the relative connectivity of mitochondrial network rather than the actual measurement of mitochondrial length.
Fatty acid degradation assay
Cells were seeded in 96-well plates at a density of 1 × 104 cells per well and incubated with PA (100 μM) for 24 h (the loading phase). Subsequently, cells were washed with PBS and cultured in low glucose DMEM supplemented with 10% lipoprotein-deficient bovine serum for additional 48 h (the unloading phase). The relative cell survival was determined by staining cells with Hoechst 33342. To determine cellular lipid contents, cells were stained with BODIPY 493/503 (1 µg/ml). The fluorescence intensity of Hoechst 33342 or BODIPY 493/503 was measured using Varioskan LUX microplate reader (Thermo Fisher). At the beginning of the unloading phase, no differences in total lipid contents were observed in different groups of cells and the fluorescence intensity for each group at this time point was normalized to 1.
Metabolite extraction and analysis
PT130 sh-control and sh-Drp1-B3 cells were treated with BSA or PA (100 µM) for 18 h. Cells were washed with PBS twice and treated with 50% methanol and incubated on ice for 10 min. Cells lysates were collected and L-norvaline was added as internal control. Polar metabolites were extracted and processed as described previously [32, 33]. For GC-MS analysis, an Agilent 7800B gas-chromatography coupled to a 7010 A triple quadrupole mass spectrometry detector equipped with a high-efficiency source was used for this study. Data analysis was performed using Aglient Masshunter software. Polar metabolites were normalized to the total biomass fraction consisting of amino acids found in the protein GC-MS quantitation. The normalized metabolite levels were uploaded to MetaboAnalyst to generate heatmap of polar metabolites across different groups. Data were normalized by Log and auto scaling for comparison.
Quantification of cellular acetate by GC-MS
Total cellular acetate levels were determined using previously published methods [32]. Briefly, PT130 sh-control and sh-Drp1-B3 cells treated with BSA or PA (100 µM) for 18 h were washed with PBS and suspended in cell lysis buffer (10 mM HEPES pH 7.5, 10 mM KCL, 0.1 mM EDTA, 1 mM DTT, 0.5% NP40, and protease/phosphatase inhibitor cocktail). After incubating on ice for 20 min, cytosolic proteins were collected by centrifugation at 12,000 g for 10 min at 4 °C. Samples were lyophilized and reconstituted in water. After chemical derivatization, acetate levels were determined using an Agilent 7890B GC system coupled to a 5977B GC/MSD. MNOVA software were employed for automated data processing using peak heights of m/z 61 ion used to quantify acetate. Raw acetate counts were normalized to protein input assessed by BCA protein assay (Thermo).
Xenograft tumorigenesis
All animal procedures were conducted using protocols approved by the University of Kentucky Animal Care and Use Committee. Six- to eight-week-old NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG, The Jackson Laboratory) mice were used. PT130 sh-control and sh-Drp1-B3 cells were resuspended in 5% Matrigel/DMEM and inoculated subcutaneously at 1 × 106 cells per injection. The tumor size was measured with a digital caliper, and the tumor volume was defined as (longest diameter) × (shortest diameter)2/2. At the end of experiments, tumors were harvested and subjected to mRNA and protein analysis. For detecting proliferating cells, fixed and paraffin-embedded tumor tissue sections were stained with the Ki67 antibody (MIB-1, Agilent). The percentage of tumor cells with positive staining of Ki67 was quantified using the HALO image analysis platform (Indica Labs). To determine the effect of adipocytes on regulating Drp1 phosphorylation in vivo, SW480 cells in 5% Matrigel suspension (1000 cells in 100 µl) were mixed with 50 µl of freshly isolated human adipocytes (contain approximately 100,000 adipocytes) or PBS and inoculated subcutaneously [8]. Tumors were harvested 3 months post injection and proteins were extracted for western blot analysis.
Bioinformatics and statistical analysis
Data from at least three independent experiments are expressed as means with SD as indicated in figure legends. Statistical analysis was performed using the Student t-test for pairwise comparisons and one-way ANOVA for multiple comparisons. For experiments with more than 10 data points, box plots were used to show the median value and data distribution. Measurements of xenograft tumor growth were summarized at each time point of follow-up and analysis was performed using longitudinal models to account for repeatedly measured tumor volume over time within each mouse. A linear mixed model was employed to compare slope of tumor volume growth curves over time between groups.
To determine the relative expression of DNM1L gene in human colon cancer patients, the mRNA expression data (478 primary tumor and 41 normal samples) in the format of fragments per kilobase of transcript per million mapped reads (FPKM) from The Cancer Genome Atlas colon cancer project (TCGA-COAD) were downloaded using the TCGAbiolinks package [34] and converted to transcripts per million (TPM). The Wilcoxon rank sum test was used to compare the expression of DNM1L in tumor vs. normal samples.
Results
FA treatment induces mitochondrial fission by activating Drp1 phosphorylation
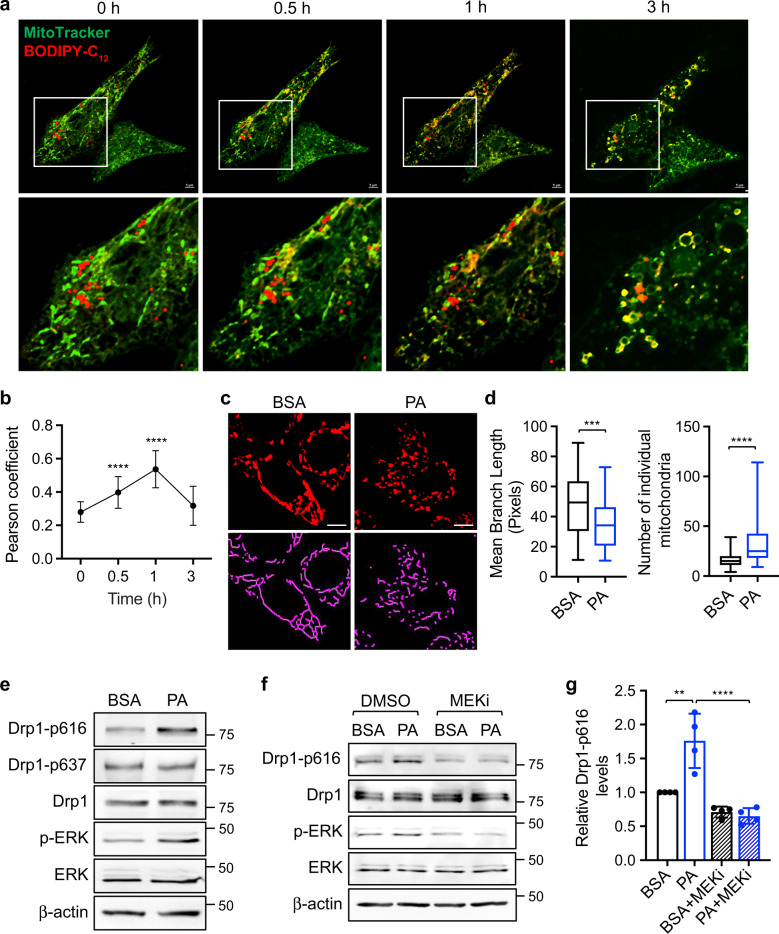
To assess the interaction between FAs and mitochondria, we monitored HCT116 cells labeled with BIODPY-C12 and MitoTracker using live cell imaging. Representative images taken at indicated time points following washout revealed that clusters of lipid droplets were initially formed in the cytoplasm with limited contact with mitochondria; subsequently, these lipid droplets dispersed and became colocalized with mitochondria (Fig. 1a). Quantitative analysis showed that the degree of colocalization first increased and then gradually decreased overtime (Fig. 1b and Supplementary Fig. S1a). Interestingly, this dynamic movement of FAs was accompanied by mitochondrial fission, in which the elongated mitochondrial network fragmentated into individual mitochondria in both HCT116 and PT130 cells (Fig. 1c and Supplementary Fig. S1b). Using the Mitochondrial Network Analysis (MiNA) toolset [31], we quantified FA-induced changes in mitochondrial morphology. As shown in Fig. 1d, the mean branch length of mitochondria decreased whereas the number of individual mitochondria increased in cells treated with PA confirming increased mitochondrial fission. Similar results were obtained in cells treated with OA (Supplementary Fig. S1c, d).
Fig. 1. Fatty acid treatment induces mitochondrial fission in colon cancer cells.
a HCT116 cells were incubated with BODIPY-C12 fluorescent fatty acid (red) and MitoTracker (green) for 1 h. After washing with PBS, the localization of BODIPY-C12 and mitochondria were monitored using live cell imaging. Representative images were taken at indicated time points. The boxed area is enlarged and shown below the corresponding image. Scale bar, 5 μm. b Colocalization of BODIPY-C12 lipid droplets with mitochondria at indicated time points were determined by Pearson coefficient as calculated using NIS-elements AR software (Nikon). Data are presented as mean ± SD (n = 30, ****p < 0.0001). c Representative confocal images of PT130 cells that were treated with BSA or PA and stained with MitoTracker (red). The skeletonized images shown below were generated by the MiNA software and used for the quantitative analysis of mitochondrial morphology. Scale bar, 10 μm. d The length of mitochondrial branch and the number of individual mitochondria were determined using MiNA with ImageJ. Results were presented as box plots (n = 20, ***p < 0.001 and ****p < 0.0001). e PT130 were treated with BSA or PA for 18 h. The levels of Drp1 phosphorylation at S616 (Drp1-p616), and S637 (Drp1-p637), Drp1, phospho-ERK (p-ERK), ERK, and β-actin were determined in cell lysates using western blot. f PT130 cells were treated with BSA or PA in combination with DMSO or trametinib (MEKi) for 18 h. The levels of Drp1-p616, Drp1, p-ERK, ERK, and β-actin were determined using western blot. g Representative western blots as shown in (f) were quantified to determine the relative Drp1-p616 levels by normalizing Drp1-p616 to Drp1. Data were presented as mean ± SD (n = 4, **p < 0.01, and ****p < 0.0001).
To investigate the mechanism underlying FA-induced mitochondria fission, we showed that PA treatment increased Drp1 phosphorylation at S616 and ERK phosphorylation (Fig. 1e). In contrast, Drp1 phosphorylation at S637, which negatively regulates Drp1 activity, was not altered by PA treatment. Similar increases in Drp1 phosphorylation at S616 were observed in HCT116 cells treated with PA and PT130 cells treated with OA (Supplementary Fig. S1e, f). However, FAs had no effects on altering the expression of total Drp1 and other regulators of mitochondrial dynamics, including MFN1, MFN2, and OPA1, both at the protein and mRNA level (Supplementary Fig. S1g, h). Previous studies have shown that increased FA levels can activate ERK signaling in obese mice [35]. To determine if PA-induced ERK activation is responsible for increased Drp1 phosphorylation, we treated colon cancer cells with trametinib (MEK inhibitor, MEKi) (Fig. 1f). Indeed, S616 phosphorylation was abolished in cells treated with MEKi confirming the involvement of ERK as the upstream kinase (Fig. 1g). Together, our results suggest that FA uptake activates mitochondrial fission by promoting ERK-dependent Drp1 phosphorylation.
FA treatment induces Drp1 mitochondrial translocation via interaction with MFF
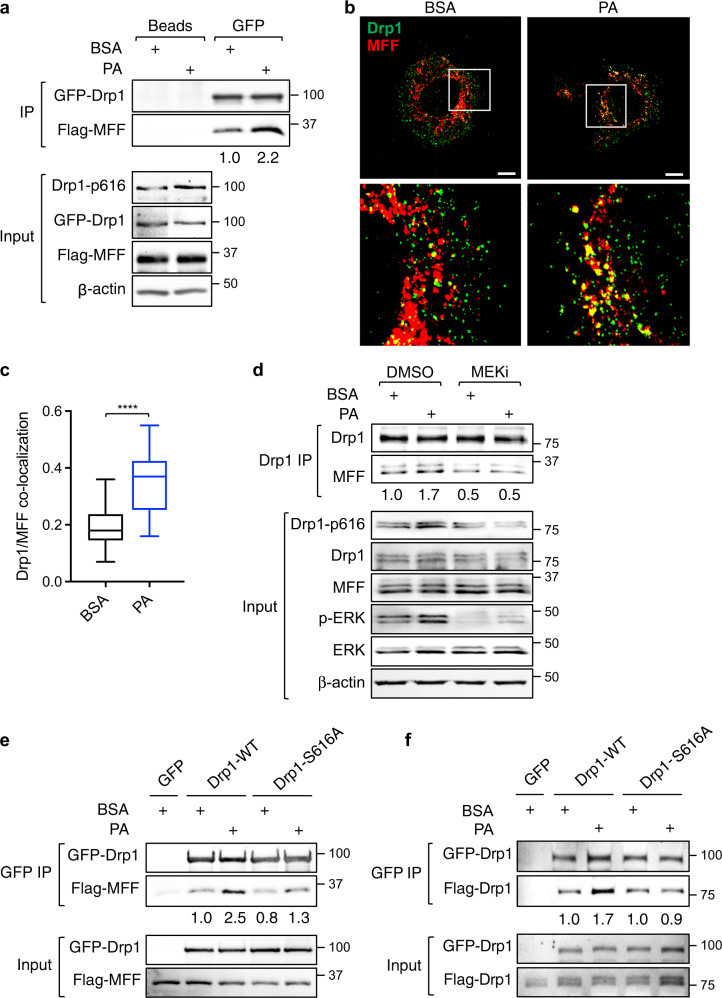
We next examined the interaction between Drp1 and its mitochondrial adapters upon FA treatment. PT130 cells transfected with GFP-Drp1 together with one of the four adapter proteins were subjected to co-immunoprecipitation experiments. Interestingly, the interaction between Drp1 and MFF was increased in cells treated with PA (Fig. 2a). In contrast, the binding of Drp1 with MiD49 or MiD51 was decreased upon PA treatment (Supplementary Fig. S2a, b), whereas the interaction between Drp1 and Fis1 was not detected in colon cancer cells (Supplementary Fig. S2c). However, future RNAi experiments are needed to validate if silencing of endogenous MiD49/51 or Fis1 affects mitochondrial translocation of Drp1. Moreover, PA increased the level of endogenous Drp1 and MFF colocalization at mitochondria as determined by IF staining (Fig. 2b, c). Consistently, the amount of Drp1 proteins localized in the mitochondrial fraction was significantly increased in PA treated cells (Supplementary Fig. S2d, e). Collectively, these results suggest that FAs promote mitochondrial translocation of Drp1 via binding to MFF.
Fig. 2. Fatty acid-induced Drp1 phosphorylation facilitates mitochondrial recruitment of Drp1 via interaction with MFF.
a PT130 cells transfected with GFP-Drp1 and Flag-MFF plasmids were treated with BSA or PA for 18 h. Cell lysates were immunoprecipitated with protein A/G agarose (beads) or GFP antibody conjugated beads. The presence of GFP-Drp1 and Flag-MFF in the immunoprecipitates was detected using Drp1 and Flag antibodies, respectively. The expression of Drp1-p616, GFP-Drp1, Flag-MFF, and β-actin in cell lysates (10% of input) was determined using western blot. The relative amount of Flag-MFF immunoprecipitated with GFP-Drp1 was obtained by normalizing MFF to Drp1 in GFP immunoprecipitates. b Representative confocal images of PT130 cells treated with BSA or PA for 6 h and stained with Drp1 (green) and MFF (red) antibodies. The boxed area is enlarged and shown below the corresponding image. Scale Bar, 10 μm. c Colocalization of Drp1 and MFF was quantitatively determined by Pearson coefficient analysis. Results were presented as a box plot (n = 20, ****p < 0.0001). d PT130 cells were treated with BSA or PA in combination with DMSO or MEKi for 18 h. Cell lysates were immunoprecipitated with the Drp1 antibody. The presence of endogenous Drp1 and MFF in immunoprecipitates were detected using Drp1 and MFF antibodies, respectively. The expression of Drp1-p616, Drp1, MFF, p-ERK, ERK, and β-actin in the input was determined using western blot. The relative amount of MFF that co-immunoprecipitated with Drp1 was determined by normalizing levels of MFF to Drp1 in the immunoprecipitates. Note that multiple endogenous MFF isoforms are detected by the MFF antibody in total cell lysates. e PT130 cells transfected with GFP, GFP-Drp1-WT, or GFP-Drp1-S616A together with Flag-MFF were treated with BSA or PA for 18 h. Cell lysates were immunoprecipitated with GFP antibody conjugated beads. The presence of GFP-Drp1 and Flag-MFF was detected using Drp1 and Flag antibodies, respectively. The relative amount of Flag-MFF that co-immunoprecipitated with GFP-Drp1 was determined by normalizing levels of MFF to Drp1 in GFP immunoprecipitates. f PT130 cells were transfected with the following combination of plasmids: (i) GFP + Flag-Drp1-WT; (ii) GFP-Drp1-WT + Flag-Drp1-WT; and (iii) GPF-Drp1-S616A + Flag-Drp1-S616A. Cell lysates prepared from transfected cells treated with BSA or PA were immunoprecipitated with GFP antibody conjugated beads. The presence of GFP-Drp1 (WT or S616A) and Flag-Drp1 (WT or S616A) was detected in the immunoprecipitates and input using GFP and Flag antibodies, respectively. The relative amount of Flag-Drp1 (WT or S616A) that co-immunoprecipitated with GFP-Drp1 (WT or S616A) was determined by normalizing Flag-Drp1 to GFP-Drp1 in GFP immunoprecipitates.
Furthermore, we determined if FA-induced Drp1-MFF interaction depends on Drp1 phosphorylation. To this end, the interaction between the endogenous Drp1 and MFF was analyzed in cells treated with PA alone or in combination with MEKi. The presence of MEKi decreased the Drp1-MFF interaction in both BSA- and PA-treated cells, which coincided with the inhibition of S616 phosphorylation (Fig. 2d). In addition, the interaction between MFF and Drp1-S616A, a phosphorylation-deficient mutant Drp1, was reduced compared to WT Drp1 in cells treated with PA (Fig. 2e), suggesting that PA-stimulated Drp1-MFF interaction relies on S616 phosphorylation. Previous studies indicate that cytosolically located Drp1 dimerizes before being recruited to mitochondria [36]. To test the hypothesis that S616 phosphorylation promotes Drp1 interaction with MFF by facilitating Drp1 dimerization, PT130 cells were co-transfected with GFP- and Flag-tagged Drp1 WT or S616A mutant. Indeed, PA increased the interaction of two WT Drp1 molecules whereas it failed to enhance the interaction of Drp1-S616A mutant proteins (Fig. 2f). Taken together, our findings suggest that FA uptake promotes S616 phosphorylation-dependent intermolecular interaction of Drp1 to enhance Drp1-MFF interaction and subsequent mitochondrial fission in colon cancer cells.
Knockdown of Drp1 disrupts FA utilization
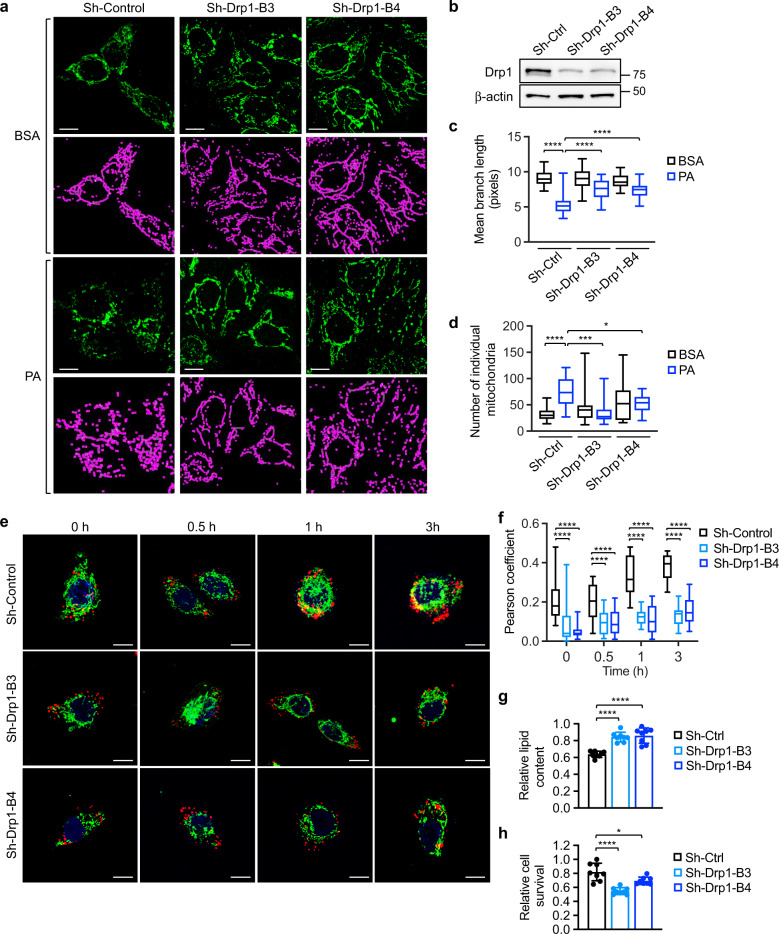
To verify the requirement of Drp1 in FA-induced mitochondrial fission, we analyzed mitochondrial morphology in stable control and sh-Drp1 colon cancer cells. Quantitative analysis revealed that downregulation of Drp1 abolished FA-induced mitochondrial fission (Fig. 3a–d). Similarly, silencing MFF alleviated the mitochondrial fragmentation phenotype induced by PA treatment (Supplementary Fig. S3a–c). Moreover, we found that knockdown of ERK blocked PA-induced mitochondrial fission as well as Drp1 phosphorylation at S616 (Supplementary Fig. S3d–g), thus confirming that ERK-dependent phosphorylation of Drp1 is required to promote mitochondrial fission. We next monitored the trafficking of lipid droplets in control and sh-Drp1 knockdown cells labeled with BODIPY-C12. Interestingly, the amount of lipid droplets that colocalized with mitochondria was significantly reduced in sh-Drp1 cells indicating impaired mitochondrial translocation (Fig. 3e, f). Similar results were obtained in cells treated with PA, in which downregulation of Drp1 prevented the association between FAs and mitochondria (Supplementary Fig. S4a, b).
Fig. 3. Knockdown of Drp1 blocks fatty acid-induced mitochondrial fission and fatty acid utilization.
a Control (sh-Ctrl) and two Drp1 knockdown (sh-Drp1-B3 and sh-Drp1-B4) PT130 cells were treated with BSA or PA for 18 h. Representative confocal images were taken of cells stained with MitoTracker Green. The skeletonized images shown below were generated by the MiNA software and used for the quantitative analysis of mitochondrial morphology. Scale bar, 10 μm. b Cell lysates of sh-Ctrl and sh-Drp1 PT130 cells were analyzed for the expression of Drp1 and β-actin using western blot. c The length of mitochondrial branch and d the number of individual mitochondria were determined using MiNA with ImageJ. Results were presented as box plots (n = 20, *p < 0.05, ***p < 0.001 and ****p < 0.0001). e Sh-Ctrl and sh-Drp1 PT130 cells were incubated with BODIPY-C12 (red) and MitoTracker (green) for 45 min and subsequently cultured in DMEM low glucose media supplemented with 10% lipoprotein-deficient serum. Representative images were taken at indicated time points. Scale bar, 10 μm. f Colocalization of BODIPY-C12 with mitochondria was determined using Pearson coefficient as calculated by NIS-elements AR software (Nikon). Results were presented as a box plot (n = 40, ****p < 0.0001). g Sh-Ctrl and sh-Drp1 PT130 cells were incubated with PA for 24 h and subsequently cultured in DMEM low glucose media supplemented with 10% lipoprotein-deficient serum for additional 48 h to allow for lipid utilization. Total cellular lipid contents were determined by BODIPY 493/503 staining. The fluorescence intensity of stained cells was measured using a fluorescence plate reader. Data were presented as mean ± SD (n = 8, ****p < 0.0001). h Sh-Ctrl and sh-Drp1 PT130 cells were loaded with PA and subsequent cultured in EBSS for 48 h. The relative cell survival for each cell line was determined by comparing to cells cultured in regular growth media. Data were presented as mean ± SD (n = 8, *p < 0.05, and ****p < 0.0001).
To determine if Drp1-mediated mitochondrial fission is necessary for cancer cells to catabolize exogenous FAs, control and sh-Drp1 cells loaded with PA were grown in low glucose media to allow for the preferential utilization of FAs. The total amount of FAs remained in sh-Drp1 cells was significantly higher after 48 h (Fig. 3g), suggesting delayed degradation of FAs. Functionally, the survival advantage provided by FA uptake was reduced in Drp1 knockdown cells when subjected to nutrient deprivation conditions (Fig. 3h). Silencing Drp1 also decreased FA degradation and cell survival in HCT116 cells (Supplementary Fig. S4c, d). Collectively, these data provide the initial evidence supporting a role of Drp1 and mitochondrial fission in regulating FA metabolism in colon cancer cells.
Drp1-dependent mitochondrial fission provides metabolic plasticity in colon cancer cells
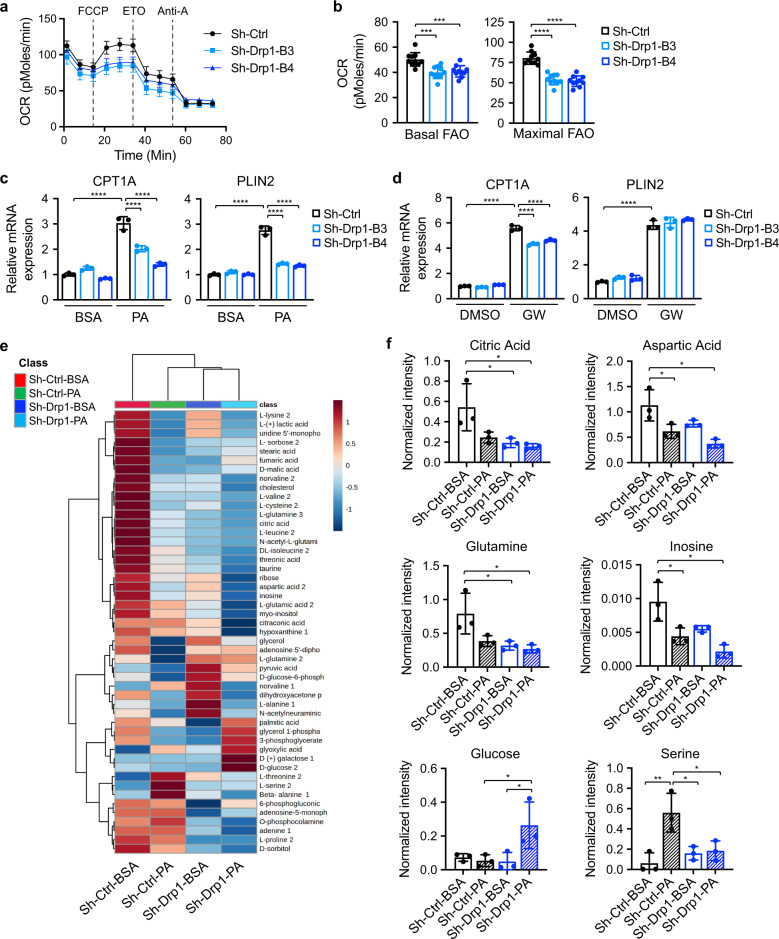
To further determine the effect of Drp1 on FA metabolism, we measured the rate of FA-driven respiration using Seahorse Extracellular Flux Analyzer. Results showed that both basal and maximal oxygen consumption rate (OCR) associated FA metabolism were significantly decreased in Drp1 knockdown PT130 (Fig. 4a, b) and HCT116 cells (Supplementary Fig. S5a). In addition, we found that silencing Drp1 decreased both basal and maximal OCR when glucose was supplied as the main metabolic substrate (Supplementary Fig. S5b). Thus, inhibition of mitochondrial fission likely attenuates the ability of mitochondria to metabolize both FAs and glucose.
Fig. 4. Knockdown of Drp1 disrupts FAO and alters cellular metabolism in colon cancer cells.
a Sh-Ctrl and sh-Drp1 PT130 cells were cultured in substrate-limited media and subsequently subjected to Seahorse FAO tests using Seahorse XF96 Extracellular Flux Analyzer as described in Materials and Methods. FCCP, ETO, and antimycin A (Anti-A) were added sequentially as indicated by dashed lines. The OCR measurements were normalized to total cell numbers. b Fatty acid-driven mitochondrial respirations were calculated based on OCR measurements and used to reflect basal and maximal levels of FAO. Results were presented as mean ± SD (n = 10, ***p < 0.001 and ****p < 0.0001). c Sh-Ctrl and sh-Drp1 PT130 cells treated with BSA or PA were analyzed for the expression of CPT1A and PLIN2 using RT-qPCR. Data were presented as mean ± SD (n = 3, ****p < 0.0001). d Sh-Ctrl and sh-Drp1 PT130 cells were treated with PPARδ agonist GW501516 (1 μM) for 24 h. The relative expression of CPT1A and PLIN2 was determined using RT-qPCR. Data were presented as mean ± SD (n = 3, ****p < 0.0001). e Sh-Ctrl and sh-Drp1 PT130 cells were treated with BSA or PA for 18 h. Polar metabolites were extracted and analyzed using GC-MS. The heatmap of polar metabolite levels was generated using Metaboanalyst. f Examples of polar metabolites that were differentially regulated in sh-Ctrl and sh-Drp1 PT130 cells. Data were presented as mean ± SD (n = 3, *p < 0.05, and **p < 0.01).
We have shown previously that FA uptake stimulates the expression of carnitine palmitoyltransferase I (CPT1A) via a PPARδ-dependent mechanism [8]. Intriguingly, FA treatment failed to induce the expression of PPARδ target genes, including CPT1A and PLIN2, in sh-Drp1 cells, whereas PPARδ agonist GW501516 was able to activate these target genes similarly in control and sh-Drp1 cells (Fig. 4c, d). To confirm the role of Drp1 in promoting FA-induced activation of PPARδ, we analyzed the expression of CPT1A and PLIN2 mRNA in control and ERK knockdown cells. Consistently, silencing ERK blocked PA-stimulated expression of both PPARδ target genes (Supplementary Fig. S5c). Thus, Drp1-dependent mitochondrial fission facilitates PPARδ activation downstream of FAs.
To better understand how mitochondrial fission regulates cellular metabolism, we measured a panel of 49 metabolites (mainly from the TCA cycle, glycolysis, amino acid, fatty acid and purine metabolism and the pentose phosphate pathway) using GC-MS. A heatmap constructed based on unsupervised hierarchical clustering analysis revealed that knockdown of Drp1 led to marked metabolic changes both basally and upon PA treatment (Fig. 4e). A large number of metabolites were reduced in cells grown in PA containing media compared to control media; and knockdown of Drp1 exacerbated PA-induced alterations (Fig. 4e, f). Consistent with reduced mitochondrial respiration as shown by Seahorse analysis, silencing Drp1 resulted in a decrease in metabolites linked to TCA cycle (such as citric acid, aspartic acid, and glutamate), nucleotide biosynthesis (such as inosine, ribose, and adenine) as well as amino acid metabolism (Fig. 4f and Supplementary Fig. S6). Intriguingly, cellular glucose levels were significantly increased in sh-Drp1 cells cultured in PA containing media. This is likely due to a compensatory upregulation of glucose uptake to overcome FAO defects in Drp1 knockdown cells. Moreover, PA preferentially increased cellular serine in control cells but not in sh-Drp1cells (Fig. 4f). Since FAO is known to produce mitochondrial reactive oxygen species (ROS) [37], increased serine production may serve as an antioxidative defensive mechanism. Taken together, our results demonstrate that Drp1-mediated regulation of mitochondrial fission is indispensable for FAs to remodel cellular metabolic pathways.
Drp1 facilitates FA-induced mitophagy
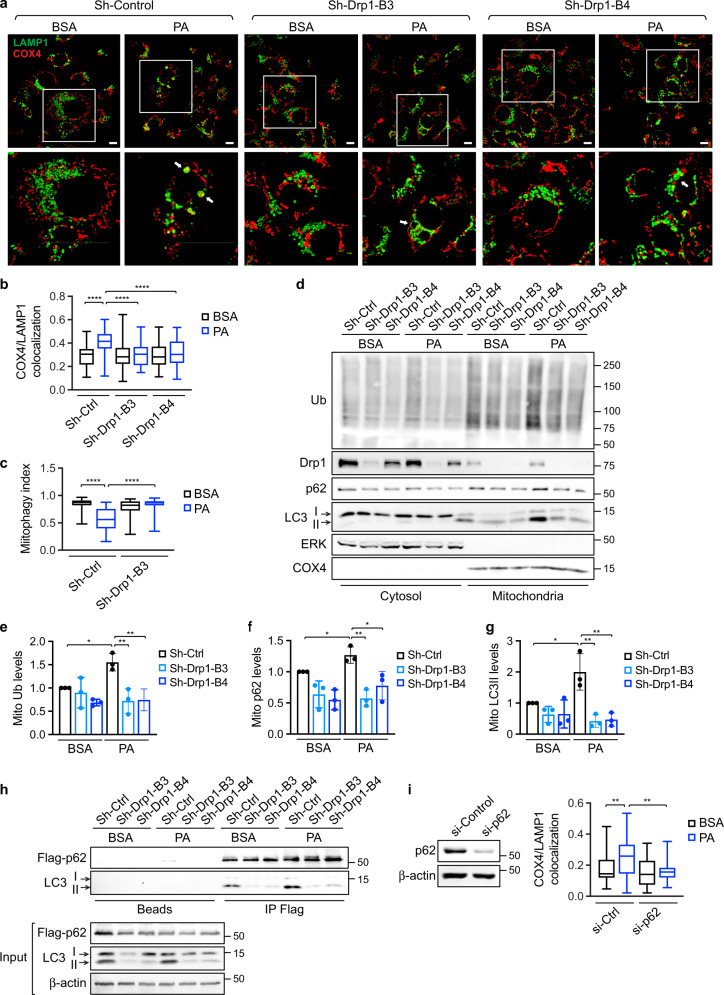
Recent studies in cardiac myocytes indicate that increased FAO induces mitophagy as a quality control mechanism [38, 39]. Here we determined whether exposure to FAs activates mitophagy in colon cancer cells. Results from IF staining showed that PA treatment enhanced the interaction between mitochondria and lysosomes in control cells as demonstrated by increased colocalization of COX4 and LAMP1 and the engulfment of fragmentated mitochondria within lysosomal compartments; however, silencing Drp1 largely inhibited PA-induced mitochondria-lysosome interaction (Fig. 5a, b). In addition, we used Su9-mCherry-GFP, a mitophagy reporter [25], to quantitatively analyze the mitophagy index by measuring mCherry and GFP colocalization. The induction of mitophagy in PA treated control cells was shown by decreased mCherry and GFP colocalization as GFP signals diminish upon mitochondria moving into the acidic environment within lysosomes. This FA-induced change in mitophagy index was not observed in sh-Drp1 cells (Fig. 5c and Supplementary Fig. S7a). Moreover, we found that silencing MFF blocked PA-induced mitophagy as determined using Su9-mCherry-GFP reporter (Supplementary Fig. S7b).
Fig. 5. Uptake of fatty acids induces mitophagy through a Drp1- and p62-dependent mechanism in colon cancer cells.
a Sh-Ctrl and sh-Drp1 PT130 cells were treated with BSA or PA for 18 h. Representative confocal images were taken from cells stained with LAMP1 (green) and COX4 (red) antibodies. The boxed areas are enlarged and shown below the corresponding images. Arrows indicate enlarged lysosomes either filled with mitochondria in sh-Ctrl cells or remained empty in sh-Drp1 cells. Scale Bar, 10 μm. b Colocalization of COX4 and LAMP1 was determined using Pearson coefficient as calculated by NIS-elements AR software. Results are presented as a box plot (n = 30, ****p < 0.0001). c Sh-Ctrl and sh-Drp1 PT130 cells transfected with Su9-mCherry-GFP reporter were treated with BSA or PA for 18 h. The degree of mCherry and GFP colocalization of was determined using Person coefficient and termed mitophagy index. Results were presented as a box plot (n = 30, ****p < 0.0001). d Sh-Ctrl and sh-Drp1 PT130 cells were treated with BSA or PA for 18 h. Cytosolic and mitochondrial fractions were prepared and analyzed for the expression of ubiquitinated proteins (Ub), Drp1, p62, and LC3 using western blot. ERK and COX4 were used as markers for cytosolic and mitochondrial fractions, respectively. e–g Relatively levels of protein ubiquitination (e), p62 (f), and LC3 (g) in the mitochondrial fraction were determined by normalizing to COX4. Data were presented as mean ± SD (n = 3, *p < 0.05 and **p < 0.01). h Sh-Ctrl and sh-Drp1 PT130 cells transfected with Flag-p62 were treated with BSA or PA for 18 h. Cell lysates were immunoprecipitated with protein A/G beads (beads) or Flag antibody conjugated beads. The presence of LC3 and Flag-p62 and in the immunoprecipitates and inputs was detected using LC3 and Flag antibodies, respectively. i PT130 cells were transfected with control or p62 siRNA. Two days following transfection, si-Ctrl and si-p62 cells were treated with BSA or PA for 18 h. Colocalization of LAMP1 and COX4 were examined using IF staining and expressed as Person coefficient. Results were presented as a box plot (n = 30, **p < 0.01). The knockdown efficiency of p62 was confirmed by western blot analysis and shown on the left.
We next investigated the mechanisms underlying FA-induced mitophagy. Results from cell fractionation experiments showed that levels of ubiquitinated proteins were increased particularly in the mitochondrial fraction of control cells upon PA treatment; and this increase in ubiquitination was accompanied by increased mitochondrial accumulation of p62 and LC3 (Fig. 5d–g). In contrast, knockdown of Drp1 attenuated the effect of PA on promoting mitochondrial protein ubiquitination and translocation of p62 and LC3 (Fig. 5d–g). Moreover, the interaction between p62 and LC3 was increased upon PA treatment in control cells but not in sh-Drp1 cells (Fig. 5h). Interestingly, the active form of LC3, LC3II, was preferentially localized to the mitochondrial fraction and interacted with p62 (Fig. 5d, h). To confirm the requirement of p62 in FA-induced mitophagy, we showed that downregulation of p62 disrupted colocalization of COX4 and LAMP1 induced by PA treatment (Fig. 5i and Supplementary Fig. S7c). Additionally, PA-induced changes in mitophagy index was blocked in p62 knockdown cells as assessed using Su9-mCherry-GFP reporter (Supplementary Fig. S7d). Together, our results suggest that FA uptake triggers mitophagy via a Drp1- and p62-dependent mechanism.
Drp1 is required for FA-induced activation of Wnt signaling
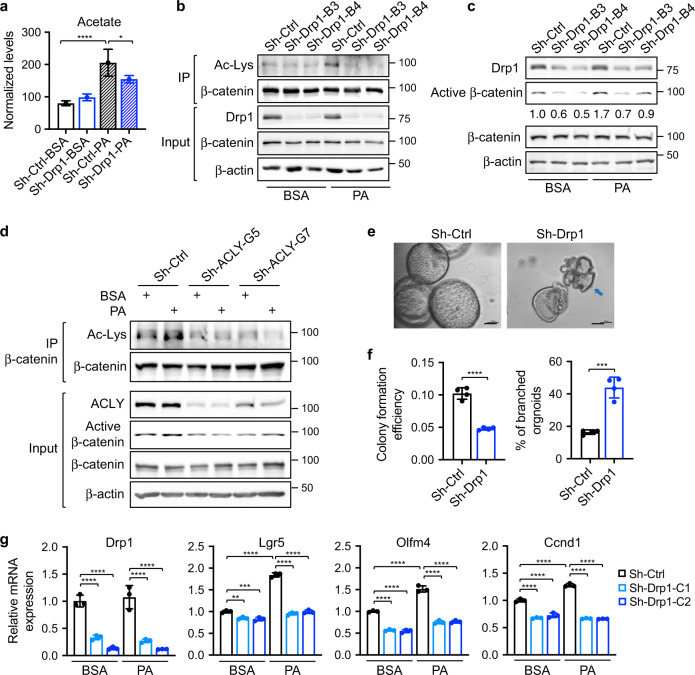
We have shown previously that FAs promote Wnt signaling by increasing β-catenin acetylation [7, 8]. Here, we investigated the role of Drp1 on regulating Wnt signaling. Consistent with the notion that FAO is one of the major metabolic pathway that provides the carbon source for the production of acetyl-CoA (Ac-CoA) and protein acetylation [8, 40], we found that PA increased cellular acetate levels in both PT130 and HCT116 cells; however, knockdown of Drp1 reduced acetate production in PA treated cells (Fig. 6a and Supplementary Fig. S8a). This change in acetate levels correlated with levels of β-catenin acetylation in that PA-stimulated β-catenin acetylation was largely inhibited in sh-Drp1 cells (Fig. 6b). In addition, silencing Drp1 also blocked FA-induced activation of β-catenin as determined using the active β-catenin antibody (Fig. 6c). Furthermore, we found that FA treatment failed to enhance β-catenin acetylation in ERK knockdown cells, suggesting that the S616 phosphorylation of Drp1 is involved in regulating FA-mediated β-catenin activation (Supplementary Fig. S8b).
Fig. 6. Downregulation of Drp1 decreases cellular levels of acetate and acetylation of β-catenin.
a Sh-Ctrl and sh-Drp1 PT130 cells were treated with BSA or PA for 18 h. Cellular acetate was extracted and analyzed using GC-MS. Data were presented as mean ± SD (n = 3 for sh-Ctrl-PA group and n = 4 for other groups, *p < 0.05 and ****p < 0.0001). b Cell lysates isolated from sh-Ctrl and sh-Drp1 PT130 cells treated with BSA or PA were immunoprecipitated using the β-catenin antibody. The acetylation of β-catenin was detected by the acetylated-lysine antibody (Ac-Lys). c Sh-Ctrl and sh-Drp1 PT130 cells were treated with BSA or PA for 18 h. GSK3 inhibitor, CHIR99021 (3 μM), was included in both groups to allow the analysis of β-catenin activation downstream of the destruction complex. The expression of Drp1, active β-catenin, total β-catenin, and β-actin were analyzed using western blot. The relative levels of active β-catenin were obtained by normalizing active β-catenin to β-actin. d Sh-Ctrl and sh-ACLY PT130 cells were treated with BSA or PA for 18 h. Cell lysates were immunoprecipitated using the anti-β-catenin antibody and the acetylation of β-catenin were detected using the Ac-Lys antibody. The expression of ACLY, active β-catenin, total β-catenin, and β-actin was detected in the 10% input using western blot. e Representative images of control and Drp1 knockdown Apc/Kras tumor organoids grown in 3D Matrigel for 6 days. Arrow indicates tumor organoid with branched phenotype. Scale bar, 100 μm. f Single cell suspensions of sh-Ctrl and sh-Drp1 tumor cells were seeded in 3D Matrigel. The colony formation efficiency and the percentage of organoids with branched phenotype were quantified. Data were presented as mean ± SD (n = 4, ****p < 0.0001). g Sh-Ctrl and sh-Drp1 tumor organoids were cultured in organoid growth media with the addition of BSA or PA for 2 days. The relative expression of Drp1 and Wnt target genes (including Lgr5, Olfm4, and Ccnd1) was determined using RT-qPCR. Data were presented as mean ± SD (n = 3, **p < 0.01 and ****p < 0.0001).
In order for FA-derived Ac-CoA produced in mitochondria to participate in protein acetylation in the cytosol, Ac-CoA is first converted to citrate via TCA cycle and subsequently transported out of mitochondria and converted back to Ac-CoA by ATP citrate lyase (ACLY) [41]. Indeed, silencing ACLY blocked PA-induced increase in β-catenin acetylation and activation (Fig. 6d). These data are consistent with previous reports that direct inhibition of FAO by silencing CPT1A impairs β-catenin acetylation and activation downstream of FA treatment [8]. Since β-catenin is a known substrate of CBP/p300 acetyltransferases [42, 43], we treated cells with SGC-CBP30, a potent inhibitor of CBP/p300, and found that inhibition of CBP/p300 attenuated PA-stimulated acetylation and activation of β-catenin (Supplementary Fig. S8c).
To determine the functional importance of Drp1-mediated regulation of β-catenin, we silenced Drp1 expression in Apc/Kras mutant mouse tumor organoids. Single cell suspensions of control and sh-Drp1 cells were seeded in 3D Matrigel and the number of tumor organoids formed was quantified (Fig. 6e). Knockdown of Drp1 significantly decreased the formation of tumor organoids and increased numbers of sh-Drp1 tumor organoids with branched phenotype suggesting potential differentiation (Fig. 6e, f). While the presence of PA in 3D growth media increased the expression of Wnt/β-catenin target genes (including Lgr5, Olfm4, and Ccnd1), silencing Drp1 significantly reduced Wnt signaling activation under both basal and PA treated conditions (Fig. 6g). Similarly, FA-stimulated Wnt target gene expression was reduced in sh-Drp1 PT130 cells (Supplementary Fig. S8d). Taken together, our results suggest that Drp1 plays an important role in regulating FA-stimulated Ac-CoA production and subsequent β-catenin activation.
Knockdown of Drp1 inhibits xenograft tumor growth in vivo
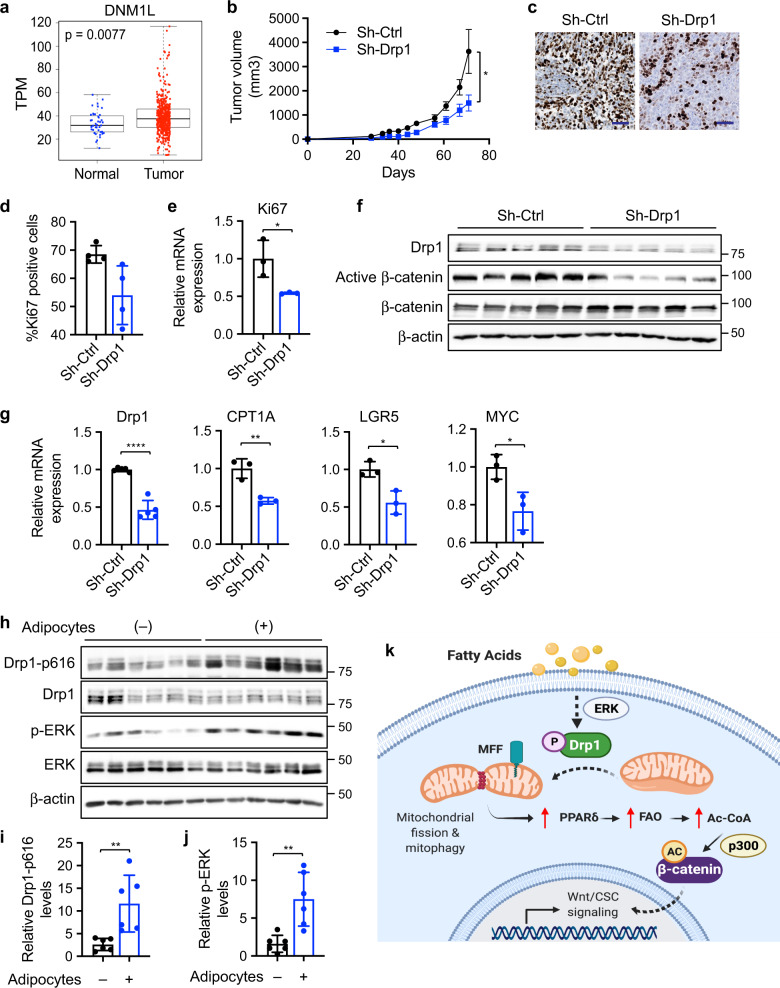
To better understand the role of Drp1 in colon cancer tumorigenesis in vivo, we analyzed the expression of Drp1 (DNM1L) mRNA in colon cancer patients using TCGA COAD dataset. Results from bioinformatic analysis showed that Drp1 is significantly elevated in colon tumors compared to normal controls (Fig. 7a). In addition, control and sh-Drp1 PT130 cells were injected subcutaneously into NSG mice and tumor formation was monitored. We found that silencing Drp1 significantly reduced the rate of xenograft tumor growth (Fig. 7b). Results from IHC staining and RT-qPCR experiments showed that the number of Ki67-positive cells and the expression of Ki67 mRNA were significantly decreased in sh-Drp1 tumors (Fig. 7c, e). Moreover, silencing Drp1 reduced levels of active β-catenin, as well as the expression of CPT1A and Wnt target genes (including LGR5 and MYC), in tumor samples indicating downregulation of Wnt signaling (Fig. 7f, g).
Fig. 7. Knockdown of Drp1 inhibits xenograft tumor growth and Wnt signaling in vivo.
a Bioinformatic analysis of TCGA-COAD dataset showed higher Drp1 (DNM1L gene) expression in tumors compared to normal samples (p = 0.0077 based on linear mixed models). b Sh-Ctrl and sh-Drp1 PT130 cells were injected subcutaneously into NSG mice. The size of tumors was measured every 3–5 days starting at the 3rd week after injection. Data were presented as mean ± SEM (n = 12, *p < 0.05). c Representative images from tumor sections stained with the Ki67 antibody. Scale bar, 25 μm. d The percentage of Ki67 positive cells were quantified in tumors from 4 mice of each group using HALO. Data were presented as mean ± SD (n = 4, *p < 0.05). e Tumor tissues from three mice of each group were analyzed for the expression of Ki67 mRNA using RT-qPCR. Data were presented as mean ± SD (n = 3, *p < 0.05). f Tumor tissues from sh-Ctrl and sh-Drp1 group were analyzed for the expression of Drp1, active β-catenin, total β-catenin, and β-actin using western blot. g Tumor tissues from three mice of each group were analyzed for the expression of Drp1, CPT1A, LGR5, and MYC mRNA using RT-qPCR. Data were presented as mean ± SD (n = 3, *p < 0.05, **p < 0.01, and ****p < 0.0001). h Suspensions of SW480 cells with or without adipocytes were injected subcutaneously into NSG mice and tumor tissues were analyzed for the expression of Drp1-p616, Drp1, p-ERK, ERK, and β-actin using western blot. i, j Relative Drp1-p616 (i) and p-ERK (j) levels were quantified by normalizing Drp1-p616 to Drp1 and p-ERK to ERK, respectively. Data were presented as mean ± SD (n = 6, **p < 0.01). k Results from our study support a model in which fatty acids activate Drp1/MFF-mediated mitochondrial fission to promote Wnt/β-catenin signaling by remodeling cellular metabolic pathways.
We have previously shown that the presence of adipocytes promotes xenograft tumor growth [7, 8]. To verify that increased availability of FAs activates ERK-dependent Drp1 phosphorylation in vivo, we analyzed the status of Drp1 and ERK phosphorylation in xenograft tumors derived from colon cancer cells that were co-injected with or without adipocytes. Consistent with our previous report that co-injection of adipocytes promotes β-catenin activation [8], the presence of adipocytes significantly increased Drp1 phosphorylation at S616 and activation of ERK in vivo (Fig. 7h–j). Together, our study provides a road map linking FA uptake with Wnt/β-catenin signaling. The presence of FAs stimulates Drp1 activity by promoting ERK-dependent phosphorylation of Drp1 at S616 and subsequent recruitment of Drp1 to mitochondria via binding to MFF. Functionally, Drp1-dependent mitochondrial remodeling enables FA metabolism to produce the necessary metabolic substrates for β-catenin acetylation and Wnt signaling activation (Fig. 7k). Therefore, Drp1 plays an essential role in maintaining metabolic plasticity to promote tumorigenesis in vivo.
Discussion
Mitochondria are well known for their ability to produce ATP. However, by controlling the production of a large number of metabolites, increasing evidence suggests that mitochondria function as signaling organelles and mitochondria can be targeted for cancer therapy [44]. Inhibition of Drp1 has been shown to prevent cell cycle progression and induce apoptosis in cancer cells [19]. However, many of the findings are obtained by using a putative Drp1 inhibitor, Mdivi-1, which has recently been re-classified as a reversible inhibitor of mitochondrial complex I [45]. Thus, more studies are needed to better understand the role of Drp1 in cancer. Here, we demonstrated that FAs promote cell proliferation and survival by stimulating Drp1-dependent mitochondrial fission in colon cancer cells. Downregulation of Drp1 prevents FA-induced mitochondrial fission, disrupts mitochondrial localization of FAs, eliminates the survival advantage provided by FAs and inhibits tumorigenesis in vivo. Mechanistically, Drp1 facilitates FA-induced activation of Wnt signaling by enabling β-catenin acetylation. However, since increased Ac-CoA production is known to stimulate the acetylation of other substrates [8], future studies are needed to determine the functional contrition of those acetylation events downstream of FA uptake.
Despite being the master regulator of mitochondrial fission, a large percentage of endogenous Drp1 is cytosolic [46]. We showed that FA uptake promotes mitochondrial localization of Drp1 and mitochondrial fission by stimulating ERK-mediated phosphorylation of Drp1 at S616. Although a number of studies have demonstrated that S616 phosphorylation activates Drp1, the underlying molecular mechanism remains largely unknown as this residue resides in a disordered region that is not visible in the crystal structure of Drp1 [17]. Our results suggest that S616 phosphorylation facilitates the intermolecular interaction of Drp1 and subsequent association with MFF. Interestingly, AMPK-mediated phosphorylation of MFF has been shown to increase Drp1-MFF interaction [47]. Since FA treatment induces AMPK activation [7], the mitochondrial fission phenotype is likely triggered by phosphorylation-mediated activation of both Drp1 and MFF.
We found in this study that exogenous FAs (BOPIDY-C12 or PA) are rapidly incorporated into lipid droplets once entered the cells; however, the initial colocalization of newly formed lipid droplets with mitochondria occurs at a relatively low level. As Drp1 activates mitochondrial fission, the colocalization of FAs with mitochondria increases. This is likely due to the transport of free FAs released from lipid droplets into mitochondria. Consistent with a recent report [48], results from our study indicated that Drp1 is required for the mitochondrial association of FAs. Interestingly, it has been shown that lipid droplet-associated mitochondria have elongated morphology but reduced FAO capacity compared to fragmentated mitochondria [49]. Thus, FA-stimulated mitochondrial recruitment of Drp1 may initiate necessary changes in the mitochondrial network in order to meet the increasing demand for FAO. However, addtional studies are needed to determine how Drp1 regulates the interaction between lipid droplets and mitochondria.
It has been shown that knockdown of Drp1 inhibits tumor growth in a mouse model of Kras-driven pancreatic cancer by disrupting the ability of mutant Kras to upregulate glycolysis. Additional defects in metabolic pathways (including FAO) are also observed in Drp1 knockout tumor cells [21]. We found that silencing Drp1 decreases mitochondrial FAO activity and alters a number of cellular metabolic pathways. Thus, Drp1 may be responsible for coordinating mitochondrial activity based on changes in nutrient status. In addition, Drp1-dependent mitochondrial fragmentation is essential for maintaining the integrity and fitness of mitochondria through mitophagy [23]. Results from our study indicate that FA uptake triggers mitophagy via a Drp1/p62/LC3-mediated molecular pathway. It is of particular interest for future studies to determine the functional interplay between the induction of mitophagy and FA metabolism.
In summary, our study identifies Drp1 as a central mediator that functions downstream of FAs to modulate cellular metabolism and cancer cell signaling. Given the importance of Wnt signaling in promoting tumor growth and maintaining cancer stem cell functions, targeting Drp1-depenent mitochondrial fission may represent a potential therapeutic strategy to eliminate the survival advantage provided by FAs.
Supplementary information
Acknowledgements
We thank Dr. Hiromi Sesaki at Johns Hopkins University for kindly providing the Su9-mCherry-GFP expression plasmid and Dr. Yekaterina Zaytseva for sharing the PT130 cell line. The diagram shown in Fig. 7k was created with BioRender.com.
Author contributions
XX, SH, and TG designed all aspects of the study; XX, SH, LEAY, DR, ATS, RM, CW, and HLW performed experiments and analysis; MSG and RCS provided critical support for metabolic analysis; XX, SH, and TG wrote the manuscript.
Funding
This work was supported by R01CA133429 (TG), R01CA208343 (TG), F31CA260840 (SH), R35NS116824 (MSG), R01AG066653 (RCS), and a pilot grant from P20GM121327 (University of Kentucky Center for Cancer and Metabolism). ATS is supported by National Science Foundation Graduate Research Fellowship Award (#1839289). The studies were conducted with support provided by the Redox Metabolism, Biospecimen Procurement and Translational Pathology, Flow Cytometry and Immune Monitoring, and Biostatistics and Bioinformatics Shared Resource Facilities of the University of Kentucky Markey Cancer Center (P30CA177558) at the University of Kentucky.
Materials availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Ethics approval
This research does not involve human subjects.
Footnotes
Edited by A. Degterev
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xiaopeng Xiong, Sumati Hasani.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-022-00974-5.
References
- 1.Bhaskaran K, Douglas I, Forbes H, dos-Santos-Silva I, Leon DA, Smeeth L. Body-mass index and risk of 22 specific cancers: a population-based cohort study of 5·24 million UK adults. Lancet. 2014;384:755–65. doi: 10.1016/S0140-6736(14)60892-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lauby-Secretan B, Scoccianti C, Loomis D, Grosse Y, Bianchini F, Straif K. Body fatness and cancer—viewpoint of the IARC working group. N. Engl J Med. 2016;375:794–8. doi: 10.1056/NEJMsr1606602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corn KC, Windham MA, Rafat M. Lipids in the tumor microenvironment: From cancer progression to treatment. Prog Lipid Res. 2020;80:101055. doi: 10.1016/j.plipres.2020.101055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ringel AE, Drijvers JM, Baker GJ, Catozzi A, García-Cañaveras JC, Gassaway BM, et al. Obesity shapes metabolism in the tumor microenvironment to suppress anti-tumor immunity. Cell. 2020;183:1848–66. doi: 10.1016/j.cell.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z, Liu H, He J, Wang Z, Yin Z, You G, et al. Acetyl-CoA synthetase 2: a critical linkage in obesity-induced tumorigenesis in myeloma. Cell Metab. 2021;33:78–93. doi: 10.1016/j.cmet.2020.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lazar I, Clement E, Dauvillier S, Milhas D, Ducoux-Petit M, LeGonidec S, et al. Adipocyte exosomes promote melanoma aggressiveness through fatty acid oxidation: a novel mechanism linking obesity and cancer. Cancer Res. 2016;76:4051–7. doi: 10.1158/0008-5472.CAN-16-0651. [DOI] [PubMed] [Google Scholar]
- 7.Wen YA, Xiong X, Harris JW, Zaytseva YY, Mitov MI, Napier DL, et al. Adipocytes activate mitochondrial fatty acid oxidation and autophagy to promote tumor growth in colon cancer. Cell Death Dis. 2017;8:e2593. doi: 10.1038/cddis.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiong X, Wen YA, Fairchild R, Zaytseva YY, Weiss HL, Evers BM, et al. Upregulation of CPT1A is essential for the tumor-promoting effect of adipocytes in colon cancer. Cell Death Dis. 2020;11:736. doi: 10.1038/s41419-020-02936-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clement E, Lazar I, Attane C, Carrie L, Dauvillier S, Ducoux-Petit M, et al. Adipocyte extracellular vesicles carry enzymes and fatty acids that stimulate mitochondrial metabolism and remodeling in tumor cells. EMBO J. 2020;39:e102525. doi: 10.15252/embj.2019102525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Attane C, Muller C. Drilling for oil: tumor-surrounding adipocytes fueling cancer. Trends Cancer. 2020;6:593–604. doi: 10.1016/j.trecan.2020.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–5. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet. 2012;46:265–87. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 13.van der Bliek AM, Shen Q, Kawajiri S. Mechanisms of mitochondrial fission and fusion. Cold Spring Harb Perspect Biol. 2013;5:a011072. [DOI] [PMC free article] [PubMed]
- 14.Kashatus JA, Nascimento A, Myers LJ, Sher A, Byrne FL, Hoehn KL, et al. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol Cell. 2015;57:537–51. doi: 10.1016/j.molcel.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem. 2007;282:11521–9. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- 16.Cribbs JT, Strack S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 2007;8:939–44. doi: 10.1038/sj.embor.7401062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalia R, Wang RY, Yusuf A, Thomas PV, Agard DA, Shaw JM, et al. Structural basis of mitochondrial receptor binding and constriction by DRP1. Nature. 2018;558:401–5. doi: 10.1038/s41586-018-0211-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang CR, Blackstone C. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J Biol Chem. 2007;282:21583–7. doi: 10.1074/jbc.C700083200. [DOI] [PubMed] [Google Scholar]
- 19.Lima AR, Santos L, Correia M, Soares P, Sobrinho-Simoes M, Melo M et al. Dynamin-related protein 1 at the crossroads of cancer. Genes. 2018;9:115. [DOI] [PMC free article] [PubMed]
- 20.Serasinghe MN, Wieder SY, Renault TT, Elkholi R, Asciolla JJ, Yao JL, et al. Mitochondrial division is requisite to RAS-induced transformation and targeted by oncogenic MAPK pathway inhibitors. Mol Cell. 2015;57:521–36. doi: 10.1016/j.molcel.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagdas S, Kashatus JA, Nascimento A, Hussain SS, Trainor RE, Pollock SR, et al. Drp1 promotes KRas-driven metabolic changes to drive pancreatic tumor growth. Cell Rep. 2019;28:1845–59. doi: 10.1016/j.celrep.2019.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashrafi G, Schwarz TL. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013;20:31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macleod KF. Mitophagy and mitochondrial dysfunction in cancer. Annu Rev Cancer Biol. 2020;4:41–60. doi: 10.1146/annurev-cancerbio-030419-033405. [DOI] [Google Scholar]
- 24.Wen YA, Xiong X, Zaytseva YY, Napier DL, Vallee E, Li AT, et al. Downregulation of SREBP inhibits tumor growth and initiation by altering cellular metabolism in colon cancer. Cell Death Dis. 2018;9:265. doi: 10.1038/s41419-018-0330-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamada T, Murata D, Adachi Y, Itoh K, Kameoka S, Igarashi A, et al. Mitochondrial stasis reveals p62-mediated ubiquitination in parkin-independent mitophagy and mitigates nonalcoholic fatty liver disease. Cell Metab. 2018;28:588–604. doi: 10.1016/j.cmet.2018.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gan T, Stevens AT, Xiong X, Wen YA, Farmer TN, Li AT, et al. Inhibition of protein tyrosine phosphatase receptor type F suppresses Wnt signaling in colorectal cancer. Oncogene. 2020;39:6789–801. doi: 10.1038/s41388-020-01472-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X, Stevens PD, Liu J, Yang H, Wang W, Wang C, et al. PHLPP is a negative regulator of RAF1, which reduces colorectal cancer cell motility and prevents tumor progression in mice. Gastroenterology. 2014;146:1301–12. doi: 10.1053/j.gastro.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, Weiss HL, Rychahou P, Jackson LN, Evers BM, Gao T. Loss of PHLPP expression in colon cancer: role in proliferation and tumorigenesis. Oncogene. 2009;28:994–1004. doi: 10.1038/onc.2008.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiong X, Li X, Wen YA, Gao T. Pleckstrin homology (PH) domain leucine-rich repeat protein phosphatase controls cell polarity by negatively regulating the activity of atypical protein kinase C. J Biol Chem. 2016;291:25167–78. doi: 10.1074/jbc.M116.740639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiong X, Wen YA, Mitov MI, Oaks CM, Miyamoto S, Gao T. PHLPP regulates hexokinase 2-dependent glucose metabolism in colon cancer cells. Cell Death Disco. 2017;3:16103. doi: 10.1038/cddiscovery.2016.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valente AJ, Maddalena LA, Robb EL, Moradi F, Stuart JA. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem. 2017;119:315–26. doi: 10.1016/j.acthis.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 32.Sun RC, Dukhande VV, Zhou Z, Young LEA, Emanuelle S, Brainson CF, et al. Nuclear glycogenolysis modulates histone acetylation in human non-small cell lung cancers. Cell Metab. 2019;30:903–16. doi: 10.1016/j.cmet.2019.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young LEA, Brizzee CO, Macedo JKA, Murphy RD, Contreras CJ, DePaoli-Roach AA, et al. Accurate and sensitive quantitation of glucose and glucose phosphates derived from storage carbohydrates by mass spectrometry. Carbohydr Polym. 2020;230:115651. doi: 10.1016/j.carbpol.2019.115651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bi L, Chiang JY, Ding WX, Dunn W, Roberts B, Li T. Saturated fatty acids activate ERK signaling to downregulate hepatic sortilin 1 in obese and diabetic mice. J Lipid Res. 2013;54:2754–62. doi: 10.1194/jlr.M039347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu R, Chan DC. The mitochondrial fission receptor Mff selectively recruits oligomerized Drp1. Mol Biol Cell. 2015;26:4466–77. doi: 10.1091/mbc.E15-08-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosca MG, Vazquez EJ, Chen Q, Kerner J, Kern TS, Hoppel CL. Oxidation of fatty acids is the source of increased mitochondrial reactive oxygen species production in kidney cortical tubules in early diabetes. Diabetes. 2012;61:2074–83. doi: 10.2337/db11-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shao D, Kolwicz SC, Jr, Wang P, Roe ND, Villet O, Nishi K, et al. Increasing fatty acid oxidation prevents high-fat diet-induced cardiomyopathy through regulating parkin-mediated mitophagy. Circulation. 2020;142:983–97. doi: 10.1161/CIRCULATIONAHA.119.043319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tong M, Saito T, Zhai P, Oka SI, Mizushima W, Nakamura M, et al. Mitophagy is essential for maintaining cardiac function during high fat diet-induced diabetic cardiomyopathy. Circ Res. 2019;124:1360–71. doi: 10.1161/CIRCRESAHA.118.314607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McDonnell E, Crown SB, Fox DB, Kitir B, Ilkayeva OR, Olsen CA, et al. Lipids reprogram metabolism to become a major carbon source for histone acetylation. Cell Rep. 2016;17:1463–72. doi: 10.1016/j.celrep.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zaidi N, Swinnen JV, Smans K. ATP-citrate lyase: a key player in cancer metabolism. Cancer Res. 2012;72:3709–14. doi: 10.1158/0008-5472.CAN-11-4112. [DOI] [PubMed] [Google Scholar]
- 42.Wolf D, Rodova M, Miska EA, Calvet JP, Kouzarides T. Acetylation of beta-catenin by CREB-binding protein (CBP) J Biol Chem. 2002;277:25562–7. doi: 10.1074/jbc.M201196200. [DOI] [PubMed] [Google Scholar]
- 43.Levy L, Wei Y, Labalette C, Wu Y, Renard CA, Buendia MA, et al. Acetylation of beta-catenin by p300 regulates beta-catenin-Tcf4 interaction. Mol Cell Biol. 2004;24:3404–14. doi: 10.1128/MCB.24.8.3404-3414.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vasan K, Werner M, Chandel NS. Mitochondrial metabolism as a target for cancer therapy. Cell Metab. 2020;32:341–52. doi: 10.1016/j.cmet.2020.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bordt EA, Clerc P, Roelofs BA, Saladino AJ, Tretter L, Adam-Vizi V, et al. The putative Drp1 inhibitor mdivi-1 is a reversible mitochondrial complex I inhibitor that modulates reactive oxygen species. Dev Cell. 2017;40:583–94. doi: 10.1016/j.devcel.2017.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–56. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toyama EQ, Herzig S, Courchet J, Lewis TL, Jr, Loson OC, Hellberg K, et al. Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science. 2016;351:275–81. doi: 10.1126/science.aab4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song JE, Alves TC, Stutz B, Sestan-Pesa M, Kilian N, Jin S, et al. Mitochondrial fission governed by Drp1 regulates exogenous fatty acid usage and storage in hela cells. Metabolites. 2021;11:322. [DOI] [PMC free article] [PubMed]
- 49.Benador IY, Veliova M, Mahdaviani K, Petcherski A, Wikstrom JD, Assali EA, et al. Mitochondria bound to lipid droplets have unique bioenergetics, composition, and dynamics that support lipid droplet expansion. Cell Metab. 2018;27:869–85. doi: 10.1016/j.cmet.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.