Abstract
We describe the utilization of a red fluorescent protein (DsRed) as an in vivo marker for Saccharomyces cerevisiae. Clones expressing red and/or green fluorescent proteins with both cytoplasmic and nuclear localization were obtained. A series of vectors are now available which can be used to create amino-terminal (N-terminal) and carboxyl-terminal (C-terminal) fusions with the DsRed protein.
Green fluorescent protein (GFP) is a powerful tool for identifying the subcellular localization of proteins and to monitor gene expression. The protein is capable of producing a strong green fluorescence when excited by blue light, without any exogenously added substrate or cofactor (3). Events inside living cell can thus be visualized in a noninvasive way (3, 12, 16). For Saccharomyces cerevisiae, a series of plasmids has been developed for the expression of N-terminal and C-terminal in-frame fusions with the protein of interest (4, 5, 16). These so-called pUG vectors have proved useful for localization and expression studies in this yeast.
Recently, fluorescent proteins have been described that emit light with a wavelength different from that of GFP. These comprise blue-, cyan-, and yellow-shifted mutants of GFP and the newly isolated DsRed (1, 6, 20). The latter is a red-emitting fluorescent protein. The longer wavelength of the emitted light minimizes problems associated with light scattering and auto-fluorescence of the cells (21, 23). Fluorescent proteins with different emission colors are valuable for in vivo multilabeling experiments, allowing comonitoring of several events (8, 22).
In this study, we show the use of the red fluorescent protein DsRed as a reporter in yeast cells. Based on the pUG plasmids, a new set of vectors expressing DsRed was constructed, allowing the production of amino-terminal (N-terminal) and carboxyl-terminal (C-terminal) fusion proteins. Our results indicate that DsRed can be expressed in S. cerevisiae, and the protein can be targeted specifically to the nucleus. Finally, we show that cells with nuclei labeled with either red or green fluorescent proteins can be used to follow mating in vivo.
The isogenic strains W303-1A (Mata) and W303-1B (Matα) of S. cerevisiae (ade2-1 his3-11,15 ura3-1 leu2-3,112 trp1) were used. Escherichia coli XL1-Blue was used as the bacterial host for plasmids (2). E. coli strains were grown in Luria-Bertani medium (LB) at 37°C (19). Yeast strains were grown in YPD (17) or, for selective purposes, in a synthetic medium (24).
The plasmid pR1 was constructed by digesting the vector pUG36 with XbaI. The product thus obtained was ligated to the 681-bp XbaI fragment of the pDsRed vector (Clontech). U. Güldener and J. H. Hegemann, Düsseldorf, Germany, kindly provided all pUG vectors used in this study. The sequences of all these vectors are available in the MIPS website (http://www.mips.biochem.mpg.de/proj/yeast/info/tools/hegemann/gfp.html).
The yEGFP3 gene was removed from the plasmids pUG34 and pUG36 by digestion with XbaI followed by self-ligation. Two plasmids were obtained pSL34 and pSL36, respectively. The DsRed gene was obtained by PCR on the vector pDsRed with the primers P1 and P2 (Table 1). After digestion with BamHI and XbaI, the fragment was ligated into pSL34 or pSL36 digested with the same enzymes. The resulting plasmids were named pUR34 and pUR36, respectively. pUR23 and pUR35 were obtained similarly by cloning a ClaI- and XhoI-digested DsRed gene PCR product obtained by using primers P3 and P4 (Table 1) into the ClaI- and XhoI-digested plasmids pUG34 and pUG36, respectively. The vectors pUR34NLS, pUR36NLS, and pUG36NLS were obtained by ligating an NLS1 fragment into the BamHI- and XhoI-digested vectors pUR34, pUR36, and pUG36, respectively. The NLS1 fragment was obtained as follows. The oligonucleotide O1 and O2 (Table 1) were mixed, boiled for 5 min, and then cooled overnight in a water bath. The vectors pUR23NLS, and pUR35NLS were obtained mainly as described above. In these constructs we used the vectors pUR23 and pUR35, the enzymes BamHI and SalI, and the oligonucleotides O3 and O4 (Table 1). DNA manipulations and transformation procedures were performed as described elsewhere (7, 10, 18, 19).
TABLE 1.
Sequences of the oligonucleotides used in the constructions of the plasmids
| Name | Sequence (5′-3′) |
|---|---|
| P1 | GCTCTAGAATGAGGTCTTCCAAGAATGTT |
| P2 | CGGGATCCAAGGAACAGATGGTGGCGTCC |
| P3 | CCGCTCGAGCATAAGGAACAGATGGTGGCGTCC |
| P4 | CCATCGATACCGTCGACATGAGGTCTTCCAAGAATGTT |
| O1 | GATCGCCAAAAAAGAAGAGAAAGGTCGTTGTTAAATAG |
| O2 | TCGACTATTTAACAACGACCTTTCTCTTCTTTTTTGGC |
| O3 | GATCCATGCCAAAAAAGAAGAGAAAGGTCGTTGTTAAAT |
| O4 | TCGAATTTAACAACGACCTTTCTCTTCTTTTTTGGCATG |
Cells collected from middle of the exponential growth phase were used for fluorescence microscopy analysis. For colocalization of DsRed, yEGFP3, and DNA, the cells were fixed as follows. Approximately 109 cells were incubated in 100 mM phosphate buffer (pH 6.5) containing 3.7% formaldehyde at room temperature during 2 h. Fixation was also performed with 70% ethanol at 4°C during 30 min. To stain DNA, the fixed cells were resuspended in 100 mM phosphate buffer containing 4′, 6′-diamidino-2-phenylindole (DAPI; 0.5 μg/ml) at room temperature for 10 min. The stained cells were washed twice and subjected to fluorescence microscopy. For fluorescence microscopy a Zeiss Axioplan microscope equipped with a mercury lamp and a 510–560 FT 580 (excitation), LP 590 (emission) filter set was used (17). The same microscope coupled to a Bio-Rad 1024 system was used for confocal laser-scanning microscopy with excitation by the 568-nm line of a krypton-argon laser and using an emission filter LP 585.
Expression and subcellular localization of the DsRed protein.
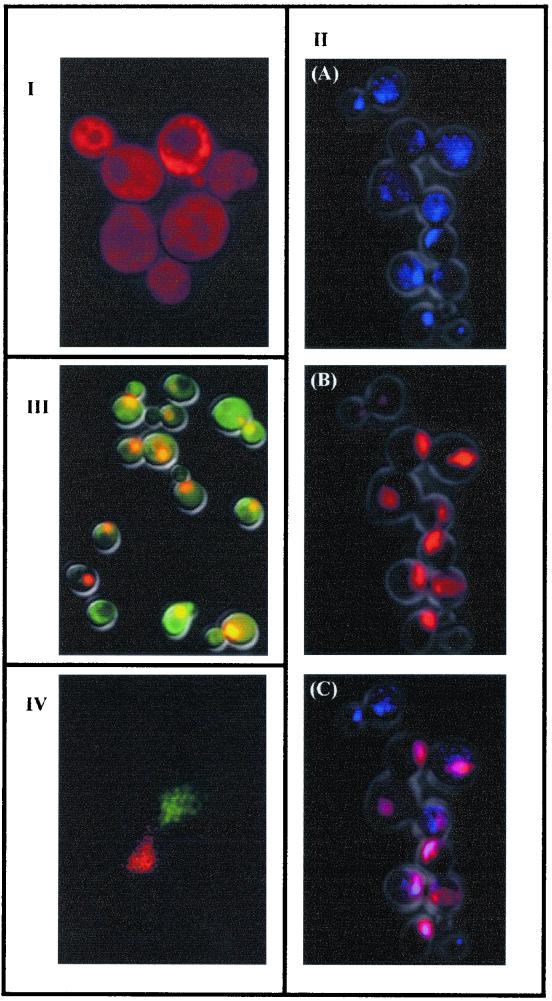
To assess whether the DsRed gene was expressed in S. cerevisiae, the yEGFP3 gene in the vector pUG36 was replaced by DsRed (4). When the resulting plasmid, pR1, was introduced into S. cerevisiae, the cells emitted a red bright fluorescence when illuminated with UV light (Fig. 1I). We developed a similar set of vectors, based on the pUG plasmids, expressing a red fluorescent protein, DsRed. pUR23 and pUR35 allow the production of C-terminal fused proteins and contain as selective markers the HIS3 and URA3 genes, respectively. The vectors for N-terminal fusions were called pUR34 (HIS3) and pUR36 (URA3). While this study was in preparation, Baird and coworkers reported the multimeric nature of DsRed (1, 9). For some purposes, this oligomerization can be troublesome since it may lead to misinterpretation of the results. Nevertheless, the new set of DsRed vectors that we described may be very useful, since genes cloned on pUG vector can be easily transferred to pUR vectors and vice versa. This allowed us to test whether DsRed oligomerization interferes with trafficking of the host protein. To use DsRed as a reporter gene, the resulting protein should (i) not carry any specific target information, (ii) result in a functional protein when fused in frame, and (iii) not be toxic when expressed in S. cerevisiae cells. The in silico analysis of the coding sequence of DsRed and yEGFP3 did not show significant differences with respect to the predicted localization (data not shown). Our data demonstrated that when DsRed was expressed in S. cerevisiae a bright red fluorescence was spread uniformly over the cell (Fig. 1I). This result is in agreement with the localization of the protein in the cytoplasm, indicating that the protein does not have its own functional targeting signal in this organism. The expression of DsRed did not appear to have any toxic effect on cell growth, as judged by specific growth rates. In addition, ca. 90% of the cell population emitted a red bright signal, and this signal was stronger in cells grown in liquid media, with high rates of oxygenation, than in cells from solid media.
FIG. 1.
Fluorescent microscopy of S. cerevisiae cells: I, transformed with the pR1 vector; II, harboring the vector pUR36NLS, showing DNA counterstained with DAPI in blue (A), red fluorescence of DsRed-NLS in red (B), and the overlap of both colors (colocalization) in violet (C); III, harboring the vectors pUR36NLS and pUG34 (IV). Confocal laser scanning microscopy of S. cerevisiae cells during mating. The different mating types are expressing yEGFP3-NLS or DsRed-NLS. The fluorescences of both yEGFP3-NLS and yEGFP3 are coded in green, and those of DsRed-NLS and DsRed are coded in red.
The NLS (PKKKRKV132) of the simian virus 40 large T antigen was used as a single cluster for nuclear targeting of DsRed in S. cerevisiae (11, 13). A dimer of oligonucleotides encoding this nuclear localization signal (NLS) was cloned in frame in all pUR, resulting in a redistribution of the fluorescent signal to a single subcellular compartment (Fig. 1IIB). As expected, counterstaining of the cells with DAPI confirmed the nuclear localization of DsRed-NLS (Fig. 1II A, -B, and -C) and yEGFP3-NLS (data not shown). Mozdy et al. have recently used the DsRed protein with a mitochondrial target in S. cerevisiae (15).
The fluorescence of the DsRed persisted after treatment of the cells with either ethanol or formaldehyde. This allows examination of preparations fixed with these chemicals and the utilization of antibodies. Clontech commercializes antibodies for DsRed protein that are specific and do not recognize yEGFP3.
Double labeling and potential applications.
Clones of S. cerevisiae expressing DsRed-NLS and yEGFP3 showed emission of both fluorescences (Fig. 1III). These cells showed green fluorescence in the cytoplasm and red, orange, or yellow fluorescence in the nuclei. The different colors of the nucleus may be attributed to colocalization of the two proteins. It has been demonstrated that some GFP can diffuse into the nuclei without any signal (22). Nevertheless, differences in the superposition of the green and red fluorescence in the cytoplasm and nuclei may occur. This fact may also explain the observed range colors of nuclei. Reinforcing this idea, cells expressing both DsRed-NLS and yEGFP3-NLS gave indeed rise to bright yellow fluorescent nuclei (data not shown).
Taking advantage of the double labeling, we followed mating in vivo. One of the mating types of the strain S. cerevisiae W303 was transformed with plasmid expressing DsRed-NLS, and the other was transformed with plasmid expressing yEGFP3-NLS, and conjugation was induced. Time-lapse digital imaging in a confocal laser-scanning microscope allowed tracking the dynamics of the mating process in S. cerevisiae. An example of the images obtained is shown in Fig. 1IV, where both nuclei have moved toward the shmoo tips. Our data demonstrated that DsRed-NLS can be used to stain the nuclei of live cells in a manner that is compatible with the use of yEGFP3 and confocal laser-scanning microscopy. This has proven to be difficult with the existing chemical fluorescent dyes for the nucleus (M. van Hemert, unpublished data). In conclusion, our data show that DsRed is well suited as a reporter gene for S. cerevisiae and is an alternative to those techniques that modify cell structure. Moreover, it allows studying dynamic processes in vivo. However, the slow maturation of DsRed (1, 9) can limit its use under the control of an inducible promoter. If we take all of the results together, DsRed should be envisaged more as an addition tool than as a substitute for yEGFP3 in expression studies.
In conclusion, DsRed, together with yEGFP3, can be used to study competition between different strains or yeast species in confined environments. This methodology also emerges as a powerful technique to investigate fusion events of organelles during mating. In fact, we are using this technique to assess whether nuclear fusion occurs during the conjugation of Zygosaccharomyces bailii prior to sporulation, since the production of spores in this species does not seem to be a product of meiosis (14).
Acknowledgments
We are especially grateful to Gerda Lamers for technical support with the confocal laser scanning microscopy experiments.
This study was supported by a research grant (contract PRAXIS XXI P/AGR/11135/98). Fernando Rodrigues was a recipient of a fellowship from PRAXIS XXI (Fundaçāo para a Ciência e Tecnologia).
REFERENCES
- 1.Baird G S, Zacharias D A, Tsien R Y. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci USA. 2000;97:11984–11989. doi: 10.1073/pnas.97.22.11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bullock W O, Fernandez J M, Short J M S. XL1-Blue: a highly efficient plasmid transforming recA Escherichia coli strain with β-galactosidase selection. BioTechniques. 1987;4:376–378. [Google Scholar]
- 3.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–803. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 4.Cormack B P, Bertram G, Egerton M, Gow N A, Falkow S, Brown A J. Yeast-enhanced green fluorescent protein (yEGFP) a reporter of gene expression in Candida albicans. Microbiology. 1997;143:303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- 5.Craven R A, Griffiths D J, Sheldrick K S, Randall R E, Hagan I M, Carr A M. Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene. 1998;221:59–68. doi: 10.1016/s0378-1119(98)00434-x. [DOI] [PubMed] [Google Scholar]
- 6.Ehrig T, O'Kane D J, Prendergast F G. Green-fluorescent protein mutants with altered fluorescence exitation spectra. FEBS Lett. 1995;367:163–166. doi: 10.1016/0014-5793(95)00557-p. [DOI] [PubMed] [Google Scholar]
- 7.Gietz R D, Schiestl R H. Transforming yeast with DNA. Methods Mol Cell Biol. 1995;5:255–269. [Google Scholar]
- 8.Haseloff J. GFP variants for multispectral imaging of living cells. Methods Cell Biol. 1999;58:139–151. doi: 10.1016/s0091-679x(08)61953-6. [DOI] [PubMed] [Google Scholar]
- 9.Heikal A A, Hess S T, Baird G S, Tsien R Y, Webb W W. Molecular spectroscopy and dynamics of intrinsically fluorescent proteins: coral red (dsRed) and yellow (Citrine) Proc Natl Acad Sci USA. 2000;97:11996–12001. doi: 10.1073/pnas.97.22.11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inune H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 11.Makkerh J P S, Dingwall C, Laskey R A. Comparative mutagenesis of nuclear localization signals reveals the importance of neutral and acidic amino acids. Curr Biol. 1996;6:1025–1027. doi: 10.1016/s0960-9822(02)00648-6. [DOI] [PubMed] [Google Scholar]
- 12.Mayer G, Launhardt H, Munder T. Application of the green fluorescent protein as a reporter for Ace1-based, two-hybrid studies. BioTechniques. 1999;27:86–84. doi: 10.2144/99271st01. [DOI] [PubMed] [Google Scholar]
- 13.Miyamoto Y, Imamoto N, Sekimoto T, Tachibana T, Seki T, Tada S, Enomoto T, Yoneda Y. Differential modes of nuclear localization signal (NLS) recognition by three distinct classes of NLS receptors. J Biol Chem. 1997;272:26375–26381. doi: 10.1074/jbc.272.42.26375. [DOI] [PubMed] [Google Scholar]
- 14.Mollapour M, Piper P W. Targeted gene deletion in Zygosaccharomyces bailii. Yeast. 2001;18:173–186. doi: 10.1002/1097-0061(20010130)18:2<173::AID-YEA663>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 15.Mozdy A D, McCaffery J M, Shaw J M. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niedenthal R K, Riles L, Johnston M, Hegemann J H. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 17.Pringle J R, Adams A E, Drubin D G, Haarer B K. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- 18.Rodrigues F, Zeeman A-M, Alves C, Sousa M J, Steensma H Y, Côrte-Real M, Leāo C. Construction of a genomic library of the food spoilage yeast Zygosaccharomyces bailii and isolation of the β-isopropylmalate dehydrogenase gene (ZbLEU2) FEMS Yeast Res. 2000;1407:1–5. doi: 10.1111/j.1567-1364.2001.tb00014.x. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1998. [Google Scholar]
- 20.Tsien R Y. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 21.Tsien R Y. Rosy down for fluorescent proteins. Nat Biotechnol. 1999;17:954–957. doi: 10.1038/13648. [DOI] [PubMed] [Google Scholar]
- 22.von Arnim A G, Deng X W, Stacey M G. Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene. 1998;221:35–43. doi: 10.1016/s0378-1119(98)00433-8. [DOI] [PubMed] [Google Scholar]
- 23.Wildt S, Deuschle U. cobA, a red fluorescent transcriptional reporter for Escherichia coli, yeast, and mammalian cells. Nat Biotechnol. 1999;17:1175–1178. doi: 10.1038/70713. [DOI] [PubMed] [Google Scholar]
- 24.Zonneveld B J M. Cheap and simple yeast media. J Microbiol Methods. 1986;4:287–291. [Google Scholar]