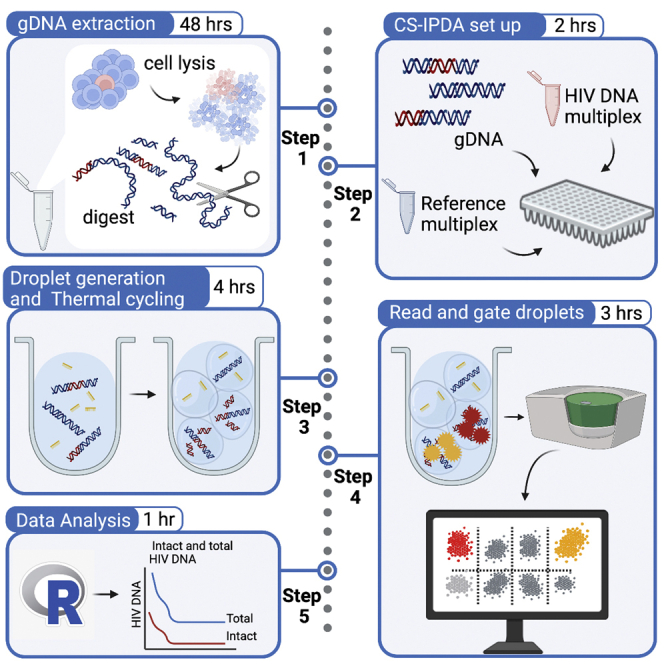
Summary
The cross-subtype intact proviral DNA assay (CS-IPDA) is a high-throughput method to quantify HIV reservoir size in populations infected with any of the dominant global HIV-1 subtypes. Our protocol includes genomic DNA isolation optimized to minimize DNA shearing, a reference droplet digital PCR (ddPCR) assay to quantify T cells and assess DNA shearing, and a multiplex ddPCR targeting three distinct regions across the HIV genome to quantify intact proviruses as an estimate of replication-competent proviruses in the reservoir.
For complete details on the use and execution of this protocol, please refer to Cassidy et al. (2022).
Subject areas: Genomics, High throughput screening, Microbiology, Molecular biology
Graphical abstract
Highlights
-
•
CS-IPDA permits high-throughput reservoir quantification across global HIV subtypes
-
•
gDNA extraction with minimal DNA shearing is crucial for detecting intact HIV provirus
-
•
Custom R code allows for high-throughput analysis of CS-IPDA experiments
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
The cross-subtype intact proviral DNA assay (CS-IPDA) is a high-throughput method to quantify HIV reservoir size in populations infected with any of the dominant global HIV-1 subtypes. Our protocol includes genomic DNA isolation optimized to minimize DNA shearing, a reference droplet digital PCR (ddPCR) assay to quantify T cells and assess DNA shearing, and a multiplex ddPCR targeting three distinct regions across the HIV genome to quantify intact proviruses as an estimate of replication-competent proviruses in the reservoir.
Before you begin
Timing: 0.5–4 h
The CS-IPDA multiplex utilizes genomic DNA (gDNA) template extracted from either fresh or cryopreserved human PBMCs collected from individuals living with HIV infection. This protocol utilizes an optimized gDNA extraction method to minimize DNA shearing (Wiegand et al., 2017; Green and Sambrook, 2012). When using cryopreserved samples, we strongly recommend using cells for gDNA extraction that have not previously been thawed, as minimal DNA shearing is critical for accurately estimating the size of the potentially intact HIV DNA reservoir. The protocol includes 2 independent multiplex droplet digital PCR (ddPCR) assays: 1) primers and probes that target multiple HIV DNA targets are pooled into a single ddPCR to quantify the intact and total HIV DNA proviruses and 2) an RPP30/deltaD multiplex reaction as a reference assay to quantify the number of cells interrogated and normalize HIV DNA copy counts.
Before you begin, we recommend you first gather the materials listed below in the key resources table and the materials and equipment section. We also suggest that you prepare gDNA positive and negative controls, plasmid controls, and HIV target and deltaD probe mixes as follows. All pre-prepared gDNA, plasmid, and probe mixes can be aliquoted and stored at −20°C until use.
Institutional permissions
This study was approved by the ethical review committees at Kenyatta National Hospital, the University of Washington, and the Fred Hutchinson Cancer Research Center. Institutional Review Board (IRB) approval to work with human specimens may be required prior to starting this protocol. Refer to your institution’s regulations and ensure that all documents are complete before collecting samples or data.
Prepare J-lat gDNA positive controls
Cell lines containing provirus, such as J-lat 5A8 or J-lat 8.4 (one copy per cell; Chan et al., 2013), serve as positive controls for gDNA extraction and HIV DNA quantification (see key resources table). To make positive control mixes for CS-IPDA and RPP30/deltaD reference assay, combine J-lat cells and HIV-negative PBMCs at a ratio of 1:1,000 cells and dilute to 2 × 10 6 cells/mL in freezing medium (90% FBS, 10% DMSO), and cryopreserve cells in 1 mL aliquots until ready for use.
Prepare HIV-negative human gDNA negative controls
Commercially prepared HIV-negative human gDNA can be conveniently purchased (ex. Promega Cat #G3041) or can be extracted from HIV-negative human PBMCs (see key resources table) using the following protocol (Step-by-step 1–28). Prior to being used as a control for CS-IPDA and RPP30/deltaD experiments, HIV-negative gDNA must be enzyme digested with BgII (NEB; see key resources table) as described below (steps 18–28).
Prepare plasmid controls
This protocol utilizes plasmids containing single or multiple CS-IPDA HIV subtype B target sequences as controls for gating the CS-IPDA data. All plasmid controls are prepared following the protocol detailed in Levy et al. (2021a, 2021b). Agar stabs of plasmids for culture and isolation can be ordered from Addgene (see Catalog numbers in key resources table) or lyophilized plasmids for resuspension can be ordered via IDT using the plasmid sequences published on Addgene.
-
1.
Linearize control plasmid stocks (resuspended or isolated from culture) using restriction enzyme AatII (key resources table) following manufacturer’s instructions for enzyme digestion (New England BioLabs).
-
2.
Gel-isolate linearized control plasmids. A variety of methods may be used; we suggest QIAquickⓇ PCR & Gel Cleanup Kit (QIAGEN) (See key resources table).
-
3.
Dilute linearized control plasmids with water to a final concentration of approximately 2,500 copies/μL for use in CS-IPDA experiments.
Note: We recommend completing a preliminary CS-IPDA on serially diluted plasmid preps to optimize results since each plasmid preparation may be variable in yield.
-
4.
Make the all-target plasmid mix control by pooling equal masses of each of the 7 linearized control plasmids and mixing 4:1 with HIV-negative human gDNA (see key resources table) that has been BgII digested and ethanol precipitated, as described in steps 18–28.
-
5.
Make the triple positive plasmid control by mixing plasmid 167347, Seattle IPDA_control_1_001 (see key resources table) 4:1 with HIV-negative human gDNA (see key resources table) that has been BgII digested and ethanol precipitated, as described in steps 18–28.
-
6.
Store all plasmid stocks and plasmid controls at −20°C in 100 μL aliquots.
Prepare CS-IPDA and RPP30 reference assay probe mixes for ddPCR
Each CS-IPDA HIV target and the deltaD target for the RPP30 reference assay has a unique probe mixture to be prepared prior to ddPCR reaction set up. Probe mixtures are prepared by pooling 100 μM stocks of custom-ordered individual primers (Thermo Fisher Scientific) and probes (IDT) (see key resources table), as detailed below. Both 5′ RPP30 and 3′ RPP30 probe mixes used in the reference assay are ordered from IDT as custom pre-mixed PrimeTimeⓇ Std qPCR stocks of lyophilized primers and probes (See key resources table).
-
7.
Prepare separate probe mixes for each of the following targets: deltaD, 5′ pol, LTR/gag, env (Tables 1 and 2). Pool probe mix reagents (Table 3) and vortex well.
-
8.
Resuspend lyophilized 5′ RPP30 and 3′ RPP30 PrimeTimeⓇ Std qPCR probe mixes to make a 40× working stock. Vortex well and incubate at 20°C–25°C for at least 30 min.
Pause point: Use probe mixes immediately for CS-IPDA experiment or aliquot (≤200 μL) and store at −20°C for ≤2 years.
Table 1.
CS-IPDA primer and probe sequences
| CS-IPDA target | Primer | Sequence | Probe dye, quencher |
|---|---|---|---|
| 5′ LTR/ gag | Forward | GACTAGCGGAGGCTAGAAGGAGAGA | |
| Probe | AT+G+GGT+GC+GAGA | HEX, BHQ | |
| Reverse | CTAATTTTCCSCCDCTTAATAYTGACG | ||
| 5′ pol | Forward | WCCYTTARYTTCCCTCARATCACTCT | |
| Probe | TTG+G+CARC+GA+CC | FAM, BHQ | |
| Reverse | TACTGTATCATCTGCTCCTGTRTCTAAKAGAGCYTC | ||
| env | Forward | TVTTCMTTGGGTTCTTRGGAGCAGCAGG | |
| Probe | A+G+CA+CKA+T+G+GG | HEX, BHQ | |
| Reverse | GCACTATRCCAGACAATAVYTGTCTGGCCTGTACC |
+ indicates a locked nucleic acid base
Table 2.
RPP30/deltaD reference assay primer and probe sequences
| RPP30/deltaD target | Primer | Sequence | Probe dye, quencher |
|---|---|---|---|
| 5′ RPP30a | Forward | GCATATCAGGGTACAGCATAGG | |
| Probe | TCTGCTCGTTGTTAGTCACCAGCT | FAM, ZEN/3′ IBFQ | |
| Reverse | CTTCCCTCACGGCATATACTTC | ||
| 3′ RPP30a | Forward | GCTGTGTTTGCTCTCTTGATTT | |
| Probe | AATGTCTGTGACTGGGTTCTGGCT | HEX, ZEN/3′ IBFQ | |
| Reverse | GGTCTGTCCATGGCATCTTAT | ||
| deltaD | Forward | GCTGGCTGTAATGGGAATGT | |
| Probe | TGTGAAGATGTCTGTAGCCATCTTAT | FAM, BHQ | |
| Reverse | TAATGGCTTGATAAAGATAAGTGATCAT |
RPP30 probe mixes ordered as pre-mixed lyophilized stocks of primers and probe from Integrated DNA Technologies (IDT) (see key resources table).
Table 3.
Recommended reagents for target probe mixes
| Reagent | Final concentration | Amount |
|---|---|---|
| Target probe (100 μM) | 5 μM | 5 μL |
| Target forward primer (100 μM) | 16.6 μM | 16.6 μL |
| Target reverse primer (100 μM) | 16.6 μM | 16.6 μL |
| Molecular-grade water | N/A | 61.8 μL |
| Total | N/A | 100 μL |
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Human Peripheral blood mononuclear cells (HIV-negative) | Bloodworks Northwest | https://www.bloodworksnw.org |
| Chemicals, peptides, and recombinant proteins | ||
| Buffer EB | QIAGEN | Cat# 19086 |
| 3 M Guanidine HCl | Thermo Fisher Scientific | Cat# 24115- 200 |
| 6 M Guanidinium isothiocyanate | Sigma | Cat# 50983 |
| Proteinase K solution, 20 mg/mL, > 318 mAu/mL at 30 C | QIAGEN | Cat# 19131 |
| Isopropanol (molecular biology grade) | Fisher Scientific | Cat# BP2618500 |
| Ethanol, 200 proof (molecular biology grade) | Fisher Scientific | Cat#BP2818500 |
| 3 M Sodium acetate | Thermo Fisher Scientific | Cat# S210 |
| NEBuffer r3.1 | New England Biolabs | Cat# B6003S |
| AatII | New England Biolabs | Cat# R0117S |
| ddPCR Supermix for probes (no dUTP) | Bio-Rad | Cat# 1863024 |
| BgII | New England Biolabs | Cat# R0143S |
| Experimental models: Cell lines | ||
| J-Lat 8.4 cell line | NIH HIV Reagent Program | Cat# 9847-427; RRID: CVCL_8284 |
| Oligonucleotides | ||
| HIV-1 probes | Integrated DNA Technologies | Table 1 |
| HIV-1 primers | Thermo Fisher Scientific | Table 1 |
| RPP30 gene PrimeTimeⓇ Std qPCR Assay primer/probe mixes. (Levy et al., 2021a, 2021b) | Integrated DNA Technologies | Table 2 |
|
TRD (deltaD) gene probe (Zoutman et al., 2017) |
Integrated DNA Technologies | Table 2 |
|
TRD (deltaD) gene primers/ probes (Zoutman et al., 2017) |
Thermo Fisher Scientific | Table 2 |
| Recombinant DNA | ||
| Seattle_IPDA_control_1_001 | Levy et al. (2021a, 2021b) | Addgene ID: 167347 |
| Seattle_IPDA_control_6_002 | Levy et al. (2021a, 2021b) | Addgene ID: 167348 |
| Seattle_IPDA_control_9_003 | Levy et al. (2021a, 2021b) | Addgene ID: 167349 |
| Seattle_IPDA_control_14_004 | Levy et al. (2021a, 2021b) | Addgene ID: 167350 |
| Seattle_IPDA_control_17_005 | Levy et al. (2021a, 2021b) | Addgene ID: 167351 |
| Seattle_IPDA_control_29_006 | Levy et al. (2021a, 2021b) | Addgene ID: 167352 |
| Seattle_IPDA_control_32_007 | Levy et al. (2021a, 2021b) | Addgene ID: 167353 |
| Software and algorithms | ||
| QuantaSoft Software (version 1.7) | Bio-Rad | Cat# 1864011 |
| QuantaSoft Analysis Pro (version 1.0.596.0525) | Bio-Rad | https://www.bio-rad.com/en-us/product/qx200-droplet-digital-pcr-system |
| R (version 4.0.0) | R project | http://www.r-project.org/ |
| Microsoft Office | Microsoft | https://www.microsoft.com/en-us/microsoft-365 |
| Other | ||
| QIAquickⓇ PCR & Gel Cleanup Kit | QIAGEN | Cat# 28506 |
| Vortex | N/A | N/A |
| Wide-bore p1000 pipette tips | Rainin | Cat# 30389166 |
| Wide-bore p200 pipette tips | Rainin | Cat# 30389241 |
| Micro-centrifuge for 1.5–2.0 mL tubes | N/A | N/A |
| Heat block for micro-tubes (1.5–2.0 mL) | N/A | N/A |
| Nanodrop 2000 Spectrophotometer | Thermo Scientific | Cat## ND-2000-US-CAN |
| ddPCR 96-well plates | Bio-Rad | Cat# 12001925 |
| DG8 gasket | Bio-Rad | Cat# 1863009 |
| DG8 Cartridge holder | Bio-Rad | Cat# 1863051 |
| DG8 Droplet generation cartridge | Bio-Rad | Cat# 1864008 |
| PCR Plate Heat Seal, foil, pierceable | Bio-Rad | Cat# 1814040 |
| Bio-Rad PX1 PCR Plate Sealer | Bio-Rad | Cat# 1814000 |
| Droplet generation oil for probes | Bio-Rad | Cat# 1863005 |
| Thermal Cycler (96 well plate compatible) | N/A | N/A |
| ddPCR droplet reader oil | Bio-Rad | Cat# 1863004 |
| QX200 Droplet generator | Bio-Rad | Cat# 1864002 |
| QX200 Droplet reader | Bio-Rad | Cat# 1864003 |
Materials and equipment
Heat Blocks: This protocol requires the use of heat blocks for genomic DNA extraction and preparation. The use of a shaking heat block during the restriction enzyme digest may be used, but is not required.
QX200 Droplet Digital PCR System from Bio-Rad: Reference manuals for the QX200 or automated QX200 AutoDG Droplet generators, PX1 PCR plate sealer, QX200 Droplet reader, and accompanying software are available from the Bio-Rad website (https://www.bio-rad.com/en-us/life-science/digital-pcr/qx200-droplet-digital-pcr-system). Note that purchase of QuantaSoft Software (version 1.7) is required to use the QX200 Droplet reader, but QuantaSoft Analysis Pro is required for analysis of .qlp files from multiplexed experiments. QuantaSoft Analysis Pro is available for free download to Bio-Rad account holders (www.bio-rad.com). Both QuantaSoft programs are compatible with Windows 7 operating systems.
Additional information about ddPCR experiment planning, data analysis, and mathematical details for applying ddPCR data can be found in the Droplet Digital PCR Applications Guide (https://www.bio-rad.com/sites/default/files/webroot/web/pdf/lsr/literature/Bulletin_6407.pdf).
Step-by-step method details
Extracting genomic DNA from human blood mononuclear cells
Timing: 4 h (2 h hands-on) if stopping prior to restriction enzyme digestion
Genomic DNA is extracted from cryopreserved human PBMCs using a protocol that reduces shearing of gDNA. Minimally sheared gDNA is critical for accurately estimating the size of the intact HIV DNA reservoir.
-
1.Thaw cryopreserved cells at 20°C–25°C. Invert tubes generously to mix cells.
-
a.While cells thaw, pre-heat two heat blocks to 42°C and 56°C.
-
b.Bring all reagents to 20°C–25°C.
-
a.
Note: If 3 M guanidine HCl or 6 M guanidine isothiocyanate contain precipitates, place in a 37°C water bath to dissolve.
-
2.
Transfer approximately ≤1 × 106 cells to 1.5 mL conical tubes (“Eppendorf”). If >1 × 106 cells have been thawed, split the cells between two conical tubes so that each tube contains ≤1 × 106 cells.
CRITICAL: Dispose of tips following local or institutional biohazardous waste procedures.
Note: We recommend using ≤2 × 106 T cells or PBMCs per extraction (2 × 106 T cells or PBMCs will yield approximately 1–2 μg gDNA). Utilizing too many cells in this gDNA extraction protocol may results in incomplete cell lysis, diminished gDNA yield, incomplete digestion by BgII enzyme, and excessively viscous gDNA samples that can cause poor droplet generation when used in ddPCR experiments. If alternative cell types are being used, we recommend testing the gDNA extraction using a variety of different cell quantities to optimize the procedure. Extraction quality can be assessed using a Nanodrop to measure yield and the RPP30 reference assay to measure gDNA shearing. We recommend a final yield of 1–5 μg gDNA with <40% shearing.
-
3.Pellet cells via centrifugation at 2,400 × g for 5 min.
-
a.Remove supernatant without disrupting the cell pellet.
-
a.
CRITICAL: Dispose of supernatant and tips following local or institutional biohazardous waste procedures.
-
4.
Resuspend the cell pellet in 100 μL of 3 M guanidine HCl and 20 μL Proteinase K solution. Mix gently by pipetting with a wide-bore tip or by inverting the tubes 10 times if wide-bore tips are unavailable.
Note: It is not crucial that the entire cell pellet is resuspended as it will dissolve during upcoming incubation.
CRITICAL: Mix gently to minimize mechanical shearing.
CRITICAL: Tips used with 3 M guanidine HCl or 6 M guanidine isothiocyanate should not be bleached prior to disposal. Dispose of tips following local or institutional chemical waste disposal procedures.
-
5.
Incubate at 56°C for 1 h.
-
6.
Add 400 μL of 6 M guanidine isothiocyanate. Mix gently by pipetting with a wide-bore tip or by inverting the tubes 10 times if wide-bore tips are unavailable.
-
7.Incubate at 42°C for 10 min.
-
a.After removing the tubes following incubation, set the heat block to 65°C.
-
a.
-
8.
Add 1 mL of 100% molecular-grade isopropanol. Invert 10 times to mix.
-
9.
Centrifuge at 21,000 × g for 10 min.
-
10.
Remove supernatant leaving behind ∼100 μL to prevent disrupting the pellet.
CRITICAL: Supernatant containing 3 M guanidine HCl or 6 M guanidine isothiocyanate should be disposed of in a designated container for chemical waste disposal. (Disposal procedures may vary by institution or local requirements.).
-
11.
Add 750 μL of 70% ethanol. Invert 10 times to mix.
-
12.
Centrifuge at 21,000 × g for 2 min.
-
13.
Remove supernatant using a p1000 pipette tip. Follow with a p20 or p10 to completely remove any liquid surrounding pellet.
-
14.
Air dry pellets for approximately 5 min or until tubes are visibly dry. Do not over-dry pellets.
Note: Check for liquid by tipping the tube slightly.
-
15.
Dissolve pellet in 300 μL Buffer EB. Vortex tubes for 5 s to mix.
-
16.
Incubate at 65°C for 1 h.
-
17.Remove tubes from the heat block and incubate at 20°C–25°C for at least 30 min and up to 24 h to ensure extraction samples return to ambient temperature prior to enzyme digestion.
-
a.Prior to continuing to enzyme digestion steps, set the heat block to 37°C.
-
a.
Pause point: This is a safe stopping point. Extraction samples can be stored beyond 24 h up to 7 days at 4°C, if desired.
Restriction enzyme digestion of genomic DNA and ethanol precipitation
Timing: Day 1: 30 min hands-on with 16–18 h incubation. Day 2: 3 h hands-on
High molecular weight gDNA is enzyme digested using a restriction enzyme with no cut-site within the HIV genome to prepare for use in ddPCR reactions. Following digest, enzyme is removed from gDNA extractions via ethanol precipitation of gDNA.
-
18.
Thaw NEBuffer r3.1 at 20°C–25°C. Once thawed, vortex well.
-
19.
Add NEBuffer r3.1 and NEB BgII to each tube (Table 4). Invert tubes 10 times to mix.
Note: If preparing multiple samples, Buffer r3.1 and NEB BgII can be made into a master mix. Dispense 48.2 μL of master mix to each extraction.
-
20.Incubate 16–18 h at 37°C.
-
a.After removing the tubes following incubation, set the heat block to 65°C.
-
a.
Note: Place 100% ethanol in freezer prior to incubation for use in step 30.
-
21.
Add 35 μL 3 M sodium acetate and 766 μL of ice-cold 100% ethanol. Invert tubes 10 times to mix.
-
22.
Incubate on wet-ice for 30 min.
-
23.
Repeat steps 9–17, as described above.
-
24.
Add 50 μL Buffer EB. Vortex tubes for 5 s to mix.
-
25.
Incubate at 65°C for 1 h.
-
26.
Remove from heat block and incubate at 20°C–25°C for 1–18 h.
Note: High molecular weight gDNA can be extremely viscous. If viscosity makes pipetting difficult, repeat BglI digestion and ethanol precipitation (steps 18–28) by bringing sample to 300 μL with Buffer EB. Highly viscous DNA can interrupt droplet formation in later ddPCR reactions.
Note: If an unknown number of cells were used for gDNA extraction, quantification of gDNA samples may be required. Input DNA for ddPCR reactions should not exceed 200 ng/μL as higher concentrations of DNA may interfere with droplet formation. Further, if DNA concentration exceeds >300 ng/μL, it is recommended that BglI digestion and ethanol precipitation are repeated (Levy et al., 2021a, 2021b).
-
27.
Prepare at least 20 μL of 1:10 diluted gDNA in molecular-grade water for use in the RPP30/deltaD reference assay.
CRITICAL: Data from the RPP30/deltaD reference assay are used to normalize the concentration of HIV DNA in the corresponding CS-IPDA sample well and infer sample quality. Ensuring sample gDNA is thoroughly mixed prior to dilution and the 1:10 diluted gDNA is thoroughly mixed prior to experiment set up is crucial.
-
28.
Genomic DNA samples and dilutions are ready for immediate use in a CS-IPDA experiment or can be stored at −20°C. Extracted gDNA can be aliquoted in small volumes to reduce number of freeze thaws that could shear gDNA.
Table 4.
Reagents for a single enzyme digest reaction of extracted gDNA
| Reagent | Final concentration | Amount |
|---|---|---|
| NEBuffer r3.1 (10×) | 1× | 35 μL |
| NEB BgII (10,000 IU/mL) | 380 IU/mL | 13.2 μL |
| gDNA extraction sample | N/A | 300 μL |
| Total | N/A | 348.2 μL |
Setting up CS-IPDA and RPP30 reference assay ddPCR
Timing: 1.5 h (for 96 reactions; requires less time for fewer reactions)
CS-IPDA and RPP30/deltaD multiplex ddPCR reactions are set-up to quantify total and intact copies of HIV DNA per million T cells using gDNA templates.
Note: Probe mixes for individual targets can be made and stored in advance of ddPCR experiments. Probes are sensitive to freeze-thaws, so we recommend probe mixes to be made and stored in small volume aliquots (≤200 μL). These aliquots can be safely stored at −20°C for ≤2 years.
Note: For HIV DNA quantification, we recommend testing samples by the CS-IPDA in triplicate and by the RPP30/deltaD reference assay in singlet (Figure 1). CS-IPDA reactions are highly reproducible; variability between replicate wells is expected to be <3-fold change.
-
29.
Thaw Supermix for probes (no dUTP) on wet ice and vortex well.
-
30.
Prepare RPP30/deltaD reference assay reaction mix (Table 5). Vortex to mix and keep on wet ice.
Note: Thaw pre-aliquoted probe mixes on wet ice and mix thoroughly.
-
31.
Prepare CS-IPDA reaction mix (Table 6). Vortex to mix and keep on wet-ice.
Note: Thaw pre-aliquoted probe mixes on wet-ice and mix thoroughly.
-
32.
Distribute 18 μL RPP30/deltaD reaction mix to designated wells.
Optional: Refer to Figure 1 for suggested plate layout.
-
33.
Add 2 μL of 1:10 diluted gDNA or RPP30/deltaD reference assay controls to wells containing RPP30/deltaD reaction mix. Pipette up and down to mix well.
Note: Try to avoid creating air bubbles while mixing reaction mix and template.
-
34.
Distribute 16 μL CS-IPDA reaction mix to designated wells.
Optional: Refer to Figure 1 for suggested plate layout.
-
35.
Add 4 μL of undiluted gDNA or CS-IPDA controls to wells containing CS-IPDA reaction mix.
Note: Try to avoid creating air bubbles while mixing reaction mix and template.
-
36.
Centrifuge plate for 10–20 s to bring contents to bottom of wells.
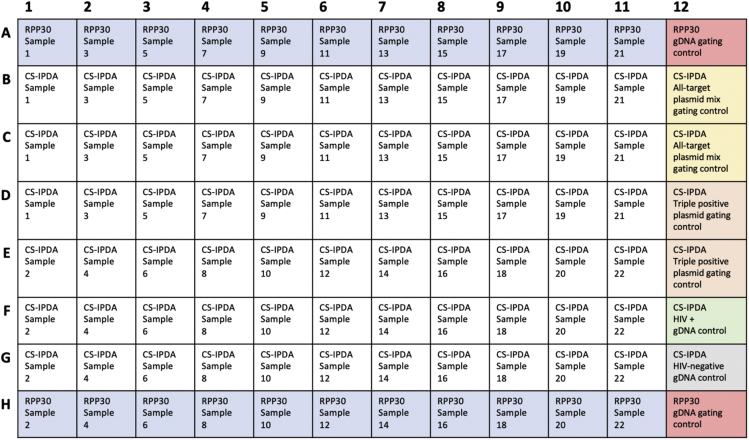
Figure 1.
Example plate layout for performing CS-IPDA and RPP30/deltaD reference assay in a single 96-well experiment
RPP30/deltaD reference assay reactions are completed in singlet (blue). CS-IPDA reactions are completed in triplicate (white). Experimental controls include all-target plasmid mix gating control (yellow), triple positive plasmid gating control (orange), HIV-positive control, such as J-lat 5A8 or 8.4 gDNA (green), HIV-negative gDNA control (gray), and RPP30 gDNA gating control, such as HIV-negative PBMCs or J-lat 5A8 or 8.4 gDNA (red).
Table 5.
Reagents of RPP30/deltaD reaction
| Probe mix (Dye)/ Reagent | Final concentration | Amount for single reaction | Amount for 96 well CS-IPDA plate |
|---|---|---|---|
| 5′ RPP30 probe mix (40×) (FAM) | 1× | 0.5 μL | 13.2 μL |
| 3′ RPP30 probe mix (40×) (HEX) | 1× | 0.5 μL | 13.2 μL |
| deltaD probe mix (FAM) | 0.25 μM | 1 μL | 26.4 μL |
| Supermix for probes (no dUTP) (2×) | 1× | 10 μL | 264 μL |
| Molecular-grade water | N/A | 6 μL | 158.4 μL |
| 1:10 diluted gDNA template | N/A | 2 μL | N/A |
| Total | N/A | 20 μL | 475.2 μL |
Table 6.
Reagents of CS-IPDA reaction
| Probe mix (Dye)/ Reagent | Final concentration | Amount for single reaction | Amount for 96 well CS-IPDA plate |
|---|---|---|---|
| env probe mix (HEX) | 0.225 μM | 0.90 μL | 67.5 μL |
| 5′ LTR/ gag probe mix (HEX) | 0.238 μM | 0.95 μL | 71.25 μL |
| 5′ pol probe mix (FAM) | 0.288 μM | 1.15 μL | 86.25 μL |
| Supermix for probes (no dUTP) (2×) | 1× | 10 μL | 750 μL |
| Molecular-grade water | N/A | 3 μL | 75 μL |
| gDNA template | N/A | 4 μL | N/A |
| Total | N/A | 20 μL | 1,050 μL |
Droplet generation and thermal-cycling incubation
Timing: 1 h hands-on to prepare for a 2.5 h incubation (for 96 reactions; requires less hands-on time for fewer reactions)
CS-IPDA and RPP30/deltaD ddPCR reactions are partitioned into droplets prior to PCR amplification of target regions.
-
37.
Prepare DG8 Droplet Generator Cartridges for droplet generation by inserting into plastic cartridge holders.
-
38.
Using a multi-channel pipette, transfer 20 μL of reaction mix from the first 8-well plate column to the middle row of wells on the cartridge.
CRITICAL: Dispense reaction mix against side of cartridge well slowly to avoid air bubbles. Air bubbles will prevent droplet formation.
-
39.
Using a multi-channel pipette, transfer 70 μL of Droplet generation oil for probes to the bottom row of wells on the cartridge.
-
40.
Apply a DG8 gasket to the cartridge and insert into QX200 Droplet Generator.
-
41.
Following droplet generation, transfer 40 μL of droplets to the first column of wells on a new ddPCR 96-well plate.
-
42.
Repeat steps 37–41 for all remaining samples, dispensing droplets into the corresponding wells on the ddPCR 96-well plate.
Note: Check wells for any droplet-generation failures. Wells containing droplets will have an opaque top layer.
-
43.
Heat-seal the droplet-containing plate with PCR Plate Heat Seal pierceable foil.
Incubate droplets on a thermal cycler using the PCR cycling protocol (Table 7).
Note: The ramp rate is set at 2°C for each step.
-
44.
Transfer droplet-containing plate from thermal cycler to QX200 Droplet reader.
Pause point: Following incubation, droplet containing plate can be stored up to 24 h at 4°C prior to droplet reading.
Table 7.
PCR cycling conditions
| Step | Temperature | Time | Cycles |
|---|---|---|---|
| Enzyme activation | 95°C | 10 min | 1 |
| Denaturing | 94°C | 30 s | 65 |
| Annealing and extension | 60°C | 60 s | |
| Enzyme deactivation | 98°C | 10 min | 1 |
| Hold | 4°C | Indefinitely | |
Droplet reading
Timing: 3.5 h (for 96 reactions; requires less time for fewer reactions). Droplet reading requires approximately 2 min per well
Droplets from PCR-amplified CS-IPDA and RPP30/deltaD reactions are analyzed individually using a droplet reader.
-
45.
Insert plate into QX200 Droplet reader. Follow manufacturer’s instructions for operating QX200 Droplet reader and QuantaSoft software (www.bio-rad.com).
-
46.Begin run by setting parameters as follows:
-
a.Define ‘Supermix’ as “Supermix (no dUTPs).”
-
b.Set ‘Experiment type’ as “ABS.”
-
c.Set ‘Dye(s) used’ as “FAM/HEX.”
-
d.Set both Channel 1 and Channel 2 to “Unknown.”
-
e.Inputting channel targets and sample names is optional.
-
a.
-
47.
Following completion of plate reading, dispose of the empty plate and droplet waste following disposal procedures as indicated by institution or local requirements.
Expected outcomes
The outcome of this gDNA extraction protocol if completed using ≤2 × 10ˆ6 cells is a 50 μL gDNA sample with a concentration of approximately 200 ng/μL (Levy et al., 2021a, 2021b). If uncertain about the number of cells used for extraction, DNA quantification by Nanodrop is suggested for optimal use of samples in RPP30/deltaD reference assay and CS-IPDA. Inputting too much DNA into ddPCR reactions can inhibit droplet formation or result in oversaturation of amplitude signals during droplet reading.
We have observed that the use of high-quality cells for gDNA extraction can result in highly “intact” gDNA samples with <10% of sample DNA having been mechanically sheared. We recommend using cells that have not been previously freeze-thawed, as this can cause shearing and will impact accurate quantification of intact HIV provirus.
Quantification and statistical analysis
Timing: 1–2 h
Overview
Analysis of data collected from the RPP30/deltaD reference assay and the CS-IPDA can be summarized into the following steps:
-
•
Gating and assigning target names to raw experiment data from the QX200 Droplet reader using QuantaSoft Analysis Pro software.
-
•Using R software,
-
○Analyze gated RPP30/deltaD reference assay data to quantify the number of cells interrogated, quantify the number of T cells interrogated, and measure DNA sample shearing.
-
○Analyze the gated CS-IPDA data to quantify both defective and potentially intact HIV DNA copies detected.
-
○Merge the RPP30/deltaD reference assay and CS-IPDA data sets to normalize HIV DNA copies to the number of interrogated cells or T cells.
-
○
CS-IPDA experiment analysis is completed using custom R code (Levy et al., 2021a, 2021b; Wickham, 2017; Hughes, 2016; Muller, 2020; Lai, 2020; Rstudio, 2015). The code and example data for analysis are publicly available (https://github.com/LehmanLab/CS-IPDA). Additionally, this protocol includes two tutorial videos to demonstrate gating raw experiment data and using the custom R code to analyze CS-IPDA experiment data (Methods videos S1 and S2).
Note: Alternatively, RPP30/deltaD reference assay and CS-IPDA experiments can be analyzed in Excel (Levy et al., 2021a, 2021b).
Note: Detailed information regarding the mathematical equations utilized for ddPCR multiplex analysis can be found in the Bio-Rad Droplet Digital PCR Applications Guide (https://www.bio-rad.com/sites/default/files/webroot/web/pdf/lsr/literature/Bulletin_6407.pdf) or as described by Levy et al. (2021a, 2021b).
Analysis of CS-IPDA and RPP30/deltaD reference assay data is completed using two scripts: CSIPDA_analysis.R and CSIPDA_helper_functions.R. Both scripts use gated data exported from QuantaSoft Analysis Pro software read in as two files: [CS-IPDA_experiment]_well_data.csv and [CS-IPDA_experiment]_ClusterData.csv file. The scripts also require a [CS-IPDA experiment]_plate_layout.csv to add sample metadata.
Data obtained through analysis with script includes:
-
•
Potentially “intact” HIV DNA copies per 1 × 106 cells or T cells.
-
•
HIV DNA single and dual target copies per 1 × 106 cells or T cells.
-
•
Total HIV copies (defective or intact) detected per 1 × 106 cells or T cells.
-
•
Samples with no intact copies of HIV DNA per 1 × 106 cells or T cells.
-
•
Samples with wells below threshold droplet count.
-
•
Total number of cells or T cells interrogated for each sample.
-
•
Concentration of cells or T cells per volume of gDNA for each sample.
-
•
DNA shearing index (DSI) and percent of unsheared DNA for all gDNA samples.
CS-IPDA and RPP30/deltaD cell reference assays can be completed in the same experiment or as separate experiments, so experiment data and metadata files may include both assays in the same file or be separated into multiple files. The R script can accommodate either scenario by permitting the input and merging of multiple experimental data files for a single analysis and output.
Note: The example CS-IPDA and RPP30/deltaD reference assay data available in our code repository are data collected for the same samples as two separate experiments.
Gating and assigning target names using QuantaSoft Analysis Pro
-
•Open the experiment .qlp data file in QuantaSoft Analysis Pro (Methods video S1).
-
○In the ‘Plate Editor’ tab,
-
-Set ‘Experiment Type’ as “Direct Quantification (DQ).”
-
-Set ‘Assay Information’ as “Amplitude Multiplex.”
-
-Assign targets (Table 8) by selecting the wells used for either RPP30/deltaD reference assay or CS-IPDA.
-
-Use the “-” button to remove “Target 4.”
-
-
-
○
-
•
Go to the “2D Amplitude” tab.
-
•Gate the RPP30/deltaD well data.
-
○Select the RPP30 control wells and select the purple “cross” tool (left panel) to show gates. Drag the purple lines to adjust.
-
○Select all RPP30 sample wells and apply the gates determined by the controls (Figure 2).
-
○
Note: Reactions completed using a high concentration of gDNA (>200 ng/uL) may result in clusters that span the amplitude plot diagonally. In this case, it is appropriate to manually determine the borders of target clusters using the “circle” gating tool. Additionally, the “Show Heat Map” option can be used to better visualize the borders of clusters when gating manually.
-
•Gate the CS-IPDA well data.
-
○Select the CS-IPDA control wells (all target plasmid mix, triple positive plasmid, HIV+ gDNA) and select the purple “cross” tool (left panel) to show gates. Drag the purple lines to adjust.
-
○Select all CS-IPDA sample wells and apply the gates determined by the controls (Figure 2).
-
○
-
•Select all wells on the plate to export gated data.
-
○Export “Well Data” to a .csv file by selecting the “options” icon in the upper right corner of the “Well Data” table. Name and save file as “[CS-IPDA_experiment]_well_data.csv.”
-
○Export “Cluster Data” to a .csv file by selecting “Analysis Tools” from the top panel of the screen, then selecting “Export Cluster Data” from the drop down menu. Name and save file as “[CS-IPDA_experiment]_ClusterData.csv”.
-
○
Note: If analyzing data in R, files must be saved in .csv format. When analyzing in Excel, files can be saved as either .csv or .xlsx formats.
Note: Exported “Cluster Data” will not contain any sample names that may have been inputted prior to droplet reading or in the ‘Plate Editor’ tab during analysis. Samples can be identified using well information.
Table 8.
Target assignments for CS-IPDA and RPP30/deltaD assays
| Assay | Target name | Signal Ch.1 | Signal Ch.2 |
|---|---|---|---|
| CS-IPDA | 5′ pol | FAM Lo | None |
| env | None | HEX Lo | |
| gag/LTR | None | Hex Hi | |
| RPP30/deltaD | 5′ RPP30 | FAM Hi | None |
| 3′ RPP30 | None | HEX Lo | |
| deltaD | FAM Lo | None |
Figure 2.
Example of ddPCR data analysis using QuantaSoft Analysis Pro software
(A) RPP30/deltaD reference assay wells are gated using extracted gDNA from provirus-containing cells mixed with HIV-negative PBMCs as a reference. Assay probe targets for each cluster are indicated by text.
(B) RPP30/deltaD gating thresholds determined by the control are applied to a clinical sample. Assay probe targets for each cluster are indicated by text.
(C) CS-IPDA gating thresholds are determined using a mixture of plasmids containing all targets.
(D) CS-IPDA gating thresholds applied to gDNA extracted from provirus containing cells (approximately 200 HIV DNA copies/reaction). Assay probe targets for each cluster are indicated by text. See also Methods video S1.
Analyzing data in R
-
•
Format a plate layout .csv file for the experiment including sample names, dilution factors, and date of experiment. Example plate layouts with appropriate formatting are available in our code repository (Methods video S2).
-
•
Save the required .csv data files (well_data.csv, plate_layout.csv, and ClusterData.csv) to a shared directory.
-
•Read data files into R and analyze all experiment data using available code (version v1.0).
-
○Refer to the README file provided with the example code and data for instructions for completing experiment analysis in R.
-
○
Note: The CS-IPDA analysis code includes quality control criteria to filter and exclude, or flag, the following:
-
•
CS-IPDA or RPP30/deltaD reaction wells with fewer than 5,000 droplets counted and interrogated. Wells containing <5,000 droplets will have inaccurate concentration estimates and are indicative of failed droplet generation (see troubleshooting, problem 3).
-
•
CS-IPDA sample replicates with calculated intact HIV copies per million cells > 3-fold different from other sample replicates. If a sample replicate is > 3-fold different, the sample will be flagged so an additional CS-IPDA replicate can be performed. If a sample replicate is >= 10-fold different from other sample replicates, replicate will be omitted from the data set and the sample will be flagged so an additional CS-IPDA replicate can be performed and used in downstream analyses.
-
•
Samples with <60% intact gDNA calculated using the RPP30/deltaD reference assay are considered “sheared” and should be omitted from downstream analyses.
-
•
Samples with no detected intact HIV copies per million T cells must have had at minimum 100,000 cells interrogated via CS-IPDA. If fewer than 100,000 PBMCs have been interrogated, additional replicates may be performed to bring the number of interrogated cells above this threshold or sample will be omitted from downstream analyses if >100,000 cells cannot be interrogated.
These criteria for data cleaning and quality control can be adapted and optimized, as needed.
Limitations
Genomic DNA sample quality and degree of shearing is dependent on cell quality prior to extraction (preservation method, number of freeze-thaws, cold-chain interruptions, long-term storage conditions). Extraction quality and subsequent accuracy of HIV DNA quantification may be less than ideal when using banked cell aliquots of cells in which any of the aforementioned conditions may be uncertain.
RPP30/deltaD reference assay and CS-IPDA replicates have shown high reproducibility with <3-fold variability in quantity of potentially “intact” HIV DNA copies per 1 × 106 T cells. Reproducibility between replicates may be diminished with the use of low quality gDNA (highly sheared, low concentration, high viscosity, high molecular weight) or when using clinical samples from individuals with low amounts of integrated provirus due to antiretroviral treatment.
Permissions and safety procedures for handling human cells may vary by institution. Please refer to your institution’s regulations and recommendations regarding safe lab practices and Institutional Review Board (IRB) approval requirements prior to using this protocol.
Troubleshooting
Problem 1
During gDNA extraction, no visible pellet is seen following centrifugation (Step-by-Step Methods steps: 3, 9–14).
Potential solution
Proceed with protocol using extra caution when removing supernatants. For extractions with low cell input/ low gDNA yield, pellets cannot always be seen by eye. In cases of low gDNA yield, extracted gDNA can often be used in downstream ddPCR reactions with success.
Problem 2
Pipetting gDNA is difficult; DNA is too viscous or clumpy (Step-by-Step Methods steps 27 and 35).
Potential solution
Bring gDNA to 300 μL by adding Buffer EB. Repeat BgII enzyme digestion and ethanol precipitation as described in Step-by-Step Methods (Step-by-Step Methods steps 18–28).
Problem 3
Droplet generation is resulting in wells without droplets or with very low droplets (no opaque layer on surface) or, during analysis, many sample wells are failing to meet the minimum droplet requirement. (Step-by-Step Methods steps 40–42, quantification and statistical analysis).
Potential solution
If droplet generation failure is consistent for a particular sample, the extracted gDNA may be too viscous. Refer to problem 2. If droplet generation is occurring inconsistently across samples or is occurring randomly, attempt to minimize air bubbles during ddPCR reaction set up and reaction mix transfer to DG8 Droplet Generator Cartridges. (For pipetting tips for sample transfer to cartridge, see the QX200 Droplet Generator Instruction Manual, page 7 (Bio-Rad).) Note that droplet generation is much more successful when ddPCR reaction mix is transferred into DG8 Droplet Generator Cartridges before Droplet generation oil is transferred. Further, the use of wide-bore pipette tips during droplet generation can be helpful to minimize air bubbles.
Problem 4
When gating, distinct target clusters are hard to distinguish. Clusters are taking on a diagonal shape at high amplitude. There are very few droplets in the expected “negative target” cluster or all “negative” droplets have a higher than anticipated amplitude intensity (quantification and statistical analysis).
Potential solution
The ddPCR reaction was run with too high a concentration of template. High amplitude cluster shifts or absence of “negative” droplets suggests the template concentration was high enough that most, or all, droplets contain a template copy. In some cases, droplets may contain multiple copies of template, resulting in high amplitude clusters. Repeat experiment using a lower concentration of template. For more guidance, see the Droplet Digital PCR Applications Guide (Bio-Rad).
Problem 5
There are no HIV DNA “intact” copies quantified for a sample prior to DSI-correction and normalization (Step-by-Step Methods step 8).
Potential solution
In some samples, HIV DNA may be present at very low levels and undetectable. If “intact” copies were anticipated, repeat the CS-IPDA experiment using a higher concentration of template (if available) or increase the number of replicates completed so as to interrogate more cells. Check sample RPP30/deltaD reference data to ensure the template is ≥60% and an adequate number of cells were used in gDNA extraction.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dara A. Lehman (dlehman@fredhutch.org).
Materials availability
The plasmid sequences used as gating controls in CS-IPDA experiments are available courtesy of Claire Levy and can be accessed via AddGene. Subtype-specific plasmid sequences generated for the development of the CS-IPDA are available from the lead contact upon request.
Acknowledgments
This work was supported by the National Institutes of Health (NIH) grants R01 HD094718 (D.A.L.), R01 DA040386 and AI116292 (F.H.), UM1 A126623 (K.R.J.), and R37 A138518 (J.O.).
Author contributions
D.L. and J.O. conceived the study. All authors contributed to the design of the study. N.C. and C.F. designed and performed experiments. N.C., C.F., C.L., S.H., and D.L. contributed analysis tools and data analysis. C.L., C.F., and N.C. developed the analysis pipeline. All authors contributed to analysis and interpretation of data. C.F. and D.L. wrote this protocol with input from all authors.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2022.101681.
Data and code availability
All original code and an example experiment for analysis are available (https://doi.org/10.5281/zenodo.6908632). Data generated from this study are available from the lead contact upon request.
References
- Cassidy N.A.J., Fish C.S., Levy C.N., Roychoudhury P., Reeves D.B., Hughes S.M., Schiffer J.T., Benki-Nugent S., John-Stewart G., Wamalwa D., et al. HIV reservoir quantification using cross-subtype multiplex ddPCR. iScience. 2022;25:103615. doi: 10.1016/j.isci.2021.103615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.K., Bhattacharyya D., Lassen K.G., Ruelas D., Greene W.C. Calcium/calcineurin synergizes with prostratin to promote NF-kappaB dependent activation of latent HIV. PLoS One. 2013;8:e77749. doi: 10.1371/journal.pone.0077749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M.R., Sambrook J. Precipitation of DNA with ethanol. Cold Spring Harb. Protoc. 2012;2012:21–25. doi: 10.1101/pdb.prot093377. [DOI] [PubMed] [Google Scholar]
- Hughes S.M. Plater: read, tidy, and display data from microtiter plates. J. Open Source Softw. 2016;1:106. doi: 10.21105/joss.00106. [DOI] [Google Scholar]
- Lai R. The Comprehensive R Archive Network; 2020. Arrangements: Fast Generators and Iterators for Permutations, Combinations, Integer Partitions and Compositions. R Package Version 1.1.9. [Google Scholar]
- Levy C.N., Hughes S.M., Roychoudhury P., Reeves D.B., Amstuz C., Zhu H., Huang M.-L., Wei Y., Bull M.E., Cassidy N.A.J., et al. A highly multiplexed droplet digital PCR assay to measure the intact HIV-1 proviral reservoir. Cell Rep. Med. 2021;2:100243. doi: 10.1016/j.xcrm.2021.100243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy C.N., Hughes S.M., Roychoudhury P., Amstuz C., Zhu H., Huang M.-L., Lehman D.A., Jerome K.R., Hladik F. HIV reservoir quantification by five-target multiplex droplet digital PCR. STAR Protoc. 2021;2:100885. doi: 10.1016/j.xpro.2021.100885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller K. The Comprehensive R Archive Network; 2020. Here: A Simpler Way to Find Your Files.https://here.r-lib.org/ R Package Version 1.0.1. ed. [Google Scholar]
- Rstudio . Rstudio, Inc; 2015. Rstudio: Integrated Development for R.http://rstudio.com/ [Google Scholar]
- Wickham H. Tidyverse: easily install and load the ’tidyverse’. 2017. https://CRAN.R-project.org/ package=tidyverse
- Wiegand A., Spindler J., Hong F.F., Shao W., Cyktor J.C., Cillo A.R., Halvas E.K., Coffin J.M., Mellors J.W., Kearney M.F. Single-cell analysis of HIV-1 transcriptional activity reveals expression of proviruses in expanded clones during ART. Proc. Natl. Acad. Sci. USA. 2017;114:E3659–E3668. doi: 10.1073/pnas.1617961114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoutman W.H., Nell R.J., Versluis M., Van Steenderen D., Lalai R.N., Out-Luiting J.J., De Lange M.J., Vermeer M.H., Langerak A.W., Van Der Velden P.A. Accurate quantification of T cells by measuring loss of germline T-cell receptor loci with generic single duplex droplet digital PCR assays. J. Mol. Diagn. 2017;19:236–243. doi: 10.1016/j.jmoldx.2016.10.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All original code and an example experiment for analysis are available (https://doi.org/10.5281/zenodo.6908632). Data generated from this study are available from the lead contact upon request.