Abstract
The regulation of stem cell survival, self-renewal, and differentiation is critical for the maintenance of tissue homeostasis. Although the involvement of signaling pathways and transcriptional control mechanisms in stem cell regulation have been extensively investigated, the role of post-transcriptional control is still poorly understood. Here, we show that the nuclear activity of the RNA-binding protein Second Mitotic Wave Missing is critical for Drosophila melanogaster intestinal stem cells and their daughter cells, enteroblasts, to maintain their progenitor cell properties and functions. Loss of swm causes intestinal stem cells and enteroblasts to stop dividing and instead detach from the basement membrane, resulting in severe progenitor cell loss. swm loss is further characterized by nuclear accumulation of poly(A)+ RNA in progenitor cells. Second Mitotic Wave Missing associates with transcripts involved in epithelial cell maintenance and adhesion, and the loss of swm, while not generally affecting the levels of these Second Mitotic Wave Missing-bound mRNAs, leads to elevated expression of proteins encoded by some of them, including the fly ortholog of Filamin. Taken together, this study indicates a nuclear role for Second Mitotic Wave Missing in adult stem cell maintenance, raising the possibility that nuclear post-transcriptional regulation of mRNAs encoding cell adhesion proteins ensures proper attachment of progenitor cells.
Keywords: swm, cheerio, rhea, intestinal stem cells, cell adhesion
Introduction
Understanding the biology and behavior of stem cells and their niches is important because of their critical role in maintaining adult tissue homeostasis throughout life. Stem cell proliferation, survival, and differentiation are tightly regulated by both cell-autonomous and noncell-autonomous factors whose mis-regulation can compromise tissue integrity (Morrison and Spradling 2008; Losick et al. 2011). Despite the interest in studying stem cell behaviors, how stem cell populations maintain cell identity and stemness to deliver proper cell and tissue functions is less well understood.
The Drosophila midgut epithelium serves as an ideal model to study stem cell behaviors because this high turnover tissue hosts a stem cell population, known as intestinal stem cells (ISCs), that gives rise to a simple cell lineage (Fig. 1a). In this lineage, ISCs are basally located and physically attached to a basement membrane that neighbors a layer of visceral muscle. ISCs are mitotically active in the midgut and often divide asymmetrically to produce a new ISC and a transient enteroblast (EB) that differentiates into an enterocyte (EC) through activation of the Notch signaling pathway (Micchelli and Perrimon 2005; Ohlstein and Spradling 2005, 2007; Biteau et al. 2011). At the same time, a small percentage of ISCs generate enteroendocrine (ee) cells through asymmetric division or direct differentiation (Amcheslavsky et al. 2014; Biteau and Jasper 2014; Guo and Ohlstein 2015; Zeng and Hou 2015). The asymmetric renewal of an ISC can generate one ISC and an intermediate ee progenitor cell (also known as a pre-ee) that divides one more time resulting in 2 differentiated ee cells (Chen et al. 2018). ISCs and EBs are collectively known as intestinal progenitors.
Fig. 1.
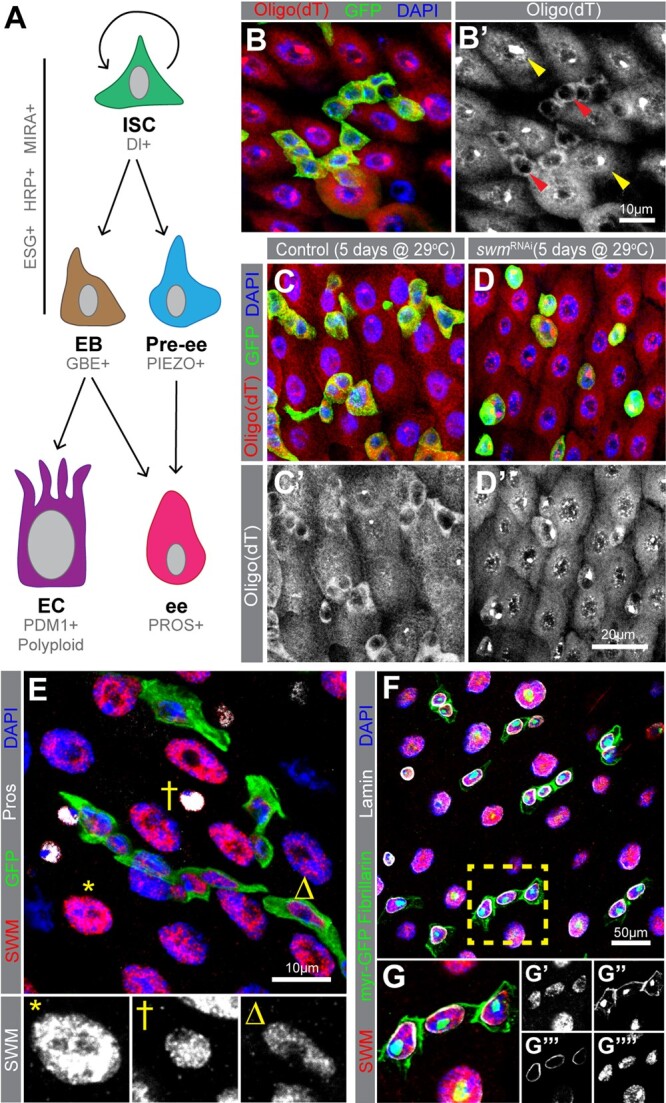
Knockdown of swm in intestinal progenitor cells leads poly(A)+ RNA accumulation in nuclei. a) A schematic of intestinal cell types and markers. In this study, we refer to ESG+ cells as progenitor cells (Ps), which include both ISCs and EBs. b) Poly(A)+ RNA distribution in cells from esgTS intestines, labeled by oligo (dT) probes (red) and Ps are labeled with GFP (anti-GFP in green) and nuclei with DAPI (blue). b′) Poly(A)+ RNA distribution is shown in gray scale in Ps (red arrowheads) and ECs (yellow arrowheads). Sections from (c) esgTS and (d) esgTS; swmRNAi PMGs after 5 days at 29°C stained with oligo (dT) (red), GFP (green), and DAPI (blue). e) PMG section from esgTS stained for Swm (anti-Swm in red). Ps are shown in GFP (green), ees are stained for Prospero (anti-Prospero in white) and nuclei for DAPI (blue). Enlarged EC (*), ee (†) and P (Δ) show Swm in gray scale. Individual channels of (e) are shown in Supplementary Fig. 1. (f and g) PMG region from UAS-myr GFP driven by esg-GAL4 stained for Swm (anti-Swm in red) and counterstained for cell membrane of Ps (myr-GFP in green), nucleoli of all cells (anti-Fibrillarin, nuclear green), nuclear membranes of all cells (anti-Lamin in white), and nuclei (DAPI in blue). (g) Enlargement of cells indicated in (e) with individual channels for Swm in red (g′), cell membranes (myr-GFP) and nucleoli (Fibrillarin) in green (g″), nuclear membranes in white (g′′′) and nuclei in blue (g′′′′). Complete genotypes are listed in Supplementary Table 1. P, progenitor cell; EC, enterocyte; ee, enteroendocrine cell; Pros, Prospero; PMG, posterior midgut.
Because the transcriptomes of the 2 progenitor cell types are largely similar, there are multiple markers that identify progenitors but few that are specific to either ISCs or EBs (Dutta et al. 2015; Hung et al. 2020). Progenitors express the transcription factor Escargot (Esg) while ISCs are identified by the expression of the Notch ligand, Delta (Dl), and EBs are identified as cells that express Notch signaling reporters. Ee cells express the transcription factor Prospero (Pros) and ECs express the transcription factor Pou domain-1 (Pdm-1) (Fig. 1a) (Micchelli and Perrimon 2005; Ohlstein and Spradling 2005; Jiang et al. 2011).
Unlike many other stem cell populations, ISCs are not associated with any specialized supporting cells making it difficult to define their exact niche. Growing evidence, however, supports the notion that the visceral muscle and basement membrane, including its extracellular matrix (ECM) proteins, serve as a basal niche to support ISCs (Lin et al. 2008; Jiang et al. 2009; Buchon et al. 2010; Biteau and Jasper 2011; O’Brien et al. 2011). A number of signaling pathways, including Wnt, Hippo, Notch, EGFR, Insulin, BMP/Dpp, and JAK/STAT, as well as environmental signals, including chemicals, nutrients, and pathogens, are involved in regulating ISC proliferation, survival and differentiation as well as maintenance of other cell types (Miguel-Aliaga et al. 2018). Recent studies have indicated that, in addition to the transcriptional regulation provided by these pathways, post-transcriptional regulation also plays critical roles in ISCs. For example, post-transcriptional gene regulation through RNA-binding proteins (RBPs) and small RNAs, such as microRNAs (miRNAs), have been shown to add important layers to regulate ISC behaviors both under homeostatic and externally challenged conditions (Foronda et al. 2014; Chen et al. 2015; Luhur et al. 2017; Buddika et al. 2020; Shanahan et al. 2020; Buddika, Huang, et al. 2021; Mukherjee et al. 2021). In other stem cell types, such as human and mouse embryonic stem cells as well as Drosophila S2R+ cells, nuclear post-transcriptional processes including alternative splicing, polyadenylation, RNA export, and nuclear RNA decay have been shown to regulate potency, self-renewal, and differentiation (Herold et al. 2003; Hurt et al. 2009; Wang et al. 2013; Chen and Hu 2017; Silla et al. 2020). However, the role of nuclear post-transcriptional processes in regulating stem cell behaviors of in vivo tissue-based stem cell models remain poorly understood.
Here, we used the Drosophila midgut as a tissue-based stem cell model to study nuclear post-transcriptional regulatory mechanisms. We identify the RBP Second Mitotic Wave Missing (Swm) as a required factor for maintaining intestinal progenitors and post-transcriptionally regulating mRNAs. Our study reveals the necessity of nuclear RBP activity for Drosophila intestinal progenitor cell survival and maintenance.
Materials and methods
Drosophila strains and husbandry
The full genotypes of all fly strains used in this study are listed in Supplementary Table 1. Age matched mated female flies were used in all experiments. All fly strains were cultured on standard Bloomington media in 18°C, 25°C, or 29°C incubators set for a 12 h light/dark schedule and 65% humidity. For temporal and regional gene expression-targeting (TARGET) experiments, flies were reared at 18°C, upon eclosion collected progeny over 2 days, aged 2 more days at 18°C and shifted to 29°C up to 10 days before being dissected. For clonal analysis using mosaic analysis with repressible cell marker (MARCM) technique, flies were grown at 25°C until eclosion, collected 0–2 days old animals and heat-shocked immediately at 38°C for 40 min in a Lauda circulating water bath. Following heat-shock, flies were reared at 25°C up to 10 days. For bleomycin feeding assays, flies were reared on standard media until 4 days after eclosion at 18°C. Then, half the flies were transferred to vials containing a chromatography paper soaked in 5% sucrose in water (control) and the other half to 5% sucrose augmented with 25 µg/ml bleomycin. Flies were then shifted to 18°C or 29°C incubators based on the experiment and dissected after 24 h of feeding.
Construction of new strains
swm RNAi-2 was generated by first predicting effective shRNAs targeting the swm 3’UTR using splashRNA (Pelossof et al. 2017) and then annealing and subcloning oligos encoding one of these shRNAs (TTAACAATTATATATCCGCGTA) into the EcoRI and NheI sites of pWalium20. The transgene was subsequently inserted into the VK33 landing site by Rainbow Genetics (Camarillo, CA).
Antibody generation
Anti-Swm antibodies were generated in rats (Cocalico Biologicals) against a 6× HIS-tagged fragment of the first 220 amino acids of Swm that was expressed and purified according to standard methods. This Swm-encoding plasmid (pNIK1384) was generated by PCR amplifying the swm coding region from cDNA LD45403 (obtained from the Drosophila Genomics Resource Center) using oligonucleotides 4,290 (acgaccgaaaacctgtattttcagggcgccATTCTGGAGAATTCGGACAAGCTCAAGGAT), 4,292 (actagttgagctcgtcgacgtaggcctttgTCAAAGACCTGCTCCGCCGGGACCACCTCC), and high-fidelity Q5 polymerase (NEB), subcloning the resulting PCR product into the NcoI and EcoRI sites of pHIS.parallel (Sheffield et al. 1999) using HiFi DNA assembly master mix (NEB), and sequence verifying the resulting plasmid to confirm the absence of any PCR-induced errors.
Dissections and immunostaining
Gastrointestinal tracts of adult female flies were dissected in 1× PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, KH2PO4, pH 7.4), fixed in 4% paraformaldehyde (Electron Microscopy Sciences, Cat. No. 15714) in 1× PBS for 45 min, washed 3 times in 1× PBT (1× PBS, 0.1% Triton X-100) and then blocked (1× PBT, 0.5% bovine serum albumin) for 45 min. Subsequently, samples were incubated at 4°C overnight with primary antibodies, including mouse anti-Prospero (MR1A, Developmental Studies Hybridoma Bank, 1:100), mouse anti-V5 (MCA1360GA, Bio-Rad, 1:250), and rabbit anti-GFP (A11122, Life Technologies, 1:1,000), rat anti-Swm (this study, 1:50), rat anti-Cheerio (Sokol and Cooley 1999) (1:1,000), mouse anti-Talin (A22A and E16B, Developmental Studies Hybridoma Bank, 1:1 mixture, 1:50 each), mouse anti-Lamin (ADL67.10, Developmental Studies Hybridoma Bank, 1:50), mouse anti-Coracle (C566.9, Developmental Studies Hybridoma Bank, 1:50), mouse anti-Discs large (4F3, Developmental Studies Hybridoma Bank, 1:100), mouse anti-Headcase (U33, Developmental Studies Hybridoma Bank, 1:3), mouse anti-Osa (Osa, Developmental Studies Hybridoma Bank, 1:20), mouse anti-Shot (mAbRod1, Developmental Studies Hybridoma Bank, 1:20), and rabbit anti-Fibrillarin (ab5821, Abcam, 1:500). The following day, samples were washed in 1× PBT and incubated for 2–3 h with secondary antibodies, including AlexaFluor 488- and 568-conjugated goat antirabbit, -mouse, -rat and -chicken antibodies (Life Technologies, 1:1,000). AlexaFluor 647-conjugated goat-HRP antibodies were used in the secondary antibody solution whenever required. Finally, samples were washed multiple times in 1× PBT, including one wash with 5 μg/ml DAPI in PBT, and mounted in Vectashield mounting medium (Vector Laboratories). An alternative staining protocol was used for mouse anti-Delta (C594.9B, Developmental Studies Hybridoma Bank, 1:500) staining as described in Buddika, Xu, et al. (2021) and these samples were mounted in ProLong Diamond mounting medium (Invitrogen, P36970). Intestines stained with anti-Delta antibody were also stained with anti-GFP antibody, since the methanol steps required in this protocol quenched GFP fluorescence. Cell death analysis was performed using the ApopTag Fluorescein in situ Apoptosis Detection Kit (Sigma-Aldrich, Cat. No. S7110) following manufacturer’s instructions.
Oligo-dT fluorescent in situ hybridization
Oligo-dT fluorescent in situ hybridization was performed as described in Buddika et al. (2020) and briefly, adult female intestines were dissected out in ice cold 1× PBS and fixed in 4% w/v paraformaldehyde (Electron Microscopy Sciences, Cat. No. 15714) in PBS for 45 min while rocking. Tissue was then washed in 1× PBT (1× PBS, 0.3% v/v Triton X-100) 3 times 5 min each. When protein immunostaining is required, samples were blocked in RNase free blocking solution (0.3% v/v PBT, 0.5% ultra-pure Bovine Serum Albumin) (Ambion, Cat. No. AM2616) for 45 min and primary and secondary antibody stainings were carried out. After secondary antibody staining, samples were washed 3× in 0.3% v/v PBT and subjected to a second sample fixation in 4% w/v paraformaldehyde (Electron Microscopy Sciences, Cat. No. 15714) in PBS for 45 min while rocking. Tissue was then washed in 1× PBT (1× PBS, 0.3% v/v Triton X-100) 3 times 5 min each. Subsequently, samples were gradually dehydrated in a series of 0.3% v/v PBT (1× PBS, 0.3% v/v Triton X-100):Methanol (7:3, 1:1, 3:7) washes and incubated in 100% Methanol for 10 min. Next, tissue was rehydrated with a series of 0.3% v/v PBT:Methanol (3:7, 1:1, 7:3) washes and finally washed in 0.3% v/v PBT for another 5 min. Then samples were rinsed once in hybridization wash buffer (20% formamide, 2× SSC, DEPC-treated water) and washed in hybridization wash buffer for 10 min. Then the wash buffer was completely removed and oligo-dT probes were added in 50 μl of ULTRAhyb-Oligo buffer (Ambion, Cat. No. AM8663). Samples were incubated overnight at 37°C. The following day, probe solution was removed, and samples were washed in hybridization wash buffer 3 times for 20 min each and DAPI was added in the second wash. Finally, samples were mounted using ProLong Diamond mounting solution (Invitrogen, Cat. No. P36971).
RNAscope in situ hybridization
RNAscope in situ hybridization was performed as described in Buddika, Huang, et al. (2021) with some changes. Briefly, adult female flies were dissected in ice cold 1× PBS and fixed in 4% w/v paraformaldehyde (Electron Microscopy Sciences, Cat. No. 15714) in PBS for 2 h while rocking. Tissue was then washed in 1× PBT (1× PBS, 0.3% v/v Triton X-100) 3 times 5 min each. Samples were gradually dehydrated in a series of 0.3% v/v PBT (1× PBS, 0.3% v/v Triton X-100):Methanol (7:3, 1:1, 3:7) washes and incubated in 100% Methanol for 10 min. Then tissue was rehydrated with a series of 0.3% v/v PBT:Methanol (3:7, 1:1, 7:3) washes and finally washed with 0.3% v/v PBT for another 5 min. For next steps, reagents from RNAscope Multiplex Fluorescent Reagent Kit v2 assay were used. Fixed tissue was transferred into 0.2 mL PCR tubes and incubated in RNAscope Protease III reagent for 5 min at 40°C (all the incubations at 40°C were done in a PCR thermal cycler). Samples were immediately washed with 1× PBS twice. RNAscope probes for cher (50×) (pre warmed to 40 °C) (ACD Bio., Cat. No. 1050721-C2) was diluted in RNAscope Probe Diluent (ACD Bio., Cat. No. 300041) to have 1× concentration and added 20 μl per sample. Next, samples were incubated at 40 °C overnight. Following day, samples were washed twice with 1× RNAscope wash buffer. Next, RNAscope Multiplex FL v2 AMP 1, RNAscope Multiplex FL v2 AMP 2, RNAscope Multiplex FL v2 AMP 3 and RNAscope Multiplex FL v2 HRP-C2 steps were done as described in Chapter 4 of Fluorescent v2 assay manual, ACD Bio, 2019. Finally, samples were incubated for 30 min at 40 °C with Opal 620 (AKOYA Biosciences, Cat. No. FP1495001KT, 1:1,500 in TSA buffer) and washed with 1× RNAscope wash buffer and counterstained with DAPI. Samples were mounted in ProLong Diamond mounting medium (Invitrogen).
Microscopy and image processing
Images of whole dissected intestines were collected on a Zeiss Axio Zoom microscope. Images of immunostained intestines were collected on a Leica SP8 Scanning Confocal microscope. Unless otherwise indicated, the posterior region R4-b and c, as defined by Buchon et al. (2013), was used for all microscopic analyses. Samples to be compared were collected under identical settings on the same day, image files were adjusted simultaneously using Adobe Photoshop CC, and figures were assembled using Adobe Illustrator CC. Image J FIJI (https://fiji.sc/; accessed 2022 July 29) was used to quantify the fluorescence intensity. For the cell detachment experiment, images of single cells (or 2 cells) of 10× magnification were collected. For each image, starting from the basal side of the cell, 40–45 Z sections of 0.3 µm were imaged, and 3D models were built using the Leica LAS software.
Cell quantification
Field of view images of 1× magnification were collected from 5 to 8 female intestines and cells were counted manually using the count tool of Adobe Photoshop CC. The percentage of progenitor cells was calculated as the number of Esg+ cells (GFP+) per field of view divided by the total number of DAPI+ cells per field of view and multiplied by 100. For the swm mutant cell clone counting, all the GFP+ cell clones of posterior midguts (R4 and R5) from 5 intestines of each genotype were imaged and used for cell quantification.
Western blot analysis
Adult female flies were used for protein isolation. Whole adult flies were lysed in I-RIPA protein lysis buffer (150 mM NaCl, 50 mM Tris–HCl pH 7.5, 1 mM EDTA, 1% Triton X-100, 1% Na Deoxycholic Acid, 1× protease inhibitor cocktail). Prepared protein extracts were resolved on a 4–20% gradient polyacrylamide gel (Bio-Rad, Cat. No. 456-1093), transferred to Immobilon-P membrane (Millipore, Cat. No. IPVH00010) and probed with rat anti-Swm (this study, 1:100), rabbit anti-GFP (ab290, Abcam, 1:10,000) or mouse anti-a-tubulin (12G10, Developmental Studies Hybridoma Bank, 1:1,000) antibodies. After washing with 1× TBST (1× TBS, 0.1% Tween-20) blots were incubated with anti-rat, -rabbit, -or -mouse conjugated HRP secondary antibodies. Subsequently, blots were washed with 1× TBST, treated with ECL-detection reagents (Thermo Scientific, Cat. No. 1859701 and 1859698) and finally exposed to chemiluminescence films (GE Healthcare, Cat. No. 28906839).
RNA immunoprecipitation, CLIP-seq library preparation, and qPCR
Gastrointestinal (GI) tracts were dissected from 200 adult female flies (per replicate) in ice-cold 1× PBS. Dissected intestines were placed on a petri dish with minimum amount of 1× PBS as a monolayer and irradiated 3 times at 2000J in a UV cross-linker, with mixing between each irradiation to maximize surface exposure. Immediately after irradiation, samples were snap frozen in liquid nitrogen and stored at −80°C until all the samples are prepared. Same day, each frozen tissue sample was lysed in 1 ml of I-RIPA protein lysis buffer (150 mM NaCl, 50 mM Tris–HCl pH 7.5, 1 mM EDTA, 1% Triton X-100, 1% Na Deoxycholic Acid, 1× protease inhibitor cocktail, RNase inhibitor). After centrifugation 50 µl of supernatant was saved as the total input and the rest was incubated with Swm antibody coated Dynabeads Protein G (Invitrogen, Cat. No. 10003D) for 2 h at 4°C while rocking. Beads were collected on a magnetic stand and washed with fresh lysis buffer containing RNase inhibitor. Subsequently, Swm-bound RNA was released from beads using Proteinase K (Ambion, Cat. No. AM2546) treatment and TRIzol LS reagent (Ambion, Cat. No. 10296028) was used to isolate immunoprecipitated RNA. The rRNA-depleted libraries were prepared using the Ovation SoLo RNA-seq system (Part No. 0502 including Parts 0407 and S02240) following the manufacturer’s instructions. The quality and the quantity of final libraries were assessed using Agilent 2200 TapeStation and KAPA Library Quantification Kit, respectively. For qPCR, isolated RNA was first treated with Turbo DNase (ThermoFisher, Cat. No. AM2239) and gDNA-free RNA was used for cDNA synthesis with Superscript III (ThermoFisher, Cat. No. 56575). qPCR was performed using the PowerUp SYBR Green Master Mix (ThermoFisher, Cat. No. A25742) in a StepOnePlus machine (ThermoFisher). Primers for all targets detected are listed in Supplementary Table 2. Transcript levels were quantified in triplicates and normalized to Gapdh1. Fold enrichment was calculated as the ratio of transcript in SWM IP vs total input.
FACS isolation of progenitor cells and RNA-seq library preparation
Gastrointestinal (GI) tracts were dissected from 80 to 100 adult female flies (per replicate) in ice-cold 1× PBS. Then cells were dissociated by treating intestines with 1 mg/ml elastase at 27°C for 1 h with agitation. Subsequently, ∼25,000–50,000 GFP+ intestinal progenitor cells were sorted using a BD FACSAria II flow cytometer equipped with a 100-µm nozzle at the IUB Flow Cytometry Core Facility. Total RNA was prepared using the TRI reagent (Molecular Research Center, Cat. No. TR118). The rRNA-depleted libraries were prepared using the Ovation SoLo RNA-seq system (Part No. 0502 including Parts 0407 and S02240) following the manufacturer’s instructions. The quality of final libraries was assessed using Agilent 2200 TapeStation and libraries were quantified using KAPA Library Quantification Kit.
CLIP-seq and RNA-seq data analysis
Swm CLIP and transcriptomic data analysis were performed as described in Buddika, Xu, et al. (2021) using a python based in-house pipeline (https://github.com/jkkbuddika/RNA-Seq-Data-Analyzer; accessed 2022 July 29). First, the quality of raw sequencing files was assessed using FastQC version 0.11.9. Then, reads with low quality were removed using Cutadapt (Martin 2011) version 2.9. Next, the remaining reads were mapped to the Berkeley Drosophila Genome Project (BDGP) assembly release 6.28 (Ensembl release 100) reference genome using STAR genome aligner (Dobin et al. 2013) version 2.7.3a and duplicated reads were eliminated using SAMtools (Li et al. 2009) version 1.10. Subsequently, the Subread (Liao et al. 2019) version 2.0.0 function featureCounts was used to count the number of aligned reads to the nearest overlapping feature. All subsequent analysis and data visualization steps were performed using custom scripts written using R. Next, differential gene expression analysis was performed with the Bioconductor package DESeq2 (https://bioconductor.org/packages/release/bioc/html/DESeq2.html; accessed 2022 July 29) (Love et al. 2014) version 1.26.0. Unless otherwise noted, significantly upregulated and downregulated genes were defined as FDR <0.05; Log2 fold change >1 and FDR <0.05; Log2 fold change <−1, respectively, and were used to identify enriched gene ontology (GO) terms using gProfiler2. A selected significantly enriched GO categories were plotted.
Statistical analysis
For all statistical analyses, GraphPad Prism, Version 9.0 was used. First, D’Agostino-Pearson test was used to test the normality of datasets. Datasets with a parametric distribution were compared with an Unpaired t-test while datasets with a nonparametric distribution were compared with a Mann–Whitney test. Three or more datasets following a parametric distribution were analyzed using an ordinary 1-way ANOVA test. Multiple comparisons of 3 or more datasets following a nonparametric distribution were analyzed using Kruskal–Wallis test. Unless otherwise noted, significance is indicated as follows: n.s., not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Results
Nuclear poly(A)+ RNA accumulates in the absence of swm
Previous analyses of RBPs in progenitor cells have focused on their cytoplasmic roles, leaving a possible role for RBPs in progenitor nuclei unexplored (Chen et al. 2015; Luhur et al. 2017; Buddika et al. 2020; Buddika, Huang, et al. 2021). To begin investigating this possibility, we first assessed the poly(A)+ RNA distribution in Drosophila intestinal cells by oligo d(T) in situ hybridization. While oligo d(T) signal was detected in the cytoplasm of all cell types as well as the nuclei of most ECs, it was noticeably absent from the nuclei of progenitor cells, defined as cells that express Esg (Fig. 1, b and b′). This prompted us to investigate factors required for poly(A)+ RNA export in progenitor cells. To address this question, we knocked down selected RNA processing and nuclear export related factors in intestinal progenitor cells via RNA interference (RNAi) and observed progenitor cell numbers as well as poly(A)+ RNA distribution detected by oligo d(T) in situ hybridization. For our screen, we focused on genes implicated in RNA processing and nuclear export and generated a candidate gene list using the GO term analysis function on Flybase (Larkin et al. 2021). From the candidate gene list, we selected 43 genes, encoding both general RNA processing and nuclear export factors as well as proteins implicated or predicted to have nuclear RNA processing functions and that were present within the Bloomington Drosophila Stock Center RNAi strain collection (see Supplementary Table 3 for the screen summary). Each RNAi strain was crossed to a strain harboring both the intestinal progenitor cell driver esg-GAL4 and the temperature-sensitive GAL4 repressor tub-Gal80[ts], hereafter referred to as esgTS (Micchelli and Perrimon 2005). RNAi expression was induced by shifting the resulting young adults to 29°C, and intestinal tissue was analyzed 7 days later. From this analysis, knockdown of 21 genes led to aberrant nuclear poly(A)+ RNA accumulation. Furthermore, knockdown of 19 genes showed progenitor cell loss, 16 of which also showed nuclear poly(A)+ RNA accumulation. Among these 16, the knockdown of second mitotic wave missing (swm) in progenitor cells led to accumulation of nuclear poly(A)+ RNA in progenitor cells (Fig. 1, c and d′). Swm is an understudied RBP that is predicted to have 4 known sequence motifs, a CCCH-type Zn+ finger domain, an RNA-binding RNA recognition motif (RRM), and 2 nuclear localization sequences (Casso et al. 2008). It is implicated in nuclear RNA adenylation and export (Hurt et al. 2009; Yamamoto-Hino et al. 2010), and consistent with our findings in progenitor cells, its knockdown in S2R+ cells also leads to nuclear poly(A)+ RNA accumulation (Farny et al. 2008).
Swm is expressed in nuclei of all cell types of the intestine
To determine where Swm was expressed in the adult intestinal epithelium, we generated a rat polyclonal antibody against the N-terminal 220 amino acids of Swm. Western blot detected a ∼115 kDa band in control tissue and tissue immunostaining detected signal in control but not cells homozygous for swmF14, a previously generated null allele of swm (Casso et al. 2008) (Supplementary Fig. 1, a–c), confirming the specificity of the antibody. We then stained dissected wildtype adult female intestines with this antibody and found that Swm protein was expressed in all 4 cell types (ISCs, EBs, ees, and ECs) (Fig. 1e; Supplementary Fig. 1d). To carefully assess the subcellular localization of Swm in progenitor cells, we analyzed Swm expression in progenitor cells counterstained for cell membranes (myristoylated GFP), nuclear membranes (anti-Lamin antibodies), and nucleoli (anti-Fibrillarin antibodies). Swm was expressed exclusively in nuclei with no apparent staining in nucleoli (Fig. 1, f–g′′′′). This nuclear localization of the protein together with the accumulation of nuclear poly(A)+ RNA after swm knockdown suggested a nuclear function for Swm.
Loss of swm leads to progenitor cell loss
To assess the effects of swm loss in progenitor cells, we crossed the UAS-RNAi line used in our screen [line HM05034 from the Transgenic RNAi Project (Ni et al. 2009), which we refer to as swmRNAi-1 for the remainder of this study] to esgTS and analyzed intestines for progenitor cell number as well as total cell number per field of view in the posterior R4 region after 1, 2, 5, and 10 days of RNAi. Progenitor cell number in swm-depleted intestines gradually declined over time and was nearly absent at the 10-day timepoint (Fig. 2a; Supplementary Fig. 2, a and b). Total cell number per field of view also declined with time (Supplementary Fig. 2c). We confirmed the loss of Swm in the swm RNAi-1 strain by staining for Swm (Supplementary Fig. 2, d–e′) and verified the Swm mediated progenitor cell loss phenotype after 7 days of RNAi using a second independently generated RNAi strain called swmRNAi-2 (Supplementary Fig. 2f).
Fig. 2.
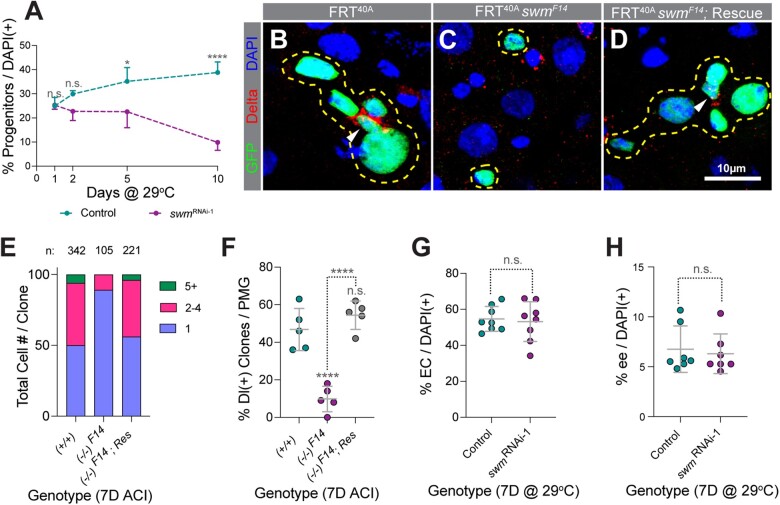
Depletion of swm results in loss of intestinal progenitor cells. a) Normalized progenitor percentage of esgTS and esgTS; swmRNAi-1 after 1, 2, 5, and 10 days at 29°C (n = 2,689, 4,268, 3,702, and 4,097 for esgTS and 13,040, 4,265, 2,811, and 2,023 for esgTS; swmRNAi-1 total cells from 7 to 8 intestines at the 1D, 2D, 5D, and 10D timepoints, respectively). Representative images of tub-GAL4, UAS-GFP- labeled (b) control, (c) swmF14, and (d) rescued swmF14 homozygous clones stained for Delta (red), GFP (green), and DAPI (blue). Clones are outlined in yellow and ISCs are indicated with white arrow heads. (e) Binned bar plot showing the proportion of total cell number per clone in tub-GAL4, UAS-GFP-labeled control, swmF14, and rescued swmF14 intestines analyzed 7 days ACI. Number of clones per genotype is indicated as n. f) Scatter dot plot of the same samples as in (e) showing the percentage of clones per PMG containing at least one Dl+ ISC. Scatter dot plots showing (g) normalized EC percentage in mex1-GAL4TS (n = 1,610) and mex1-GAL4TS; swmRNAi-1 (n = 1,363) and (h) normalized ee percentage in prosV1-GAL4TS (n = 1,870) and prosV1-GAL4TS; swmRNAi-1 (n = 1,859) intestines analyzed after 7 days at 29°C (n = total cells counted from 6 to 8 intestines). Error bars on plots show mean ± SD and asterisks denote statistical significance from Kruskal–Wallis test (a), ordinary one-way ANOVA (f), and unpaired t-test (g and h). *P < 0.05; **P < 0.01; ***P < 0.001; **** P < 0.0001; n.s., not significant. Complete genotypes are listed in Supplementary Table 1. ACI, after clone induction; EC, enterocyte; ee, enteroendocrine cell; Dl, delta; PMG, posterior midgut.
To further validate these results, we generated swmF14 mutant cell clones with the Mosaic Analysis with a Repressible Cell Marker (MARCM) technique (Lee and Luo 1999) and analyzed intestines 7 days after clone induction. Consistent with the swmRNAi-1 RNAi results, 89% of swmF14 mutant cell clones were single-cell clones and the rest of the clones contained 2 or 3 cells while 50% of control clones contained single cells, 44% contained 2–5 cells, and 6% contained 6, or more cells. In addition, while 47% of control clones contained at least 1 cell stained for the ISC marker Dl (Ohlstein and Spradling 2007), only 10% of swmF14 mutant contained an ISC (Fig. 2, b, c, e, and f). These defects were completely rescued by a transgenic insertion of a GFP-tagged ∼48 kb fosmid clone that contained the entire swm locus (fTRG01287.sfGFP-TVPTBF) (Fig. 2, d–f) (Sarov et al. 2016). In contrast to depletion of swm in progenitor cells, knocking down swm in either ECs using the EC driver midgut expression1-GAL4 (mex1-GAL4) (Phillips and Thomas 2006) or in ee cells using prosV1-GAL4, an enhancer trap generated by the insertion of P{GawB} upstream of the transcription start site of pros (Balakireva et al. 1998) did not affect the respective differentiated cell number (Fig. 2, g and h; Supplementary Fig. 2, g–j′). Taken together, our data suggested that swm is specifically required in intestinal progenitor cells for their maintenance and normal tissue homeostasis.
Loss of swm leads to loss of ISC/EB properties
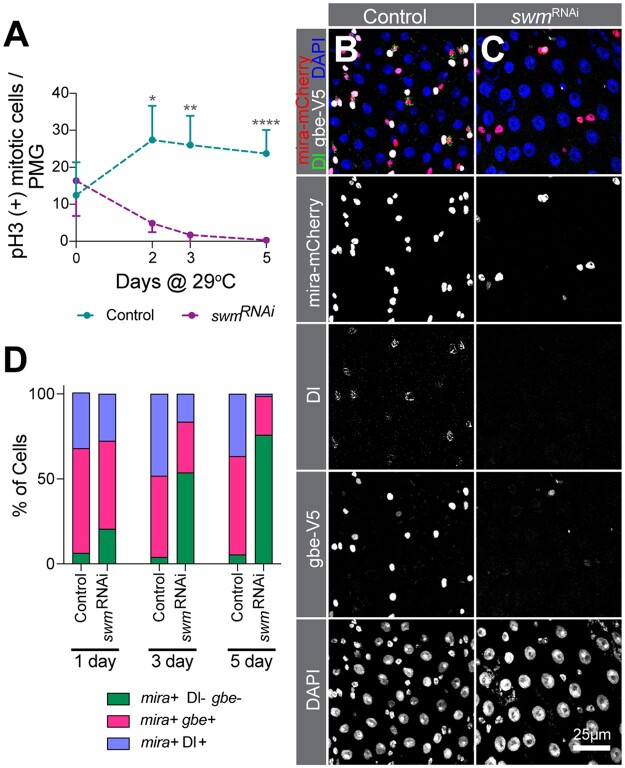
To determine whether ISCs can proliferate in the absence of swm, we induced ISC proliferation by feeding flies bleomycin, a DNA-damaging agent known to trigger ISC division (Amcheslavsky et al. 2009). EsgTS; swmRNAi-1 flies were reared at the nonpermissive temperature (18°C) until 4 days after eclosion, at which point flies were shifted to the permissive temperature (29°C) for an additional 2, 3, or 5 days. Flies were fed with bleomycin for the 24 h prior to dissection. The number of mitotically active ISCs was then counted after immunostaining for phosphorylated-Histone 3 (pH3), a highly specific marker of condensed chromosomes during mitosis. Consistent with the swm mutant cell phenotype of single cell clones with very low-to-no Dl+ cells, knockdown of swm caused a significant reduction in the number of pH3 positive cells over time (Fig. 3a). This could be due to the loss of the ability of ISCs to divide or the loss of ISCs themselves. To distinguish between these possibilities, we examined the cell identities of GFP+-progenitor cells expressing swmRNAi-1 for 1, 3, and 5 days that were stained for both the Dl antibody, which labeled ISCs, as well as 2 previously described transgenic reporters that labeled EBs with gbe-smGFP.V5.nls and all progenitors with mira-His2A.mCherry.HA (Buddika, Xu, et al. 2021). Interestingly, we found that after 5 days of swmRNAi-1 expression, nearly 80% of GFP+ cells did not express either the ISC-marker, Dl, or the EB-marker, gbe-smGFP.V5.nls, but did express the progenitor marker, mira-His2A.mCherry.HA, suggesting at least partial loss of ISC and EB cell identities (Fig. 3, b–d; Supplementary Fig. 3, a–d).
Fig. 3.
Loss of swm leads to loss of ISC/EB activity and identity. a) pH3 (+) cell number per PMG (n = 7 or 8 intestines) of bleomycin fed esgTS and esgTS; swmRNAi-1 intestines after 0, 2, 3, and 5 days at 29°C. PMG section from control (b) and swmRNAi-1 (c) after 5 days at 29°C stained for Ps (anti-RFP for mira-mCherry, in red), ISCs (anti-Dl, in green), EBs (anti-V5 for gbe-smGFP.V5.nls, in white) and all nuclei (DAPI, in blue). Individual channels are shown in gray scale. (d) Binned bar plot showing the quantification of percentage of mira+Dl-gbe- (no defined identity), mira+gbe+ (EB), and mira+Dl+ (ISC) of intestines from genotypes shown in (b) and (c) after 1, 3, and 5 days at 29°C. See Supplementary Fig. 3, a–d for additional statistics. Error bars on plots show mean±SD and asterisks denote statistical significance from Kruskal–Wallis test (a). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; n.s., not significant. Complete genotypes are listed in Supplementary Table 1. P, progenitor cell; ISC, intestinal stem cell; EB, enteroblast; Dl, delta; PMG, posterior midgut.
One possible reason for the progenitor cell number reduction could be due to accelerated differentiation into ECs or ee cells. To test whether swm depleted progenitors differentiate into ECs and ee cells, we took advantage of the Repressible Dual Differential stability cell Marker (ReDDM) cell-lineage tracing system (Antonello et al. 2015). This system uses 2 reporters, a short-lived GFP and a longer-lived, highly stable histone tagged RFP, both of which are under the regulation of the esg-GAL4 driver. Undifferentiated progenitors are labeled with both GFP and RFP but, since GFP degrades as cells differentiate, the newly differentiated ECs and ee cells are only labeled with RFP. ReDDM analysis after 10 days of swm RNAi revealed that, in the absence of swm, the percentage of newborn ECs and ee cells was largely reduced (4%) compared with the control (41%) (Supplementary Fig. 3, e–i), suggesting that swm depleted progenitor cells did not produce differentiated ECs and ee cell types. Taken together, these data indicated that swm is required to maintain progenitor cell properties of ISC and EB cells and thereby homeostasis of differentiated cell numbers.
swm depleted progenitor cells detach from the basement membrane
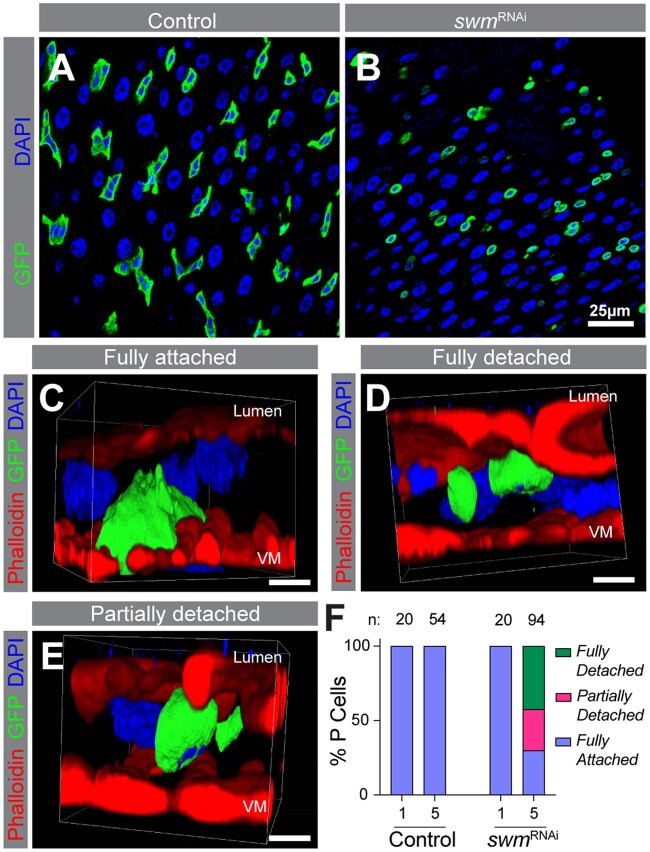
Why are progenitor cells lost in the absence of swm? One possible cause of cell loss is cell death. To test whether swm depleted progenitor cells undergo apoptotic cell death, we used ApopTag assay and found that progenitor cells were not ApopTag positive (Supplementary Fig. 3j). Furthermore, the cell loss phenotype was not rescued by overexpression of the apoptosis inhibitor P35, further suggesting that apoptosis was not the cause of cell loss (Supplementary Fig. 3k). During this analysis, we noticed that swm depleted progenitor cells appeared small and round in comparison to the control progenitor cells, which were mostly triangular (Fig. 4, a and b). One possible reason for the altered cell shape is due to disruption of the physical attachment of progenitor cells to the underlying basement membrane; this basal location of progenitor cells is likely important for proper reception of signals for their maintenance and function (Lin et al. 2013). To determine whether the progenitor cell-basement membrane attachment was affected, we took a confocal microscopy-based approach where we labeled the Filamentous-actin (F-actin) of intestinal tissue with phalloidin and progenitors with GFP, collected images of Z sections spaced 0.3 µm from one another that spanned the basal-to-lumen sides of the intestine, and reconstructed 3D models from them. After 5 days of swmRNAi-1 expression, within a total population of 94 cells, we observed 3 different GFP+ progenitor cell populations: (1) one that was fully detached from the basement membrane and shifted toward the lumen side (42% of cells); (2) one that was partially detached from the basement membrane (28% of cells); and (3) one that was fully attached (30% of cells). In comparison, 100% of control progenitor cells (n = 54) were fully attached to the basement membrane (Fig. 4, c–f; Supplementary Videos 1–3). We defined a cell as fully detached if we could see space between it and the basement membrane, as fully attached if it displayed the embedded appearance of a progenitor with broad attachment to the visceral muscle, and as partially detached if it was not fully attached but still had some contact with the basement membrane. Taken together, our data indicated that in the absence of swm, progenitors detached from the basement membrane and shifted toward the lumenal side of the gut tissue, likely leading to progenitor cell elimination from the epithelium.
Fig. 4.
In the absence of swm, Intestinal progenitors detach from the basement membrane. PMG section from esgTS (a) and esgTS; swmRNAi-1 (b) after 5 days at 29°C. Ps are labeled by GFP (anti-GFP in green) and nuclei with DAPI (blue). Representative snapshot of 3D reconstructed confocal images of fully attached cell from esgTS (c), fully detached cell from esgTS; swmRNAi-1 (d), and partially detached cell from esgTS; swmRNAi-1 (e) after 5 days at 29°C labeled F-actin (phalloidin-TRITC in red), Ps (anti-GFP in green), and nuclei (DAPI in blue). Apical lumen side is on top and basal VM is on the bottom of images as labeled. f) Binned bar plot showing the quantification of fully detached, fully attached, and partially detached P cell percentages in esgTS control (left) and esgTS; swmRNAi-1 (right) after 1 and 5 days at 29°C. n indicates the number of individual cells used for the quantification across up to 5 intestines in each time point. Complete genotypes are listed in Supplementary Table 1. VM, visceral muscle; P, progenitor cell. Three-dimensional arrangement of fully attached, fully detached, and partially detached cells are shown in Supplementary Videos 1–3.
Swm physically associates with epithelial cell maintenance and adhesion related transcripts
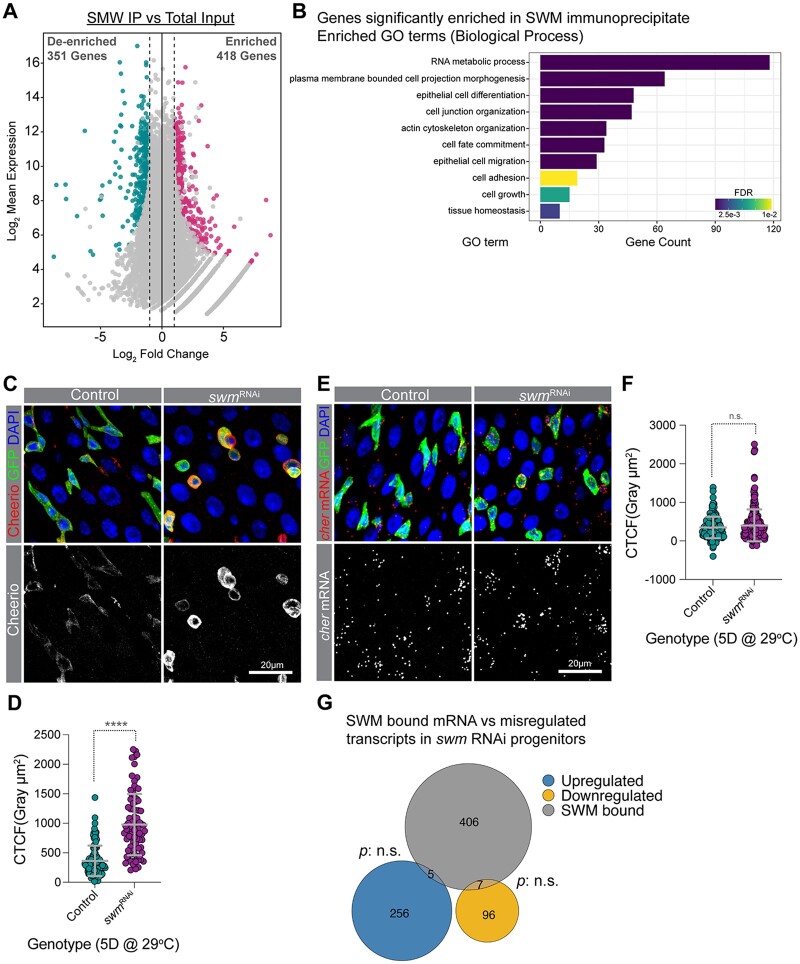
To begin to understand the mechanism of Swm function, we set out to identify the mRNAs that were bound by Swm using crosslinking immunoprecipitation (CLIP) combined with sequencing (CLIP-seq). Using the anti-Swm antibody, endogenous Swm was immunoprecipitated from lysates prepared from UV-crosslinked, dissected intestines of w1118 females (Supplementary Fig. 4a). We prepared libraries from the RNA extracted from 2 independent immunoprecipitates (SWM-IPs) and, in parallel, a third library was prepared from RNA extracted from one “input” lysate, which we used as our normalization control. Differential gene expression analysis revealed that 418 genes were enriched in the SWM-IP samples (Fig. 5a; Supplementary Table 4). We validated 3 SWM-IP enriched targets (cheerio [cher], headcase [hdc], and CG12194) by performing qPCR on SWM-IPs, thus demonstrating that our results were reliable (Supplementary Fig. 4b). Interestingly, GO analysis showed that epithelial cell differentiation, actin cytoskeleton organization, cell fate commitment, and cell adhesion-related genes were enriched in SWM-IPs (Fig. 5b). Genes such as cher, coracle (cora), discs large 1 (dlg1), futsch, molecule interacting with CasL (mical), Myosin 10A (Myo10A), rhea, scribble (scrib), short stop (shot), split ends (spen), Tenascin accessory (Ten-a), and Tenascin major (Ten-m) were found among SWM-IP enriched genes (Supplementary Table 4). Taken together, our identification of RNAs associated with endogenous Swm in intestinal tissue suggested a role for Swm in regulating genes involved in epithelial cell adhesion, cell fate commitment, and maintenance.
Fig. 5.
Swm binds and post-transcriptionally regulates transcripts related to cell adhesion. a) A scatter dot plot visualizing differentially enriched genes in SWM IP vs total input. Each dot represents a single gene. Cyan and pink dots indicate genes with a false discovery rate (FDR) adjusted P-value <0.05 and a Log2 fold change <−1 or >1, respectively. b) A bar plot showing a selected set of significantly enriched GO terms (biological processes) for genes that are physically associated with Swm. c) Representative confocal micrographs from PMGs of esgTS control (left) and esgTS; swmRNAi-1 (right) after 5 days at 29°C stained for Cheerio (red) and DAPI (blue). Ps are labeled with GFP in green. Bottom panel shows Cheerio staining in gray scale. d) Scatter dot plot of normalized Cheerio fluorescence intensity (corrected total cell fluorescence, CTCF) of progenitor cells from esgTS (n = 95 cells, 8 intestines) and esgTS; swmRNAi-1 (n = 84 cells, 7 intestines) after 5 days at 29°C. e) Intestinal progenitors from PMGs of esgTS control (left) and esgTS; swmRNAi-1 (right) after 5 days at 29°C labeled cheerio mRNA (red), Ps stained for GFP (anti-GFP in green) and nuclei with DAPI (blue). Bottom panel shows cheerio mRNA in gray scale. f) Scatter dot plot of normalized cheerio mRNA fluorescence intensity (CTCF) of progenitor cells from genotypes in F (n = 167 cells, 7 intestines for esgTS and n = 241 cells, 10 intestines for esgTS; swmRNAi-1). g) Venn diagram showing the overlap between genes enriched in SWM-CLIP (gray) and genes that are upregulated (blue) or downregulated (yellow) in swm depleted progenitor cells identified from RNAseq. Error bars on plots show mean ± SD and asterisks denote statistical significance from Mann–Whitney test (d, f). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; n.s., not significant. Complete genotypes are listed in Supplementary Table 1. PMG, posterior midgut; P, progenitor cell; CTCF, corrected total cell fluorescence.
Swm post-transcriptionally regulates target mRNAs
To begin to understand how Swm binding to transcripts impacted expression of genes involved in epithelial differentiation, actin cytoskeleton organization, cell fate commitment, and cell adhesion, we knocked down swm in progenitor cells using RNAi for 5 days and immunostained for proteins encoded by Swm-bound transcripts using available antibodies. These included a total of 9 proteins encoded by Cadherin-N (CadN), cher, cora, cut (ct), dlg1, hdc, osa, rhea, and shot. Among the proteins for which we stained, Cheerio (encoded by cher), Talin (encoded by rhea), and Shortstop (encoded by shot) were noticeably upregulated, and Cheerio showed the most obvious increase in progenitor cells upon swm loss (Fig. 5, c and d). Cheerio is a Drosophila member of the Filamin family of actin-binding proteins (Sokol and Cooley 1999). Filamins bind to F-actin to crosslink actin filaments into parallel bundles and connect F-actin to cell membranes, and have been shown to play key roles in modulating cell shape, cell adhesion, cell motility, and differentiation (Sokol and Cooley 1999; Lamsoul et al. 2020). In addition to Cheerio, Talin protein was also increased in swm depleted progenitor cells (Supplementary Fig. 4, c–e). Talin is a large adaptor protein, encoded by the gene rhea, which links ECM-bound integrins to the actin cytoskeleton and thereby regulates adhesion of cells to the ECM (Brown et al. 2002).
Given the upregulated protein levels, to begin to understand how Swm was mediating effects on these bound RNAs, we next asked whether transcript level or distribution were affected. To address this question, we analyzed the subcellular localization of cher transcripts in swm depleted intestinal progenitors using RNAscope in situ hybridization probes after first verifying the specificity of cher probes in progenitors expressing cher RNAi (Supplementary Fig. 5, a–d′) (Wang et al. 2012). From the analysis, we found that cher transcript level was not significantly changed in GFP+ cells after 5 days of swm RNAi in progenitor cells (Fig. 5, e and f) even though our previous analysis indicated that Cher protein was elevated at this timepoint. In addition, we did not observe an obvious change in cellular distribution of the transcript. To investigate the steady state mRNA levels of SWM-IP targets after swm depletion, we performed transcriptomic profiling (RNA-seq) of FACS (fluorescent activated cell sorting) isolated swm RNAi and control intestinal progenitor cells after 5 days of RNAi. In agreement with the RNAscope in situ hybridization results, swm knock down in progenitor cells did not significantly alter the abundance of SWM-IP target transcripts, including cher, and, despite the loss of progenitor cell properties and functions we had observed, the transcriptome of swmRNAi-1-expressing progenitor cells showed only slight changes compared with control cells (Fig. 5g; Supplementary Table 5). Altogether, the analysis of the cher protein and transcript levels in swm-depleted progenitor cells indicated that Swm post-transcriptionally regulated Cher expression and likely other Swm-bound transcripts as well.
Discussion
Here, we report the results of a candidate RNAi screen that identified the RBP Swm as critically important to maintain ISC and EB cell identities and thereby progenitor cell function. This conclusion is based on observations that Swm is nuclearly localized; that nuclear poly(A)+ RNA accumulates in the absence of swm; that swm depleted progenitor cells lose physical contact with the basement membrane and defined cell identities and are lost from the epithelium; that Swm physically associates with mRNAs related to epithelial cell maintenance and adhesion including cher; and, finally, that swm loss leads to upregulation of proteins encoded by Swm-bound targets, including Cher, without affecting transcript levels.
Based on our results, we propose a model where Swm binds and regulates the translation of mRNAs involved in maintaining epithelial progenitor cell properties and function. An open question is how exactly Swm regulates the expression of its bound RNA targets. One possibility is that Swm regulates the poly(A) tail length of the mRNAs, that these mRNAs are hyperadenylated in the absence of swm, and that hyperadenylation of targeted mRNAs leads to enhanced protein translation in intestinal cells. Supporting this, longer poly(A) tails can improve protein translation in context dependent manner in both Drosophila and vertebrate models (Eichhorn et al. 2016; Passmore and Tang 2021; Xiang and Bartel 2021). In addition, Swm has been reported to be a physical interactor of Drosophila Zinc finger CCCH domain-containing protein 3 (dZC3H3), and depletion of dZC3H3 in Drosophila S2R+ cells resulted in hyperadenylation of mRNAs (Hurt et al. 2009). Importantly, dZC3H3 also interacts with Polyadenylate-Binding Protein 2 (Pabp2), the Drosophila nuclear poly(A)-binding protein, which controls poly(A) tail length. Our RNAi screen revealed that progenitor specific knock down of dZC3H3, just like swm, resulted in severe nuclear poly(A)+ RNA accumulation and cell loss (Supplementary Table 1), supporting the notion that these genes also functionally interact in intestinal tissue. The 2 human orthologs of Swm, RNA Binding Motif protein 26 and 27 (RBM26 and RBM27), are also known to bind to ZC3H3 in HEK293 cells (Silla et al. 2020), indicating that the functional interaction between these proteins is evolutionarily conserved. Of course, there are other possibilities for how Swm might regulate mRNA, including via transcript biogenesis, export, localization, and/or translation.
According to our observations, wildtype progenitors do not display nuclear poly(A) RNA accumulation while ECs do. One possible reason for this differential localization of poly(A) RNA between cell types is that nuclear export of poly(A) is more efficient in progenitor cells compared to differentiated EC cells. Previous studies show that intestinal progenitor cells show high protein synthesis (Buddika et al. 2020) and this may demand an efficient nuclear poly(A) RNA export in progenitor cells.
One of the Swm mRNA targets that we identified encodes Cher, a member of the Filamin family of actin-binding proteins. Mammalian Filamin A is known to control the architecture and mechanics of the actin cytoskeleton in a dosage-dependent manner. Tighter F-actin bundles were observed at high Filamin A concentration, leading to reduced flexibility of cells. In contrast, at lower Filamin-A levels, the F-actin cytoskeleton was more dynamic. Filamin A is also known to negatively regulate cell adhesion by binding to the cytoplasmic domains of integrins (Lamsoul et al. 2020). It is possible that modulation of Cher levels could adjust progenitor cell adhesion properties that contribute to the maintenance of stem cell characteristics and function. Given the number of Swm-bound mRNAs we identified, however, we suspect, that Swm must regulate multiple mRNA targets coordinately to adjust stem cell adhesion. Consistently, we failed to rescue the swm loss-of-function phenotype by reducing Cher levels via RNAi (data not shown), indicating that elevated Cher levels are not the only cause of swm phenotypes. In addition, despite the nuclear poly(A)+ RNA accumulation in swm depleted progenitor cells, we did not observe a nuclear accumulation of cher mRNA by in situ hybridization. One possible reason for this could be that one RNA target is not enough to visualize the accumulation, but rather the accumulation is a result of multiple poly(A)+ RNAs.
Even though Swm is expressed in all 4 cell types of the intestinal epithelium, only progenitor cells showed a cell loss phenotype in the absence of swm. One likely reason for this differential requirement of Swm for cell survival could be the differential requirement of Swm-bound target mRNAs for the viability of the different cell types of the intestine. Among SWM-IP targets, genes encoding cell adhesion-related molecules, including integrin linker molecules and actin cytoskeleton binding molecules, were abundant. Adult stem cells are typically associated with a specific microenvironment or niche and this stem cell attachment is frequently mediated via integrins, integrin binding linker molecules, and the cellular actin cytoskeleton (Xi 2009; Chen et al. 2013). Recent single cell transcriptomic analyses, tissue immunostaining, and mutant studies have shown that cell adhesion-related molecules are differentially expressed and required for intestinal progenitors, including both integrins, encoded by myospheroid (mys), multiple edematous wings (mew) and scab (scb), as well as integrin linker molecules, such as talin/rhea, Laminin A (lanA), and integrin-linked kinase (ilk) (Lin et al. 2013; Hung et al. 2020).
We found that swm depleted progenitor cells do not express Dl or gbe, indicating a loss of ISC and EB cell identities. Identities in the Drosophila intestinal cell lineage is specified by Notch signaling (Micchelli and Perrimon 2005; Ohlstein and Spradling 2005, 2007). ISCs express the Notch ligand, Dl and thereby activate Notch signaling in neighboring ISC daughter cells, EBs. Downregulation of Dl in swm-depleted ISCs disrupts Notch signaling and that might compromise EB cell fate and subsequent cell differentiation. However, whether this swm-mediated Dl downregulation is the cause, or the effect of progenitor cell detachment is an open question. Furthermore, although other studies have shown that loss of Dl leads to loss of Notch signaling and induces ee-like tumors (Ohlstein and Spradling 2007), swm loss did not result in a tumor-like cell mass. This could be simply due to loss of ISC stem cell properties including impaired cell proliferation when swm is absent. A similar phenotype was reported for terribly reduced optic lobes mutant clones, which lost Dl but did not display an overproliferation phenotype (You et al. 2014).
Another open question raised by our study is how swm mutant progenitor cells are lost from the epithelium. Previous studies have found that stem cells that detach from the ECM undergo anoikis, an apoptotic condition (Gilmore 2005). However, we found that detached progenitors do not undergo apoptosis, a finding consistent with other studies in the Drosophila intestines that showed that the progenitor cell loss that was associated with the loss of integrins, integrin linker molecules, or ECM components was not caused by apoptosis (Lin et al. 2013; You et al. 2014). These observations indicate that intestinal progenitors are not generally eliminated by anoikis. However, we cannot rule out the possibility that detached progenitor cells are eliminated via a different cell death mechanism. As an alternative, we hypothesize that detached or extruded progenitor cells are shed into the lumen and thereby get eliminated from the epithelium. Intestinal progenitor cell extrusion is not previously reported, but live imaging of the midgut has revealed EC extrusion from the epithelium (Martin et al. 2018), and it has also been reported that ISC tumors promote apoptotic EC extrusion to create space on the basement membrane (Patel et al. 2015).
Nuclear post-transcriptional gene regulatory mechanisms in stem cell biology are largely understudied (Wang et al. 2013) and our study suggests active nuclear post-transcriptional mechanisms in intestinal cells. Future work including identifying molecular and genetic interactors of Swm will dissect the Swm mediated nuclear post-transcriptional regulatory mechanisms.
Supplementary Material
Acknowledgments
We thank Claire Thomas, Jean-Francois Ferveur, the Bloomington Drosophila Stock Center (supported by grant NIHP4OOD018537), the Vienna Drosophila Resource Center (VDRC), the Drosophila Genome Resource Center (supported by grant NIH2P40OD010949), and the Developmental Studies Hybridoma Bank (created by the NICHD of the NIH) for reagents; the FlyBase for resources and information; the Light Microscopy Imaging Center (supported by grant NIH1S10OD024988-01) for access to the SP8 confocal; and the Flow Cytometry Core Facility at Indiana University, Bloomington for access to the BD FACSAria II flow cytometer;
Funding
This work was supported by the National Institute of General Medical Sciences (Award R01GM124220 to NSS and transferred to BRC).
Conflicts of interest
None declared.
Contributor Information
Ishara S Ariyapala, Department of Biology, Indiana University, Bloomington, IN 47405, USA.
Kasun Buddika, Department of Biology, Indiana University, Bloomington, IN 47405, USA.
Heather A Hundley, Medical Sciences Program, Indiana University School of Medicine, Bloomington, IN 47405, USA.
Brian R Calvi, Department of Biology, Indiana University, Bloomington, IN 47405, USA.
Nicholas S Sokol, Department of Biology, Indiana University, Bloomington, IN 47405, USA.
Data Availability
Strains and plasmids are available upon request. RNA-seq data has been submitted to the Gene Expression Omnibus and is available under accession number GSE206632. Supplementary Table 1 lists the full genotypes used in each figure. Supplementary Table 2 lists the sequence of all primers used in the study. Supplementary Table 3 lists genes screened by RNAi. Supplementary Table 4 lists genes enriched in SWM-IPs. Supplementary Table 5 lists differentially expressed genes identified by RNA-seq.
Supplemental material is available at GENETICS online.
Literature cited
- Amcheslavsky A, Jiang J, Ip YT.. Tissue damage-induced intestinal stem cell division in Drosophila. Cell Stem Cell. 2009;4(1):49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amcheslavsky A, Song W, Li Q, Nie Y, Bragatto I, Ferrandon D, Perrimon N, Ip YT.. Enteroendocrine cells support intestinal stem cell-mediated homeostasis in Drosophila. Cell Rep. 2014;9(1):32–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonello ZA, Reiff T, Ballesta-Illan E, Dominguez M.. Robust intestinal homeostasis relies on cellular plasticity in enteroblasts mediated by miR-8–Escargot switch. EMBO J. 2015;34(15):2025–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakireva M, Stocker RF, Gendre N, Ferveur J-F.. Voila, a new Drosophila courtship variant that affects the nervous system: behavioral, neural, and genetic characterization. J Neurosci. 1998;18(11):4335–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Hochmuth CE, Jasper H.. Maintaining tissue homeostasis: dynamic control of somatic stem cell activity. Cell Stem Cell. 2011;9(5):402–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Jasper H.. EGF signaling regulates the proliferation of intestinal stem cells in Drosophila. Development. 2011;138(6):1045–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Jasper H.. Slit/Robo signaling regulates cell fate decisions in the intestinal stem cell lineage of Drosophila. Cell Rep. 2014;7(6):1867–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NH, Gregory SL, Rickoll WL, Fessler LI, Prout M, White RAH, Fristrom JW.. Talin is essential for integrin function in Drosophila. Dev Cell. 2002;3(4):569–579. [DOI] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Kuraishi T, Lemaitre B.. DrosophilaEGFR pathway coordinates stem cell proliferation and gut remodeling following infection. BMC Biol. 2010;8(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N, Osman D, David FPA, Fang HY, Boquete J-P, Deplancke B, Lemaitre B.. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep. 2013;3(5):1725–1738. [DOI] [PubMed] [Google Scholar]
- Buddika K, Ariyapala IS, Hazuga MA, Riffert D, Sokol NS.. Canonical nucleators are dispensable for stress granule assembly in Drosophila intestinal progenitors. J Cell Sci. 2020;133:jcs243451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buddika K, Huang Y-T, Ariyapala IS, Butrum-Griffith A, Norrell SA, O’Connor AM, Patel VK, Rector SA, Slovan M, Sokolowski M, et al. Coordinated repression of pro-differentiation genes via P-bodies and transcription maintains Drosophila intestinal stem cell identity. Curr Biol. 2021;32:386–397.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buddika K, Xu J, Ariyapala IS, Sokol NS.. I-KCKT allows dissection-free RNA profiling of adult Drosophila intestinal progenitor cells. Development. 2021;148:dev196568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casso DJ, Liu S, Iwaki DD, Ogden SK, Kornberg TB.. A screen for modifiers of Hedgehog signaling in Drosophila melanogaster identifies swm and mts. Genetics. 2008;178(3):1399–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Hu G.. Post-transcriptional regulation of the pluripotent state. Curr Opin Genet Dev. 2017;46:15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Lewallen M, Xie T.. Adhesion in the stem cell niche: biological roles and regulation. Development. 2013;140(2):255–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-H, Luhur A, Sokol N.. Lin-28 promotes symmetric stem cell division and drives adaptive growth in the adult Drosophila intestine. Development. 2015;142(20):3478–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Xu N, Wang C, Huang P, Huang H, Jin Z, Yu Z, Cai T, Jiao R, Xi R.. Transient scute activation via a self-stimulatory loop directs enteroendocrine cell pair specification from self-renewing intestinal stem cells. Nat Cell Biol. 2018;20(2):152–161. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D, Dobson AJ, Houtz PL, Gläßer C, Revah J, Korzelius J, Patel PH, Edgar BA, Buchon N.. Regional cell-specific transcriptome mapping reveals regulatory complexity in the adult Drosophila midgut. Cell Rep. 2015;12(2):346–358. [DOI] [PubMed] [Google Scholar]
- Eichhorn SW, Subtelny AO, Kronja I, Kwasnieski JC, Orr-Weaver TL, Bartel DP.. mRNA poly(A)-tail changes specified by deadenylation broadly reshape translation in Drosophila oocytes and early embryos. eLife. 2016;5:e16955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farny NG, Hurt JA, Silver PA.. Definition of global and transcript-specific mRNA export pathways in metazoans. Genes Dev. 2008;22(1):66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foronda D, Weng R, Verma P, Chen Y-W, Cohen SM.. Coordination of insulin and Notch pathway activities by microRNA miR-305 mediates adaptive homeostasis in the intestinal stem cells of the Drosophila gut. Genes Dev. 2014;28(21):2421–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore AP. Anoikis. Cell Death Differ. 2005;12(S2):1473–1477. [DOI] [PubMed] [Google Scholar]
- Guo Z, Ohlstein B.. Bidirectional Notch signaling regulates Drosophila intestinal stem cell multipotency. Science (80) 2015;350(6263):927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold A, Teixeira L, Izaurralde E.. Genome-wide analysis of nuclear mRNA export pathways in Drosophila. EMBO J. 2003;22(10):2472–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung R-J, Hu Y, Kirchner R, Liu Y, Xu C, Comjean A, Tattikota SG, Li F, Song W, Sui SH, et al. A cell atlas of the adult Drosophila midgut. Proc Natl Acad Sci U S A. 2020;117(3):1514–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt JA, Obar RA, Zhai B, Farny NG, Gygi SP, Silver PA.. A conserved CCCH-type zinc finger protein regulates mRNA nuclear adenylation and export. J Cell Biol. 2009;185(2):265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA.. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137(7):1343–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Grenley MO, Bravo MJ, Blumhagen RZ, Edgar BA.. EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell. 2011;8(1):84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamsoul I, Dupré L, Lutz PG.. Molecular tuning of filamin A activities in the context of adhesion and migration. Front Cell Dev Biol. 2020;8:591323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin A, Marygold SJ, Antonazzo G, Attrill H, dos Santos G, Garapati PV, Goodman JL, Gramates LS, Millburn G, Strelets VB, et al. ; FlyBase Consortium. FlyBase: updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res. 2021;49(D1):D899–D907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L.. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22(3):451–461. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W.. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019;47(8):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, Xu N, Xi R.. Paracrine Wingless signalling controls self-renewal of Drosophila intestinal stem cells. Nature. 2008;455(7216):1119–1123. 2008 4557216 [DOI] [PubMed] [Google Scholar]
- Lin G, Zhang X, Ren J, Pang Z, Wang C, Xu N, Xi R.. Integrin signaling is required for maintenance and proliferation of intestinal stem cells in Drosophila. Dev Biol. 2013;377(1):177–187. [DOI] [PubMed] [Google Scholar]
- Losick VP, Morris LX, Fox DT, Spradling A.. Drosophila stem cell niches: a decade of discovery suggests a unified view of stem cell regulation. Dev Cell. 2011;21(1):159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhur A, Buddika K, Ariyapala IS, Chen S, Sokol N.. Opposing post-transcriptional control of InR by FMRP and LIN-28 adjusts stem cell based tissue growth. Cell Rep. 2017;21(10):2671–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17(1):10. [Google Scholar]
- Martin JL, Sanders EN, Moreno-Roman P, Jaramillo Koyama LA, Balachandra S, Du X, O'Brien LE.. Long-term live imaging of the Drosophila adult midgut reveals real-time dynamics of division, differentiation and loss. eLife. 2018;7:e36248. doi: 10.7554/eLife.36248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli CA, Perrimon N.. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2005;439:475–479. [DOI] [PubMed] [Google Scholar]
- Miguel-Aliaga I, Jasper H, Lemaitre B.. Anatomy and physiology of the digestive tract of Drosophila melanogaster. Genetics. 2018;210(2):357–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Spradling AC.. Stem cells and Niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132(4):598–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Paricio N, Sokol NS.. A stress-responsive miRNA regulates BMP signaling to maintain tissue homeostasis. Proc Natl Acad Sci U S A. 2021;118:e2022583118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J-Q, Liu L-P, Binari R, Hardy R, Shim H-S, Cavallaro A, Booker M, Pfeiffer BD, Markstein M, Wang H, et al. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics. 2009;182(4):1089–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien LE, Soliman SS, Li X, Bilder D.. Altered modes of stem cell division drive adaptive intestinal growth. Cell. 2011;147(3):603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A.. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2005;439:470–474. [DOI] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A.. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science. 2007;315(5814):988–992. [DOI] [PubMed] [Google Scholar]
- Passmore LA, Tang TT.. The long and short of it. Elife. 2021;10:e70757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PH, Dutta D, Edgar BA.. Niche appropriation by Drosophila intestinal stem cell tumours. Nat Cell Biol. 2015;17(9):1182–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelossof R, Fairchild L, Huang C-H, Widmer C, Sreedharan VT, Sinha N, Lai D-Y, Guan Y, Premsrirut PK, Tschaharganeh DF, et al. Prediction of ultra-potent shRNAs with a sequential classification algorithm. Nat Biotechnol. 2017;35(4):350–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MD, Thomas GH.. Brush border spectrin is required for early endosome recycling in Drosophila. J Cell Sci. 2006;119(Pt 7):1361–1370. [DOI] [PubMed] [Google Scholar]
- Sarov M, Barz C, Jambor H, Hein MY, Schmied C, Suchold D, Stender B, Janosch S, KJ VV, Krishnan R, et al. A genome-wide resource for the analysis of protein localisation in Drosophila. eLife. 2016;5:e12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanahan MT, Kanke M, Singh AP, Villanueva JW, McNairn AJ, Oyesola OO, Bonfini A, Hung Y-H, Sheahan B, Bloom JC, et al. Single cell analysis reveals multi-faceted miR-375 regulation of the intestinal crypt. bioRxiv; 2020. doi: 10.1101/2020.10.01.321612. [DOI] [Google Scholar]
- Sheffield P, Garrard S, Derewenda Z.. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr Purif. 1999;15(1):34–39. [DOI] [PubMed] [Google Scholar]
- Silla T, Schmid M, Dou Y, Garland W, Milek M, Imami K, Johnsen D, Polak P, Andersen JS, Selbach M, et al. The human ZC3H3 and RBM26/27 proteins are critical for PAXT-mediated nuclear RNA decay. Nucleic Acids Res. 2020;48(5):2518–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol NS, Cooley L.. Drosophila Filamin encoded by the cheerio locus is a component of ovarian ring canals. Curr Biol. 1999;9(21):1221–1230. [DOI] [PubMed] [Google Scholar]
- Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, Luo Y.. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14(1):22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Miao YL, Zheng X, Lackford B, Zhou B, Han L, Yao C, Ward JM, Burkholder A, Lipchina I, et al. The THO complex regulates pluripotency gene mRNA export and controls embryonic stem cell self-renewal and somatic cell reprogramming. Cell Stem Cell. 2013;13(6):676–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi R. Anchoring stem cells in the niche by cell adhesion molecules. Cell Adh Migr. 2009;3(4):396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang K, Bartel DP.. The molecular basis of coupling between poly(A)-tail length and translational efficiency. eLife. 2021;10:e66493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto-Hino M, Kanie Y, Awano W, Aoki-Kinoshita KF, Yano H, Nishihara S, Okano H, Ueda R, Kanie O, Goto S.. Identification of genes required for neural-specific glycosylation using functional genomics. PLoS Genet. 2010;6(12):e1001254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J, Zhang Y, Li Z, Lou Z, Jin L, Lin X.. Drosophila perlecan regulates intestinal stem cell activity via cell-matrix attachment. Stem Cell Rep. 2014;2(6):761–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Hou SX.. Enteroendocrine cells are generated from stem cells through a distinct progenitor in the adult Drosophila posterior midgut. Development. 2015;142(4):644–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains and plasmids are available upon request. RNA-seq data has been submitted to the Gene Expression Omnibus and is available under accession number GSE206632. Supplementary Table 1 lists the full genotypes used in each figure. Supplementary Table 2 lists the sequence of all primers used in the study. Supplementary Table 3 lists genes screened by RNAi. Supplementary Table 4 lists genes enriched in SWM-IPs. Supplementary Table 5 lists differentially expressed genes identified by RNA-seq.
Supplemental material is available at GENETICS online.