Abstract
Despite the fast development of vaccines, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) still circulates through variants of concern (VoC) and escape the humoral immune response. SARS-CoV-2 has provoked over 200,000 deaths/months since its emergence and only a few antiviral drugs showed clinical benefit up to this moment. Thus, chemical structures endowed with anti-SARS-CoV-2 activity are important for continuous antiviral development and natural products represent a fruitful source of substances with biological activity. In the present study, agathisflavone (AGT), a biflavonoid from Anacardium occidentale was investigated as a candidate anti-SARS-CoV-2 compound. In silico and enzymatic analysis indicated that AGT may target mainly the viral main protease (Mpro) and not the papain-like protease (PLpro) in a non-competitive way. Cell-based assays in type II pneumocytes cell lineage (Calu-3) showed that SARS-CoV-2 is more susceptible to AGT than to apigenin (APG, monomer of AGT), in a dose-dependent manner, with an EC50 of 4.23 ± 0.21 μM and CC50 of 61.3 ± 0.1 μM and with a capacity to inhibit the level of pro-inflammatory mediator tumor necrosis factor-alpha (TNF-α). These results configure AGT as an interesting chemical scaffold for the development of novel semisynthetic antivirals against SARS-CoV-2.
Keywords: SARS-CoV-2, COVID-19, Biflavonoid
1. Introduction
Members of the Coronaviridae family include seasonal and highly pathogenic viruses [1]. In the last two years and a half, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that emerged in China was spread globally [2], causing over 581 million confirmed cases and 6.41 million deaths worldwide of 2019 coronavirus disease (COVID-19) until the end of July 2022 [3]. Coronaviruses (CoV) are enveloped positive-sense RNA viruses that infect humans and zoonotic hosts [4]. The CoVs genome encodes two polycistronic polyproteins that are further cleaved by its own proteases to 16 non-structural proteins (nsp) [5], thus proteolytic processing is a critical step in CoV life cycle. Two viral proteases are required to cleave the CoV polyprotein into functional units: the main protease (Mpro; a chymotrypsin-like cysteine protease, also known as 3CLpro), which cleaves in about eleven sites [5], [6], and the papain-like protease (PLpro), which is responsible for cleavage three sites [7], [8]. Additionally, PLpro has the function of stripping ubiquitin and interferon-stimulated gene 15 (ISG15) from host-cell proteins to aid CoV in their evasion of the host innate immune responses [9].
Both Mpro and PLpro enzymes have been considered as interesting targets for drug development and were proposed to be targeted by clinically approved antiviral drugs [10], [11], [12], [13]. The importance of Mpro as a target has been reinforced by the drug PF-07321332 (PAXLOVID™; Pfizer), which reduced COVID-19-associated hospitalization by 80 % [14]. Despite that, continuous drug development against COVID-19 is necessary, due to the virus ability to escape humoral response induced by vaccines, as evidenced by the emergence of variants of concern (VoC) [15], [16], [17], [18]. Thus, antiviral resistance could also occur in the future and must be anticipated by a sustained effort in drug development.
Natural products are fruitful sources of active compounds with a wide variety of chemicals and biochemical arrangements [19]. It has been described that natural product present activity against CoVs, impairing mainly the viral replication [20]. In this context, polyphenolic metabolites, such as flavonoids, represent a class of molecules that includes the largest source of substances with antiviral activity [19]. Indeed, numerous flavonoids, e.g., fisetin, myricetin, quercetin, and kaempferol, have been described with antiviral activity, targeting proteases (Mpro or PLpro) of SARS-CoV, MERS-CoV, and SARS-CoV-2 [19], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30]. On the other hand, recent reports indicated that natural dimers of flavonoids, known as biflavonoids, frequently reported in plants used in traditional and modern medicine, have attracted more attention than flavonoids due to their highest anti-inflammatory, antioxidant, antibacterial, antidiabetic, antitumor, and antiviral (including anti-SARS-CoV and anti-SARS-CoV-2) properties [31], [32], [33], [34]. Additionally, biflavonoids are more promising candidates than flavonoids in either pre- or clinical stages due to their larger physical-chemical stability during pharmaceutical preparation and their better pharmacokinetics profile [32].
Severe cases of COVID-19 have been characterized by developing cytokine storm [35], therefore, the discovery of antivirals that also might inhibit the hyperinflammatory response and/or regulate the immune responses is an interesting strategy to better attenuate illness in patients infected with SARS-CoV-2. Different reports previously indicated flavonoids, e.g., flavones, isoflavones, and flavonols, as natural sources with the capacity to decrease both releasing of proinflammatory mediators and transcription of proinflammatory genes [36], [37], [38], [39], [40], as example, pre-treatment of pre-inflamed human macrophage with apigenin (APG) shows significant inhibition not only of pro-inflammatory interleukin-6 (IL-6) secretion but also the inflammatory chemokines (CCL5) and adhesion molecules (ICAM1 and VCAM1) [41], reinforcing the importance to evaluate flavonoids/biflavonoids against COVID-19 [42].
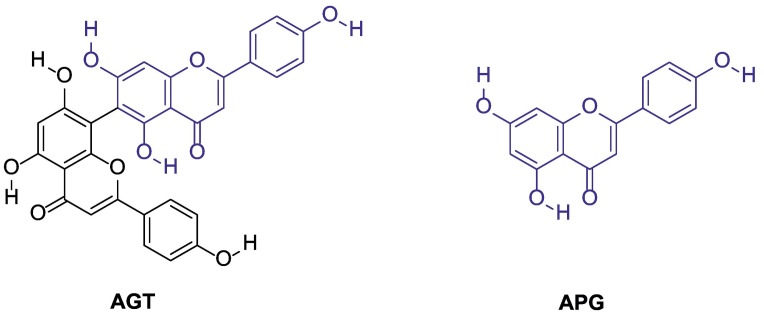
Agathisflavone (AGT, 8-[5,7-dihydroxy-2-(4-hydroxyphenyl)-4-oxochromen-6-yl]-5,7-dihydroxy-2-(4- hydroxyphenyl) chromen-4-one, Fig. 1 ) is a plant-derived biflavonoid that has been spotlight due to its diverse biological activities, e.g., antioxidant, antiviral, antiparasitic, cytotoxic, and neuroprotective [43], [44], [45]. From a structural point of view, AGT is a dimer of flavonoid APG (Fig. 1) [46]. Recently, aiming to explore the antiviral activity of biflavonoid, our group studied the anti-influenza action of AGT obtained from ethanolic extracts of Anacardium occidentale leaves [47], [48]. In addition to the anti-influenza activity, AGT has been described with anti-inflammatory properties [49], [50]. Considering that both COVID-19 and influenza trigger cytokine storm in critically ill patients [51] and the importance of finding novel antivirals hits that not only target both acute respiratory infections, but also act as anti-inflammatory is an interesting approach to the design of novel leads, the present work investigate the anti-SARS-CoV-2 and anti-inflammatory activity of AGT via cell-based and enzymatic assays combined with in silico calculations.
Fig. 1.
Chemical structure of agathisflavone (AGT) and apigenin (APG).
2. Materials and methods
2.1. Chemicals
Agathisflavone (AGT) was isolated from Anacardium occidentale leaves and characterized according to De Freitas et al. [47]. Apigenin (APG), remdesivir (RDV), 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT), N-methyl dibenzopyrazine methyl sulfate (PMS), dimethyl sulfoxide (DMSO), carboxymethyl cellulose (CMC), formaldehyde, phosphate buffer solution (PBS), crystal violet, bovine serum albumin (BSA), and methanol were purchased from Sigma-Aldrich/Merck (St. Louis, MO, USA). Atazanavir sulfate (ATV) was kindly donated by Instituto de Tecnologia de Fármacos, Farmanguinhos, Rio de Janeiro, Brazil.
2.2. Cells and virus
African green monkey kidney (Vero, subtype E6), human lung carcinoma (A549), and human lung epithelial (Calu-3) cells were cultured in high-glucose Dulbecco's modified Eagle medium (DMEM - HyClone, Logan, Utah) supplemented with 10 % fetal bovine serum (FBS - HyClone, Logan, Utah), 100 U/mL penicillin, and 100 μg/mL streptomycin (P/S - Thermo Fisher Scientific®, Massachusetts, USA). All cells were incubated at 37 °C in 5 % of carbon dioxide (CO2).
The SARS-CoV-2 virus used in the experiments was isolated in Vero-E6 cells from nasopharyngeal swab of a confirmed case from Rio de Janeiro, Brazil. The virus strain was sequenced to confirm the virus identity and its complete genome was deposited in GenBank under the identification number #MT710714 and received the approval number 30650420.4.1001.0008 from the Institutional Review Broad. All procedures related to virus culture were handled at biosafety level 3 (BSL3) multi-user facility at Fundação Oswaldo Cruz (FIOCRUZ), Rio de Janeiro, Brazil, according to World Health Organization (WHO) guidelines [52].
2.3. Citotoxicity assays
Vero-E6 cells (2.0 × 104 cells/well) in 96-well plates (Nalge Nunc Int, Rochester, New York, USA) were treated with different concentrations of AGT or APG (ranging from 1 to 800 μM) and observed for 3 days to subsequent addition of 5 mg/mL of XTT in DMEM in the presence of 0.01 % PMS. After incubation of 4 h at 37 °C, the plates were read in a spectrophotometer at excitation and emission wavelengths of 492 and 620 nm, respectively. The mean ± standard deviation (SD) of 50 % cytotoxic concentration (CC50) was calculated by a non-linear regression analysis of the dose–response curves.
2.4. Yield-reduction and virus titration assays
Cells were infected with a multiplicity of infection (MOI) 0.1. Firstly, a screening in Vero-E6, A549, and Calu-3 cells was conducted with AGT, ATV (positive control), or RDV (positive control) in a concentration of 1.25 and 10 μM, however, the screening of APG was only conducted for infected-Calu-3 cells also in a drug concentration of 1.25 and 10 μM. The dose-response curve was conducted only in Calu-3 cells for AGT in concentrations of 0.03, 0.1, 0.3, 1.0, 3.16, and 10 μM, for APG and ATV in concentrations of 0.63, 1.25, 2.50, 5.00, and 10.0 μM, and for RDV in concentrations of 0.0001, 0.001, 0.01, 0.1, 0.5, 1.0, 5.0, and 10 μM. The cells were infected at densities of 2.0 × 104 cells/well (Vero-E6 and A549) and 5.0 × 105 cells/well (Calu-3), performed in 96-well plates (Nalge Nunc Int, Rochester, New York, USA) for 1 h at 37 °C. The cells were washed, and various concentrations of the compounds were added in DMEM with 2 % FBS for Vero-E6 and A549, while 10 % FBS was used for Calu-3 cells. After 48 h, the supernatants were harvested, and infectious titers were quantified by plaque-based assays previously described [13]. All experiments were carried out at least three independent times, including a minimum of two technical replicates in each assay, and each data was analyzed with Prism GraphPad 8.0 (Windows GraphPad Software, San Diego, California USA). The selectivity index (SI) was calculated through the ratio between CC50 and EC50 values.
2.5. Measurements of inflammatory mediators
The levels of interleukin-6 (IL-6) and tumor necrosis factor-alpha (TNF-α) were quantified from the supernatant of uninfected Calu-3 cells (MOCK), infected cells without treatment (NIL), and infected and AGT or APG treated (10 μM) cells, using specific kits following the manufacturer's instructions (code #DY206 and #DY210 for IL-6 and TNF- α, respectively, from R&D Systems® Inc., Minneapolis, USA). All experiments were carried out at least three independent times and analyzed with Prism GraphPad 8.0 (Windows GraphPad Software, San Diego, California USA).
2.6. Enzymatic assays
The AGT capacity to inhibit the SARS-CoV-2 PLpro and Mpro processability was determined by the commercial kit provided by BPS Bioscience® company (catalog number: #79995-1 and #79955-1, respectively) following the procedure and recommendations from literature and manufacturer [53], [54], [55]. Basically, 100 nM PLpro was incubated in 50 mM HEPES pH 7.4, 0.01 % Triton X-100 (v/v), 0.1 mg/mL bovine serum albumin (BSA), 2 mM dithiothreitol (DTT) with 25 μM of its substrate (modified peptide Z-RLRGG-AMC with CAS number 167698-69-3) in the presence of different concentrations of inhibitors (0–10 μM of AGT or GRL0617 as positive control) for 45–60 min at 37 °C. On the other hand, 88.8 nM Mpro was incubated overnight in reaction buffer (20 mM Tris pH 7.3, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, and 1 μM BSA) containing 25 μM of substrate (modified peptide DABCYL-KTSAVLQSGFRKME-EDANS with CAS number 730985-86-1) and AGT or GC376 (positive control), at concentrations ranging from 0 to 10 μM at 25 °C. Fluorescence signal was measured at excitation and emission wavelengths of 360 nm and 460 nm, respectively, in a GloMax® (Promega) plate reader. The quadratic equation describing tight-binding behavior, known commonly as the Morrison equation (one of the widely used approximation mainly due to its high accuracy for this type of inhibitor) [56], [57] was applied to treat the experimental enzymatic data. Therefore, the Morrison's inhibitory constant (K i) was calculated by this non-linear regression using Prism GraphPad 8.0 (Windows GraphPad Software, San Diego, California USA). The Michaelis-Menten plots were obtained for 100 nM PLpro or 88.8 nM Mpro in assay buffer with substrate concentrations varying from 0 to 100 μM with and without 2.5 μM of AGT, following the procedure described above. After fluorescence quantification, the Michaelis-Menten constant (K m) and maximum velocity (V max) were calculated by non-linear regression also using Prism GraphPad software 8.0.
2.7. In silico simulations
Models obtained by X-ray diffraction from crystal structures of Mpro (PDB ID: 6LU7, dimeric structure) and PLpro (PDB ID: 6W9C, trimeric structure) [58] were obtained with resolutions of 2.16 Å and 2.70 Å, respectively.
2.7.1. Molecular docking
Ligand docking was performed with DOCK v6.9 [59] together with sphgen [60] to generate a maximum of 100 spheres within 10.0 Å of the dyad and catalytic triad used as reference position. Then, it was built a box of 10.0 Å edges around the spheres with showbox. Finally, was computed the energy interactions of each ligand and receptor atom at a 0.375 Å resolution inside the box with grid [61].
The substrate of Mpro (SMpro, modified peptide DABCYL-KTSAVLQSGFRKME-EDANS with CAS number 730985-86-1) and substrate of PLpro (SPLpro, modified peptide Z-RLRGG-AMC with CAS number 167698-69-3) was docked in the active site of chain A of the corresponding protease, and the best result was replicated for the other chains. On the other hand, AGT docking in Mpro was performed with SMpro in the active site, as well as to the allosteric sites of Mpro. The active site of this protease is characterized by the presence of the amino acids His41 and Cys145, which form a catalytic dyad. In addition to its active site, two allosteric sites were proposed in previous studies: (i) a binding site formed by Glu288, Asp289 and Phe290-291 (named as ALST1 below), which is a region at the interface between both monomers and (ii) a binding site formed by Lys12, Cys 16, and Lys97 (named as ALST2 below) on each chain A and B [62], [63], [64]. Finally, the AGT docking in PLpro was performed with and without SPLpro in the active site, which is characterized by the catalytic triad Cys108, His269, and Asp283.
Ligands and amino acid side chains at the selected sites of protein targets were treated as flexible [65]. The docking parameters were as follow: (i) groups with at least five atoms were considered as rigid segments of the molecule and treated as anchors. The maximum number of anchor orientations (max_orientations) was 10,000; (ii) 10,000 anchor orientations (pruning_max_orients) were retained to maximize the docking score according to a cut-off heuristic of 100 that combines conformational classification and mean squared deviation (RMSD); (iii) recursive cycles of ligand conformation based on energy minimization to search the best docking pose; (iv) rejection of conformers with a score >100.0 kcal/mol (pruning_conformer_score_cutoff) after energy minimization; (v) maximum number of 10,000.00 scored conformers for each optimization run; and (vi) grouping of the best conformers with a 2.0 Å RMSD threshold [59], [65].
The conformer that performed the largest number of hydrogen bonds and had the best gridscore was used for further analyses.
2.7.2. Ligand parameterization
The substrate of Mpro (SMpro) and PLpro (SPLpro) as well as AGT structure corresponding to CID numbers: 145707754, 101438889, and 5281599, respectively, were retrieved from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). The structures were converted to 3D topologies and their electrical charges calculated using the Ambertools 1.5 package [66]. Finally, it was used the force field Amber99SB [67], [68] and the General AMBER Force Field (GAFF) [69] to define bonds, angles, and twists.
2.7.3. Molecular dynamics (MD)
The MD simulations of Mpro/SMpro and PLpro/SPLpro were performed using Amber 18 [70]. The systems were solvated with a cubic water box TIP3P of 12 Å [71], [72]. The energy of MD systems was minimized in two steps: (i) steepest descent method and (ii) conjugate gradient in constant volume for 5000 cycles each.
The energy minimized systems were gradually heated (i) from −273 to 25 °C for Mpro and (ii) from −273 to 37 °C for PLpro during 1 ns with data collection in each 1 ps. The backbone and ligands were constrained with a constant force of 1.5 kcal/mol.Å2 during heating. From this step on, all bonds involving hydrogen atoms were constrained with the SHAKE algorithm and the PME distance cutoff was set to 12 Å. In addition, during all MD steps, temperature was controlled with a Langevin thermostat whose collision frequency was set to 1 ps−1.
Afterward, the heated systems were equilibrated for 1 ns and their evolution was recorded by collecting data each 1 ps at constant temperature (25 °C and 37 °C, for Mpro and PLpro, respectively) and pressure (1 bar). The backbone and ligands were constrained with a constant force of 0.5 kcal/mol·Å2 during the equilibration step. Pressure was controlled with isotropic position scaling, setting the relaxation time to 1 ps.
For each system, the final snapshot of the equilibration process was used as input to MD simulations for 200 ns of simulation for each protease complex. Following MD simulations, temperature trajectories were joined to identify the most representative cluster using (i) K-means clustering [73], (ii) hierarchical agglomerative (bottom-up) [74], and (iii) DBScan clustering [75]; all these methods were parameterized with a maximum cluster equal to 10 and a minimum distance of 4 Å between clusters to be reached at the end of the clustering process. This cut-off was set to 4 Å for all methods and temperatures investigated. The cluster whose 3D-structure presented the lowest RMSD compared to the initial structure among the three methods, was the cluster used for the subsequent docking and MD analyzes.
3. Results
3.1. SARS-CoV-2 is susceptible to AGT in Vero-E6, A549, and Calu-3 cells
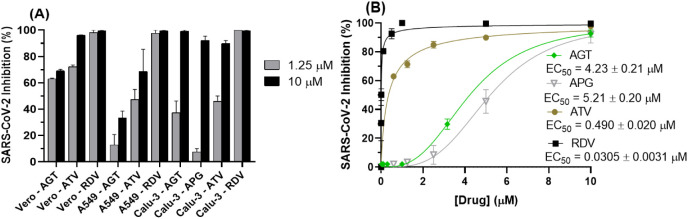
Cell-based assays in Vero-E6, A549, and Calu-3 cells were performed with AGT and two controls: atazanavir (ATV), a human immunodeficiency virus (HIV) protease inhibitor repurposed against SARS-CoV-2 (targeting SARS-CoV-2 Mpro), and remdesivir (RDV) an approved drug by the U.S. Food and Drug Administration (FDA) against COVID-19 (targeting SARS-CoV-2 RNA-dependent RNA polymerase, RdRp) [13], [76], [77]. Fig. 2A shows that SARS-CoV-2 replication is susceptible to AGT, comparable to ATV, in Vero-E6 and Calu-3 lineages, while RDV was the most effective drug in inhibiting SARS-CoV-2 replication in all cell lines, during initial antiviral screening. The APG (monomer of AGT) was less effective in inhibiting SARS-CoV-2 replication than AGT in Calu-3 cells (Fig. 2A). Fig. 2B depicts the dose-dependent inhibitory effect of AGT, APG, ATV, and RDV in the SARS-CoV-2 replication in Calu-3 cells, and the corresponding 50 % effective concentration (EC50) value. Since the 50 % cytotoxic concentration (CC50) value for AGT, APG, ATV, and RDV was 61.3 ± 0.1, 282 ± 4 (Fig. S1), 312 ± 8 [12], and 512 ± 30 [78] μM, respectively, were obtained the corresponding selective indeces (SI) of 14.5, 54.1, 637, and 1.68 × 104.
Fig. 2.
Susceptibility of SARS-CoV-2 replication to different concentrations of AGT, APG, ATV, and RDV. (A) Inhibitory capacity of AGT, ATV or RDV (1.25 and 10 μM) on SARS-CoV-2 replication in Vero-E6, A549, and Calu-3 cells. Screening for APG was conducted only in infected-Calu-3 cells. (B) Dose-dependent drug concentration for SARS-CoV-2 inhibition by AGT, APG, ATV or RDV in Calu-3 cells (5.0 × 105 cells/well). ATV and RDV are positive controls. [AGT] = 0.03, 0.1, 0.3, 1.0, 3.16, and 10 μM, [ATV] = [APG] = 0.63, 1.25, 2.50, 5.00, and 10.0 μM, and [RDV] = 0.0001, 0.001, 0.01, 0.1, 0.5, 1.0, 5.0, and 10 μM.
3.2. Anti-inflammatory profile of AGT in Calu-3 infected with SARS-CoV-2
COVID-19 frequently leads to fatal inflammatory responses and acute lung injury in critically ill patients. In this sense, monocytes/macrophages from patients with severe COVID-19 may be the main source of uncontrolled levels of the pro-inflammatory mediators TNF-α and IL-6 in the peripheral blood of the respiratory tract [51]. Fig. 3 depicts the AGT and APG capacity (in the concentration of 10 μM, close to their corresponding EC90 value of 9.24 ± 0.46 and 8.94 ± 0.51 μM, respectively) to reduce inflammatory markers upon SARS-COV-2 infection, in which TNF-α levels in infected Calu-3 cells were reduced by both natural products.
Fig. 3.
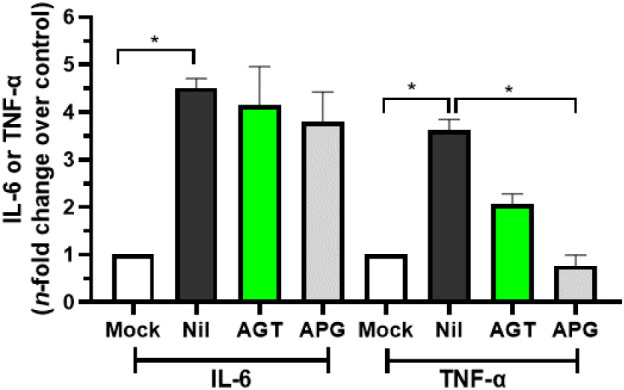
The anti-inflammatory profile of AGT and APG assessed by the IL-6 and TNF-α levels from uninfected Calu-3 cells supernatant (MOCK), SARS-CoV-2 infected cells without treatment (NIL), and infected and treated cells with 10 μM of AGT or APG. *p < 0.05.
3.3. Enzymatic assays and in silico calculations of the inhibitory capacity of AGT to SARS-CoV-2 proteases
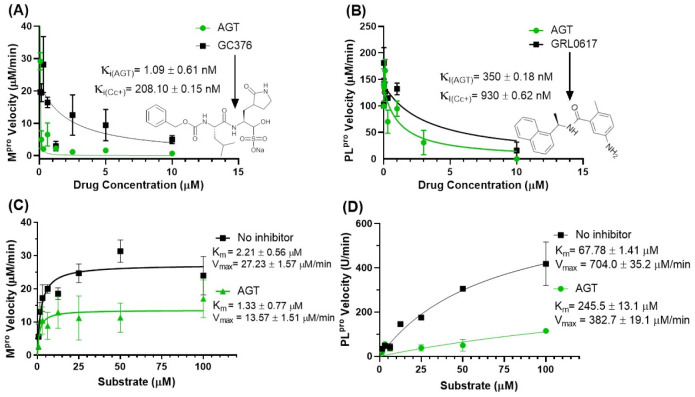
Considering previous literature that flavonoids inhibit SARS-CoV-2 proteases [79], Mpro and PLpro were assayed in the presence of AGT – better SARS-CoV-2 inhibitor than APG from the cell-based data. Fig. 4A and B depict the influence of AGT in the enzymatic parameters of Mpro and PLpro, respectively. Morrison's inhibitory constant (K i) for Mpro was 321-fold lower than that for PLpro (Fig. 4A and B). Additionally, AGT showed superior inhibitory capacity against Mpro and PLpro than the corresponding positive controls GC376 and GRL0617. Although these results suggest that Mpro is a better target than PLpro, K i values for both enzymes are below the in vitro antiviral potency displayed in Fig. 2B (EC50).
Fig. 4.
Enzymatic parameters for the inhibition of Mpro and PLpro by AGT. (A) The activity of 0–10 μM AGT and/or GC376 on the velocity of 88.8 nM Mpro (protease assay kit #79955-1, BPS Biosciences, USA). (B) The activity of 0–10 μM AGT and/or GRL0617 on the velocity of 100 nM PLpro (protease assay kit #79995-1, BPS Biosciences, USA). Michaelis-Menten plot for (C) Mpro and (D) PLpro incubated with substrate concentrations from 0 to 100 μM with or without 2.5 μM of AGT.
Next, either Mpro and PLpro were assayed in the presence of various substrate concentrations (0 to 100 μM) under a fixed concentration of AGT (2.5 μM). For Mpro, the Michaelis-Menten constant (K m) value with and without AGT was the same inside the experimental error, while the maximum velocity (V max) value decreased, indicating the possibility of a non-competitive inhibitory mechanism (Fig. 4C), which suggests that AGT might interact Mpro/substrate complex and/or with the allosteric site(s). For PLpro, both K m and V max values changed from the absence to the presence of AGT, indicating the possibility of a mixed inhibitory mechanism (Fig. 4D), therefore, AGT might interact with the catalytic site of PLpro without or in the presence of its native substrate.
To offer a molecular level explanation of the inhibitory capacity of AGT for the two SARS-CoV-2 proteases in silico calculations were performed based on the experimental inhibitory mechanism. Firstly, the proteases' substrate SMpro and SPLpro were docked in the active site of the center of mass of the catalytic dyad (Mpro) and triad (PLpro) (Fig. S2) The best docking pose for the systems Mpro/SMpro and PLpro/SPLpro was subsequently used as input for MD simulations (Fig. S3), observing that chain B of Mpro was more stable than chain A, while for PLpro the chain C remained stable throughout the simulation. Finally, the analysis of MD clusters showed that the DBScan method released structures with the lowest RMSD, i.e., 1.378 Å and 3.154 Å for Mpro and PLpro, respectively (Fig. S4). These structures were those used for the docking studies of AGT.
The same coordinates used to dock SMpro and SPLpro in the active site of Mpro and PLpro, respectively, were applied for AGT with the corresponding substrate. Additionally, for Mpro, the allosteric sites suggested in previous studies [62], corresponding to amino acid residues Lys12, Cys16, and Lys97 of chains A and B, and Glu288, Asp289, Phe290, Phe291, and Cys300 of the chain A were also investigated.
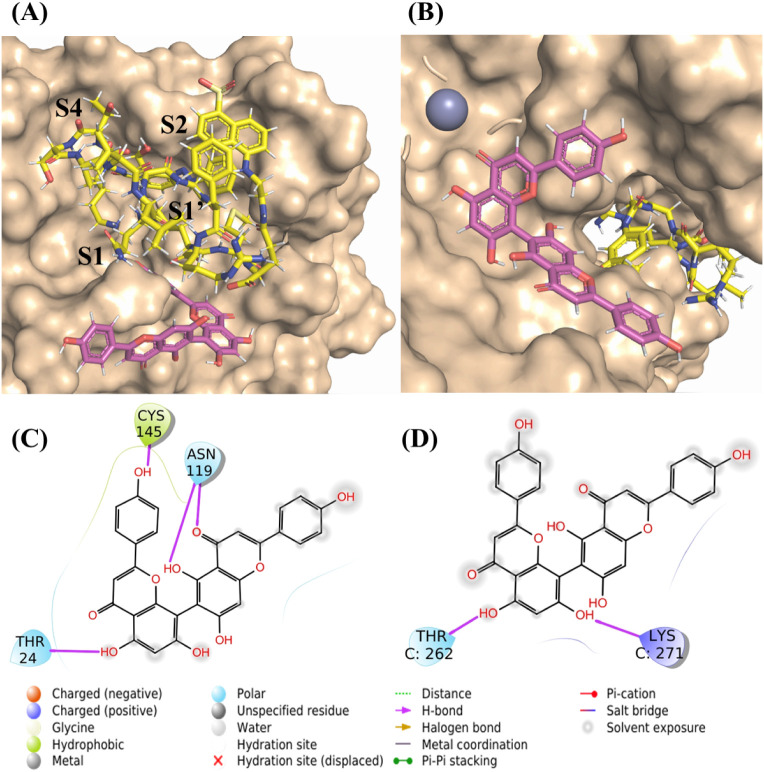
The gridscores of AGT at the active sites of Mpro and PLpro complexed with their substrates were −54.696 kcal·mol−1 and −4.824 kcal·mol−1, respectively (Fig. 5A and B), which are consistent with higher potency of AGT over Mpro than PLpro. In this case, AGT's interacted with residues Thr24, Asn119 and Cys145 for Mpro (Table 1 , Fig. 5C) and Thr262 and Lys271 for PLpro (Table 2 , Fig. 5D). It is worth noting that the hydrogen from the enol group of the ring C of AGT is a potential donor for hydrogen bonding with the carboxylic acid from SMpro structure (2.70 Å). In comparison, AGT docked in the part of PLpro most exposed to the solvent and the carbonyl group of ring C from AGT structure is at 7.50 Å away from the central Zn(II) ion (violet in Fig. 5B, O7–Zn(II)), which suggests some role for AGT-Zn(II).
Fig. 5.
The in silico results for AGT in the catalytic site of Mpro and PLpro. (A) AGT (pink) positioned at the catalytic site of Mpro complexed with the substrate SMpro (yellow) in the chain B. (B) AGT (pink) positioned at the catalytic site of PLpro complexed with the substrate SPLpro (yellow) in the chain C. In this case, Zn(II) is represented as violet sphere on the top left. (C) 2D-representation of AGT interactions at the catalytic site of the Mpro/SMpro complex. (D) 2D-representation of AGT interactions at the catalytic site of the PLpro/SPLpro complex.
Table 1.
Interaction of AGT with the amino acid (AA) residues of the Mpro/SMpro complex in the active site (chain B).
| AGT | AA | Distance (Å) |
|---|---|---|
| H50 | Thr24 – O | 2.56 |
| O8 | Asn119 – HD1 | 2.02 |
| H45 | Asn119 – OD1 | 1.78 |
| H57 | Cys145 – SG | 1.98 |
Table 2.
Interaction of AGT with the amino acid (AA) residues of the PLpro/SPLpro complex in the active site (chain C).
| AGT | AA | Distance (Å) |
|---|---|---|
| H50 | Thr262 – OG1 | 2.67 |
| O5 | Lys271 – HD3 | 2.31 |
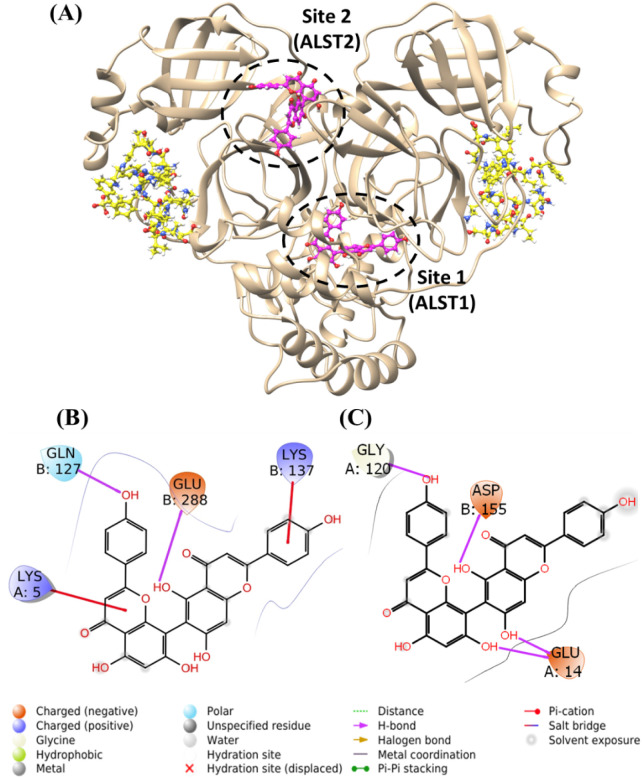
The gridscore of AGT onto the putative allosteric site was −72.946 kcal.mol−1 for ALST1 and − 66.853 kcal.mol−1 for ALST2 (Fig. 6A), which is more favorable than its predictive effect on the Mpro's active site; and thus, more consistent with the kinetic data assayed at different substrate concentrations. The interactions of AGT with ALST1 occurred following two modes: (i) hydrogen bonds with Gln127 and Glu288 in the chain B and (ii) π-cation bonds with Lys5 in the chain A and with Lys137 in the chain B (Table 3 , Fig. 6B). The interactions of AGT with ALST2 occurred through hydrogen bonds with Glu14, Gly120 in the chain A and Asp155 in the chain B (Table 4 , Fig. 6C).
Fig. 6.
Docking of AGT at the allosteric sites of Mpro. (A) Superposition of the best docking pose of AGT (pink) in the two allosteric sites of Mpro (ellipse) in the system Mpro/SMpro (substrate in yellow). (B) The 2D-representation of AGT interactions at the putative ALST1 of Mpro. (C) The 2D-representation of AGT interactions at the putative ALST2 of Mpro.
Table 3.
Interaction of AGT with the amino acid (AA) residues of the Mpro/SMpro complex in ALST1.
| AGT | AA | Distance (Å) |
|---|---|---|
| O9 | Gln127 – H | 2.64 |
| H45 | Glu288 – OE1 | 2.75 |
| π | Lys5 – HZ1 | 6.32 |
| π | Lys137 HZ1 | 3.12 |
Table 4.
Interaction of AGT with the amino acid (AA) residues of the Mpro/SMpro complex in ALST2.
| AGT | AA | Distance (Å) |
|---|---|---|
| H46 | Glu14-O1 | 2.10 |
| H47 | 1.70 | |
| H57 | Gly120 | 1.97 |
| H45 | Asp155 | 2.57 |
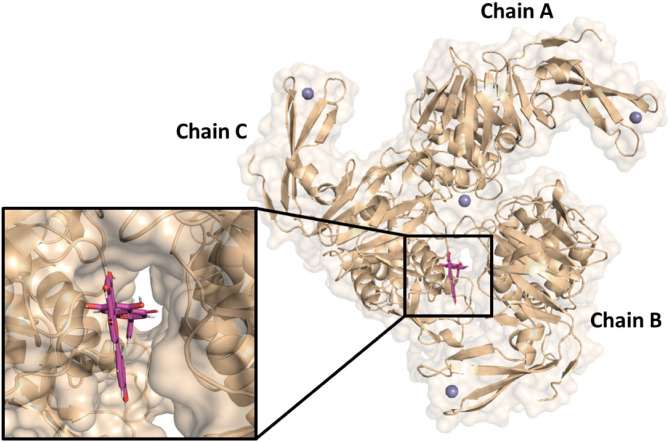
Finally, the gridscore of AGT at the active site of PLpro (a mixed inhibitory mechanism) without its substrate is −33.152 kcal·mol−1 (Fig. 7 ) without the detection of any hydrogen bonding or covalent interactions with the amino acid (AA) residues at the active site of PLpro, even occupying the same docking region as SPLpro.
Fig. 7.
Molecular docking and the corresponding zoom representation of AGT (pink) in the active site of PLpro without the substrate SPLpro in the chain C.
4. Discussion
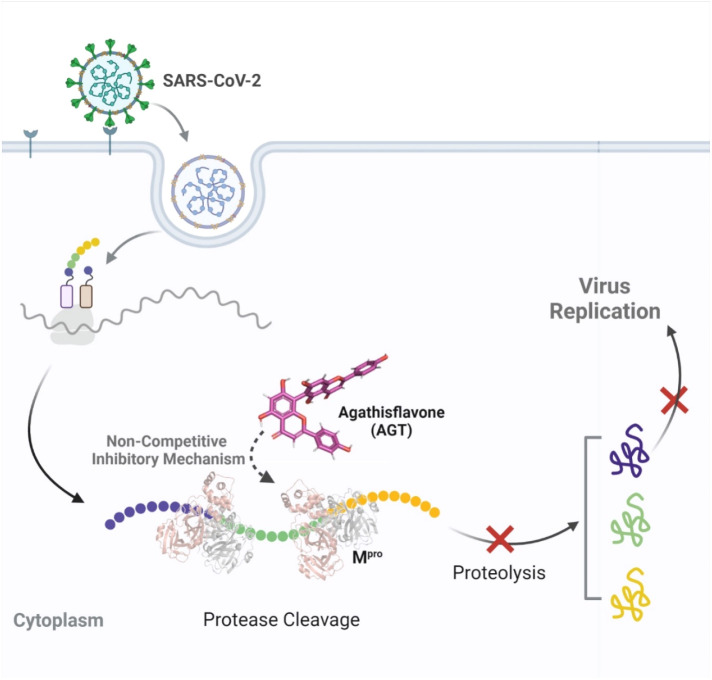
Biflavonoids have been widely explored as potential drugs and scaffolds for the design of semisynthetic drugs with wide-spectrum activity, e.g., anti-inflammatory, antioxidant, antibacterial, and antiviral [32], [79]. Recently, Lokhande and coworkers reported by in silico calculations that AGT interacts strongly with the catalytic residues of SARS-CoV-2 Mpro [79], however, this inference was not experimentally validated either by enzymatic or cell-based assays. Thus, here, it was investigated AGT inhibitory activity against SARS-CoV-2 and its anti-inflammatory profile in Calu-3-based assays (a physiologically relevant cell lineage) [80] and evaluated experimentally the AGT capacity in inhibiting the two proteases of SARS-CoV-2. Finally, since the rational design has entered the state of the art for the discovery of new SARS-COV-2 inhibitors [6], was also mimicked the lab enzymatic experiments by in silico modeling to better shed light on the molecular interactions of AGT with Mpro and PLpro.
The cell-based screening in three different lineages confirmed the AGT activity as anti-SARS-CoV-2. Although the biflavonoid did not show comparable potency and SI value with the positive controls ATV (a FDA-approved HIV-protease inhibitor that targets SARS-CoV-2 Mpro via competitive mechanism) [12], [13] and RDV (a FDA-approved inhibitor of SARS-CoV-2 RdRp) [76], there is still a difference of over 10-fold between its potency and cytotoxicity values, configuring a safety range for eventual clinical applications. Additionally, AGT was more potent than APG (the natural monomer of AGT) in inhibiting SARS-CoV-2 replication, with an EC50 value 4.6-fold lower than APG, reinforcing our preliminary hypothesis that biflavonoid might increase the antiviral capacity of flavonoids. Finally, AGT might reduce in the same way of APG the severity of COVID-19 symptoms by significantly decreasing the production of the pro-inflammatory mediator TNF-α in the peripheral blood of the respiratory tract which has been associated with patients with severe COVID-19 [51]. Therefore, the cell-based results indicated AGT as a dual activity against COVID-19 (antiviral and anti-inflammatory), corroborating with other studies that also identified some flavonoids, e.g., luteolin, fisetin, kaempferol, and myricetin with dual biological activity [81].
The experimental K i value for the AGT-Mpro complex is over 10-fold lower than that of the AGT-PLpro one, demonstrating a higher selectivity of AGT for Mpro compared with PLpro. Indeed, numerous flavonoids have been described with antiviral activity due to their inhibitory capacity for the proteases of SARS- and MERS-CoV, e.g., the flavonols quercetin and kaempferol have potency values in the range of 52.7–116.3 and 8.6–16.3 μM for Mpro and PLpro, respectively [22], [23], [82], that are not so good compared with AGT for the proteases of SARS-CoV-2, probably due to the chemical nature of biflavonoid compared with flavonoid. Morrison's approximation was used to treat the enzymatic data due to the tight-binding inhibitor capacity of AGT (K i ≈ 1.1 nM) [56], [57], reinforced by the using Mpro's substrate at proportion >10-fold above the K m value.
The experimental enzymatic assays agree with the in silico calculation of interaction energy that showed a difference also over 10-fold between both protease-AGT complexes. Based on the AGT structure, this biflavonoid showed a better interaction with protease than other flavonoids, such as quercetin and myricetin [83], [84], [85], [86], which is presumably due to the number of hydrophobic contacts provided by the largest number of aromatic rings in AGT. The experimental K m and V max values for Mpro indicated that AGT might interact not into the active site as a competitive inhibitor but close to the active site in a non-competitive way via the allosteric region or in the protease-substrate complex, while for PLpro a mixed inhibitory mechanism was identified. Comparing the enzymatic data of AGT with those previously described for ATV [13], the commercial HIV-protease inhibitor showed a competitive inhibitory mechanism to SARS-CoV-2, suggesting that the insertion of amino acid moieties in the AGT scaffold during drug design could lead this compound to similar mechanism of action and probably increase the anti-SARS-CoV-2 activity - semisynthetic compounds that might improve both EC50 and SI values.
The in silico simulations of SMpro by MD within the active site of Mpro suggested that the S2 region was not initially occupied by the SMpro, but the sulfonic group of its C-terminal side moved to this active pocket throughout the simulation, which increased the RMSD value. Upon reaching the S2 region, the SMpro stabilized throughout the simulation time. The SMpro's sulfonic group moved into the S2 region and liberate a space for AGT in the S1’ pocket, which is close to the catalytic amino acid residue Cys145. The AGT interaction with Cys145, even in the presence of the substrate occupying the S1-S4 binding regions, may disable the proton transfer from Cys145 to His41, which is possibly triggered by substrate binding or occurring in a transition state during the attack of sulfur on the carbonyl carbon atom of the scissile peptide bond [87]. Thus, the AGT interaction with Cys145 can break the interaction with His41 and, consequently, lead to the Mpro inactivation [88], [89].
An alternative form of Mpro inhibition is via its allosteric site that is located at the dimeric interface near the C-terminal region [63]. The impediment of the Mpro dimeric formation should lead to its lack of activity since it is essentially functional in its dimeric form. In general, the enzymatic activity of Mpro relies on the architecture of the active site, which critically depends on the dimerization of the enzyme and the correct relative orientation of its subdomains. Ligands binding outside of the active site in ALST1 could lead to monomer misalignment and affect enzyme activity. In fact, Günther and coworkers [63] identified two such allosteric binding sites for Mpro. As outlined above, AGT presented the highest negative gridscore for ALST1 and ALST2 compared to the active site in the presence of substrate, which suggests that Mpro inhibition at the allosteric sites occurs preferentially, mainly at ALST1, and should be better explored. Overall, our enzymatic and in silico data for AGT-Mpro (Fig. 8 ) invalidate the in silico results previously reported by Lokhande and coworkers [79]. Additionally, it is important to highlight that despite the difference in the experimental potency between AGT and APG, both compounds achieve the same experimental efficacy, e.g., the same EC90 value inside the experimental error, probably, following the in silico trend for AGT, due to the excess of the monomer of AGT in the biological assay might interact with the allosteric binding pocket of SARS-CoV-2 Mpro via dimerization of the inhibitor achieving the same inhibitory capacity of AGT.
Fig. 8.
Proposed mechanism of AGT to inhibit SARS-CoV-2 replication.
By contrast to Mpro, the structure of PLpro is characterized by a very narrow catalytic site, which requires a flexible ligand. Docking results showed that SPLpro can insert into the narrow active site of the protease between Leu73 and Gly76, which is consistent with the high local specificity for flexible molecules [90]. In this case, the mode of inhibition of PLpro can occur in two ways: (i) by interaction with the Zn(II) ion that binds the three PLpro monomers and that is responsible for the correct folding of the protein as well as its structural stability [91]; and (ii) by interaction with the catalytic triad [92]. Since AGT presented the lowest negative gridscore for PLpro in the absence of SPLpro than in its presence, one may conclude that the PLpro inhibition at the catalytic site by AGT occurs preferentially without the substrate.
Overall, our results revealed that AGT is a better SARS-CoV-2 inhibitor than its monomer with the capacity to inhibit mainly SARS-CoV-2 Mpro than PLpro via a non-competitive fashion, probably interacting with the Mpro's allosteric site(s) (Fig. 8). Considering the anti-inflammatory profile of AGT and APG, both reduced the TNF-α levels in infected Calu-3 cells. This first approach combining experimental and in silico calculations to identify biflavonoid capacity to inhibit SARS-CoV-2 proteases offers an optimistic scenario regarding the continuous effort to identify natural products as a hit for future development of leads based on their phytochemical core, however, additional assays, including in vivo experiments are necessary to ensure the efficacy of AGT as antiviral and/or anti-inflammatory, as well as understanding the pharmacokinetic and pharmacodynamic of the biflavonoid.
Moreover, we understand that the finding of pan-inhibitors of proteases (compounds able to inhibit different proteases) is a good strategy to find therapeutic for the treatment of different viral diseases, however, for example, SARS-CoV-2 Mpro/PLpro, dengue virus NS2B-NS3 protease, and HIV-protease are cysteine, serine, and aspartic proteases, respectively, belonging to different families of proteases [93], [94], [95], making difficult the correlation between the inhibitory capacity of AGT with different viral proteases, even for the two proteases of SARS-CoV-2 this correlation is also difficult since Mpro (dimer) and PLpro (trimer) have different composition and structural arrangements. This limitation is feasible considering the reactive pocket of the proteases, however, considering the allosteric site is not ruled out the possibility to identify, in future works, some structural motifs that make AGT a pan-inhibitor of proteases.
5. Conclusion
The SARS-CoV-2 replication is susceptible to treatment with AGT showing EC50 and CC50 values of 4.23 ± 0.21 and 61.3 ± 0.1 μM, respectively, however, although the biflavonoid did not show a comparable potency and SI value with ATV and RDV controls, there is still a difference of over 10-fold between its potency and cytotoxicity values, which configure a safety range for clinical applications and an interesting scaffold to improve its anti-SARS-CoV-2 potential by drug design. Additionally, AGT was more potent than its natural monomer APG in inhibiting SARS-CoV-2 replication, with an EC50 value 4.6-fold lower than APG, indicating that biflavonoid might increase the antiviral capacity of flavonoids, however, both natural compounds achieved the same experimental efficacy - probably the excess of APG in the biological assay might interact with the allosteric binding pocket of SARS-CoV-2 Mpro via dimerization of the inhibitor achieving the same inhibitory capacity of AGT. Finally, both AGT and APG decreased the pro-inflammatory mediator TNF-α levels in infected cells. The experimental enzymatic data indicated a non-competitive and mixed inhibitory mechanism of AGT for Mpro and PLpro, respectively. The K i of Mpro in the presence of AGT was 321-fold lower than that of PLpro, which was corroborated by in silico calculations mainly due to the gridscore of AGT interaction at the allosteric site of the Mpro/SMpro complex.
CRediT authorship contribution statement
Otávio Augusto Chaves: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing, Visualization. Carlyle Ribeiro Lima: Methodology, Software, Investigation, Writing – original draft. Natalia Fintelman-Rodrigues: Methodology, Validation, Formal analysis, Investigation, Writing – review & editing. Carolina Q. Sacramento: Methodology, Validation, Formal analysis, Investigation. Caroline S. de Freitas: Investigation, Data curation, Writing – original draft. Leonardo Vazquez: Methodology, Formal analysis, Investigation, Writing – original draft. Jairo R. Temerozo: Methodology, Formal analysis, Investigation. Marco E.N. Rocha: Investigation. Suelen S.G. Dias: Investigation. Nicolas Carels: Methodology, Software, Investigation, Resources, Writing – review & editing, Funding acquisition. Patrícia T. Bozza: Validation, Resources, Funding acquisition. Hugo Caire Castro-Faria-Neto: Validation, Investigation, Resources, Writing – review & editing, Funding acquisition. Thiago Moreno L. Souza: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Resources, Data curation, Writing – review & editing, Visualization, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare no conflict of interest and the funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Acknowledgments
The authors acknowledge the following Brazilian agencies for the finantial support: Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Finance Codes 441019/2020-0, 307162/2017-6) and Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ, Finance Codes E-26/210.182/2020, E-26/201.067/2021, E-26/210.112/2020). This study was financed in part by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Brazil, Finance Code 88887.506989/2020-00). Funding was also provided by CNPq, CAPES, and FAPERJ through the National Institutes of Science and Technology Program (INCT) on Diseases of Neglected Populations (INCT-IDPN, Finance Code 465313/2014-0). O.A.C. also thanks Dumith Chequer Bou-Habib and Fundação para o Desenvolvimento Científico e Tecnológico em Saúde (FIOTEC) both from Oswaldo Cruz Foundation (IOC, Rio de Janeiro, Brazil) for the grant VPPCB-005-FIO-20. C.R.L. also thanks CAPES-Fiocruz for the fellowship number 88887.319299/2019-00. Finally, the authors thank André C. Ferreira and Mayara Mattos from the Laboratory of Immunopharmacology, Oswaldo Cruz Institute (IOC), Rio de Janeiro, Brazil, for the assistance in the anti-inflammatory assays.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijbiomac.2022.09.204.
Appendix A. Supplementary data
Supplementary material
Data availability
Data will be made available on request.
References
- 1.Guo Y.-R., Cao Q.-D., Hong Z.-S., Tan Y.-Y., Chen S.-D., Jin H.-J., Tan K.-S., Wang D.-Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak- An update on the status. Mil. Med. Res. 2020;7:1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronavirus disease 2019. 2022. https://www.who.int/emergencies/diseases/novel-coronavirus-2019
- 3.2022. COVID-19 Dashboardby the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU)https://coronavirus.jhu.edu/map.html [Google Scholar]
- 4.Khailany R.A., Safdar M., Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;19 doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bzówka M., Mitusińska K., Raczyńska A., Samol A., Tuszyński J.A., Góra A. Structural and evolutionary analysis indicate that the SARS-CoV-2 Mpro is a challenging target for small-molecule inhibitor design. Int. J. Mol. Sci. 2020;21:3099. doi: 10.3390/ijms21093099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 7.Klemm T., Ebert G., Calleja D.J., Allison C.C., Richardson L.W., Bernardini J.P., Lu B.G.C., Kuchel N.W., Grohmann C., Shibata Y., Gan Z.Y., Cooney J.P., Doerflinger M., Au A.E., Blackmore T.R., van der Heden van Noort G.J., Geurink P.P., Ova H., Newman J., Riboldi-Tunnicliffe A., Czabotar P.E., Mitchell J.P., Feltham R., Lechtenberg B.C., Lowes K.N., Dewson G., Pellegrini M., Lessene G., Komander D. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 2020;39 doi: 10.15252/embj.2020106275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freitas B.T., Durie I.A., Murray J., Longo J.E., Miller H.C., Crich D., Hogan R.T., Tripp R.A., Pegan S.D. Characterization and noncovalent inhibition of the deubiquitinase and deISGylase activity of SARS-CoV-2 papain-like protease. ACS Infect. Dis. 2020;6:2099–2109. doi: 10.1021/acsinfecdis.0c00168. [DOI] [PubMed] [Google Scholar]
- 9.Báez-Santos Y.M., St S.E., John A.D.Mesecar. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X., Wang X.-J. Potential inhibitors for 2019-nCoV coronavirus M protease from clinically approved medicines. J. Genet. Genomics. 2020;47:119–121. doi: 10.1016/j.jgg.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campos D.M.O., Oliveira C.B.S., Andrade J.M.A., Oliveira J.I.N. Fighting COVID-19. Braz. J. Biol. 2020;80:698–701. doi: 10.1590/1519-6984.238155. [DOI] [PubMed] [Google Scholar]
- 12.Fintelman-Rodrigues N., Sacramento C.Q., Lima C.R., da Silva F.S., Ferreira A.C., Mattos M., de Freitas C.S., Soares V.C., Dias S.S.G., Temerozo J.R., Miranda M., Matos A.R., Bozza F.A., Carels N., Alves C.R., Siqueira M.M., Bozza P.T., Souza T.M.L. Atazanavir, alone or in combination with ritonavir, inhibits SARS-CoV-2 replication and proinflammatory cytokine production. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00825-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaves O.A., Sacramento C.Q., Ferreira A.C., Mattos M., Fintelman-Rodrigues N., Temerozo J.R., Pinto D.P., da Silveira G.P.E., da Fonseca L.B., Pereira H.M., Carlos A.S., d’Ávila J.C.P., Viola J.P.B., Monteiro R.Q., Vazquez L., Bozza P.T., Castro-Faria-Neto H.C., Souza T.M.L. Atazanavir is a competitive inhibitor of SARS-CoV-2 Mpro, impairing variants replication in vitro and in vivo. Pharmaceuticals. 2021;15:21. doi: 10.3390/ph15010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.2022. Pfizer's Novel COVID-19 Oral Antiviral Treatment Candidate Reduced Risk of Hospitalization or Death by 89% in Interim Analysis of Phase 2/3 Epic-HR Study.https://www.pfizer.com/news/press-release/press-release-detail/pfizers-novel-covid-19-oral-antiviral-treatment-candidate [Google Scholar]
- 15.Pedro S.A., Ndjomatchoua F.T., Jentsch P., Tchuenche J.M., Anand M., Bauch C.T. Conditions for a second wave of COVID-19 due to interactions between disease dynamics and social processes. Front. Phys. 2020;8 [Google Scholar]
- 16.Ali I., Alharbi O.M.L. COVID-19: disease, management, treatment, and social impact. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tregoning J.S., Flight K.E., Higham S.L., Wang Z., Pierce B.F. Progress of the COVID-19 vaccine effort: viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat. Rev. Immunol. 2021;21:626–636. doi: 10.1038/s41577-021-00592-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venkatesan P. Repurposing drugs for treatment of COVID-19. Lancet. 2021;9:E63. doi: 10.1016/S2213-2600(21)00270-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parvez M.K., Arbab A.H., Al-Dosari M.S., Al-Rehaily A.J. Antiviral natural products against chronic hepatitis B: recent developments. Curr. Pharm. Des. 2016;22:286–293. doi: 10.2174/1381612822666151112152733. [DOI] [PubMed] [Google Scholar]
- 20.Das P., Majumder R., Mandal M., Basak P. In-silico approach for identification of effective and stable inhibitors for COVID-19 main protease (Mpro) from flavonoid based phytochemical constituents of Calendula officinalis. J. Biomol. Struct. Dyn. 2021;39:6265–6280. doi: 10.1080/07391102.2020.1796799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaves O.A., Fintelman-Rodrigues N., Wang X., Sacramento C.Q., Temerozo J.R., Ferreira A.C., Mattos M., Pereira-Dutra F., Bozza P.T., Castro-Faria-Neto H.C., Russo J.J., Ju J., Souza T.M.L. Commercially available flavonols are better SARS-CoV-2 inhibitors than isoflavone and flavones. Viruses. 2022;14:1458. doi: 10.3390/v14071458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park J.-Y., Yuk H.J., Ryu H.W., Lim S.H., Kim K.S., Park K.H., Ryu Y.B., Lee W.S. Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. J. Enzym. Inhib. Med. Chem. 2017;32:504–515. doi: 10.1080/14756366.2016.1265519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muchtaridi M., Fauzi M., Ikram N.K.K., Gazzali A.M., Wahab H.A. Natural flavonoids as potential angiotensin-converting enzyme 2 inhibitors for anti-SARS-CoV-2. Molecules. 2020;25:3980. doi: 10.3390/molecules25173980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vijayakumar B.G., Ramesh D., Joji A., Prakasan J.J., Kannan T. In silico pharmacokinetic and molecular docking studies of natural flavonoids and synthetic indole chalcones against essential proteins of SARS-CoV-2. Eur. J. Pharmacol. 2020;886 doi: 10.1016/j.ejphar.2020.173448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maroli N., Bhasuran B., Natarajan J., Kolandaivel P. The potential role of procyanidin as a therapeutic agent against SARS-CoV-2: a text mining, molecular docking and molecular dynamics simulation approach. J. Biomol. Struct. Dyn. 2022;40:1230–1245. doi: 10.1080/07391102.2020.1823887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Omotuyi I.O., Nash O., Ajiboye B.O., Olumekun V.O., Oyinloye B.E., Osuntokun O.T. Aframomum melegueta secondary metabolites exhibit polypharmacology against SARS-CoV-2 drug targets: in vitro validation of furin inhibition. Phytother. Res. 2021;35:908–919. doi: 10.1002/ptr.6843. [DOI] [PubMed] [Google Scholar]
- 27.Báez-Santos Y.M., John S.E.S., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen T.T.H., Woo H.-J., Kang H.-K., Nguyen V.D., Kim Y.-M., Kim D.-W., Ahn S.-A., Xia Y., Kim D. Flavonoid mediated inhibition of SARS coronavirus 3C-like protease expressed in Pichia pastoris. Biotechnol. Lett. 2012;34:831–838. doi: 10.1007/s10529-011-0845-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jo S., Kim S., Shin D.H., Kim M.-S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzyme Inhib. Med. Chem. 2020;35:145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.da Silva F.M.A., da Silva K.P.A., de Oliveira L.P.M., Costa E.V., Koolen H.H.F., Pinheiro M.L.B., de Souza A.Q.L., de Souza A.D.L. Flavonoid glycosides and their putative human metabolites as potential inhibitors of the SARS-CoV-2 main protease (Mpro) and RNA-dependent RNA polymerase (RdRp) Mem. Inst. Oswaldo Cruz. 2020;115 doi: 10.1590/0074-02760200207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Šamec D., Karalija E., Dahija S., Hassan S.T.S. Biflavonoids: important contributions to the health benefits of ginkgo (Ginkgo biloba L.) Plants. 2022;11:1381. doi: 10.3390/plants11101381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He X., Yang F., Huang X. Proceedings of chemistry, pharmacology, pharmacokinetics and synthesis of aúdeiaoids. Molecules. 2021;26:6088. doi: 10.3390/molecules26196088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryu Y.B., Jeong H.J., Kim J.H., Kim Y.M., Park J.-Y., Kim D., Nguyen T.T.H., Park S.-J., Chang J.S., Park K.H., Rho M.-C., Lee W.S. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CL(pro) inhibition. Bioorg. Med. Chem. 2010;18:7940–7947. doi: 10.1016/j.bmc.2010.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartini Y., Saputra B., Wahono B., Auw Z., Indayani F., Adelya L., Namba G., Hariono M. Biflavonoid as potential 3-chymotrypsin-like protease (3Clpro) inhibitor of SARS-coronavirus. Res. Chem. 2021;3 doi: 10.1016/j.rechem.2020.100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahmudpour M., Roozbeh J., Keshavarz M., Farrokhj S., Nabipour I. COVID19 cytokine storm: the anger of inflammation. Cytokine. 2020;133 doi: 10.1016/j.cyto.2020.155151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan M.-H., Lai C.-S., Ho C.-T. Anti-inflammatory activity of natural dietary flavonoids. Food Funct. 2010;1:15–31. doi: 10.1039/c0fo00103a. [DOI] [PubMed] [Google Scholar]
- 37.Hanakova Z., Hosek J., Kutil Z., Temml V., Landa P., Vanek T., Schuster D., Dall’Acqua S., Cvacka J., Polansky O., Šmejkal K. Anti-inflammatory activity of natural geranylated flavonoids: Cyclooxygenase and lipoxygenase inhibitory properties and proteomic analysis. J. Nat. Prod. 2017;80:999–1006. doi: 10.1021/acs.jnatprod.6b01011. [DOI] [PubMed] [Google Scholar]
- 38.Al-Khayri J.M., Sahana G.R., Nagella P., Joseph B.V., Alessa F.M., Al-Mssallem M.Q. Flavonoids as potential anti-inflammatory molecules: a review. Molecules. 2022;27:2901. doi: 10.3390/molecules27092901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ginwala R., Bhavsar R., Chigbu D.G.I., Jain P., Khan Z.K. Potential role of flavonoids in treating chronic inflammatory diseases with a special focus on the anti-inflammatory activity of apigenin. Antioxidants. 2019;8:35. doi: 10.3390/antiox8020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maleki S.J., Crespo J.F., Cabanillas B. Anti-inflammatory Effects of Flavonoids. Vol. 299. 2019. p. 125124. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X., Wang G., Gurley E.C., Zhou H. Flavonoid apigenin inhibits lipopolysaccharide-induced inflammatory response through multiple mechanisms in macrophages. PloS One. 2014;9 doi: 10.1371/journal.pone.0107072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alzaabi M.M., Hamdy R., Ashmawy N.S., Hamoda A.M., Alkhayat F., Khademi N.N., Al Joud S.M.A., El-Keblawy A.A., Soliman S.S.M. Flavonoids are promising safe therapy against COVID-19. Phytochem. Rev. 2022;21:291–312. doi: 10.1007/s11101-021-09759-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Islam M.T., Zihad S.M.N.K., Rahman M.S., Sifat N., Khan M.R., Uddin S.J., Rouf R. Agathisflavone: botanical sources, therapeutic promises, and molecular docking study. IUBMB Life. 2019;71:1192–1200. doi: 10.1002/iub.2053. [DOI] [PubMed] [Google Scholar]
- 44.Andrade A.W.L., Machado K.C., Machado K.C., Figueiredo D.D.R., David J.M., Islam M.T., Uddin S.J., Shilpi J.A., Costa J.P. In vitro antioxidant properties of the bifavonoid agathisfavone. Chem. Cent.J. 2018;12:75. doi: 10.1186/s13065-018-0443-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.do Nascimento R.P., de Jesus L.B., Oliveira-Junior M.S., Almeida A.M., Moreira E.L.T., Paredes B.D., David J.M., Souza B.S.F., Costa M.F.D., Butt A.M., Silva V.D.A., Costa S.L. Agathisflavone as a single therapy or in association with mesenchymal stem cells improves tissue repair in a spinal cord injury model in rats. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.858190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bahia M.V., dos Santos J.B., David J.P., David J.M. Biflavonoids and other phenolics from Caesalpinia pyramidalis (Fabaceae) J. Braz. Chem. Soc. 2005;16:1402–1405. [Google Scholar]
- 47.de Freitas C.S., Rocha M.E.N., Sacramento C.Q., Marttorelli A., Ferreira A.C., Rocha N., de Oliveira A.C., Gomes A.M.O., dos Santos P.S., da Silva E.O., da Costa J.P., Moreira D.L., Bozza P.T., Silva J.L., Barroso S.P.C., Souza T.M.L. Agathisflavone, a biflavonoid from Anacardium occidentale L., inhibits influenza virus neuraminidase. Curr. Top. Med. Chem. 2019;20:111–120. doi: 10.2174/1568026620666191219150738. [DOI] [PubMed] [Google Scholar]
- 48.2022. Secretaria de Saúde do Estado de Pernambuco. Cartilha de Plantas Medicinais e Medicamentos Fitoterápicos.http://farmacia.saude.pe.gov.br/sites/aúdeia.saude.pe.gov.br/files/cartilha.pdf [Google Scholar]
- 49.Velagapudi R., Ajileye O.O., Okorji U., Jain P., Aderogba M.A., Olajide O.A. Agathisflavone isolated from Anacardium occidentale suppresses SIRT1-mediated neuroinflammation in BV2 microglia and neurotoxicity in APPSwe-transfected SH-SY5Y cells. Phyther. Res. 2018;32:1957–1966. doi: 10.1002/ptr.6122. [DOI] [PubMed] [Google Scholar]
- 50.Islam M.T., Zihad S.M.N.K., Rahman M.S., Sifat N., Khan M.R., Uddin S.J., Rouf R. Agathisflavone: botanical sources, therapeutic promises, and molecular docking study. IUBMB Life. 2019;71:1192–2000. doi: 10.1002/iub.2053. [DOI] [PubMed] [Google Scholar]
- 51.Ferreira A.C., Soares V.C., de Azevedo-Quintanilha I.G., Dias S.S.G., Fintelman-Rodrigues N., Sacramento C.Q., Mattos M., de Freitas C.S., Temerozo J.R., Teixeira L., Hottz E.D., Barreto E.A., Pão C.R.R., Palhinha L., Miranda M., Bou-Habib D.C., Bozza F.A., Bozza P.T., Souza T.M.L. SARS-CoV-2 engages inflammasome and pyroptosis in human primary monocytes. Cell Death Discov. 2021;7:43. doi: 10.1038/s41420-021-00428-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bain W., Lee J.S., Watson A.M., Stitt-Fischer M.S. Practical guidelines for collection, manipulation and inactivation of SARS-CoV-2 and COVID-19 clinical specimens. Curr. Protoc. Cytom. 2020;93 doi: 10.1002/cpcy.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weglarz-Tomczak E., Tomczak J.M., Talma M., Burda-Grabowska M., Giurg M., Brul S. Identifcation of ebselen and its analogues as potent covalent inhibitors of papain-like protease from SARS-CoV-2. Sci. Rep. 2021;11:3640. doi: 10.1038/s41598-021-83229-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu W., Morse J.S., Lalonde T., Xu S. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochem. 2020;21:730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murphy D.J. Determination of accurate KI values for tight-binding enzyme inhibitors: an in silico study of experimental error and assay design. Anal. Biochem. 2004;327:61–67. doi: 10.1016/j.ab.2003.12.018. [DOI] [PubMed] [Google Scholar]
- 57.Yang H., Li X., Li G., Huang H., Yang W., Jiang X., Sen M., Liu J., Liu Y., Pan Y., Wang G. Accurate quantitative determination of affinity and binding kinetics for tight binding inhibition of xanthine oxidase. Biomed.Pharmacother. 2021;139 doi: 10.1016/j.biopha.2021.111664. [DOI] [PubMed] [Google Scholar]
- 58.Berman H., Henrick K., Nakamura H. Announcing the worldwide protein data bank. Nat. Struct. Mol. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 59.Allen W.J., Balius T.E., Mukherjee S., Brozell S.R., Moustakas D.T., Lang P.T., Case D.A., Kuntz I.D., Rizzo R.C. DOCK 6: impact of new features and current docking performance. J. Comp. Chem. 2015;36:1132–1156. doi: 10.1002/jcc.23905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DesJarlais R.L., Sheridan R.P., Seibel G.L., Dixon J.S., Kuntz I.D., Venkataraghavan R. Using shape complementarity as an initial screen in designing ligands for a receptor binding site of known three-dimensional structure. J. Med. Chem. 1988;31:722–729. doi: 10.1021/jm00399a006. [DOI] [PubMed] [Google Scholar]
- 61.Meng E.C., Shoichet B.K., Kuntz I.D. Automated docking with grid-based energy evaluation. J. Comp. Chem. 1992;13:505–524. [Google Scholar]
- 62.Yuce M., Cicek E., Inan T., Dag A.B., Kurkcuoglu O., Sungur F.A. Repurposing of FDA-approved drugs against active site and potential allosteric drug-binding sites of COVID-19 main protease. Proteins: Struct. Funct. Bioinf. 2021;89:1425–1441. doi: 10.1002/prot.26164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Günther S., Reinke Patrick Y.A., Fernández-García Y., Lieske J., Lane Thomas J., Ginn Helen M., Koua Faisal H.M., Ehrt C., Ewert W., Oberthuer D., Yefanov O., Meier S., Lorenzen K., Krichel B., Kopicki J.-D., Gelisio L., Brehm W., Dunkel I., Seychell B., Gieseler H., Norton-Baker B., Escudero-Pérez B., Domaracky M., Saouane S., Tolstikova A., White Thomas A., Hänle A., Groessler M., Fleckenstein H., Trost F., Galchenkova M., Gevorkov Y., Li C., Awel S., Peck A., Barthelmess M., Schlünzen F., Lourdu Xavier P., Werner N., Andaleeb H., Ullah N., Falke S., Srinivasan V., França Bruno A., Schwinzer M., Brognaro H., Rogers C., Melo D., Zaitseva-Doyle J.J., Knoska J., Peña-Murillo G.S., Mashhour Aida R., Hennicke V., Fischer P., Hakanpää J., Meyer J., Gribbon P., Ellinger B., Kuzikov M., Wolf M., Beccari Andrea R., Bourenkov G., von Stetten D., Pompidor G., Bento I., Panneerselvam S., Karpics I., Schneider T.R., Garcia-Alai M.M. X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease. Science. 2021;372:642–646. doi: 10.1126/science.abf7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sztain T., Amaro R., McCammon J.A. Elucidation of cryptic and allosteric pockets within the SARS-CoV-2 main protease. J. Chem. Inform. Model. 2021;61:3495–3501. doi: 10.1021/acs.jcim.1c00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mukherjee S., Balius T.E., Rizzo R.C. Docking validation resources: protein family and ligand flexibility experiments. J. Chem. Inf. Model. 2010;50:1986–2000. doi: 10.1021/ci1001982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Case D.A., Cheatham T.E., Darden T., Gohlke H., Luo R., Merz K.M., Jr., Onufriev A., Simmerling C., Wang B., Woods R.J. The Amber biomolecular simulation programs. J. Comp. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lindorff-Larsen K., Piana S., Dror R.O., Shaw D.E. How fast-folding proteins fold. Science. 2011;334:517–520. doi: 10.1126/science.1208351. [DOI] [PubMed] [Google Scholar]
- 68.Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O., Shaw D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Prot. Struct. Funct. Bioinf. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general amber force field. J. Comp. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 70.Case D.A., Ben-Shalom I.Y., Brozell S.R., Cerutti D.S., Cheatham T.E., Cruzeiro V.W.D., Darden T.A., Duke R.E., Ghoreishi D., Gilson M.K., Gohlke H., Goetz A.W., Greene D., Harris R., Homeyer N., Izadi S., Kovalenko A., Kurtzman T., Lee T.S., LeGrand S., Li P., Lin C., Liu J., Luo R., Mermelstein D.J., Merz K.M., Miao Y., Monard G., Nguyen C., Nguyen H., Omelyan I., Onufriev A., Pan F., Qi R., Roe D.R., Roitberg A., Luchko T., Sagui C., Schott-Verdugo S., Shen J., Simmerling C.L., Smith J., Salomon-Ferrer R., Swails J., Walker R.C., Wang J., Wei H., Wolf R.M., Wu X., Xiao L., Kollman D.M.York, Kollman P.A. Vol. 810. University of California; San Francisco: 2018. AMBER 2018. [Google Scholar]
- 71.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 72.Martínez J.M., Martínez L. Packing optimization for automated generation of complex system's initial configurations for molecular dynamics and docking. J. Comp. Chem. 2003;24:819–825. doi: 10.1002/jcc.10216. [DOI] [PubMed] [Google Scholar]
- 73.Bremer P.L., De Boer D., Alvarado W., Martinez X., Sorin E.J. Overcoming the heuristic nature of k-means clustering: identification and characterization of binding modes from simulations of molecular recognition complexes. J. Chem. Inf. Model. 2020;60:3081–3092. doi: 10.1021/acs.jcim.9b01137. [DOI] [PubMed] [Google Scholar]
- 74.Noé F., Horenko I., Schütte C., Smith J.C. Hierarchical analysis of conformational dynamics in biomolecules: transition networks of metastable states. J. Chem. Phys. 2007;126 doi: 10.1063/1.2714539. [DOI] [PubMed] [Google Scholar]
- 75.Lemke O., Keller B.G. Density-based cluster algorithms for the identification of core sets. J. Chem. Phys. 2016;145 doi: 10.1063/1.4965440. [DOI] [PubMed] [Google Scholar]
- 76.Coronavirus (COVID-19) 2022. Drugs.https://www.fda.gov/drugs/emergency-preparedness-drugs/coronavirus-covid-19-drugs [Google Scholar]
- 77.The Nitazoxanide Plus Atazanavir for COVID-19 Study 2022. https://clinicaltrials.gov/ct2/show/NCT04459286
- 78.Sacramento C.Q., Fintelman-Rodrigues N., Temerozo J.R., da Silva A.P.D., Dias S.S.G., da Silva C.S., Ferreira A.C., Mattos M., Pão C.R.R., de Freitas C.S., Soares V.C., Hoelz L.V.B., Fernandes T.V.A., Branco F.S.C., Bastos M.M., Boechat N., Saraiva F.B., Ferreira M.A., Jockusch S., Wang X., Tao C., Chien M., Xie W., Patel D., Garzia A., Tuschl T., Russo J.J., Rajoli R.K.R., Pedrosa C.S.G., Vitória G., Souza L.R.Q., Goto-Silva L., Guimarães M.Z., Rehen S.K., Owen A., Bozza F.A., Bou-Habib D.C., Ju J., Bozza P.T., Souza T.M.L. In vitro antiviral activity of the anti-HCV drugs daclatasvir and sofosbuvir against SARS-CoV-2, the aetiological agent of COVID-19. J. Antimicrob. Chemother. 2021;76:1874–1885. doi: 10.1093/jac/dkab072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lokhande K., Nawani N., Venkateswara S.K., Pawar S. Biflavonoids from Rhus succedanea as probable natural inhibitors against SARSCoV-2: a molecular docking and molecular dynamics approach. J. Biomol. Struct. Dyn. 2020;1–13 doi: 10.1080/07391102.2020.1858165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sacramento C.Q., Fintelman-Rodrigues N., Dias S.S.G., Temerozo J.R., da Silva C.S., Blanco C., Ferreira A.C., Mattos M., Soares V.C., Pereira-Dutra F., Miranda M.D., Barreto-Vieira D.F., da Silva M.A.N., Santos S.S., Torres M., Chaves O.A., Rajoli R.K.R., Paccanaro A., Owen A., Bou-Habib D.C., Bozza P.T., Souza T.M.L., Da Silva A.D.P.D. Unlike chloroquine, mefloquine inhibits SARS-CoV-2 infection in physiologically relevant cells. Viruses. 2022;14:374. doi: 10.3390/v14020374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zaragozá C., Villaescusa L., Monserrat J., Zaragozá F., Álvarez-Mon M. Potential therapeutic anti-inflammatory and immunomodulatory effects of dihydroflavones, flavones, and flavonols. Molecules. 2020;25:1017. doi: 10.3390/molecules25041017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Solnier J., Fladerer J.-P. Flavonoids: a complementary approach to conventional therapy of COVID-19? Phytochem. Rev. 2021;20:773–795. doi: 10.1007/s11101-020-09720-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.di Pierro F., Iqtadar S., Khan A., Mumtaz S.U., Chaudhry M.M., Bertuccioli A., Derosa G., Maffioli P., Togni S., Riva A., Allegrini P., Khan S. Potential clinical benefits of quercetin in the early stage of COVID-19: results of a second, pilot, randomized, controlled and open-label clinical trial. Int. J. Gen. Med. 2021;14:2807–2816. doi: 10.2147/IJGM.S318949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Biancatelli R.M.L.C., Berrill M., Catravas J.D., Marik P.E. Quercetin and vitamin C: an experimental, synergistic therapy for the prevention and treatment of SARS-CoV-2 related disease (COVID-19) Front. Immunol. 2020;11:1451. doi: 10.3389/fimmu.2020.01451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xiao T., Cui M., Zheng C., Wang M., Sun R., Gao D., Bao J., Ren S., Yang B., Lin J., Li X., Li D., Yang C., Zhou H. Myricetin inhibits SARS-CoV-2 viral replication by targeting mpro and ameliorates pulmonary inflammation. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.669642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kaul R., Paul P., Kumar S., Busselberg D., Dwivedi V.D., Chaari A. Promising antiviral activities of natural flavonoids against SARS-CoV-2 targets: systematic review. Int. J. Mol. Sci. 2021;22:11069. doi: 10.3390/ijms222011069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kneller D.W., Phillips G., O’Neill H.M., Jedrzejczak R., Stols L., Langan P., Joachimiak A., Coates L., Kovalevsky A. Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography. Nat.Commun. 2020;11:3202. doi: 10.1038/s41467-020-16954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dai W., Zhang B., Jiang X.-M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L.-K., Xu Y., Yang H., Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Qiao J., Li Y.-S., Zeng R., Liu F.-L., Luo R.-H., Huang C., Wang Y.-F., Zhang J., Quan B., Shen C., Mao X., Liu X., Sun W., Yang W., Ni X., Wang K., Xu L., Duan Z.-L., Zou Q.-C., Zhang H.-L., Qu W., Long Y.-H.-P., Li M.-H., Yang R.-C., Liu X., You J., Zhou Y., Yao R., Li W.-P., Liu J.-M., Chen P., Liu Y., Lin G.-F., Yang X., Zou J., Li L., Hu Y., Lu G.-W., Li W.-M., Wei Y.-Q., Zheng Y.-T., Lei J., Yang S. SARS-CoV-2 mpro inhibitors with antiviral activity in a transgenic mouse model. Science. 2021;371:1374–1378. doi: 10.1126/science.abf1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rut W., Lv Z., Zmudzinski M., Patchett S., Nayak D., Snipas S.J., El Oualid F., Huang T.T., Bekes M., Drag M., Olsen S.K. Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: a framework for anti–COVID-19 drug design. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abd4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gil-Moles M., Basu U., Büssing R., Hoffmeister H., Türck S., Varchmin A., Ott I. Gold metallodrugs to target coronavirus proteins: inhibitory effects on the spike-ACE2 interaction and on PLpro protease activity by auranofin and gold organometallics. Chem. Eur. J. 2020;26:15140–15144. doi: 10.1002/chem.202004112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lv Z., Cano K.E., Jia L., Drag M., Huang T.T., Olsen S.K. Targeting SARS-CoV-2 proteases for COVID-19 antiviral development. Front. Chem. 2022;9 doi: 10.3389/fchem.2021.819165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.de Sousa L.R.F., Wu H., Nebo L., Fernandes J.B., da Silva M.F.G.F., Kiefer W., Kanitz M., Bodem J., Diederich W.E., Schirmeister T., Vieira P.C. Flavonoids as noncompetitive inhibitors of dengue virus NS2B-NS3 protease: inhibition kinetics and docking studies. Bioorg. Med. Chem. 2015;23:466–470. doi: 10.1016/j.bmc.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 94.Weber I.T., Wang Y.-F., Harrison R.W. HIV protease: historical perspective and current research. Viruses. 2021;13:839. doi: 10.3390/v13050839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Narayanan A., Narwal M., Majowicz S.A., Varricchio C., Toner S.A., Ballatore C., Brancale A., Murakami K.S., Jose J. Identification of SARS-CoV-2 inhibitors targeting Mpro and PLpro using in-cell-protease assay. Commun. Biol. 2022;5:169. doi: 10.1038/s42003-022-03090-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available on request.