Abstract
The European earwig Forficula auricularia is an important model for studies of maternal care, sexual selection, sociality, and host–parasite interactions. However, detailed genetic investigations of this species are hindered by a lack of genomic resources. Here, we present a high-quality hybrid genome assembly for Forficula auricularia using Nanopore long-reads and 10× linked-reads. The final assembly is 1.06 Gb in length with 31.03% GC content. It consists of 919 scaffolds with an N50 of 12.55 Mb. Half of the genome is present in only 20 scaffolds. Benchmarking Universal Single-Copy Orthologs scores are ∼90% from 3 sets of single-copy orthologs (eukaryotic, insect, and arthropod). The total repeat elements in the genome are 64.62%. The MAKER2 pipeline annotated 12,876 protein-coding genes and 21,031 mRNAs. Phylogenetic analysis revealed the assembled genome as that of species B, one of the 2 known genetic subspecies of Forficula auricularia. The genome assembly, annotation, and associated resources will be of high value to a large and diverse group of researchers working on dermapterans.
Keywords: Forficula auricularia, hybrid genome assembly, repeatome, genome annotation
Introduction
Insects have been at the forefront of genetic research for various biological questions (Wilson-Sanders 2011; Mukherjee et al. 2015; Simons and Tibbetts 2019). However, most of the genetic studies are carried out on a small number of holometabolous insects that undergo true metamorphosis. In contrast to Holometabola, hemimetabolous insects undergo incomplete metamorphosis with a series of nymphal molts that increasingly resemble the adult form (Truman 2019). It is widely accepted that Holometabola branched out from hemimetabolous ancestors during the Permian 300 Mya (Labandeira and Phillips 1996; Yang 2001). Yet the conserved mode of development, embryonic organization, and the adult body plan of hemimetabolous insects offer a unique model for the study of developmental and evolutionary mechanisms. However, even with the increasing number of sequenced genomes, the majority belong to the Holometabola (Ylla et al. 2021). This has been a bottleneck for the exploration of the diverse biology and life history of hemimetabolous insects. To address this paucity, we report a high-quality annotated genome of the European earwig, Forficula auricularia (Dermaptera: Forficulidae).
The European earwig F. auricularia is widely distributed, comprising 2 recognized subspecies, A and B (Wirth et al. 1998). They are native to the western Eurasian region and were introduced to North America, Australia, and New Zealand, where they have quickly adapted and became abundant throughout the regions (Quarrell et al. 2018; Tourneur and Meunier 2019). The 2 subspecies A and B differ through mitochondrial divergence and in their reproductive life histories (Guillet, Guiller, et al. 2000; Guillet, Josselin, et al. 2000). Subspecies A is found in relatively colder climates and is univoltine with a long gregarious phase, whereas subspecies B is found in temperate and oceanic climates and is bivoltine (Lamb and Wellington 1974). In laboratory conditions, they fail to produce offspring by cross mating (Wirth et al. 1998). Their propensity to dwell on flower and kitchen gardens can cause significant damage to crops, flowers, and commercial vegetables and make them important agricultural pests (Campos et al. 2011; Hill et al. 2019).
They have been of particular interest for many researchers not just because of their importance in the agricultural ecosystem (Binns et al. 2021) but also their importance as a research model for various biological and evolutionary phenomena like sexual selection, maternal care, family interactions, reproductive strategy, and social behavior (Forslund 2000; Falk et al. 2014; Kramer et al. 2015; Van Meyel and Meunier 2020). They have been extensively studied by behavioral ecologists for the early evolution of group-living and family life (Falk et al. 2014). The male earwigs also show an unusual bias in their use of lateral left and right sexual organs without any conspicuous anatomical differentiation (Kamimura 2006). Like the right-handedness in humans, 90% of males of giant earwig Labidura riparia show a preference for the right penis for copulation, providing insights into the evolutionary origin of lateralization (Kamimura et al. 2021). Similarly, they are an excellent lab model to study extended phenotypes as they exhibit strange suicidal water-seeking behavior during the late stages of infection by mermithid nematodes (Herbison et al. 2019). However, their use as a genetic model has been severely limited by the lack of a reference genome.
Here, we have sequenced, assembled, annotated, and analyzed the genome of the European Earwig, F. auricularia and confirmed the subspecies identity of the individuals we used. This genome will help researchers study multiple facets of this insect’s exciting biology and evolutionary characters and broaden our understanding of insect and genome evolution.
Methods and materials
Sample collection and preparation
Earwigs (F. auricularia) were field collected from the Dunedin Botanic Garden (−45°51′27.59″S, 170°31′15.56″E) and reared in a temperature-controlled room (temperature: cycling from 15 to 12°C, day/night; photoperiod of L:D 16:8) in the Department of Zoology, University of Otago, Dunedin. Earwigs were snap-frozen in liquid nitrogen and stored at −80°C before dissection and subsequent nucleotide extraction. Earwigs were dissected in 1× PBS buffer under a dissection microscope to check for nematode parasites, and only nonparasitized individuals were used in this study. The head, wings and muscles from the thorax region were used for DNA extraction to avoid the gut microbiota. Juvenile instars required for RNA extraction were obtained directly from the field.
DNA extractions
DNA was extracted using either the Nanobind Tissue Big DNA kit (Circulomics, USA) for high molecular weight DNA or DNeasy Blood & Tissue Kit (Qiagen, Germany) by following the manufacturer’s protocol. Tissues from a single individual were used for each extraction. After the extraction, RNase treatment was performed using 4 µl of RNase A (10 mg/ml) per 200 µl of DNA elute. DNA was quantified in a Qubit 2.0 Fluorometer (Life Technologies, USA) and quality analyzed using Nanodrop. Low-quality DNA samples were further cleaned with 1.8× by volume AMPure XP beads (Beckman Coulter, USA), wherever applicable, following the manufacturer’s instructions and eluted in 55 µl of molecular grade water. High-quality DNA samples were stored at −20°C and were used within a week of extraction.
Linked-read library preparation and sequencing
Linked-read library was prepared at the Genetic Analysis Services (GAS), University of Otago (Dunedin, New Zealand). DNA from an adult male was extracted using the Nanobind kit and size-selected for fragments over 40 kbp using Blue Pippin (Sage Science, USA). A 10× linked-reads (10× Genomics, USA) library was prepared following the manufacturer’s instructions. The library was sequenced on the Illumina Nova-seq platform to generate 2 × 151-bp paired-end reads (Garvan Institute, Australia).
Long-read library preparation and sequencing
Five long-read sequencing libraries for Oxford Nanopore MinION were prepared using the Ligation Sequencing Kit (SQK-LSK109) (Oxford Nanopore Technologies, Oxford, UK) following the manufacturer’s instructions. To increase the raw Nanopore read N50, the first and the second libraries were prepared using 1.75 and 0.75 µg of DNA extracted via a Circulomics kit from 2 adult male earwigs. Both libraries were sequenced in a single Minion flow cell, flushing the flow cell to remove remains of the first library before loading the second library with a Flow Cell Wash Kit (EXP-WSH004) (Oxford Nanopore Technologies, Oxford, UK).
To increase the total raw output, the third and the fourth libraries were prepared with DNA from 2 adult female earwigs, both extracted with a DNeasy Blood & Tissue, Qiagen kit followed by the AMPure XP beads clean-up step. Input DNA for these 2 libraries were 2.6 and 3.2 µg. These were each sequenced on an individual minion flow cell. The fifth library was prepared using 3.0 µg of DNA from an adult male earwig. As before DNA was extracted using a DNeasy Blood & Tissue, Qiagen kit followed by AMPure XP beads clean-up. However, before library preparation, the DNA was sheared 5 times using a 26 G × 0.5″ needle (Terumo, Japan). Since the sample type and the extraction method can impact the molecular weight of extracted DNA and the nanopore sequencing output, we tried samples from both sexes and different extraction protocols to optimize our sequencing output. All prepared libraries were sequenced with R9 chemistry MinION flow cell (FLO-MIN106) (Oxford Nanopore Technologies, UK) on a MinION connected to a laptop and operated with MinKNOW (v.2.0) interface.
RNA extraction and sequencing
Total RNA from the different developmental stages, sex, and tissues was extracted using a Direct-zol RNA MicroPrep kit (Zymo Research, USA) with an on-filter DNAse treatment following the manufacturer’s instructions. Samples included: whole body (gut removed) of juvenile instars 1–2 and juvenile instars 3–4, dissected tissues (antennae, head, thorax, abdomen, legs, and gonads) of adult males and females. RNA from each individual and tissue type was extracted separately. RNA was quantified on a Qubit 2.0 Fluorometer (Life Technologies, USA) and initially quality checked using a nanodrop. Only high-quality extracts were further processed and were stored at −80°C until use.
RNA integrity was evaluated on a Fragment Analyzer (Advanced Analytical Technologies Inc., USA) at the Otago Genomics Facility (OGF), University of Otago, Dunedin, New Zealand. As with most of the insect RNA extracts (Winnebeck et al. 2010) RNA quality number (RQN) values ranged from 2.5 to 10 due to the collapsing of the 28S peak; quality was thus determined via the trace rather than RQN. Four pools of samples at equimolar concentration underwent library preparation. Pools consisted of: 8 whole body extractions for juvenile instar 1–2, 8 whole body extractions for juvenile instar 3–4, individual body tissues from 5 adult males, and individual body tissues from 5 adult females. TruSeq stranded mRNA libraries were prepared and sequenced as 2 × 100-bp paired-end reads across 2 lanes of HiSeq 2500 Rapid V2 flowcell at the OGF.
Genome size estimation
Flow cytometry and k-mer-based approach with short-read data were used to estimate the genome size. Flow cytometry analysis was performed on a single head of earwig with 2 biological replicates at Flowjoanna (Palmerston North, NZ, USA). Briefly, the earwig’s head was dissociated with a pestle in 500 µl of the stock solution containing 0.1% w/v trisodium citrate dihydrate, 0.1% v/v IGEPAL, 0.052% w/v spermine tetrahydrochloride, and 0.006% sigma 7–9 (all Sigma-Aldrich, USA). Rooster red blood cells (RRBC) derived from the domestic chicken (Gallus gallus), stored in citrate buffer, were used as reference samples. Test samples were filtered through a 35-µl filter cap and further dissociated by adding 100 µl of 0.21 mg/ml trypsin followed by 75 µl of 2.5 mg/ml trypsin inhibitor (both Sigma-Aldrich) for 10 min at 37°C. Nuclei were stained using 100 µl of prestain (containing 416 mg/ml propidium iodide with 500 mg/ml RNAse in-stock solution). Two sample tubes, 1 prepared with RRBC and 1 prepared without, were then processed on a FACSCalibur (BD Biosciences, USA). The instrument was equipped with a 488-nm laser to produce fluorescence collected using the FL-2-Area signal (585/42 BP), along with forward scatter and side scatter signals that enabled RRBC nuclei to be resolved from earwig nuclei. Data were analyzed using Flowjo (BD Biosciences, USA) and the pg/nuclei of the sample calculated.
For k-mer-based genome size estimation, we used the paired-end linked-read sequences. Reads were processed with the scaff_reads script from Scaff10x (v.5.0) (https://github.com/wtsi-hpag/Scaff10X) to remove the 10× link adapters. Quality control was carried out with Trimmomatic (v.0.39) (Bolger et al. 2014) (options: SLIDINGWINDOW:4:15 LEADING:5 TRAILING:5 MINLEN:35). We used KMC (v.3.1.1) (Kokot et al. 2017) with a k-mer size of 21 to count the k-mers, the histogram produced was then visualized in Genomescope (v.2.0) (Ranallo-Benavidez et al. 2020) web browser.
Phylogenetic analysis
Two sibling species of F. auricularia have been described (Wirth et al. 1998). To assess which of these we sequenced, nucleotide sequences covering the COI and COII region from 34 isolates of F. auricularia were downloaded from NCBI. Those included 15 isolates reported by Wirth et al. (1998) originally used to infer sibling species A and B and other isolates from Belgian orchards submitted to NCBI. Nucleotide sequence covering COI and COII regions from the assembled genome was extracted through BLAST hits. To ensure that a single subspecies was sequenced across all the individuals, raw reads from each run were blasted back to this sequence to ensure the presence of a single haplotype. The same genomic region extracted from the mitochondrial genome of Euborellia arcanum was used as an outgroup. Nucleotide sequences were aligned using Clustal Omega (v1.2.3) (Goujon et al. 2010). The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei 1987) with 1,000 bootstrap replicates (Felsenstein 1985). The evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al. 2004) and are in the units of the number of base substitutions per site. All ambiguous positions were removed for each nucleotide sequence pair (pairwise deletion option). There were a total of 799 positions in the final dataset. The optimal tree is presented and the evolutionary analyses were conducted in MEGA11 (Tamura et al. 2021).
Bioinformatic pipeline
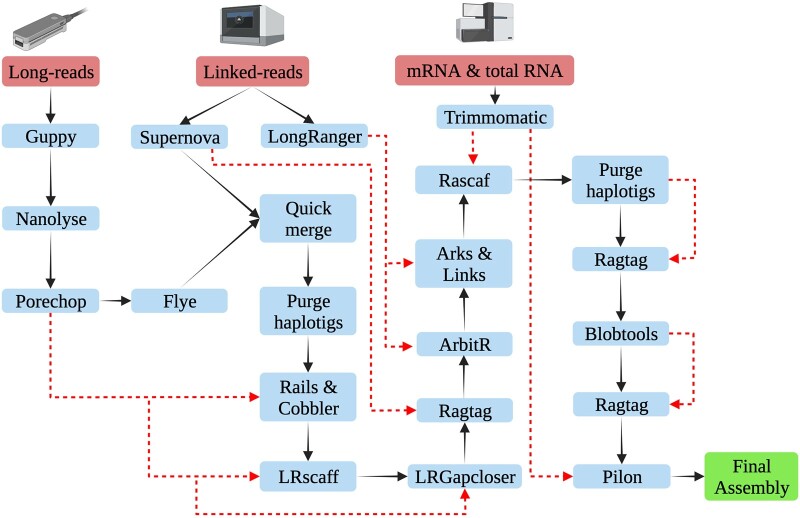
All the scripts used for genome assembly, de novo repeat library construction, and annotation are available on GitHub (https://github.com/upendrabhattarai/Earwig_Genome_Project). The bioinformatics software and packages were run in New Zealand eScience Infrastructure. Below is a description of the pipeline (Fig. 1).
Fig. 1.
Schematic representation of the assembly pipeline for the F. auricularia genome. The solid black arrow represents the workflow and the red dotted lines represent the additional input data in the pipeline (created with BioRender.com).
Genome assembly
Paired-end Illumina reads from the Chromium library were assembled using Supernova (v.2.1.1) (Weisenfeld et al. 2017). Assembly metrics such as N50 values and contig/scaffold number were assessed using Quast (v.5.0.2) (Gurevich et al. 2013) and the presence of the single-copy ortholog genes was assessed using the insecta_odb10 database in BUSCO (v.5.1.3) (Simão et al. 2015). BUSCO score from Quast analysis wherever mentioned used BUSCO version 3.0.2 and the eukaryote_odb9 database. Based on several trial assemblies, we down-sampled the total input to 660 million paired-end reads using “—maxreads” option with “supernova run” to produce an assembly with better completeness and contiguity. The assembled fasta sequence was obtained with “pseudohap” style of the supernova “mkoutput” function.
Nanopore reads were basecalled using Guppy (v.5.0.7) (Wick et al. 2019) and processed with Nanolyse (v.1.2.0) (De Coster et al. 2018) and Porechop (v.0.2.4) (Wick et al. 2017) to remove lamda DNA and adapters from the raw reads. The reads were then assembled using Flye (v.2.7.1) (Kolmogorov et al. 2019) with default parameters. The Flye assembly had higher N50 and BUSCO scores compared to the Supernova assembly so we used the Flye assembly as a reference and supernova assembly as a query assembly in Quickmerge (v.0.3) (Chakraborty et al. 2016) to improve the contiguity and completeness of the assembly. The resulting assembly was processed with Purgehaplotigs (v.1.0.0) (Roach et al. 2018) to remove unpaired allelic contigs.
The purged genome underwent further scaffolding and gap-closing steps using Rails (v.1.5.1) and Cobbler (v.0.6.1) (Warren 2016), Lrscaf (v.1.1.11) (Qin et al. 2019), and Lrgapcloser (Xu et al. 2019) with the raw Nanopore long-read data. The resulting assembly was scaffolded with Ragtag (v.2.1.0) (Alonge et al. 2021) using the Supernova assembly. The raw linked-read data was aligned to the assembly with Long Ranger (v.2.0) (Ott et al. 2018) and used to further scaffold with ArbitR (v.0.2) (Hiltunen et al. 2021), Arks (v.1.0.4) (Coombe et al. 2018), and Links (v.1.8.7) (Warren et al. 2015). mRNA-seq reads sequenced for genome annotation purposes, and total RNA-seq reads sequenced for another project (manuscript under preparation) were also used for scaffolding the assembly with Rascaf (v.1.0.2) (Song et al. 2016). Duplicated and redundant haplotigs were again removed using Purgehaplotigs (Roach et al. 2018), and discarded haplotigs were used for scaffolding the assembly using Ragtag.
BlobTools2 (Laetsch and Blaxter 2017) was used to remove small (<1,000 bp) and low coverage contigs (<5× coverage). We followed the tutorial provided by the developers of the BlobTools2 in the genomehubs website for creating, updating, filtering, and generating plots (see more at: https://blobtoolkit.genomehubs.org/blobtools2/blobtools2-tutorials/). Contigs that were filtered out were used for re-scaffolding the assembly with Ragtag (v.1.0.2) (Alonge et al. 2021). Finally, we used 1 iteration of Pilon (v.1.24) (Walker et al. 2014) to polish the exonic region of the assembly using mRNA-seq data.
Repeat content analysis
To assist with annotation a custom repeat library was generated for the Earwig genome using different de novo repeat and homology-based identifiers, including LTRharvest (v.1.5.10) (Ellinghaus et al. 2008), LTRdigest (v.1.5.10) (Steinbiss et al. 2009), RepeatModeler (v.2.0) (Flynn et al. 2020), TransposonPSI (v.1.0.0) (Haas 2010), and SINEBase (v.1.1) (Vassetzky and Kramerov 2013). We concatenated the individual libraries, and sequences with more than 80% similarity were merged to remove redundancy using usearch (v.11.0.667) (Edgar 2010). The library was then classified with RepeatClassifier (v.2.0) (Flynn et al. 2020). Sequences with unknown categories in the library were mapped against the UniProtKB/Swiss-Prot database (e-value <1e−01); if sequences were not annotated as repeat sequences they were removed from the library. The final repeat library was used in RepeatMasker (v.4.1.2) (Chen 2004) to generate a report for genome repeat content and provided to the MAKER2 pipeline to mask the genome.
Genome annotation
Genome annotation was carried out with 3 iterations of the MAKER2 (v.2.31.9) (Holt and Yandell 2011) pipeline combining evidence-based and ab initio gene models. The first round of MAKER2 used evidence-based models and the other 2 rounds were run using ab initio gene models. For the first round, we provided the MAKER2 pipeline with 180,119 mRNA transcripts denovo assembled via the Trinity pipeline (v.2.13.2) (Grabherr et al. 2011) along with 26,414 mRNA and 1,529 protein sequences of dermapterans from NCBI and 779 dermapteran protein sequences from the Uniprot database.
Augustus was trained using BRAKER (v.2.16) (Hoff et al. 2019) and SNAP was trained after each round of MAKER2 to use for ab initio gene model prediction. For the functional annotation, we ran InterProScan (v.5.51-85.0) (Jones et al. 2014) for the predicted protein sequences obtained from MAKER2 and retrieved InterPro ID, PFAM domains, and Gene Ontology (GO) terms. Furthermore, we ran BLASTp (Altschul et al. 1990) with the Uniprot database to assign gene descriptors to each transcript based on the best BLAST hit.
Results and discussion
Genome size estimates
The flow cytometer estimated the genome size of 968.22 ± 20.747 Mb (mean ± SD) for the earwig genome. Similarly, the k-mer-based approach using adapter removed paired-end data from linked-read sequencing estimated the male earwig to be 988 Mb. Whereas an earlier estimation of an unknown dermapteran (earwig) species genome size was 1.4 Gb (Gregory 2005) showing a variable genome size within the order.
Phylogenetic analysis
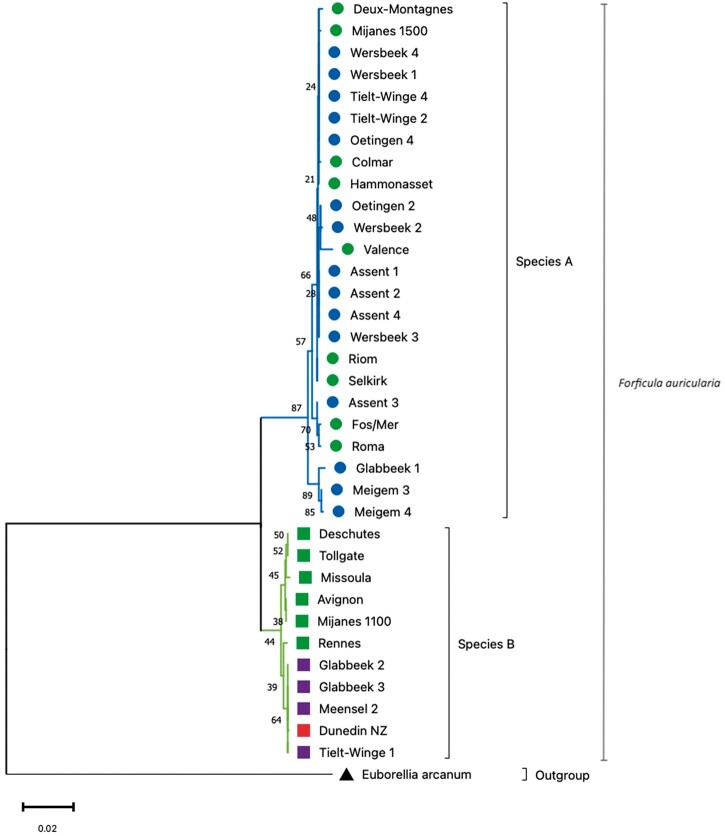
The phylogenetic analysis showed 2 distinct subspecies groups within F. auricularia (Fig. 2), in agreement with Wirth et al. (1998). One clade includes 24 individuals including 9 originally identified as species A (green circle labels, Fig. 2). While the remaining 11 individuals cluster into a separate clade that include the assembled genome herein (red square label, Fig. 2) and 6 individuals originally identified as species B (green square labels, Fig. 2). The analysis confirmed that the genome reported in this article (Dunedin, NZ) belongs to the subspecies B of F. auricularia. This is also in accordance with the report from Quarrell et al. (2018) where 2 isolates from New Zealand were reported as subspecies B of F. auricularia.
Fig. 2.
The phylogenetic relationships of F. auricularia obtained from different geographic regions inferred from COI and COII using a Neighbour-Joining method and Maximum Composite Likelihood approach in MEGA11. All ambiguous positions were removed for each nucleotide sequence pair (pairwise deletion). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Species labeled with the colored squares are subspecies B. The red square (Dunedin NZ) is the one for which the genome is reported in this article. Green squares are the species categorized as subspecies B by Wirth et al. (1998) and the purple squares are others for which the nucleotide sequences were downloaded from NCBI. Species labeled with colored circles belong to subspecies A. Green circles represent subspecies A inferred by Wirth et al. (1998) and blue are other species for which nucleotide sequences were downloaded from NCBI. E. arcanum is the outgroup labelled with a black triangle.
Genome assembly
A total of 799.6 million paired-end reads was generated from 10× linked-read sequencing. Downsampled to 660 million paired-end reads, Supernova estimated the genome size of 1.22 Gb, raw coverage of 82.02%, effective coverage of 39.50% and weighted mean molecule size of 22.45 kb. The Supernova assembly was 1.15 Gb in size and had 145,055 contigs, with an N50 of 0.03 Mb and L50 of 7,500. Quast reported a complete BUSCO of 64.69% and a partial BUSCO of 9.24% from the eukaryotic database.
The Nanopore sequencing yielded approximately 10.7 Gb of data, consisting of over 3 million reads. The median read length was 897 bp with an N50 length of 11,986 bp (Supplementary Table 1). The median read Phred quality was 13.34. Flye produced an assembly of 1.1 Gb, comprised of 18,766 contigs with N50 of 0.18 Mb and L50 of 1,832. Quast reported a complete BUSCO of 82.18% and a partial BUSCO of 9.24%. The long-read assembly was more complete based on the BUSCO scores and demonstrated better contiguity, so we merged the 2 assemblies using the Flye assembly as the primary assembly (Table 1).
Table 1.
Assembly statistics at different stages of assembly for the genome of the European earwig F. auricularia.
| Assembly length | No. scaffolds | N50 | L50 | Ns per 100 kbp | BUSCO % (Quast) |
||
|---|---|---|---|---|---|---|---|
| Complete | Partial | ||||||
| Supernova assembly | 1,145,470,221 | 145,055 | 30,358 | 7,500 | 3,677.89 | 64.69 | 9.24 |
| Flye assembly | 1,118,374,848 | 18,766 | 180,737 | 1,832 | 0.35 | 82.18 | 9.24 |
| Final hybrid assembly | 1,062,210,345 | 919 | 12,548,649 | 20 | 846.85 | 87.13 | 2.97 |
The Supernova and the Flye assembly statistics are for the assembly right after the assembler and no further processing, whereas the Final hybrid assembly shows the statistics of the assembly through all the assembly process as described in this article. Quast scores are to its default Eukaryota database.
The BlobTools2 filtering produced a clean assembly with only 215 contigs out of 2.7 K assigned as no-hits and all other contigs with blast hits to the Arthropoda database (Supplementary Fig. 1).The final hybrid assembly has a size of 1.06 Gb. It has 919 scaffolds with an N50 of 12.55 Mb, which shows that the assembly is highly contiguous. Half of the genome is present in just 20 scaffolds, as denoted by the L50 number (Table 1). Assembly has 846.85 “N’s” per 100kbp. The BUSCO score from the insect database (n = 1,367) for the assembly is 87.1% complete, among which 4.1% were duplicated, and 3.1% fragmented BUSCO (Supplementary Fig. 2). Improvement in assembly statistics after each processing step is given in Supplementary Table 2.
The only other whole-genome sequence publicly available from the Dermaptera order is of the earwig Anisolabis maritima [GenBank assembly accession: GCA_010014785.1, available to download from InsectBase (v.2)]. The A. maritima genome assembly is 649.7 Mb with a N50 of 1.4 Mb, (Mei et al. 2022), while its BUSCO score is 83.4% complete and 10.8% fragmented using the insect database (n = 1,367). In comparison, the F. auricularia genome assembly has a better gene model and contiguity.
Genome repeat contents
Repeat analysis of the assembly showed that interspersed repeats comprised 686.43 Mb (64.62%) of the F. auricularia genome. This includes 248.24 Mb of retroelements (23.37% of the genome), 178.33 Mb of DNA transposons (16.79% of the genome), 35.83 Mb of rolling circles (3.28% of the genome) and 260.87 Mb of unclassified elements (Table 2). Unusually large and variable genome sizes characterize Hemimetabolans (Wu et al. 2017). Comparative analysis in 6 species of Gomphocerine grasshoppers showed a strong positive correlation between repeat content and genome size. Genome size ranged from 8.2 to 13.7 Gb in these 6 species with a repeat content ranging from 79% to 87%, with the exception of Stauroderus scalaris whose genome is 96% repetitive DNA and the second-largest insect genome documented. Our estimation of genome size for F. auricularia does not show gigantism (968.22 Mb, flow cytometer estimate). However, its repeatome (64.62%) is almost twice that of other hemimetabolous insects like Gryllus bimaculatus (33.69%) and Laupala kohalensis (35.51%) (Ylla et al. 2021). This fold increase in the repeatome is surprising given both G. bimaculatus and L. kohalensis have bigger genomes (1.6 Gb) than F. auricularia.
Table 2.
Repeat content analysis in the European earwig Forficula auricularia genome.
| No. sequences | 919 | ||
|---|---|---|---|
| Total length (bp) | 1,062,210,345 | ||
| GC level | 31.03% | ||
| Bases masked | 722,769,501 bp (68.04%) |
||
| Numbers | Length (bp) | Percentage | |
| Retroelements | 1,385,007 | 248,236,495 | 23.37 |
| SINEs | 41,157 | 5,138,497 | 0.48 |
| Penelope | 50,409 | 10,372,837 | 0.98 |
| LINEs | 660,178 | 124,985,146 | 11.77 |
| CRE/SLACS | 0 | 0 | 0.00 |
| L2/CR1/Rex | 112,418 | 20,654,321 | 1.94 |
| R1/LOA/Jockey | 167,317 | 22,277,052 | 2.10 |
| R2/R4/NeSL | 23,348 | 4,271,189 | 0.40 |
| RTE/Bov-B | 136,406 | 28,799,096 | 2.71 |
| L1/CIN4 | 10,079 | 1,892,539 | 0.18 |
| LTR elements | 683,672 | 118,112,852 | 11.12 |
| BEL/Pao | 60,561 | 12,114,300 | 1.14 |
| Ty1/Copia | 97,132 | 14,352,992 | 1.35 |
| Gypsy/DIRS1 | 521,467 | 91,083,363 | 8.57 |
| Retroviral | 3,701 | 443,583 | 0.04 |
| DNA transposons | 1,040,870 | 178,326,460 | 16.79 |
| hobo-Activator | 362,395 | 59,188,939 | 5.57 |
| Tc1-IS630-Pogo | 355,781 | 66,331,225 | 6.24 |
| En-Spm | 0 | 0 | 0.00 |
| MuDR-IS905 | 0 | 0 | 0.00 |
| PiggyBac | 21,153 | 2,726,812 | 0.26 |
| Tourist/Harbinger | 5,541 | 1,187,174 | 0.11 |
| Other (Mirage, P-element, Transib) | 10,240 | 1,580,945 | 0.15 |
| Rolling circles | 174,964 | 34,830,487 | 3.28 |
| Unclassified | 1,563,937 | 259,874,747 | 24.47 |
| Total interspersed repeats | 686,437,702 | 64.62 | |
| Small RNA | 9,913 | 1,406,877 | 0.13 |
| Satellites | 1,110 | 495,561 | 0.05 |
| Simple repeats | 0 | 0 | 0.00 |
| Low complexity | 0 | 0 | 0.00 |
Genome annotation
Combining evidence-based and ab initio gene models in the MAKER2 pipeline, we identified 12,876 genes and 21,031 mRNAs in the genome assembly. The mean gene length is 12,096 bp and the total gene length is 155.75 Mb, which makes 14.7% of the whole assembly. The longest gene annotated is 412,198 bp and the longest CDS is 19,035 bp (Table 3). 61.35% of total predicted mRNAs and 59.53% of predicted proteins are also functionally annotated through either 1 or more of InterPro, GO, and Pfam databases (Supplementary Table 3). The annotated transcriptome and proteome had a complete BUSCO score of 73.4% and 70% respectively using the insect database (Supplementary Fig. 3). 98.3% of the gene models have AED score of 0.5 or less, assuring highly confident gene prediction (Supplementary Fig. 4).
Table 3.
Genome annotation summary for the European earwig Forficula auricularia.
| Total sequence length | 1,062,210,345 |
|---|---|
| Number of genes | 12,876 |
| Number of mRNAs | 21,031 |
| Number of exons | 145,003 |
| Number of introns | 123,973 |
| Number of CDS | 21,030 |
| Total gene length | 155,753,058 |
| Total mRNA length | 271,884,000 |
| Total exon length | 32,584,454 |
| Total intron length | 239,538,939 |
| Total CDS length | 23,936,568 |
| Longest gene | 412,198 |
| Longest mRNA | 412,198 |
| Longest exon | 10,240 |
| Longest intron | 319,382 |
| Longest CDS | 19,035 |
| Mean gene length | 12,096 |
| Mean mRNA length | 12,928 |
| Mean exon length | 225 |
| Mean intron length | 1,932 |
| Mean CDS length | 1,138 |
| % of genome covered by genes | 14.7 |
| % of genome covered by CDS | 2.3 |
| Mean mRNAs per gene | 2 |
| Mean exons per mRNA | 7 |
| Mean introns per mRNA | 6 |
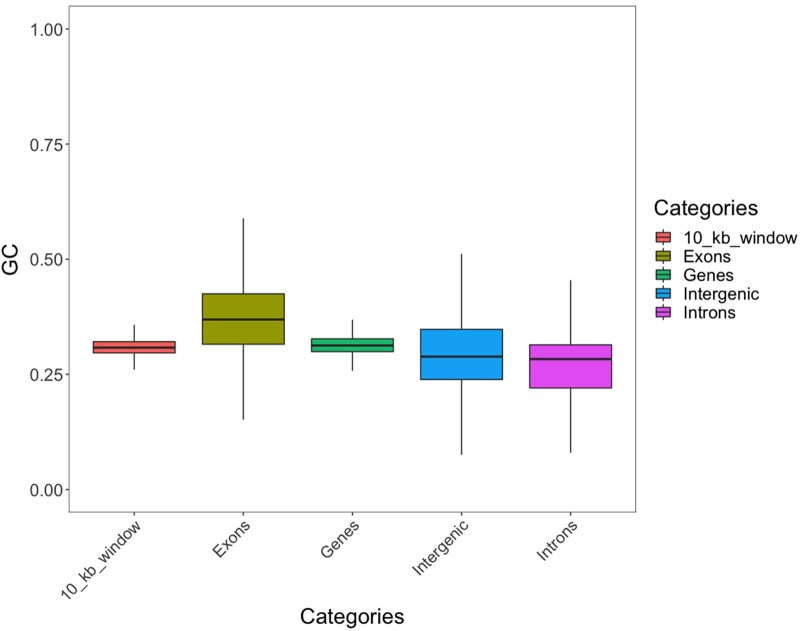
The GC content of the F. auricularia genome is 31.03%, far greater than the 19.3% GC in the genome of the earwig A. maritima reported in InsectBase2 database (Mei et al. 2022). So we compared the GC content between different regions of F. auricularia genome to see if there are any abnormal distributions. Our analysis showed that exons have higher GC content (0.372 ± 0.087) (mean ± SD) and introns have lower (0.267 ± 0.075) when compared between intergenic regions (N = 823,037), genes (N = 12,876), exons (N = 145,003), introns (N = 123,973), and nonoverlapping 10-kb windows throughout the genome (N = 106,686) (Fig. 3). GC content for 10-kb windows was 0.308 ± 0.032, which resembles the mean GC content of the whole genome (0.310). This finding is not unexpected as a higher GC content in exons vs. introns is common across the animal and plant kingdom because of the evolutionary selection of exon regions (Amit et al. 2012). There was a significant difference for each pairwise comparison using ANOVA followed by Tukey HSD with P < 0.0001.
Fig. 3.
GC percentage in different genomic features of the F. auricularia genome. GC content for 10-kb windows was generated without regard to any genomic features. Whiskers extend to 25th and 75th percentiles. GC content in exons is higher and in introns is lower compared to the genome average.
Recently there has been a growing interest in hemimetabolous insects for use as genetic research models and hence sequencing and analyzing their genomes (Adamski et al. 2019; Ylla et al. 2021). Because of their primitive yet successful biology, the evolutionary insights they can offer for various biological traits are enormous. Genomes of milkweed bug (Oncopeltus fasciatus) (Panfilio et al. 2019) and field cricket (G. bimaculatus) (Ylla et al. 2021) have been instrumental for developmental biology research. Similarly, the genome of Rhodnius prolixus, a medically important hemimetabolous insect vector, provides key insights into the genetic re-organization contributing to the evolution of a blood-feeding lifestyle (Mesquita et al. 2015). Furthermore, the genome of Halyomorpha halys has informed research on polyphagy and insecticide resistance and contributed to advances in research on insect–pest control strategies (Sparks et al. 2020). In this context, we believe that, the genome of F. auricularia will be a key resource to develop this important insect species as a genetic model. We anticipate this will enhance the genetic study on various aspects of its biology, including developmental biology, sociality, and evolutionary characteristics.
Supplementary Material
Acknowledgments
We thank the support team at New Zealand eScience Infrastructure (NeSI) for their help running various software on their HPC platforms and the staff at Dunedin Botanical Garden, Dunedin, New Zealand, for their help during earwig collection in the field.
Funding
This study was funded by the Royal Society Te Apārangi Marsden Fund grant (16-UOO-152).
Conflicts of interest
None declared.
Contributor Information
Upendra R Bhattarai, Department of Anatomy, University of Otago, Dunedin 9016, New Zealand.
Mandira Katuwal, Department of Anatomy, University of Otago, Dunedin 9016, New Zealand.
Robert Poulin, Department of Zoology, University of Otago, Dunedin 9016, New Zealand.
Neil J Gemmell, Department of Anatomy, University of Otago, Dunedin 9016, New Zealand.
Eddy Dowle, Department of Anatomy, University of Otago, Dunedin 9016, New Zealand.
Data Availability
The genome assembly and annotation of F. auricularia are available through FigShare https://doi.org/10.6084/m9.figshare.19092044. The raw sequencing reads are deposited in NCBI with accession number PRJNA800435.
The scripts used for genome assembly, repeat library preparation and masking, and genome annotation are available at GitHub under GNU GPLv3 license (https://github.com/upendrabhattarai/Earwig_genome_project).
Supplemental material is available at G3 online.
Literature cited
- Adamski Z, Bufo SA, Chowański S, Falabella P, Lubawy J, Marciniak P, Pacholska-Bogalska J, Salvia R, Scrano L, Słocińska M, et al. Beetles as model organisms in physiological, biomedical and environmental studies—a review. Front Physiol. 2019;10:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonge M, Lebeigle L, Kirsche M, Aganezov S, Wang X, Lippman ZB, Schatz MC, Soyk S. Automated assembly scaffolding elevates a new tomato system for high-throughput genome editing. bioRxiv 2021.11.18.469135, 2021. 10.1101/2021.11.18.469135 [DOI] [PMC free article] [PubMed]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Amit M, Donyo M, Hollander D, Goren A, Kim E, Gelfman S, Lev-Maor G, Burstein D, Schwartz S, Postolsky B, et al. Differential GC content between exons and introns establishes distinct strategies of splice-site recognition. Cell Rep. 2012;1(5):543–556. [DOI] [PubMed] [Google Scholar]
- Binns M, Hoffmann AA, van Helden M, Heddle T, Hill MP, Macfadyen S, Nash MA, Umina PA.. Lifecycle of the invasive omnivore, Forficula auricularia, in Australian grain growing environments. Pest Manag Sci. 2021;77(4):1818–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos MR, Picanço MC, Martins JC, Tomaz AC, Guedes RNC.. Insecticide selectivity and behavioral response of the earwig Doru luteipes. Crop Prot. 2011;30(12):1535–1540. [Google Scholar]
- Chakraborty M, Baldwin-Brown JG, Long AD, Emerson JJ.. Contiguous and accurate de novo assembly of metazoan genomes with modest long read coverage. Nucleic Acids Res. 2016;44:gkw654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinforma. 2004;Chapter 4:Unit 4.10. [DOI] [PubMed] [Google Scholar]
- Coombe L, Zhang J, Vandervalk BP, Chu J, Jackman SD, Birol I, Warren RL.. ARKS: chromosome-scale scaffolding of human genome drafts with linked read kmers. BMC Bioinformatics. 2018;19(1):234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Coster W, D'Hert S, Schultz DT, Cruts M, Van Broeckhoven C.. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics. 2018;34(15):2666–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. [DOI] [PubMed] [Google Scholar]
- Ellinghaus D, Kurtz S, Willhoeft U.. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics. 2008;9:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk J, Wong JWY, Kölliker M, Meunier J.. Sibling cooperation in earwig families provides insights into the early evolution of social life. Am Nat. 2014;183(4):547–557. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution (N Y). 1985;39(4):783. [DOI] [PubMed] [Google Scholar]
- Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, Smit AF.. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci U S A. 2020;117(17):9451–9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund P. Male–male competition and large size mating advantage in European earwigs, Forficula auricularia. Anim Behav. 2000;59(4):753–762. [DOI] [PubMed] [Google Scholar]
- Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R.. A new bioinformatics analysis tools framework at EMBL–EBI. Nucleic Acids Res. 2010;38(Web Server issue):W695–W699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory TR. Genome size evolution in animals. In: Gregory TR, editor. The Evolution of the Genome. Burlington: Elsevier; 2005. p. 3–87. [Google Scholar]
- Guillet S, Guiller A, Deunff J, Vancassel M.. Analysis of a contact zone in the Forficula auricularia L. (Dermaptera: Forficulidae) species complex in the Pyrenean Mountains. Heredity (Edinb). 2000; 85(5):444–449. [DOI] [PubMed] [Google Scholar]
- Guillet S, Josselin N, Vancassel M.. Multiple introductions of the Forficula auricularia species complex (Dermaptera: Forficulidae) in eastern North America. Can Entomol. 2000; 132(1):49–57. [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G.. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. TransposonPSI: An Application of PSI-Blast to Mine (Retro-)Transposon ORF Homologies; Cambridge, MA, USA: Broad Institute, 2010.
- Herbison REH, Evans S, Doherty J-F, Poulin R.. Let’s go swimming: mermithid-infected earwigs exhibit positive hydrotaxis. Parasitology. 2019;146(13):1631–1635. [DOI] [PubMed] [Google Scholar]
- Hill MP, Binns M, Umina PA, Hoffmann AA, Macfadyen S.. Climate, human influence and the distribution limits of the invasive European earwig, Forficula auricularia, in Australia. Pest Manag Sci. 2019;75(1):134–143. [DOI] [PubMed] [Google Scholar]
- Hiltunen M, Ryberg M, Johannesson H.. ARBitR: an overlap-aware genome assembly scaffolder for linked reads. Bioinformatics. 2021;37(15):2203–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff KJ, Lomsadze A, Borodovsky M, Stanke M.. Whole-genome annotation with BRAKER. Methods Mol Biol. 2019;1962:65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C, Yandell M.. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 2011;12:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura Y. Right-handed penises of the earwig Labidura riparia (Insecta, Dermaptera, Labiduridae): evolutionary relationships between structural and behavioral asymmetries. J Morphol. 2006;267(11):1381–1389. [DOI] [PubMed] [Google Scholar]
- Kamimura Y, Matsumura Y, Yang C-CS, Gorb SN.. Random or handedness? Use of laterally paired penises in Nala earwigs (Insecta: Dermaptera: Labiduridae). Biol J Linn Soc. 2021;134(3):716–731. [Google Scholar]
- Kokot M, Dlugosz M, Deorowicz S.. KMC 3: counting and manipulating k-mer statistics. Bioinformatics. 2017;33(17):2759–2761. [DOI] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, Pevzner PA.. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37(5):540–546. [DOI] [PubMed] [Google Scholar]
- Kramer J, Thesing J, Meunier J.. Negative association between parental care and sibling cooperation in earwigs: a new perspective on the early evolution of family life? J Evol Biol. 2015;28(7):1299–1308. [DOI] [PubMed] [Google Scholar]
- Labandeira CC, Phillips TL.. A Carboniferous insect gall: insight into early ecologic history of the Holometabola. Proc Natl Acad Sci U S A. 1996;93(16):8470–8474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laetsch DR, Blaxter ML.. BlobTools: interrogation of genome assemblies. F1000Research. 2017;6:1287. [Google Scholar]
- Lamb RJ, Wellington WG.. Techniques for studying the behavior and ecology of the European earwig, Forficula auricularia (Dermaptera: Forficulidae). Can Entomol. 1974;106(8):881–888. [Google Scholar]
- Mei Y, Jing D, Tang S, Chen X, Chen H, Duanmu H, Cong Y, Chen M, Ye X, Zhou H, et al. InsectBase 2.0: a comprehensive gene resource for insects. Nucleic Acids Res. 2022;50(D1):D1040–D1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesquita RD, Vionette-Amaral RJ, Lowenberger C, Rivera-Pomar R, Monteiro FA, Minx P, Spieth J, Carvalho AB, Panzera F, Lawson D, et al. Genome of Rhodnius prolixus, an insect vector of Chagas disease, reveals unique adaptations to hematophagy and parasite infection. Proc Natl Acad Sci U S A. 2015;112(48):14936–14941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meyel S, Meunier J.. Filial egg cannibalism in the European earwig: its determinants and implications in the evolution of maternal egg care. Anim Behav. 2020;164:155–162. [Google Scholar]
- Mukherjee K, Twyman RM, Vilcinskas A.. Insects as models to study the epigenetic basis of disease. Prog Biophys Mol Biol. 2015;118(1–2):69–78. [DOI] [PubMed] [Google Scholar]
- Ott A, Schnable JC, Yeh C-T, Wu L, Liu C, Hu H-C, Dalgard CL, Sarkar S, Schnable PS.. Linked read technology for assembling large complex and polyploid genomes. BMC Genomics. 2018;19(1):651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panfilio KA, Vargas Jentzsch IM, Benoit JB, Erezyilmaz D, Suzuki Y, Colella S, Robertson HM, Poelchau MF, Waterhouse RM, Ioannidis P, et al. Molecular evolutionary trends and feeding ecology diversification in the Hemiptera, anchored by the milkweed bug genome. Genome Biol. 2019;20(1):64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin M, Wu S, Li A, Zhao F, Feng H, Ding L, Ruan J.. LRScaf: improving draft genomes using long noisy reads. BMC Genomics. 2019;20(1):955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarrell SR, Arabi J, Suwalski A, Veuille M, Wirth T, Allen GR.. The invasion biology of the invasive earwig, Forficula auricularia in Australasian ecosystems. Biol Invasions. 2018;20(6):1553–1565. [Google Scholar]
- Ranallo-Benavidez TR, Jaron KS, Schatz MC.. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat Commun. 2020;11(1):1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach MJ, Schmidt SA, Borneman AR.. Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinformatics. 2018;19(1):460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M.. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Simons M, Tibbetts E.. Insects as models for studying the evolution of animal cognition. Curr Opin Insect Sci. 2019;34:117–122. [DOI] [PubMed] [Google Scholar]
- Song L, Shankar DS, Florea L.. Rascaf: improving genome assembly with RNA sequencing data. Plant Genome. 2016;9. doi: 10.3835/plantgenome2016.03.0027. [DOI] [PubMed] [Google Scholar]
- Sparks ME, Bansal R, Benoit JB, Blackburn MB, Chao H, Chen M, Cheng S, Childers C, Dinh H, Doddapaneni HV, et al. Brown marmorated stink bug, Halyomorpha halys (Stål), genome: putative underpinnings of polyphagy, insecticide resistance potential and biology of a top worldwide pest. BMC Genomics. 2020;21(1):227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbiss S, Willhoeft U, Gremme G, Kurtz S.. Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res. 2009;37(21):7002–7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Nei M, Kumar S.. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101(30):11030–11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Kumar S.. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol. 2021;38(7):3022–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourneur J-C, Meunier J. Thermal regimes, but not mean temperatures, drive patterns of rapid climate adaptation at a continent-scale: evidence from the introduced European earwig across North America. bioRxiv 550319, 2019. 10.1101/550319 [DOI]
- Truman JW. The evolution of insect metamorphosis. Curr Biol. 2019;29(23):R1252–R1268. [DOI] [PubMed] [Google Scholar]
- Vassetzky NS, Kramerov DA.. SINEBase: a database and tool for SINE analysis. Nucleic Acids Res. 2013;41(Database issue):D83–D89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren RL. RAILS and Cobbler: scaffolding and automated finishing of draft genomes using long DNA sequences. JOSS. 2016;1(7):116. [Google Scholar]
- Warren RL, Yang C, Vandervalk BP, Behsaz B, Lagman A, Jones SJM, Birol I.. LINKS: scalable, alignment-free scaffolding of draft genomes with long reads. Gigascience. 2015;4:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenfeld NI, Kumar V, Shah P, Church DM, Jaffe DB.. Direct determination of diploid genome sequences. Genome Res. 2017;27(5):757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR, Judd LM, Gorrie CL, Holt KE.. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb. genomics. 2017;3(10):e000132. e000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR, Judd LM, Holt KE. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome biol. 2019;20:129. https://doi.org/10.1186/s13059-019-1727-y. [DOI] [PMC free article] [PubMed]
- Wilson-Sanders SE. Invertebrate models for biomedical research, testing, and education. ILAR J. 2011;52(2):126–152. [DOI] [PubMed] [Google Scholar]
- Winnebeck EC, Millar CD, Warman GR.. Why does insect RNA look degraded? J Insect Sci. 2010;10(159):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T, Le Guellec R, Vancassel M, Veuille M.. Molecular and reproductive characterization of sibling species in the European earwig (Forficula auricularia). Evolution (N Y). 1998;52(1):260. [DOI] [PubMed] [Google Scholar]
- Wu C, Twort VG, Crowhurst RN, Newcomb RD, Buckley TR.. Assembling large genomes: analysis of the stick insect (Clitarchus hookeri) genome reveals a high repeat content and sex-biased genes associated with reproduction. BMC Genomics. 2017;18(1):884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G-C, Xu T-J, Zhu R, Zhang Y, Li S-Q, Wang H-W, Li J-T.. LR_Gapcloser: a tiling path-based gap closer that uses long reads to complete genome assembly. Gigascience. 2019;8(1): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang AS. Modularity, evolvability, and adaptive radiations: a comparison of the hemi- and holometabolous insects. Evol Dev. 2001;3(2):59–72. [DOI] [PubMed] [Google Scholar]
- Ylla G, Nakamura T, Itoh T, Kajitani R, Toyoda A, Tomonari S, Bando T, Ishimaru Y, Watanabe T, Fuketa M, et al. Insights into the genomic evolution of insects from cricket genomes. Commun Biol. 2021;4(1):733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome assembly and annotation of F. auricularia are available through FigShare https://doi.org/10.6084/m9.figshare.19092044. The raw sequencing reads are deposited in NCBI with accession number PRJNA800435.
The scripts used for genome assembly, repeat library preparation and masking, and genome annotation are available at GitHub under GNU GPLv3 license (https://github.com/upendrabhattarai/Earwig_genome_project).
Supplemental material is available at G3 online.