Abstract
The metabolic characteristics of Lactococcus lactis IL1403 were examined on two different growth media with respect to the physiological response to two sugars, glucose and galactose. Analysis of specific metabolic rates indicated that despite significant variations in the rates of both growth and sugar consumption, homolactic fermentation was maintained for all cultures due to the low concentration of either pyruvate-formate lyase or alcohol dehydrogenase. When the ionophore monensin was added to the medium, flux through glycolysis was not increased, suggesting a catabolic flux limitation, which, with the low intracellular concentrations of glycolytic intermediates and high in vivo glycolytic enzyme capacities, may be at the level of sugar transport. To assess transcription, a novel DNA macroarray technology employed RNA labeled in vitro with digoxigenin and detection of hybrids with an alkaline phosphatase-antidigoxigenin conjugate. This method showed that several genes of glycolysis were expressed to higher levels on glucose and that the genes of the mixed-acid pathway were expressed to higher levels on galactose. When rates of enzyme synthesis are compared to transcript concentrations, it can be deduced that some translational regulation occurs with threefold-higher translational efficiency in cells grown on glucose.
Lactococcus lactis is generally recognized as the model organism for the study of lactic acid bacteria, and the complete genome sequence of the IL1403 strain (3, 4) will undoubtedly consolidate this position. When growing on rapidly metabolized sugars, this species shows homolactic metabolism in which more than 90% of metabolized sugar is converted to lactic acid. However, under certain conditions, this homolactic metabolism is replaced by a shift in pyruvate metabolism towards alternative fermentation pathways. Under anaerobic conditions, this shift involves an increased flux through the pyruvate-formate lyase reaction and leads to mixed acid fermentation and the accumulation of formate, acetate, and ethanol. These products accumulate in only low quantities during homolactic metabolism but have been shown to account for the majority of carbon flux under circumstances in which the rate of glycolysis of sugars is significantly diminished.
This shift in pyruvate metabolism has been correlated with the NADH/NAD ratio (11). When flux through glycolysis is high, the high NADH/NAD ratio favors lactate dehydrogenase activity and provokes the inhibition of glyceraldehyde-3P dehydrogenase activity upstream of pyruvate. Under such conditions, metabolite pools upstream of glyceraldehyde-3P dehydrogenase (notably triose-Ps) increase within the cell, due to the controlling influence of this enzyme on glycolytic flux (10). Triose-Ps have a negative allosteric effect on pyruvate-formate lyase activity, thereby greatly diminishing the in vivo activity of the enzyme. Sugars metabolized at diminished rates lead to the relaxation of this coordinated control and facilitate the shift towards the more energetically favorable mixed-acid fermentation with an additional gain in ATP linked to the acetate kinase reaction.
Recently, this control has been shown to be a complex mechanism taking into account both the intrinsic rate of catabolism of specific sugars and the energy requirement for biomass synthesis (12). Furthermore, a number of lactococcal strains have been shown to obey this general control structure, though the sequenced strain (IL1403) has not been examined.
However, despite significant progress in understanding the allosteric mechanisms controlling carbon flux, the mechanisms leading to the observed modification of enzyme concentrations have received little attention. Catabolite repression mechanisms have been postulated to govern gene expression in gram-positive bacteria (20) via the fixation of activated CcpA-HPr protein complex to catabolite response element (CRE) sites of certain genes, though the extent of this phenomenon within the catabolic gene network is not yet known.
In this study, the physiological behavior of L. lactis IL1403 has been studied and the transcriptional analysis of all genes encoding enzymes of glycolysis and lactate and mixed-acid fermentative pathways has been performed. This analysis was performed under various defined physiological conditions: two different substrates (glucose and galactose) and two media of varying nutritional complexity (complete MCD medium [19] and a simplified medium, MS10R [8]). Furthermore, rate modeling has been used to identify the extent to which transcriptional phenomena control the concentration of each enzyme.
MATERIALS AND METHODS
Organism and growth conditions.
The bacterium used throughout this work was Lactococcus lactis subsp. lactis IL1403, which lacks the lactose plasmid. The strain was grown on complete MCD medium or simplified synthetic medium, MS10R. The MS10R medium, compared to MCD medium, lacks nucleotidic bases and contains a diminished composition of oligoelements and vitamins. These two synthetic media were supplemented with glucose or galactose (10 g/liter) as a carbon source.
Cultures were grown under anaerobic conditions, under N2 atmosphere, in butyl rubber-stoppered tubes or in a 2-liter fermentor (Setric Genie Industriel, Toulouse, France) at a temperature of 30°C, pH 6.6, and an agitation speed of 300 rpm. The cultures in the fermentor were maintained at pH 6.6 by automatic addition of KOH (10 N). Inoculation was with cells from precultures grown on the same medium, harvested during the exponential phase, and concentrated to obtain an initial optical density at 580 nm of 0.1 to 0.2 in the fermentor.
Fermentation analysis.
Bacterial growth was monitored spectrophotometrically at 580 nm and calibrated against cell dry weight measurements. A change of 1 U was shown to be equivalent to 0.3 g of dry matter per liter. Sugars (glucose and galactose) and fermentation products (formate, acetate, ethanol, and lactate) were determined by high-pressure liquid chromatography as previously described (8).
Preparation of crude extract and enzyme assays.
A volume of culture corresponding to 115 mg (dry weight) of cells was centrifuged (4°C; 10 min at 6,000 × g) and washed twice with 0.2% (vol/vol) KCl. The cells were resuspended in Tris (45 mM)-tricarballylate (15 mM) buffer (pH 7.2) containing glycerol (20%), MgCl2 (4.5 mM), and dithiothreitol (1 mM). Cell disruption by sonication (five cycles of 30 s with 1-min cooling periods) was followed by the removal of cell debris by centrifugation for 10 min at 6,000 × g and 4°C. The supernatant was used for all enzyme assays. The protein concentration of enzymatic extracts was determined by the method of Lowry et al. (17) with bovine serum albumin as the standard. Specific enzyme activities (nmol/min · mg of protein) were converted to whole-cell activities (nmol/min · mg of dried cells) using the measured protein content (42% [dry weight]) of L. lactis.
All enzymes were assayed immediately after cell disruption at 30°C and pH 7.2. Enzyme assays were based on the coupling of the enzyme activity with the consumption or production of NADH, monitored at 340 nm (ɛ340 = 6.22 · 103 M−1 · cm−1), except for P-transacetylase, where dithionitrobenzoic acid (DTNB) was used (ɛ405 = 13.6 · 103 M−1 · cm−1).
Glucokinase was assayed as previously described (26). Glucose-6P isomerase was measured using a modified method derived from that of Gracy and Tylley (13). The reaction mixture contained Tris-HCl buffer (100 mM; pH 7.2), MgCl2 (5 mM), NADH (0.3 mM), ATP (5 mM), phosphofructokinase (1 U), fructose-1,6P2 aldolase (1 U), glycero-P dehydrogenase (2 U), triose-P isomerase (5 U), and glucose-6P (10 mM), which was used to initiate the reaction. Phosphofructokinase and fructose-1,6P2 aldolase were assayed by the method previously described by Le Bloas et al. (15) and modified as follows: the reaction mixture for phosphofructokinase contained triethanolamine-HCl buffer (100 mM; pH 7.2), MgCl2 (5 mM), KCl (10 mM), NADH (0.3 mM), ATP (5 mM), fructose-1,6P2 aldolase (1 U), glycero-P dehydrogenase (2 U), triose-P isomerase (5 U) and fructose-6P (20 mM), which was used to initiate the reaction. The reaction mixture for fructose-1,6P2 aldolase contained triethanolamine-HCl buffer (100 mM; pH 7.2), KCl (200 mM), NADH (0.3 mM), glycero-P dehydrogenase (2 U), and triose-P isomerase (5 U) and was initiated by the addition of fructose-1,6P2 (30 mM). Triose-P isomerase and 3P-glycerate kinase were assayed as previously described (9). The reaction mixture for triose-P isomerase contained triethanolamine-HCl buffer (125 mM; pH 7.2), NADH (0.3 mM), glycero-P dehydrogenase (1 U), and glyceraldehyde-3P (6 mM). The reaction mixture for 3P-glycerate kinase contained triethanolamine-HCl buffer (125 mM; pH 7.2), NADH (0.3 mM), ATP (5 mM), glyceraldehyde-3P dehydrogenase (2 U), EDTA (1 mM), and 3P-glycerate (10 mM) to initiate the reaction. Glyceraldehyde-3P dehydrogenase was assayed as previously described (10) but without the reactivation step, which did not increase measurable activity for protein extracts of the L. lactis IL1403 strain. P-glycerate mutase and enolase were assayed using optimized methods based upon those described previously (14, 23). The enolase activity was measured in a reaction mixture containing Tris-HCl buffer (100 mM; pH 7.2), MgCl2 (5 mM), KCl (10 mM), NADH (0.3 mM), ADP (3 mM), pyruvate kinase (3 U), and lactate dehydrogenase (10 U). The reaction was initiated by the addition of 2P-glycerate (5 mM). The P-glycerate mutase reaction mixture was similar, but triethanolamine-HCl (125 mM; pH 7.2) was used, enolase (2 U) was added, and initiation of the reaction was by addition of 3P-glycerate (5 mM). Pyruvate kinase was assayed by the method described by Thomas (27) and was optimised as follows: the reaction mixture consisted of Tris-HCl buffer (100 mM; pH 7.2), MnSO4 (5 mM), KCl (10 mM), NADH (0.3 mM), lactate dehydrogenase (10 U), GDP (3 mM), and phosphoenolpyruvate (PEP) (6 mM). Lactate dehydrogenase was assayed as previously described (11). P-Transacetylase, acetate kinase, and alcohol dehydrogenase were assayed by the method of Vasconcelos et al. (28), modified as follows. The reaction mixture for P-transacetylase contained phosphate buffer (100 mM; pH 7.2), acetyl-coenzyme A (0.4 mM), and DTNB (0.08 mM). The reaction was initiated by the addition of crude extract. Acetate kinase was measured in a reaction mixture containing Tris-HCl buffer (100 mM; pH 7.2), MgCl2 (5 mM), NADP (0.5 mM), ADP (3 mM), hexokinase (2 U), glucose-6P dehydrogenase (2 U), glucose (2 mM), and acetyl-P (5 mM) to initiate the reaction. The alcohol dehydrogenase assay mixture contained phosphate buffer (100 mM; pH 7.2), dithiothreitol (2 mM), NADH (0.3 mM), and acetaldehyde (20 mM).
Cell extracts for assaying pyruvate-formate lyase activity were prepared in an anaerobic chamber maintained under N2-H2-CO2 (80:10:10%) gas phase. The activity was assayed in the anaerobic chamber as described by Takahashi et al. (25).
Estimation of intracellular metabolites and cellular coenzyme concentrations.
Intracellular metabolites, inorganic phosphate, and cellular coenzymes were extracted, and their concentrations were determined as previously described (11). All measurements were obtained relative to cell dry weight but were expressed as aqueous molar values, using the average intracellular volume of 1.7 ml/g reported previously (24).
Handling of RNA and transcript labeling.
A volume of culture (corresponding to 6 mg [dry weight] of cells) harvested during the exponential growth phase was centrifuged (4°C; 5 min at 8,000 × g), washed with 1 ml of TE buffer (Tris-HCl [10 mM; pH 8], EDTA [1 mM]), resuspended in 500 μl of TE buffer, and frozen immediately in liquid nitrogen. The cells were stored at −80°C until extraction.
Total RNA was extracted as previously described (21). Cells were disrupted with glass beads (three cycles of 1 min on a minibeadbeater [Biospec Products] with 2-min cooling periods) in a tube containing glass beads (0.6 g), 50 μl of sodium dodecyl sulfate (SDS) (10%), 500 μl of phenol (pH 4.7), and 170 μl of macaloid (2% in TE; pH 8), used as an RNase inhibitor (21). After centrifugation (4°C; 30 min at 13,000 rpm), the aqueous phase was extracted with phenol-chloroform. The RNA was precipitated with ethanol and redissolved in TE buffer for quantification at 260 and 280 nm.
Four micrograms of total L. lactis IL1403 RNA was chemically labeled on purine residues with digoxigenin and stored at −20°C until hybridization. The RNA labeling and detection system with digoxigenin and AttoPhos (see below) was that previously used for DNA hybridization (22) except that digoxigenin was chemically incorporated in RNA using the DIG-Chem-Link labeling set (Roche).
Primer design and PCR amplification.
The sequences of the genes glk, pgi, fba, pgk, pgm, eno, pta, and ack were kindly provided by the principal investigators of the L. lactis sequencing project prior to publication (4). PCR primer pairs (Table 1) were designed to amplify 500-bp probes corresponding to glycolysis and mixed-acid metabolism genes. Each primer contained 23 or 24 bases and was designed to achieve a melting temperature of 72 (calculated with the relation 4 GC + 2 AT).
TABLE 1.
Oligonucleotide primers for PCRs
| Gene | Oligonucleotide | Reference |
|---|---|---|
| glk | 5′-GAATTATCGCTTCAGGTGAGCTCG-3′ | Personal communication |
| 5′-TTAAGAGCAAGACTTGCGGCACCG-3′ | INRAa | |
| pgi | 5′-GGCTGACCTTGTAGATTACGTTGC-3′ | Personal communication |
| 5′-TCTTTCCCTTCTGATTCACCAGCC-3′ | INRA | |
| pfk | 5′-TCTTGTACTCAGCACGTTACCCAG-3′ | 16 |
| 5′-CTTCACCAGTCATAACGCCTTCAG-3′ | GenBank L 07920 | |
| fba | 5′-AGTTGCTATTCACCTTGACCACGG-3′ | Personal communication |
| 5′-ACGAATTTACGAGTAGCAGCTGCG-3′ | INRA | |
| tpi | 5′-TTTTCCTCGCTCCTATGGCTTACC-3′ | 6 |
| 5′-TCCACCGTATTGGATACGTACAGC-3′ | GenBank U 07640 | |
| gap | 5′-ATAGTTTAGCACCAATGGCCGACG-3′ | 7 |
| 5′-TGTCCACCGTCTTTTAGGTCAGTC-3′ | GenBank L 36907 | |
| pgk | 5′-CACTCGTTTTGAAGACATCGACGG-3′ | Personal communication |
| 5′-TTGCTACGTCAAGTCCCATGAAGC-3′ | INRA | |
| pgm | 5′-CCTATGACGATATGGATGTTCCGC-3′ | Personal communication |
| 5′-GAAAGAGAAGTACCATGTGTTCCG-3′ | INRA | |
| eno | 5′-TGAAGCGGTTGAACTTCGTGATGG-3′ | Personal communication |
| 5′-GTTTCTACACCGTCTTCAGTTCCG-3′ | INRA | |
| pyk | 5′-TGACGGTGCTGATTCAATCAGCGT-3′ | 16 |
| 5′-TCATGATACCATCAGCTGCTTCG-3′ | GenBank L 07920 | |
| ldh | 5′-AATCTTCCTCGTTGCTGCTAACCC-3′ | 16 |
| 5′-AACACCTTCAGCACCAACTACAGC-3′ | GenBank L 07920 | |
| pfl | 5′-GCGATAAAGCAAGCGTTACTCGCT-3′ | 1 |
| 5′-AAGGTAATCAGCACCGTAAAGGGC-3′ | GenBank AJ 000326 | |
| pta | 5′-AGGTGCTGCAAATCGTCTTAACGC-3′ | Personal communication |
| 5′-GGCTGAAGCTACAGCAATATCGG-3′ | INRA | |
| ack | 5′-ATCACTGCTCATATCGGGAATGGC-3′ | Personal communication |
| 5′-TGTTGAAATTTCTCCCTCGACGCC-3′ | INRA | |
| adhE | 5′-ACCACAAGTTGCCATTGTTGACCC-3′ | 2 |
| 5′-TTACCAGCAAATCCCATGAAGCG-3′ | GenBank AJ 001007 | |
| ccpA | 5′-ACTTACTTCGCAGCGATTACTCGC-3′ | GenBank AF 106673 |
| 5′-TACTACTGCAGAAGTTGCTCCTCG-3′ | GenBank AF 106673 |
INRA, Institut National de la Recherche Agronomique.
Amplification reactions were performed in a 100-μl reaction volume containing L. lactis IL1403 genomic DNA template (1 to 2 μg), each primer (1 μM), deoxynucleoside triphosphates (200 μM), and 1 U of Taq (Sigma). Reactions were cycled 45 times as follows: 94°C for 30 s, 56°C for 30 s, and 70°C for 1 min, with a final cycle of 70°C for 10 min. All PCR products were purified by electrophoresis on 2% agarose gels in 0.5× Tris-acetate (20 mM)–EDTA (0.5 mM). The probes were extracted and purified using the QIAquick gel extraction kit (Qiagen). The probes were analyzed by two different enzymatic digestions to confirm the success of the PCR amplification.
Preparation of macroarrays and hybridization.
A 0.2-μg portion of each probe was denatured at 95°C for 5 min in 250 μl of 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and was blotted on a nylon membrane, positively charged (Roche), using a slot blot filtration manifold (PR 6458; Hoefer Scientific Instruments). DNA was fixed on the membrane by UV exposure. The membranes were briefly washed in sterile water before being dried at room temperature and stored at 4°C. The concentration of target DNA was in excess compared to the hybridized RNA concentration (no increase of signal was observed when the amount of target DNA increased).
Prior to hybridization, membranes were prehybridized for 5 h at 68°C in roller bottles containing 20 ml of 1× hybridization buffer (5× SSC, 1% [wt/vol] blocking reagent [Roche], 0.02% [wt/vol] N-lauroylsarcosine, 0.02% [wt/vol] NaCl, 0.02% [wt/vol] SDS). The membranes were then hybridized (68°C for 15 h) with 1× hybridization buffer containing labeled total RNA (previously denatured [95°C for 10 min]) and washed twice at room temperature (5 min with 2× SSC and 0.1% SDS) and twice at 68°C (15 min with 0.5× SSC and 0.1% SDS) prior to detection.
Array detection and analysis.
All incubation steps were performed at room temperature with agitation. After a brief wash (1 to 5 min) in the washing buffer (maleic acid buffer [0.1 M; pH 7.5], NaCl [0.15 M], Tween 20 [0.3% {vol/vol}]), the membranes were incubated for 30 min in the blocking solution (maleic acid buffer [0.1 M; pH 7.5], NaCl [0.15 M], blocking reagent [1% {wt/vol}]). The blocking solution was removed and replaced with blocking solution containing the antidigoxigenin conjugate coupled to the alkaline phosphatase (75 mU/ml) (Roche) for a further incubation of 30 min. The membrane was then washed twice with the washing buffer for 15 min, equilibrated for 5 min in alkaline phosphatase buffer (Tris-HCl [0.1 M; pH 9.5], NaCl [0.1 M]), and sealed in polypropylene bags to avoid drying. AttoPhos (Amersham Pharmacia Biotech), the alkaline phosphatase substrate, was then added directly, and the membranes were scanned at different times with a phosphofluoroimager (STORM 860; Molecular Dynamics) tuned at 100-μm pixel resolution. Images were stored electronically and analyzed using ImageQuant version 5.1 analysis software (Molecular Dynamics). The signal intensity of each spot was calculated using the volume quantification method of ImageQuant. A grid of individual squares corresponding to each of the DNA spots was laid down on the image to designate the spots to be quantified. The background was subtracted automatically by the software using the local median background function of ImageQuant. Quantification was performed at different times after the addition of AttoPhos, leading to the calculation of the alkaline phosphatase activity for each slot. Alkaline phosphatase activity correctly calculated in its linearity domain is directly correlated with the mRNA hybridized on the slot probe. Since the total RNA amount was normalized in the assay (4 μg), the alkaline phosphatase activity revealed the abundance of a specific messenger in the total RNA population. This value was corrected by the cellular RNA concentrations (see Results) in order to arrive at the cellular concentration of a specific messenger.
The linearity of the method (measured signal/amount of mRNA) was verified by diluting RNA of L. lactis with baker's yeast tRNA, the total RNA amount being unchanged (6 μg). The reproducibility of the method was estimated in four identical cell samples, and the standard deviation for the mRNA cellular concentrations was evaluated to be ±30%. Since the results were expressed as expression ratios, only those ratios differing by at least 2 standard deviations were analyzed (i.e., <0.6 or >1.6).
RESULTS
Growth kinetics.
Despite significant variations in specific rates of exponential growth, sugar consumption, and product formation (Table 2) as a function of medium composition (sugar and/or nutritional charge), L. lactis IL1403 retained a homolactic fermentation profile (>90% carbon recovery as lactate). Mixed-acid fermentation products, formate, acetate, and ethanol, were produced in only small quantities (<10% of the total products) and in the proportion 2:1:1 necessary to regenerate NAD. Irrespective of the growth medium used, the specific rates were at all times higher on glucose than on galactose, and for both sugars the rates were higher in the more complete MCD medium.
TABLE 2.
Specific rates of growth, substrate consumption, and product formation during growth of L. lactis IL1403 on two different synthetic media (MCD and MS10R) with glucose or galactose as carbon source
| Parameter | Specific rate
|
|||
|---|---|---|---|---|
| MS10R
|
MCD
|
|||
| Galactose | Glucose | Galactose | Glucose | |
| Growth (h−1) | 0.17 ± 0.01 | 0.55 ± 0.06 | 0.26 ± 0.03 | 0.81 ± 0.03 |
| Sugar consumption (mmol/g · h) | 7.1 ± 0.1 | 17.8 ± 1.1 | 10.6 ± 1.2 | 21.1 ± 1.5 |
| Lactate production (mmol/g · h) | 13.1 ± 0.7 | 32.9 ± 2.2 | 18.4 ± 1.7 | 40.9 ± 3.0 |
| Formate production (mmol/g · h) | 1.2 ± 0.1 | 1.3 ± 0.3 | 1.9 ± 0.1 | 1.4 ± 0.3 |
| Acetate production (mmol/g · h) | 0.6 ± 0.3 | 0.6 ± 0.3 | 1.1 ± 0.3 | 0.7 ± 0.3 |
| Ethanol production (mmol/g · h) | 0.5 ± 0.2 | 0.6 ± 0.2 | 1.2 ± 0.2 | 0.6 ± 0.2 |
In order to determine the maximum rates of sugar catabolism, monensin (0.5 μM), a K+-Na+ ionophore previously used by Bond et al. (5), was added to exponentially growing cultures. In the case of Streptococcus bovis, this ionophore led to an increase in glycolytic flux when catabolic capacity was not limiting to compensate for ATP wastage (5). However, in the case of L. lactis IL1403, sugar consumption rates were not increased for either glucose or galactose, though a strong decline in the growth rate was observed. This indicates that the rates of sugar consumption observed (without monensin) were effectively maximal rates, suggesting that flux through the catabolic pathways could not be further increased.
Intracellular metabolites.
The relative abundance of intracellular metabolites is an efficient way to identify reactions that might be operating close to their physiological maxima and hence have a controlling effect on pathway flux. In all cases, glucose-6P and fructose-1,6P2 were measured at high concentrations, higher for glucose than galactose (Table 3). The intracellular concentration of glucose-1P was low on glucose but high during growth on galactose, as would be expected from the metabolic pathways specific to galactose. Other metabolite concentrations were low (notably the triose-Ps), conditions which would normally be expected to favor mixed-acid fermentation (11). The intracellular NAD and NADH concentrations were also measured, but no significant differences were detected, leading to a constant NADH/NAD ratio of approximately 0.08. Likewise, the concentrations of intracellular inorganic phosphate were shown to be identical during growth on glucose and on galactose.
TABLE 3.
Intracellular concentrations of glycolytic intermediaries, coenzymes, and Pi during exponential growth of L. lactis IL1403 on two different synthetic media with glucose or galactose as carbon source
| Metabolite | Concn (mM)
|
|||
|---|---|---|---|---|
| MS10R
|
MCD
|
|||
| Galactose | Glucose | Galactose | Glucose | |
| Glucose-1P | 8.1 ± 1.3 | <0.4 | 3.7 ± 0.5 | <0.4 |
| Glucose-6P | 6.7 ± 1.3 | 15.4 ± 1.2 | 6.0 ± 0.7 | 10.7 ± 1.1 |
| Fructose-6P | 0.7 ± 0.4 | <0.4 | 0.6 ± 0.2 | <0.4 |
| Fructose-1,6P2 | 9.2 ± 2.4 | 17.8 ± 1.8 | 22.3 ± 1.4 | 23.5 ± 1.7 |
| Dihydroxyacetone-P | <0.4 | 0.7 ± 0.7 | 0.5 ± 0.9 | 0.7 ± 0.3 |
| Glyceraldehyde-3P | <0.4 | <0.4 | <0.4 | <0.4 |
| 1,3P2-glycerate | <0.4 | <0.4 | <0.4 | <0.4 |
| 3P-glycerate | <0.4 | <0.4 | <0.4 | <0.4 |
| PEP | 0.8 ± 0.5 | <0.4 | 2.6 ± 0.9 | 1.0 ± 0.7 |
| NAD | 5.8 ± 0.7 | 5.8 ± 0.7 | 5.1 ± 0.4 | 5.2 ± 0.6 |
| NADH | 0.45 ± 0.19 | 0.44 ± 0.11 | 0.47 ± 0.15 | 0.35 ± 0.11 |
| NADH/NAD | 0.08 ± 0.04 | 0.08 ± 0.03 | 0.09 ± 0.03 | 0.07 ± 0.03 |
| Pi | 60 ± 7 | 56 ± 5 | NDa | ND |
ND, not determined.
Enzyme activities.
In vitro specific activities of all enzymes of glycolysis and the lactate and mixed-acid pathways were assayed in exponentially growing cells (Table 4). No obvious correlation can be observed between specific activity measurements and pathway flux under the different growth conditions, and with the exception of galactokinase, which was significantly induced during growth on galactose, other enzymes were present at fairly constant levels, with variations of specific activity rarely exceeding a factor of 2. This is perhaps not that surprising in view of the importance of this central pathway. Indeed, the specific activities of glucokinase and glyceraldehyde-3P dehydrogenase remained constant in cells grown on either glucose or galactose on both media. While triose-P isomerase, P-glycerate mutase, and enolase activities were somewhat higher on glucose and P-transacetylase and alcohol dehydrogenase activities were higher on galactose, irrespective of the medium used, other enzyme activities were as dependent on the medium composition as on the sugar content. Indeed, the activity of glucose-6P isomerase, phosphofructokinase, fructose-1,6P2 aldolase, pyruvate kinase, and lactate dehydrogenase were higher on glucose only on MCD medium, while 3P-glycerate kinase and acetate kinase activities were higher on galactose only on MS10R medium. Other changes in specific activity were correlated with medium composition other than the carbon source, since glucose-6P isomerase and acetate kinase activities were higher on MS10R medium than on MCD medium, as were phosphofructokinase, pyruvate kinase, and lactate dehydrogenase activities, though only on galactose. The activity values obtained for pyruvate-formate lyase activity were at all times extremely low, despite the use of strictly anaerobic conditions and a protocol previously shown to be effective in other lactococcal strains. The reasons for this lack of enzyme stability are not known.
TABLE 4.
Enzyme specific activities in cell extracts of L. lactis IL1403 grown on two different synthetic media with glucose or galactose as carbon sourcea
| Enzyme | Sp act (nmol/min · mg of dried cells)
|
|||
|---|---|---|---|---|
| MS10R
|
MCD
|
|||
| Galactose | Glucose | Galactose | Glucose | |
| Galactokinase | 560 ± 30 (5) | 13 ± 15 | 720 ± 60 (4) | 24 ± 3 |
| Glucokinase | 100 ± 20 | 150 ± 30 | 130 ± 10 | 130 ± 15 |
| Glucose-6P isomerase | 1,500 ± 200 (13) | 1,300 ± 100 (4) | 995 ± 5 (6) | 1,180 ± 30 (3) |
| Phosphofructokinase | 430 ± 20 (4) | 450 ± 100 (2) | 290 ± 20 (2) | 360 ± 10 (1) |
| Fructose-1,6P2 aldolase | 1,800 ± 100 (15) | 2,000 ± 300 (7) | 1,270 ± 50 (7) | 1,600 ± 200 (5) |
| Triose-P isomerase | 22,100 ± 600 | 31,500 ± 1500 | 25,000 ± 2,000 | 35,600 ± 30 |
| Glyceraldehyde-3P DH | 5,800 ± 900 (25) | 5,400 ± 500 (9) | 6,130 ± 50 (17) | 6,075 ± 5 (9) |
| 3P-glycerate kinase | 1,700 ± 100 | 1,400 ± 400 | 1,260 ± 40 | 1,380 ± 20 |
| P-glycerate mutase | 1,500 ± 100 (6) | 2,900 ± 500 (5) | 1,200 ± 100 (3) | 1,900 ± 100 (3) |
| Enolase | 2,000 ± 200 (8) | 2,500 ± 200 (4) | 1,940 ± 70 (5) | 2,400 ± 200 (3) |
| Pyruvate kinase | 990 ± 40 (4) | 880 ± 80 (3b) | 580 ± 30 (2) | 780 ± 70 (2b) |
| Lactate DH | 6,500 ± 300 (30) | 6,600 ± 1200 (12) | 3,790 ± 60 (12) | 5,900 ± 600 (9) |
| P-transacetylase | 1,200 ± 100 (115) | 860 ± 70 (86) | 2,800 ± 200 (150) | 1,400 ± 300 (118) |
| Acetate kinase | 2,600 ± 300 (261) | 1,300 ± 400 (129) | 800 ± 100 (44) | 760 ± 90 (65) |
| Alcohol DH | 52 ± 2 (7) | 20 ± 8 (2) | 63 ± 3 (3) | 30 ± 10 (3) |
All values represent averages of several samples taken throughout exponential growth and measured in triplicate (standard deviations are given). The figures in parentheses indicate the ratio between enzymatic activity (when measured in the physiological direction) and the flux through the enzyme estimated from rates of sugar and product accumulation. DH, dehydrogenase.
Flux in the pyruvate kinase was calculated with no flux through glucokinase (100% phosphotransferase system) in the following manner: overall flux of PEP − flux through PTS = 2 · qs − qs
Key enzymes and major regulations.
The substrate affinities of glyceraldehyde-3P dehydrogenase of the IL1403 strain were 1 mM for d-glyceraldehyde-3P and 0.25 mM for NAD. The Km values of the pyruvate kinase reaction were 4 mM for ADP and 1 mM for PEP. Taking into account the experimentally determined value of inorganic phosphate, the Km value for glucose-6P was 5 mM in the glucose-6P isomerase reaction (1.5 mM in the absence of Pi), while the affinity of the fructose-1,6P2 aldolase reaction for fructose-1,6P2 was 9 mM (3 mM in the absence of Pi). The three dehydrogenases (glyceraldehyde-3P dehydrogenase, lactate dehydrogenase, and alcohol dehydrogenase) were all sensitive to the NADH/NAD ratio, which at the in vivo value of 0.08, led to diminished activities of approximately 50% of the maximal flux capacity under otherwise optimal in vitro conditions.
Gene expression.
The abundance of glycolytic and fermentative pathway gene transcripts was estimated using DNA macroarrays in exponentially growing cells (Table 5). The total RNA concentration in the cells varied in the different cultures. Indeed, RNA concentrations were generally higher on glucose than on galactose and on MCD medium rather than MS10R medium but were not linearly correlated (Table 5). Hence, mRNA abundance was corrected by the cellular RNA concentrations to obtain the cellular concentration of each specific messenger, and the ratio of expression between glucose and galactose was calculated for each medium (Table 5). Once the intrinsic experimental error had been taken into account, three groups of genes could be identified based on the influence of the sugar on their expression. The first group consisted of those genes expressed at higher levels on a specific sugar. This group consisted of pfk, fba, tpi, pyk, and ldh for glucose and glk, pta, ack, and adhE for galactose. The effect of the sugar was more or less pronounced on each of the two media, and in some cases the differences in expression fell into the zone of intrinsic error of the method. A second group of genes consisting of pgi, gap, pgk, pgm, and eno showed similar levels of expression on all media. Finally, a third group of genes consisting of pfl, the gene involved in the mixed-acid pathway, and the regulatory gene ccpA were expressed at higher levels on galactose, but only on one of the media.
TABLE 5.
Transcript abundance and total RNA concentrations in exponentially growing cells of L. lactis IL1403 grown in two different culture media with glucose or galactose as carbon source
| Gene | Transcript abundancea
|
|||||
|---|---|---|---|---|---|---|
| MS10R
|
MCD
|
|||||
| Galactose | Glucose | Ratiob | Galactose | Glucose | Ratiob | |
| glk | 28,700 | 11,900 | 0.6 | 21,400 | 11,300 | 0.6 |
| pgi | 31,600 | 30,400 | 1.3 | 40,500 | 31,500 | 0.9 |
| pfk | 32,500 | 42,000 | 1.8 | 39,600 | 62,800 | 1.8 |
| fba | 143,300 | 160,900 | 1.5 | 120,700 | 248,000 | 2.3 |
| tpi | 41,500 | 44,100 | 1.5 | 31,900 | 67,400 | 2.3 |
| gap | 130,700 | 59,900 | 0.6 | 86,000 | 69,500 | 0.9 |
| pgk | 74,800 | 56,600 | 1.0 | 44,000 | 59,800 | 1.5 |
| pgm | 9,500 | 8,500 | 1.2 | 11,600 | 7,200 | 0.7 |
| eno | 134,100 | 148,000 | 1.5 | 79,200 | 105,200 | 1.5 |
| pyk | 47,300 | 78,500 | 2.3 | 48,600 | 104,800 | 2.4 |
| ldh | 74,700 | 94,700 | 1.7 | 51,300 | 109,900 | 2.4 |
| pfl | 35,000 | 23,500 | 0.9 | 20,300 | 9,700 | 0.5 |
| pta | 21,300 | 8,800 | 0.6 | 16,100 | 6,100 | 0.4 |
| ack | 30,600 | 12,000 | 0.5 | 25,300 | 16,400 | 0.7 |
| adhE | 99,000 | 25,400 | 0.4 | 60,700 | 36,400 | 0.7 |
| ccpA | 25,300 | 7,800 | 0.4 | 8,800 | 12,400 | 1.6 |
| Total RNAc | 5.2 ± 0.1 | 7.1 ± 0.1 | 9.9 ± 0.3 | 11.0 ± 1 | ||
Expressed as phosphatase alkaline activity (PA) (pixel density/min).
Ratio of the intracellular concentrations of a transcript in glucose and galactose calculated as follows: PAglucose × [RNA]glucose/PAgalactose × [RNA]galactose. Only expression ratios of >1.6 or <0.6 are significant, taking into account the precision of the method (30%) (see Materials and Methods).
Percent dry weight.
DISCUSSION
Metabolism.
Growth of L. lactis IL1403 on various media led to significantly different kinetic characteristics. Despite this, the shift from homolactic to mixed-acid fermentation observed under similar conditions for other strains of L. lactis (11) did not occur. The lack of a stimulatory effect on glycolytic flux when monensin was added to the culture indicates a catabolic rate limitation and suggests that carbon flux was limited by an enzymatic step(s). However, when the enzyme specific activities (other than sugar transport) are compared with pathway flux (estimated directly from rates of sugar consumption and/or product accumulation), it can be seen that all glycolytic enzymes plus lactate dehydrogenase were capable of supporting an increased flux (Table 4). Even when the negative effect of the NADH/NAD ratio on dehydrogenase reactions and various known allosteric phenomena were taken into account, the flux capacity remained in excess of experimental rates. This is consistent with the observation that metabolite concentrations were relatively low and in all cases lower than would be expected if one or more enzymes were functioning close to substrate saturation. In the cases of glucose-6P isomerase and fructose-1,6P2 aldolase, the pools of glucose-6P and fructose-1,6P2, respectively, were relatively large. However, when the effect of phosphate on the enzymes' substrate affinities was taken into account, the metabolite pools were close to the experimental Km values, indicating that these enzymes were functioning, in vivo, at considerably lower rates than those theoretically feasible. In view of this, it is tempting to suggest that sugar transport was probably a major rate-controlling limitation in this strain under the conditions used.
Rapid rates of sugar catabolism provoke, in other strains of L. lactis, the coordinated control of enzyme activity and homolactic fermentation characteristics. This has been attributed to a controlling influence of glyceraldehyde-3P dehydrogenase activity and knock-on effects to inhibit pyruvate-formate lyase activity (11). Diminished rates relax this control, enabling carbon flux to be redirected into the mixed-acid pathway. This appears not to be the case for the IL1403 strain, since homolactic metabolism was maintained without apparent saturation of the glyceraldehyde-3P reaction and the associated increase in triose-P levels. It should be noted that glycolytic activities were generally higher in the IL1403 strain than those reported for other model strains. Thus, some other metabolic phenomena must play a role in the sustained homolactic characteristics. One possibility is that only very low pyruvate-formate lyase activity was measured in the cells. The level of gene expression would tend to suggest that this is attributable to a problem in enzyme stability, since the activity is known to be rather difficult to measure in certain strains while the level of pfl transcript abundance was comparable to those of other genes of the central pathways. The enzymes involved in acetate production (P-transacetylase and acetate kinase) were present in large amounts compared to the flux (Table 4) and could certainly sustain an increased flux. However, alcohol dehydrogenase was present in amounts similar to the flux capacity once NADH-NAD effects on activity (50% inhibition) were allowed for. This enzyme is then a plausible candidate for explaining the relatively low flux through the mixed-acid pathway. A more detailed biochemical investigation of the enzyme will substantiate the validity of this hypothesis. Of course, the high lactate dehydrogenase activity and the adequate NADH availability would also explain a maintained flux through this enzyme.
Transcriptional analysis.
The relative cellular concentration of transcripts, estimated under different conditions, enabled a number of differences to be observed. In most cases, the profile could be attributed to the presence of one of the two sugars. However, in a number of cases this effect was more or less pronounced on one of the two media, suggesting that the growth rate or anabolic efficiency might also have an effect on transcriptional regulation. Modifications in gene expression were in general not particularly high (a factor always lower than 3). This low degree of transcriptional control is perhaps logical for essential genes of the major catabolic pathway of this bacterium. Despite this, most of the differences in transcript abundance were consistent with some degree of control via CcpA, the catabolite repression regulator. Genes known to be positively regulated by CcpA, such as those of the las operon (pfk, pyk, and ldh) (18), increased on glucose, while those postulated to be negatively controlled by CcpA (e.g., ack [G. Perez Martinez, personal communication]) decreased on glucose. Indeed, all genes of the mixed-acid pathway showed significantly diminished expression on glucose, suggesting a common mechanism of regulation. Other genes, whose regulation has not been clearly determined, showed expression profiles similar to those of the las operon (e.g., fba and tpi), suggesting a catabolite repression control. The genome sequence when fully annotated will probably facilitate the proposal of a putative control strategy exerted by this bacterium over catabolic genes in response to sugar specificity.
It should be observed that the expression of the regulatory gene (ccpA) also responded to sugar. While ccpA expression was constant on MCD medium, increased expression was observed in cells grown on galactose on MS10R medium. This suggests that ccpA is to some extent under the control of catabolite repression.
From gene to enzyme.
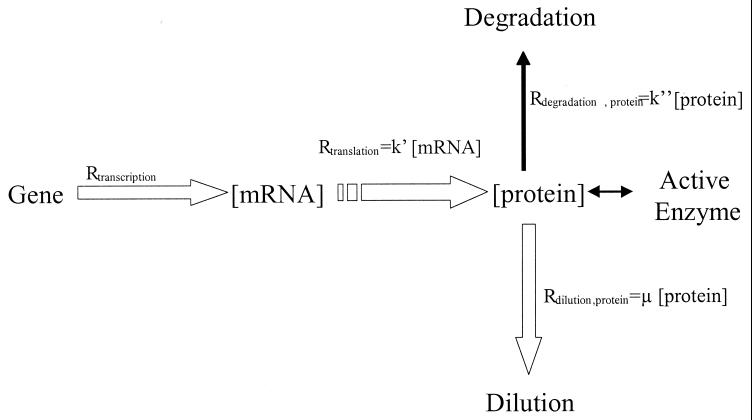
The comparison of transcript analysis and specific activity measurements for the corresponding enzymes indicates that some further degree of control was exerted, presumably at the translational level. However, to assess this correctly it is necessary to compare the cellular concentrations of transcripts to the rate of enzyme synthesis and not to the concentration of enzyme more commonly used (18). To attain this level of comparison, a number of kinetic phenomena need to be taken into account (Fig. 1), notably the factors influencing cellular enzyme concentrations. The enzyme concentration results from the rate of protein synthesis but is subject to dilution by protein turnover (normally insignificant except under specific stress conditions) and dilution resulting from the rate of cell division (at each division, the cellular content of enzymes will be halved). The rate of enzyme synthesis at steady state can therefore be obtained by multiplying the enzyme concentration by the specific growth rate (Rtranslation = Rdilution = μ · [enzyme], where μ is the specific growth rate [Table 2] and [enzyme] is the in vitro maximum specific activity [Table 4]). Thus, cells growing at different rates will have substantially different rates of protein synthesis even though the specific activities may remain similar.
FIG. 1.
Kinetic phenomena influencing cellular concentrations and rate of protein synthesis. μ, growth rate; k′, translation efficiency; k", enzyme turnover.
The ratio between the rate of enzyme synthesis and the cell transcript concentration enabled the translational efficiency to be calculated (k′ = Rtranslation/[mRNA], where [mRNA] is the phosphatase alkaline activity · [total RNA] [Table 5]). For all the genes, an average of threefold increase in translational efficiency was observed for glucose-grown cells compared to that of galactose-grown cells on both media. This observation suggests that translation was regulated by a sugar-dependent mechanism. This translation efficiency was not correlated to a proportional change in rRNA concentration. Indeed, the total RNA concentrations (Table 5), the great majority of which consist of rRNA, were similar for both sugars when MCD medium was used, although the k′ differed significantly. Other factors governing rates of translation (regulation of ribosome activity, availability of charged tRNA, etc.) may therefore be suspected.
Though the mechanism involved remains obscure, the consequences of the phenomenon can be seen. Transcriptional control will be subject to amplification or attenuation, depending upon the response and the sugar being consumed. Those genes expressed at identical levels on the two sugars or expressed at higher levels on glucose will be translated more rapidly on glucose, thereby amplifying the extent of positive catabolite activation phenomena. In view of the generally higher rates of growth obtained on glucose, this will enable adequate concentrations of the enzymes to be maintained. On the other hand, those genes subject to negative catabolite repression by glucose will see this effect attenuated, since the lower level of expression will be to some extent compensated for by the higher degree of translational efficiency. Again, the rates of cell division will further modify the concentration of the enzyme. Thus, predicting phenotypic behavior without integrating quantitative analysis of transcript abundance into the general physiological characteristics of the cells will tend to generate erroneous hypotheses. When the analysis is scaled up to the full genome, this type of coordinated treatment of experimental data will be essential.
ACKNOWLEDGMENTS
We thank the principal investigators of the L. lactis IL1403 sequencing project, Alexander Bolotin, Alexei Sorokin, and Dusko Ehrlich (INRA, Jouy en Josas, France) and Patrick Wincker, Olivier Jaillon, and Jean Weissenbach (CNS Genoscope, Evry, France), for providing the sequences of certain genes prior to publication as well as P. Renault and E. Jamet (INRA Jouy-en-Josas, France) for useful discussions.
REFERENCES
- 1.Arnau J, Jørgensen F, Madsen S M, Vrang A, Israelsen H. Cloning, expression and characterization of the Lactococcus lactis pfl gene, encoding pyruvate formate lyase. J Bacteriol. 1997;179:5884–5891. doi: 10.1128/jb.179.18.5884-5891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnau J, Jørgensen F, Madsen S M, Vrang A, Israelsen H. Cloning of the Lactococcus lactis adhE gene, encoding multifunctional alcohol dehydrogenase, by complementation of a fermentative mutant of Escherichia coli. J Bacteriol. 1998;180:3049–3055. doi: 10.1128/jb.180.12.3049-3055.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolotin A, Mauger S, Marlarme K, Ehrlich D S, Sorokin A. Low-redundancy sequencing of the entire L. lactis IL 1403 genome. Antonie Leeuwenhoek. 1999;76:27–76. [PubMed] [Google Scholar]
- 4.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Marlarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. The complete genome sequence of the lactic acid bacterium Lactococcus lactis IL 1403. Genome Res., in press. [DOI] [PMC free article] [PubMed]
- 5.Bond D R, Tsai B M, Russell J B. The diversion of lactose carbon through the tagatose pathway reduces the intracellular fructose-1,6-bisphosphate and growth rate of Streptococcus bovis. Appl Microbiol Biotechnol. 1998;49:600–605. doi: 10.1007/s002530051220. [DOI] [PubMed] [Google Scholar]
- 6.Cancilla M R, Davidson B E, Hillier A J, Nguyen N Y, Thompson J. The Lactococcus lactis triosephosphate isomerase gene, tpi, is monocistronic. Microbiology. 1995;141:229–238. doi: 10.1099/00221287-141-1-229. [DOI] [PubMed] [Google Scholar]
- 7.Cancilla M R, Hillier A J, Davidson B E. Lactococcus lactis glyceraldehyde-3-phosphate dehydrogenase gene, gap: further evidence for strongly biased codon usage in glycolytic pathway genes. Microbiology. 1995;141:1027–1036. doi: 10.1099/13500872-141-4-1027. [DOI] [PubMed] [Google Scholar]
- 8.Cocaign-Bousquet M, Garrigues C, Novak L, Lindley N D, Loubière P. Rational development of a simple synthetic medium for the sustained growth of Lactococcus lactis. J Appl Bacteriol. 1995;79:108–116. [Google Scholar]
- 9.Dominguez H, Rollin C, Guyonvarch A, Guerquin-Kern J-L, Cocaign-Bousquet M, Lindley N D. Carbon-flux distribution in the central metabolic pathways of Corynebacterium glutamicum during growth on fructose. Eur J Biochem. 1998;254:96–102. doi: 10.1046/j.1432-1327.1998.2540096.x. [DOI] [PubMed] [Google Scholar]
- 10.Even S, Garrigues C, Loubière P, Lindley N D, Cocaign-Bousquet M. Pyruvate metabolism in Lactococcus lactis is dependent upon glyceraldehyde-3-phosphate deydrogenase activity. Metabol Eng. 1999;1:198–205. doi: 10.1006/mben.1999.0120. [DOI] [PubMed] [Google Scholar]
- 11.Garrigues C, Loubière P, Lindley N D, Cocaign-Bousquet M. Control of the shift from homolactic to mixed acid fermentation in Lactococcus lactis: predominant role of the NADH/NAD+ ratio. J Bacteriol. 1997;179:5282–5287. doi: 10.1128/jb.179.17.5282-5287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrigues, C., M. Mercade, M. Cocaign-Bousquet, N. D. Lindley, and P. Loubière. Regulation of pyruvate metabolism in Lactococcus lactis depends upon the imbalance between catabolism and anabolism. Biotech. Bioeng., in press. [PubMed]
- 13.Gracy R W, Tylley B E. Phosphoglucose isomerase of human erythrocytes and cardiac tissue. Methods Enzymol. 1975;41:392–400. doi: 10.1016/s0076-6879(75)41086-2. [DOI] [PubMed] [Google Scholar]
- 14.Kulbe K D, Foellmer H, Fuchs J. Simultaneous purification of glyceraldehyde-3-phosphate dehydrogenase, 3-phosphoglycerate kinase, and phosphoglycerate mutase from pig liver and muscle. Methods Enzymol. 1982;90:498–499. doi: 10.1016/s0076-6879(82)90177-x. [DOI] [PubMed] [Google Scholar]
- 15.Le Bloas P, Guilbert N, Loubière P, Lindley N D. Growth inhibition and pyruvate overflow during glucose metabolism of Eubacterium limosum are related to a limited capacity to reassimilate CO2 by the acetyl-CoA pathway. J Gen Microbiol. 1993;139:1861–1868. [Google Scholar]
- 16.Llanos R M, Harris C J, Hillier A J, Davidson B E. Identification of a novel operon in Lactococcus lactis encoding three enzymes for lactic acid synthesis: phosphofructokinase, pyruvate kinase and lactate dehydrogenase. J Bacteriol. 1993;175:2541–2551. doi: 10.1128/jb.175.9.2541-2551.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowry O H, Rosebrough N J, Farr A L, Randa P P. Protein measurement with the Folin reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 18.Luesink E J, van Herpen R E M A, Grossiord B P, Kuipers O P, de Vos W M. Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactococcus lactis are mediated by the catabolite control protein CcpA. Mol Microbiol. 1998;30:789–798. doi: 10.1046/j.1365-2958.1998.01111.x. [DOI] [PubMed] [Google Scholar]
- 19.Poolman B, Konings W N. Relation of growth of Streptococcus cremoris to amino transport. J Bacteriol. 1988;170:700–707. doi: 10.1128/jb.170.2.700-707.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saier M H, Jr, Chauvaux S, Deutscher J, Reizer J, Ye J J. Protein phosphorylation and regulation of carbon metabolism in Gram-negative versus Gram-positive bacteria. Trends Biochem Sci. 1995;20:267–271. doi: 10.1016/s0968-0004(00)89041-6. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Scholler P, Heber S, Hoheisel J D. Optimisation and automation of fluorescence-based DNA hybridization for high-throughput clone mapping. Electrophoresis. 1998;19:504–508. doi: 10.1002/elps.1150190409. [DOI] [PubMed] [Google Scholar]
- 23.Siciliano M J, White B F. Isozyme identification of chromosomes in interspecific somatic cell hybrids. Methods Enzymol. 1987;151:169–194. doi: 10.1016/s0076-6879(87)51018-7. [DOI] [PubMed] [Google Scholar]
- 24.Sjoberg A, Hahn-Hagerdal B. β-Glucose-1-phosphate, a possible mediator for polysaccharide formation in maltose-assimilating Lactococcus lactis. Appl Environ Microbiol. 1989;55:1549–1554. doi: 10.1128/aem.55.6.1549-1554.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi S, Abbe K, Yamada T. Purification of pyruvate formate-lyase from Streptococcus mutans and its regulatory properties. J Bacteriol. 1982;149:1034–1040. doi: 10.1128/jb.149.3.1034-1040.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas T D, Crow V L. Selection of galactose-fermenting Streptococcus thermophilus in lactose-limited chemostat cultures. Appl Environ Microbiol. 1984;48:186–191. doi: 10.1128/aem.48.1.186-191.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas T D. Activator specificity of pyruvate kinase from lactic Streptococci. J Bacteriol. 1976;125:1240–1242. doi: 10.1128/jb.125.3.1240-1242.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vasconcelos I, Girbal L, Soucaille P. Regulation of carbon and electron flow in Clostridium acetobutylicum grown in chemostat culture at neutral pH on mixtures of glucose and glycerol. J Bacteriol. 1994;176:1443–1450. doi: 10.1128/jb.176.5.1443-1450.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]