Abstract
Evidence is presented that, in Methanosarcina barkeri oxaloacetate synthesis, an essential and major CO2 fixation reaction is catalyzed by an apparent α4β4-type acetyl coenzyme A-independent pyruvate carboxylase (PYC), composed of 64.2-kDa biotinylated and 52.9-kDa ATP-binding subunits. The purified enzyme was most active at 70°C, insensitive to aspartate and glutamate, mildly inhibited by α-ketoglutarate, and severely inhibited by ATP, ADP, and excess Mg2+. It showed negative cooperativity towards bicarbonate at 70°C but not at 37°C. The organism expressed holo-PYC without an external supply of biotin and, thus, synthesized biotin. pycA, pycB, and a putative bpl gene formed a novel operon-like arrangement. Unlike other archaeal homologs, the putative biotin protein ligases (BPLs) of M. barkeri and the closely related euryarchaeon Archaeoglobus fulgidus appeared to be of the Escherichia coli-type (bifunctional, with two activities: BirA or a repressor of the biotin operon and BPL). We found the element Tyr(Phe)ProX5Phe(Tyr) to be fully conserved in biotin-dependent enzymes; it might function as the hinge for their “swinging arms.”
The oxaloacetate (OAA) synthesis step is an essential physiological component and a major CO2 fixation site in a methanarchaeon (38), for it primes therein an incomplete tricarboxylic acid cycle reaction sequence that generates intermediates for the synthesis of amino acids (via OAA, α-ketoglutarate [α-KG], and succinate) and tetrapyrroles (via α-KG) (38). Methanococcus jannaschii and Methanococcus maripaludis use a pyruvate carboxylase (PYC) for OAA synthesis (28, 37). Methanobacterium thermoautotrophicum possesses two OAA-generating enzymes: PYC and phosphoenolpyruvate carboxylase (PPC) (18, 22, 31). The PYC and PPC reactions take the following forms:
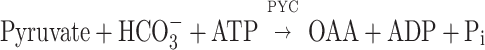 |
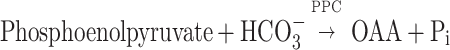 |
The avenue for OAA biosynthesis in Methanosarcinaspecies is unknown. Weimer and Zeikus (42) showed that Methanosarcina barkeri is devoid of PPC activity. We describe below PYC as an OAA-synthesizing activity in M. barkeri and present a biochemical and molecular genetic characterization of the enzyme.
Synthesis of biotin and a biotinylated protein.
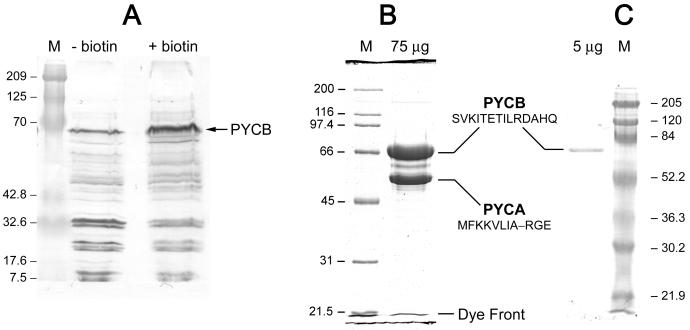
An avidin blot (29) with extracts of M. barkeri cells grown in a medium lacking biotin exhibited several bands (Fig. 1A). Hence, the organism was capable of synthesizing biotin and of biotinylating a candidate polypeptide without any external source of biotin. The intense band at ∼65 kDa was typical of the biotin-carrying subunit (PYCB) of an arcaheal PYC (28, 31). Thus, we purified and characterized the corresponding protein. As shown below, it possessed PYC activity and the above-mentioned avidin-reacting ∼65-kDa band indeed corresponded to the PYCB subunit of the enzyme (Fig. 1B and C). For the cells grown with a supply of biotin, the relative intensity of the ∼65-kDa band increased. Thus, M. barkeri exhibited a less stringent version of the control on PYC synthesis seen in Methanobacterium thermoautotrophicum (31); the latter requires exogenously supplied biotin to express holo-PYC (31), although it makes biotin (33), and a similar phenomenon exists in several bacteria (reviewed in reference 31). Whether other bands in the blot (Fig. 1A) represented certain biotinylated proteins or originated from charge-charge interactions between certain negatively charged nonbiotinylated polypeptides and avidin was not determined; the biotin-independent interactions with avidin have been seen with Methanobacterium thermoautotrophicum and Methanococcus jannaschii cell extracts (28, 31).
FIG. 1.
SDS-PAGE and avidin blot analyses. The methods were as described previously (24, 29) with modifications. The acrylamide concentrations in SDS gels were 12% for the results shown in panels A and C and 10% for panel B. For the blots phosphatase-conjugated avidin was used at a concentration of 0.1 mg liter−1. The molecular masses for the standards (in lanes marked M) are given in kilodaltons. (A) Avidin blot analysis of M. barkeri cell extracts. + biotin, cells were grown in a medium containing several vitamins (9), including d-biotin to a final concentration of 200 nM; − biotin, growth medium lacked added biotin. In each case 60 μg of cell extract protein was analyzed. Lane M, Bio-Rad Laboratories' Kaleidoscope prestained standards. (B) SDS-PAGE for purified PYC. (C) Avidin blot for purified PYC. Lane M, prestained standards.
Purification and molecular characterization of PYC.
M. barkeri strain Fusaro (20) was grown at 37°C on methanol (125 mM) in single-cell morphology. A bicarbonate-CO2-buffered medium (27) with the following modifications was used. The trace metals were supplied from a stock (30), which was modified to have Fe(NH4)2(SO4)2 · 6H2O as the Fe source and to provide an Fe level of 20 μM in the final medium. The vitamin solution was that of Bryant et al. (9). The cultures were raised according to the method of Balch and Wolfe (4). The harvested cells were stored frozen at −70°C.
For a typical enzyme purification experiment, 10 g (wet weight) of cell pellets was resuspended in a 10-ml solution containing 100 mM Tris-HCl buffer (pH 8), 10% inositol, 10 mM MgCl2, 4 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride. The cell suspension was passed three times through a French pressure cell at 1,240 atm. The resulting broken-cell slurry was supplemented with 0.4 mg of bovine pancreal DNase I (from a 5-mg/ml stock solution) (Sigma Chemical Co., St. Louis, Mo.) and held on ice for 30 min. The thinned slurry was mixed with a 10-ml solution of 3 M KCl, 50 mM Tris-HCl buffer (pH 8), 5% inositol, 5 mM MgCl2, 2 mM dithiothreitol, and 0.5 mM phenylmethylsulfonyl fluoride. The mixture was centrifuged for 30 min at 27,000 × g. The supernatant collected from this stage was recentrifuged for 1 h at 100,000 × g to obtain a KCl-enriched high-spin extract. From this extract, PYC was purified as described previously (31). The entire procedure was completed in 6 to 7 h, yielding about 0.16 mg of enzyme (in 6 ml of eluent) with a specific activity value (at 70°C) in the range of 72 to 118 U mg−1; protein was assayed according to the method of Bradford (8), and the activity was determined as described below. The product exhibited a Coomassie blue staining smear in a native-polyacrylamide gel (data not shown), indicating structural changes during electrophoresis. A similar observation has been made for the PYC from Methanobacterium thermoautotrophicum (31).
The sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (24) pattern in Fig. 1B shows that the M. barkeri PYC (MsbPYC) was composed of two polypeptides with apparent molecular masses of 66 and 52 kDa, and the avidin blot in Fig. 1C shows that the larger polypeptide was biotinylated. A matrix-assisted laser desorption ionization–time of flight mass spectrometric analysis (29) provided the following more precise estimates for the subunit molecular mass values (daltons): 64,169 for the larger biotinylated subunit (PYCB) and 52,930 for the smaller nonbiotinylated subunit (PYCA). From Edman degradation experiments, the NH2-terminal sequences for the PYCA and PYCB polypeptides were determined to be MFKKVLIA–RGE and SVKITETILRDAHQ, respectively. Size exclusion chromatography was performed as described previously (29), but a flow rate of 0.8 ml min−1 and the following mobile phase were used: 100 mM sodium phosphate buffer (pH 7.0), 5 mM MgCl2, 10% inositol, and either 100 mM NaCl or 1 M KCl. From the elution volume data, two values for the Stokes radius of the native PYC were obtained: 77.3 Å with 100 mM NaCl as a buffer component and 75.16 Å when 1 M KCl was used in place of NaCl. The same set of data yielded the following values for the apparent native molecular masses: 476.4 kDa in the presence of 100 mM NaCl and 433.8 kDa with 1 M KCl. Thus, it is plausible that MsbPYC is an α4β4-type enzyme, where α is the 52.9-kDa PYCA subunit (A is for ATP motif possession [31]) and β is the 64.2-kDa PYCB subunit (B is for biotinylated [31]). However, molecular mass data derived solely from gel filtration data are unreliable (12). Hence, a final conclusion on the number of αβ units per native molecule must await an accurate determination of the native molecular mass for the protein by use of a more appropriate method.
Similar to PYC of Methanobacterium thermoautotrophicum, the M. barkeri enzyme was found to be relatively unstable. When the enzyme was stored in the elution buffer for 24 h, the following values for the residual activities were recorded: 70% (4°C or room temperature), 8% (−20°C), and 65% (−80°C). The corresponding values for a 48-h storage were 62% at 4°C, 57% at room temperature, 4% at −20°C, and 54% at −80°C. Removal of either KCl, inositol, or Mg2+ lowered the activity further (data not shown). The gel filtration data (see above) showed that the hydrodynamic radius of the enzyme decreased in the presence of 1 M KCl. Thus, it is plausible that by inducing a more compact structure and allowing certain specific interactions, KCl enhanced both activity (see below) and stability.
The kinetic characteristics.
The PYC activity was assayed as described previously (31) but with modifications. Unless otherwise indicated, the assay temperature was 37°C and the reaction mixture had the following composition: 100 mM Tris-HCl (pH 8), 250 mM KCl, 4 mM MgCl2, 20 mM sodium pyruvate, 20 mM potassium bicarbonate, 4 mM disodium ATP, 0.2 mM disodium NADH, and 2 U of thermophilic malate dehydrogenase from Thermus flavus (Sigma Chemical Co.) per ml. For pH experiments the Tris-HCl buffer was adjusted to the desired pH (6 to 9.5) with HCl. The initial-rate data were analyzed according to the method of Cleland (15).
The purified enzyme was absolutely dependent on pyruvate, ATP, Mg2+, and HCO3− for activity. Phosphoenolpyruvate could not replace pyruvate, and GTP or ADP did not substitute for ATP. Acetyl coenzyme A was not required and it (at a 50 μM level) did not enhance or inhibit the activity of the enzyme. Incubation of purified enzyme for 5 min with avidin at a 100 molar excess (with respect to PYC-bound biotin) completely inhibited activity, establishing the typical dependence of PYC activity on protein-bound biotin for this enzyme. No activity was lost if avidin was incubated for 10 min with a 10-fold molar excess of biotin prior to its addition to the enzyme. If an avidin-inactivated enzyme preparation was incubated for 10 min with biotin, 23% of the original enzymatic activity was restored.
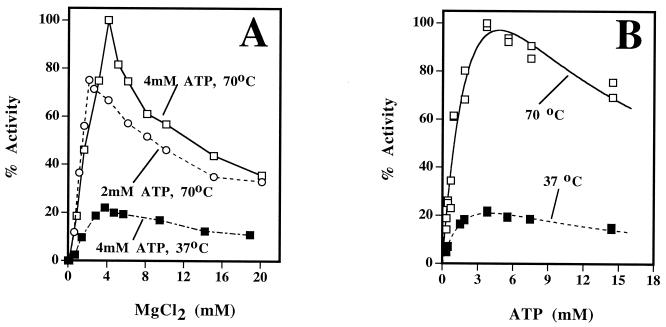
For MsbPYC, the optimum pH was found to be 8.0. This enzyme from a mesophilic host (optimum temperature for growth, 35 to 42°C [6]) was found to be thermophilic, exhibiting maximum activity at 70°C. From the linear portion of the Arrhenius plot (20 to 70°C), a value of 37.6 kJ/mol was obtained for the activation energy. A thermophilic nature is also typical for several well-characterized methanogenic enzymes from mesophilic Methanosarcina (17, 21, 23, 26, 40), indicating a possible thermophilic ancestry for this organism. At low concentrations, KCl stimulated MsbPYC, and at high concentrations, it was inhibitory; the maximum specific activity was obtained at 0.25 M KCl. A similar pattern was seen with NaCl, but the extent of stimulation was less than that recorded with KCl. Although it was absolutely required for both stability and activity, Mg2+ was inhibitory when it was present in the assay at a concentration higher than that for ATP (Fig. 2A). Maximum activity was obtained only when the concentration of Mg2+ was equal to that of ATP. In assays with 4 mM divalent cation and 4 mM ATP, Mn2+ and Co2+ provided, respectively, 9 and 42% of the activity recorded with Mg2+; the superiority of Co2+ over Mn2+ has been observed also with other archaeal PYCs (28, 31).
FIG. 2.
Inhibition of the activity of M. barkeri PYC by MgCl2 and ATP. The standard assay mixture was used but with modifications as indicated. Each data set (A or B) was obtained with an independent enzyme preparation, and thus the maximum activity value for one is different from that for the other. (A) Effect of MgCl2. Two or 4 mM disodium ATP and a desired concentration of MgCl2 were used. Each activity value is reported as a percentage of that obtained at 70°C with 4 mM disodium ATP and 4 mM MgCl2, which was 57 U/mg of protein. (B) Effect of ATP. The concentration of MgCl2 was equal to that of disodium ATP. Each activity value is reported as a percentage of the maximum (118 U/mg of protein), which was attained at an ATP concentration of 4 mM. A solid or dashed line indicates a fit of the data to the following substrate inhibition relationship: v = VmS/[Km + S + (S2/Ki)].
The initial-velocity data over a range of pyruvate concentrations (0.23 to 11.6 mM pyruvate, 4 mM ATP, 4 mM Mg2+, and 20 mM HCO3−) fit the Henri-Michaelis-Menten relationship well, and from these fits the apparent Km values for pyruvate were determined to be 0.5 ± 0.026 mM at 37°C and 0.56 ± 0.061 mM at 70°C. A similar fit was seen for the initial-velocity data collected at 37°C and at HCO3− concentrations of 0.23 to 11.6 mM (4 mM ATP, 4 mM Mg2+, and 20 mM pyruvate), and it provided an apparent Km value of 5.3 ± 0.59 mM for HCO3−. However, at 70°C the enzyme showed negative cooperativity towards HCO3−; an Eadie-Hofstee or v/S versus v plot (v, initial velocity; S, substrate concentration) was nonlinear. Hence, these data were fitted to a 2/1 function given by the following formula (15): v = Vm (S2 + DS)/(S2 + BS + C) where Vm is maximum velocity, B, C, and D are constants, and Km is 0.5B − D + [(0.5B − D)2 + C]1/2. This fit yielded the following values for bicarbonate: an apparent Km of 2.1 ± 1.8 mM, a B of 12.2 ± 10.8 mM, a C of 1.6 ± 2.6 mM2, and a D of 5.4 ± 4.8 mM. Methanobacterium thermoautotrophicum PYC shows negative cooperativity towards bicarbonate at 60°C (31). Thus, it would be interesting to examine whether such a response is seen only when a PYC acts at higher temperatures.
ATP inhibited the enzyme at high concentrations. The initial-velocity–versus–ATP concentration data (0.23 to 14 mM ATP, 20 mM pyruvate, 20 mM HCO3−, and a concentration of Mg2+ equal to that of ATP) were fitted to the substrate inhibition relationship v = VmS/[Km + S + (S2/Ki)] (Fig. 2B). These fits provided the following values: apparent Kms for ATP of 2.5 ± 0.43 mM at 37°C and 3.6 ± 1.88 mM at 70°C, and apparent Kis for ATP of 6.7 ± 4.24 mM at 37°C and 6.3 ± 7.57 mM at 70°C.
At a concentration of 9 mM in the assay, aspartate and glutamate reduced the PYC activity by only 5 and 4%, respectively, and α-KG reduced the PYC activity by 15%. GTP (4 mM) reduced the activity by 50%, but AMP (4 mM) had no effect. ADP acted as a competitive inhibitor with respect to ATP, and the corresponding Ki value was 1.2 ± 0.24 mM. In these tests AMP, ADP, or GTP was accompanied by an equimolar amount of MgCl2.
Cloning of the pycA, pycB, and bpl/birA genes and DNA sequence analysis.
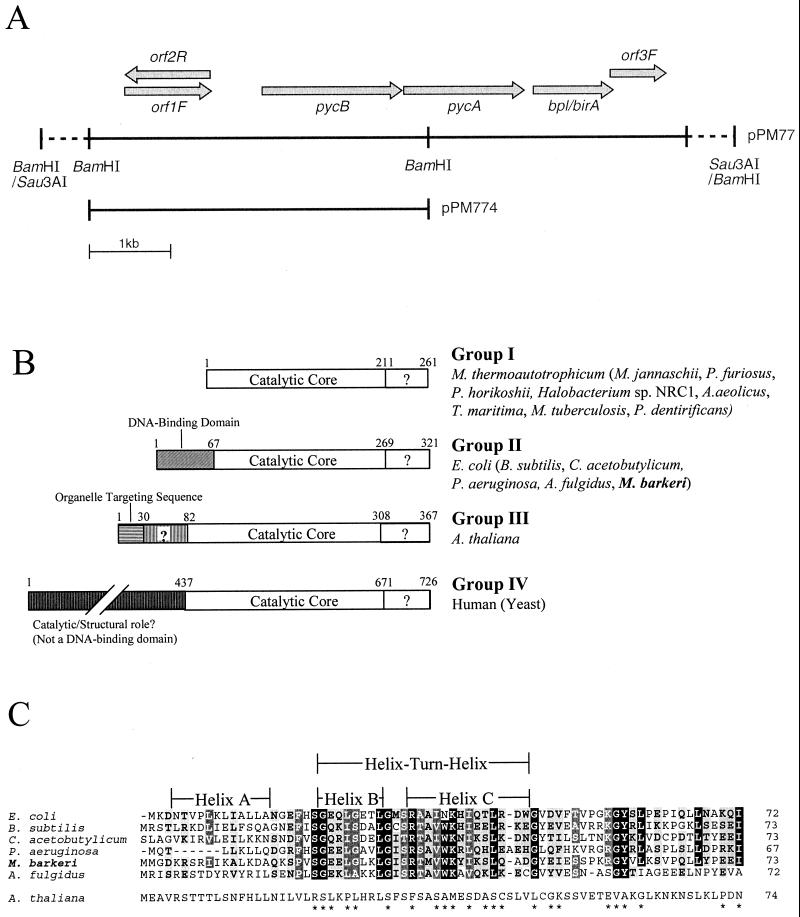
The chromosomal DNA from M. barkeri strain Fusaro was isolated as described before (32), except that the proteinase K and SDS treatments were conducted at room temperature. The DNA was partially digested with Sau3AI, and the resulting fragments were used to generate a cosmid library in E. coli XL-1 Blue MR (19) by using plasmid SuperCos1 (Stratagene, Inc., La Jolla, Calif.). This library was screened by using the colony hybridization technique. The degenerate oligonucleotide 5′ GTN AAR ATH ACN GAR ACN AT 3′, which was designed based on the determined NH2-terminal sequence of the biotinylated subunit PYCB (residues 2 to 8), was used as the probe. Prehybridization and hybridization were conducted at 50°C, and posthybridization washes were at room temperature (22°C). This screen provided a strongly hybridizing colony, from which the strain E. coli PM77 was isolated. The corresponding cosmid was designated pPM77 (Fig. 3A). A 4-kb BamHI fragment of the insert in pPM77 was found to hybridize to the above-mentioned oligonucleotide. This fragment was cloned into the BamHI site of plasmid pBluescriptII SK(+) (Stratagene, Inc.), generating plasmid pPM774 (Fig. 3A).
FIG. 3.
pyc-bpl/birA clone and selected features of BPL/BirA. (A) Clones. pPM77 is the comid clone, and pPM774 is a subclone. pycA and pycB are genes for PYC subunits, bpl/birA is the gene for a putative BPL with a repressor function (BirA). A solid line indicates that the sequence has been determined; a dashed line indicates that the section is not to scale and that there is no sequence information available. (B) Classification of BPLs. In each case the sequence numbers correspond to the organism listed first (before the parenthesis). The points of demarcation between various domains are according to reference 13. ?, the function of this region is unknown. BPLs shown (organism, accession number) are as follows: M. thermoautotrophicum strain ΔH, AAB86376; M. jannaschii, AAB99640; Pyrococcus furiosus, AAC25558; Pyrococcus horikoshii, A71236; Halobacterium sp. strain NRC-1, AAG19158; Thermotoga maritima, C72401; Mycobacterium tuberculosis, H70979; E. coli, M10123; B. subtilis, P42975; Clostridium acetobutylicum, ORF CAC0266 (http://www.genomecorp.com/genesequences/clostridium/clospage.html); Pseudomonas aeruginosa, AAG07668; A. fulgidus, AAB91153; M. barkeri, AF317651; Arabidopsis thaliana, AAC49706; Saccharomyces cerevisiae, P48445; and Homo sapiens P50747. (C) Alignment of the NH2-terminal regions of several bifunctional or group II BPLs. Each helix location and designation are according to reference 43. Arabidopsis thaliana BPL (a group III member) is shown for comparison. An ∗ indicates a departure (in the Arabidopsis thaliana BPL) from a character typical of a group II member. White letters in a black background indicate identical residues, white letters in a shaded background indicate strongly conserved residues (very small, A, G, C, T, S; aliphatic, V, I, L; aromatic, F, Y, W, H; positively charged, H, K, R; negatively charged, D, E), and bold letters in a shaded background indicate weakly conserved residues (charged, H, K, R, D, E; polar, N, Q, S, T, W, Y, and charged residues; nonpolar, A, F, I, L, M, P, V).
The DNA sequences for the insert in pPM774 and the corresponding adjoining regions in pPM77 were determined. From sequence analysis it was found that an ∼4.3-kb region of the clone in pPM77 harbored the genes for the PYC subunits (pycA and pycB) and for a putative biotin protein ligase-biotin operon repressor (bpl/birA) in that order and in a head-to-tail operon-like configuration (Fig. 3A). The pycA and pycB genes were identified by comparing the corresponding deduced amino acid sequences with the determined NH2-terminal sequences for the respective subunits of the purified enzyme; the matches were perfect except that the matured PYCB polypeptide was devoid of the initiator methionine. The open reading frame (ORF) for the bpl/birA was identified from homology searches (2); a justification for the composite designation bpl/birA is given below. The initiation codons of pycB, pycA, and bpl/birA were preceded, respectively, by the following ribosome binding site-type elements (location with respect to the initiation codon): AAGAGG (−13 to −8), AGGTGG (−14 to −9), and ATGGGGT (−9 to −4). Three ORFs were found in the vicinity of the pyc-bpl/birA region (Fig. 3A); ORF1F and ORF2R overlapped each other. None of these ORFs showed significant similarities to protein sequences in the databases. M. barkeri presented the only known example in all three domains of life, where the genes for a biotin-dependent enzyme and the biotinylating enzyme BPL are juxtaposed in an operon-like arrangement (Fig. 3A). In Methanobacterium thermoautotrophicum strain ΔH, only pycA and bpl form such an arrangement (31, 39), and in Methanococcus jannaschii, pycB and pycA are present in close proximity (10, 31). However, in each of these cases data from a transcriptional analysis are needed to determine whether a polycistronic message is indeed synthesized.
Sequence-derived molecular properties for the PYC subunits.
The calculated molecular mass for the biotinylated subunit (MsbPYCB) was 63,735 Da (+226 for the biotin prosthetic group), and that for the nonbiotinylated subunit (MsbPYCA) was 53,684 Da. These values agreed well with the data from mass spectrometry with the purified enzyme (see above). At the primary-structure level, MsbPYCA showed 64% identity and 16% strong similarity to the Methanobacterium thermoautotrophicum PYCA (31, 39), and the corresponding values with respect to the Methanococcus jannaschii PYCA (10, 28, 31) were 59 identity and 19% similarity. The MsbPYCB was 64% identical and 19% strongly similar to Methanobacterium thermoautotrophicum PYCB (31, 39) and 60% identical and 20% strongly similar to Methanococcus jannaschii PYCB (10, 28, 31).
MsbPYCB clearly possessed the structural elements that are characteristics of the carboxytransferase and biotin carboxyl carrier functions, and MsbPYCA bore the features typical of the biotin carboxylase function (reviewed in references 25 and 31). But, a few significant deviations were observed. A PROSITE (University of Geneva) search was unable to identify a putative serine/threonine dehydratase-type pyridoxal-phosphate (PLP) attachment site (accession number PS00165) in MsbPYCB. However, a direct comparison with the mycobacterial PYCB domains (29) and other archaeal PYCBs (31) showed that a PLP attachment site was indeed present in MsbPYCB. (163EELECDSICIKDMAG177, where K is the proposed PLP-binding residue). It is thought that, in biotin-dependent (de)carboxylases, a hinge allows the biotinylated domain to move between the biotin carboxylation (or carboxy-biotin decarboxylation) and the carboxytransfer sites (16, 36). Most α4-type PYCs possess the proposed hinge sequence PX(P/A) at a location ∼29 residues upstream of the canonical biotinylation site (31, 36); proline residues are often found in the hinge regions of proteins (5). However, this element was absent in MsbPYCB. Such a site is also absent in the biotin carrier subunit of acetyl coenzyme A carboxylase or biotin carboxyl carrier protein (BCCP) of E. coli. On the other hand, we found the Tyr(Phe)ProX5Phe(Tyr) element, corresponding to Pro433 of MsbPYCB (106 residues upstream of the biotinylated Lys) to be fully conserved in the biotin-dependent enzymes across the phylogenetic and reaction-type boundaries, including in E. coli BCCP. We suggest that this motif may form a part of the hinge for the “swinging arm” of a biotin-dependent enzyme. E. coli BCCP has been the model for studies of the biotinylation reaction (13, 14). However, the NH2-terminal region of this protein (bearing the proposed hinge element, residues 41 to 48) has not been studied. Since no structure information for this region is available, for the X-ray-crystallographic and nuclear magnetic resonance experiments with BCCP, two clipped versions of this subunit (lacking the first 76 and 69 residues [3, 35]) were used. Our proposal brings this unstudied region into focus. In this context, we note that if E. coli BCCP is excluded from comparison, many other proline residues are found to be conserved in the region NH2 terminal to our proposed hinge element.
A putative E. coli-type bifunctional biotin protein ligase (BPL/BirA) in M. barkeri strain Fusaro.
An alignment with several BPLs by use of CLUSTAL W (41) showed that the putative BPL of M. barkeri belonged to the E. coli class or group II, as described for Fig. 3B. Unlike other putative archaeal BPLs (except that from Archaeoglobus fulgidus) and most bacterial BPLs, M. barkeri BPL possessed an NH2-terminal extension over the catalytic core. Figure 3C shows a multiple-sequence alignment for the NH2-terminal regions of several putative and known BPLs, which revealed several fully conserved residues and several positions with conservative replacements. In E. coli BPL the NH2-terminal extension has been proposed to form a DNA-binding domain (43), which enables this enzyme to act as BirA or the repressor for the biotin operon (13, 14). Such a bifunctional nature (BPL/BirA) is also true for Bacillus subtilis BPL (7). The importance of Ser32, Arg33, and Ala34 of E. coli BPL/BirA and Glu23, Trp38, and Gly59 of the B. subtilis enzyme in repressor activity has been demonstrated (7, 11). Figure 3C shows that these residues as well as several other residues of E. coli and B. subtilis BPLs are conserved in other putative group II enzymes. Thus, it can be conjectured that an E. coli-type (BirA-mediated) control on biotin biosynthesis exists in M. barkeri and A. fulgidus. Interestingly, the above-described conserved residues belong to the B and C helices and not to helix A of E. coli BPL/BirA (43). Helices B and C constitute a helix-turn-helix motif similar to those found in many DNA-binding proteins (43). Helix C probably binds to the operator site of the bio operon in E. coli. Some of the conserved residues shown in Fig. 3C resembled in type and relative locations the DNA-interacting residues of the phage proteins Cro and the λ repressor (34). The identification of the additional conserved residues in the helix-turn-helix motif of E. coli BPL/BirA (Fig. 3C) would now allow us to build more refined models for analyzing this very interesting repressor-target interaction system (14, 43). The NH2 terminus of Arabidopsis thaliana BPL showed significant similarity to the corresponding region of the group II enzymes (Fig. 3C) but lacked most of the conserved residues. This difference might explain why it fails to function as BirA in E. coli (1). It is noteworthy that, in the sequence databases, almost every BPL is called BirA, thus implying a repressor function for the protein, regardless of whether it possesses or lacks the DNA-binding domain.
Conclusion.
The results from this investigation established an apparent α4β4-type PYC as an OAA-biosynthesizing activity in M. barkeri. Thus, this work in conjunction with previously reported information (38, 42) made our understanding of the synthesis of α-KG from the primary substrates (H2-CO2, CH3OH, methylamines, or acetate) in this organism complete. It also showed that PYC is at least a widespread, if not universally present, OAA-synthesizing activity in methanarchaea (28, 31, 37). A structure composed of two types of subunits, a lack of inhibition by aspartate or glutamate, a mild inhibition by α-KG, and inhibition by ATP and excess Mg2+ made MsbPYC similar to other methanarchaeal PYCs (28, 31). The strong competitive (with respect to ATP) inhibition offered by ADP would provide Methanosarcina with control over OAA synthesis if ATP synthesis slows down. This work also provided hypotheses concerning the control of biotin biosynthesis in M. barkeri and A. fulgidus, the hinge sequence for the swinging arm of biotin-dependent enzymes, and functionally important residues in the DNA-binding domains of bifunctional biotin-protein ligases.
Note.
A copy of the alignment establishing a conservation of the proposed hinge element, Tyr(Phe)ProX5Phe(Tyr), in the biotin-dependent enzymes is available upon request.
Nucleotide sequence accession number.
The nucleotide sequence described in this report has been submitted to GenBank and assigned accession number AF317651.
Acknowledgments
We thank Bryce V. Plapp for help in kinetic analysis, John Cronan, Jr., for discussions, and Edward M. Concar for help in preparing the figures.
This work was supported by Department of Energy grant DE-FG02-87ER13651 and National Institutes of Health grant GM 51334.
REFERENCES
- 1.Alban C. Is plant biotin holocarboxylase synthetase a bifunctional enzyme? C R Acad Sci Ser III. 2000;323:681–688. doi: 10.1016/s0764-4469(00)01223-3. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Athappilly F K, Hendrickson W A. Structure of the biotinyl domain of acetyl-coenzyme A carboxylase determined by MAD phasing. Structure. 1995;3:1407–1419. doi: 10.1016/s0969-2126(01)00277-5. [DOI] [PubMed] [Google Scholar]
- 4.Balch W E, Wolfe R S. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium. Appl Environ Microbiol. 1976;32:781–791. doi: 10.1128/aem.32.6.781-791.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergdoll M, Remy M H, Cagnon C, Masson J M, Dumas P. Proline-dependent oligomerization with arm exchange. Structure. 1997;5:391–401. doi: 10.1016/s0969-2126(97)00196-2. [DOI] [PubMed] [Google Scholar]
- 6.Boone D R, Whitman W B, Rouvière P. Diversity and taxonomy of methanogens. In: Ferry J G, editor. Methanogenesis: ecology, physiology, biochemistry, and genetics. New York, N.Y: Chapman & Hall; 1993. pp. 35–80. [Google Scholar]
- 7.Bower S, Perkins J, Yocum R R, Serror P, Sorokin A, Rahaim P, Howitt C L, Prasad N, Ehrlich S D, Pero J. Cloning and characterization of the Bacillus subtilisbirA gene encoding a repressor of the biotin operon. J Bacteriol. 1995;177:2572–2575. doi: 10.1128/jb.177.9.2572-2575.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford M M. A rapid and sensitive approach for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Bryant M P, Tzeng S F, Robinson I M, Joyner A E, Jr, et al. Nutrient requirements of methanogenic bacteria. Adv Chem Ser. 1971;105:23–40. [Google Scholar]
- 10.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Venter A J C, et al. Complete genome sequence of the methanogenic archaeon Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 11.Buoncristiani M R, Howard P K, Otsuka A J. DNA-binding and enzymatic domains of the bifunctional biotin operon repressor (BirA) of Escherichia coli. Gene. 1986;44:255–261. doi: 10.1016/0378-1119(86)90189-7. [DOI] [PubMed] [Google Scholar]
- 12.Cantor C R, Schimmel P R. Biophysical chemistry. II. New York, N.Y: W. H. Freeman Co.; 1980. [Google Scholar]
- 13.Chapman-Smith A, Cronan J E., Jr In vivo enzymatic protein biotinylation. Biomol Eng. 1999;16:119–125. doi: 10.1016/s1050-3862(99)00046-7. [DOI] [PubMed] [Google Scholar]
- 14.Chapman-Smith A, Cronan J E., Jr The enzymatic biotinylation of proteins: a post-translational modification of exceptional specificity. Trends Biochem Sci. 1999;24:359–363. doi: 10.1016/s0968-0004(99)01438-3. [DOI] [PubMed] [Google Scholar]
- 15.Cleland W W. Statistical analysis of enzyme kinetic data. Methods Enzymol. 1979;63:103–138. doi: 10.1016/0076-6879(79)63008-2. [DOI] [PubMed] [Google Scholar]
- 16.Dimroth P. Primary sodium ion translocating enzymes. Biochim Biophys Acta. 1997;1318:11–51. doi: 10.1016/s0005-2728(96)00127-2. [DOI] [PubMed] [Google Scholar]
- 17.Enssle M, Zirngibl C, Linder D, Thauer R K. Coenzyme F420-dependent N5, N10-methylenetetrahydromethaopterin dehydrogenase in methanol grown Methanosarcina barkeri. Arch Microbiol. 1991;155:483–490. [Google Scholar]
- 18.Jansen K, Stupperich E, Fuchs G. Carbohydrate synthesis from acetyl-CoA in the autotroph from Methanobacterium thermoautotrophicum. Arch Microbiol. 1982;132:355–364. [Google Scholar]
- 19.Jerpseth B, Greener A, Short J M, Viola J, Kretz P L. XL-1 Blue MRF′ E. coli cells: McrA−, McrB−, McrF−, Mrr−, HsdR−derivative of XL-1 Blue cells. Strat Mol Biol. 1992;5:81–83. [Google Scholar]
- 20.Kandler O, Hippe H. Lack of peptidoglycan in the cell walls of Methanosarcina barkeri. Arch Microbiol. 1977;113:57–60. doi: 10.1007/BF00428580. [DOI] [PubMed] [Google Scholar]
- 21.Karrasch M, Borner G, Enssle M, Thauer R K. The molybdoenzyme formylmethanofuran dehydrogenase from Methanosarcina barkericontains a pterin cofactor. Eur J Biochem. 1990;194:367–372. doi: 10.1111/j.1432-1033.1990.tb15627.x. [DOI] [PubMed] [Google Scholar]
- 22.Kenealy W R, Zeikus J G. Characterization and function of phosphoenol-pyruvate carboxylase in Methanobacterium thermoautotrophicum. FEMS Microbiol Lett. 1982;14:7–10. [Google Scholar]
- 23.Kunow J, Shima S, Vorholt J A, Thauer R K. Primary structure and properties of the formyltransferase from the mesophilic Methanosarcina barkeri: comparison with the enzymes from thermophilic and hyperthermophilic methanogens. Arch Microbiol. 1996;165:97–105. doi: 10.1007/s002030050303. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Lim F, Morris C P, Occhiodoro F, Wallace J C. Sequence and domain structure of yeast pyruvate carboxylase. J Biol Chem. 1988;263:11493–11497. [PubMed] [Google Scholar]
- 26.Ma K, Thauer R K. N5, N10-Methylenetetrahydromethaopterin reductase from Methanosarcina barkeri. FEMS Microbiol. 1990;70:119–124. [Google Scholar]
- 27.Metcalf W W, Zhang J K, Shi X, Wolfe R S. Molecular, genetic, and biochemical characterization of the serC gene of Methanosarcina barkeriFusaro. J Bacteriol. 1996;178:5797–5802. doi: 10.1128/jb.178.19.5797-5802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mukhopadhyay B, Patel V J, Wolfe R S. A stable archaeal pyruvate carboxylase from the hyperthermophile Methanococcus jannaschii. Arch Microbiol. 2001;174:406–414. doi: 10.1007/s002030000225. [DOI] [PubMed] [Google Scholar]
- 29.Mukhopadhyay B, Purwantini E. Pyruvate carboxylase from Mycobacterium smegmatis: stabilization, rapid purification, molecular and biochemical characterization and regulation of the cellular level. Biochim Biophys Acta. 2000;1475:191–206. doi: 10.1016/s0304-4165(00)00064-7. [DOI] [PubMed] [Google Scholar]
- 30.Mukhopadhyay B, Johnson E F, Wolfe R S. Reactor-scale cultivation of the hyperthermophilic methanarchaeon Methanococcus jannaschiito high cell densities. Appl Environ Microbiol. 1999;65:5059–5065. doi: 10.1128/aem.65.11.5059-5065.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukhopadhyay B, Stoddard S F, Wolfe R S. Purification, regulation, and molecular and biochemical characterization of pyruvate carboxylase from Methanobacterium thermoautotrophicumstrain ΔH. J Biol Chem. 1998;273:5155–5166. doi: 10.1074/jbc.273.9.5155. [DOI] [PubMed] [Google Scholar]
- 32.Mukhopadhyay B, Purwantini E, Pihl T D, Reeve J N, Daniels L. Cloning, sequencing, and transcriptional analysis of the coenzyme F420-dependent methylene-5,6,7,8-tetrahydromethanopterin dehydrogenase gene from Methanobacterium thermoautotrophicum strain Marburg and functional expression in Escherichia coli. J Biol Chem. 1995;270:2827–2832. doi: 10.1074/jbc.270.6.2827. [DOI] [PubMed] [Google Scholar]
- 33.Noll K M, Barber T S. Vitamin contents of archaebacteria. J Bacteriol. 1988;170:4315–4321. doi: 10.1128/jb.170.9.4315-4321.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ptashne M. A genetic switch: gene control in phage λ. Oxford, United Kingdom: Blackwell; 1986. [Google Scholar]
- 35.Roberts E L, Shu N, Howard M J, Broadhurst R W, Chapman-Smith A, Wallace J C, Morris T, Cronan J E, Jr, Perham R N. Solution structures of apo and holo biotinyl domains from acetyl coenzyme A carboxylase of Escherichia colidetermined by triple-resonance nuclear magnetic resonance spectroscopy. Biochemistry. 1999;38:5045–5053. doi: 10.1021/bi982466o. [DOI] [PubMed] [Google Scholar]
- 36.Samols D, Thornton C G, Murtif V L, Kumar G K, Haase F C, Wood H G. Evolutionary conservation among biotin enzymes. J Biol Chem. 1988;263:6461–6464. [PubMed] [Google Scholar]
- 37.Shieh J, Whitman W B. Pathway of acetate assimilation in autotrophic and heterotrophic methanococci. J Bacteriol. 1987;169:5327–5329. doi: 10.1128/jb.169.11.5327-5329.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simpson P G, Whitman W B. Anabolic pathways in methanogens. In: Ferry J G, editor. Methanogenesis: ecology, physiology, biochemistry, and genetics. New York, N.Y: Chapman & Hall; 1993. pp. 445–472. [Google Scholar]
- 39.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Reeve J N, et al. Complete genome sequence of Methanobacterium thermoautotrophicumΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.te Brommelstroet B W, Hensgens C M, Geerts W J, Keltjens J T, van der Drift C, Vogels G D. Purification and properties of 5,10-methenyltetrahydro-methanopterin cyclohydrolase from Methanosarcina barkeri. J Bacteriol. 1990;172:564–571. doi: 10.1128/jb.172.2.564-571.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weimer P J, Zeikus J G. Acetate assimilation pathway of Methanosarcina barkeri. J Bacteriol. 1979;137:332–339. doi: 10.1128/jb.137.1.332-339.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson K P, Shewchuk L M, Brennan R G, Otsuka A J, Matthews B W. Escherichia coli biotin holoenzyme synthetase/biorepressor crystal structure delineates the biotin- and DNA-binding domains. Proc Natl Acad Sci USA. 1992;89:9257–9261. doi: 10.1073/pnas.89.19.9257. [DOI] [PMC free article] [PubMed] [Google Scholar]