Summary
Tumor vessel co-option, a process in which cancer cells “hijack” pre-existing blood vessels to grow and invade healthy tissue, is poorly understood but is a proposed resistance mechanism against anti-angiogenic therapy (AAT). Here, we describe protocols for establishing murine renal (RENCA) and breast (4T1) cancer lung vessel co-option metastases models. Moreover, we outline a reproducible protocol for single-cell isolation from murine lung metastases using magnetic-activated cell sorting as well as immunohistochemical stainings to distinguish vessel co-option from angiogenesis.
For complete details on the use and execution of this protocol, please refer to Teuwen et al. (2021).
Subject areas: Antibody, Cancer, Cell isolation, Model organisms, Single cell
Graphical abstract
Highlights
-
•
Murine RENCA and 4T1 cancer lung vessel co-option metastases models
-
•
High purity and quality single-cell isolation from murine lung metastases
-
•
Immunohistochemical stainings to distinguish vessel co-option from angiogenesis
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Tumor vessel co-option, a process in which cancer cells “hijack” pre-existing blood vessels to grow and invade healthy tissue, is poorly understood but is a proposed resistance mechanism against anti-angiogenic therapy (AAT). Here, we describe protocols for establishing murine renal (RENCA) and breast (4T1) cancer lung vessel co-option metastases models. Moreover, we outline a reproducible protocol for single-cell isolation from murine lung metastases using magnetic-activated cell sorting as well as immunohistochemical stainings to distinguish vessel co-option from angiogenesis.
Before you begin
This protocol describes the material and procedure to generate murine sunitinib-induced (RENCA) and spontaneous (4T1) vessel co-option lung metastasis models for single-cell isolation of metastases-derived cells and endothelial cells (ECs). These models were established to investigate tumor vessel co-option, a process in which cancer cells “hijack” pre-existing blood vessels (in contrast to inducing the formation of new blood vessels starting from pre-existing blood vessels (vessel sprouting angiogenesis)) in order to grow and invade healthy tissue, which may occur spontaneously or which can be induced by vascular endothelial growth factor (VEGF)-blockade (Kuczynski et al., 2019). To study the heterogeneity of metastases-derived cells and co-opted ECs in tumors growing via vessel co-option, isolation of high purity single cells is required.
Here, we describe the methods how to establish the models, to perform immunohistochemical stainings to differentiate vessel co-option from vessel sprouting and to isolate single ECs, cancer cells, immune cells and perivascular cells. For EC isolation, the protocol is based on magnetic bead sorting with the MACS system (Miltenyi Biotec) to deplete immune cells (CD45)/epithelial cells (EpCAM) and to enrich for ECs (CD31). Further steps/assays in which these isolated cells can be used, such as single-cell RNA sequencing (scRNA-seq), are not addressed in this protocol but are described in more detail by Teuwen et al. (Teuwen et al., 2021). Besides scRNA-seq, isolated cells can also be used for bulk RNA-seq or for cell culture, etc.
Institutional permissions
All experimental procedures were approved by the Institutional Animal Ethics Committee of the KU Leuven (Belgium) under protocol number P084/2016.
Culturing "RENCA" renal cancer cell line and "4T1" breast cancer cell line
Timing: ∼1 week
-
1.
Culture RENCA cells and 4T1 cells at 37°C, 5% CO2 in RPMI 1640 (Thermo Fisher Scientific) supplemented with 2 mM L-glutamine (Thermo Fisher Scientific), 10% fetal bovine serum (Merck - Biochrom), 100 IU/mL penicillin and 100 μg/mL streptomycin (Thermo Fisher Scientific).
-
2.
Keep the RENCA and 4T1 cells in culture for at least two passages.
-
3.
Check the cell lines for mycoplasma on a regular basis.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Hamster anti-mouse PDPN (1/500) | DSHB | Cat# 8.1.1; RRID: AB_531893 |
| Rat anti-mouse CD31 (1/50) | BD Biosciences | Cat# 550274; RRID: AB_393571 |
| Rat anti-mouse CD34 (1/25) | BD Biosciences | 553731; RRID: AB_395015 |
| Goat anti-mouse ESM1 (1/50) | R&D | Cat# AF 1999; RRID: AB_2101810 |
| Prolong Gold Antifade Mountant | Thermo Fisher Scientific | Cat# P36934 |
| Chemicals, peptides, and recombinant proteins | ||
| Antibiotic-antimycotic | Thermo Fisher Scientific | 15240062 |
| Benzyl alcohol | Sigma-Aldrich | 402834 |
| Bovine serum albumin (BSA) | Sigma-Aldrich | A2058 |
| Carboxymethylcellulose sodium | Sigma-Aldrich | C9481 |
| Click-iTTM Plus EdU Cell Proliferation Kit for Imaging | Thermo Fisher Scientific | C10338 |
| Collagenase type II | Thermo Fisher Scientific | 17101015 |
| Collagenase type IV | Worthington Biochemical | LS004188 |
| DNase I | Sigma-Aldrich | D4527-10KU |
| Endothelial cell growth factor supplements (ECGS/Heparin) | PromoCell | C-30120 |
| EDTA | Sigma-Aldrich | ED2P-500G |
| EdU (5-ethynyl-2′-deoxyuridine) | Thermo Fisher Scientific | A10044 |
| Fetal bovine serum (FBS) | Merck - Biochrom | S 0415 |
| Glutamine | Thermo Fisher Scientific | 25030149 |
| Hoechst 33258 | Sigma-Aldrich | B2261 |
| KnockOutTM DMEM | Thermo Fisher Scientific | 10829018 |
| MEM NEAA | Thermo Fisher Scientific | 11140035 |
| Nimatek (100 mg/mL); Ketamine | Dechra | N/A |
| Penicillin/streptomycin | Thermo Fisher Scientific | 15140122 |
| Phosphate buffered saline (DPBS) | Thermo Fisher Scientific | 14190094 |
| RBC lysis buffer | Sigma-Aldrich | R7757 |
| ROTI® HISTOFIX 4% | Carl Roth | P087.1 |
| RPMI 1640 | Thermo Fisher Scientific | 21875-034 |
| Sodium pyruvate | Thermo Fisher Scientific | 11360070 |
| Sunitinib, Malate Salt | LC Laboratories | S-8803 |
| Thimerosal | Sigma-Aldrich | 71230-50G |
| Triton X-100 | Merck - Millipore | 1.08603 |
| TSA Cyanine 3 | PerkinElmer | NEL704A001KT (Cy3) |
| TSA Cyanine 5 | PerkinElmer | NEL705A001KT (Cy5) |
| TSA Fluorescein | PerkinElmer | NEL701A001KT (FT) |
| Tween 80 | Sigma-Aldrich | P1754 |
| Xylazine | VMD | XYL-M 2% |
| Critical commercial assays | ||
| CD31 MicroBeads, mouse | Miltenyi Biotec | 130-097-418 |
| CD45 MicroBeads, mouse | Miltenyi Biotec | 130-052-301 |
| CD326 (EpCAM) MicroBeads, mouse | Miltenyi Biotec | 130-105-958 |
| Deposited data | ||
| RNA-sequencing raw data mouse EC | (Teuwen et al., 2021) | ArrayExpress: (https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-9227) |
| Experimental models: Cell lines | ||
| Luciferase-tagged 4T1 cells | A. Reynolds | N/A |
| Luciferase-tagged RENCA cells | A. Reynolds | N/A |
| Experimental models: Organisms/strains | ||
| BALB/c mice (8-10 week-old; female; immunocompetent) | KU Leuven animal facility | N/A |
| BALB/c mice (8-10 week-old; female; immunocompetent) | Charles River | N/A |
| Other | ||
| 24G AGANI needle (25 × 0.55 mm) | Terumo | AN∗2425R1 |
| 29G needle | Terumo | 51908 |
| 40 μm cell strainer Nylon (blue) | Sigma-Aldrich | CLS431750-50EA |
| Cell culture Flask T-75 | Corning | 3290 |
| Centrifuge tube, conical, HDPE CentriStar™, PP, 15 mL | VWR | 734-1867 |
| Centrifuge tube, conical, HDPE CentriStar™, PP, 50 mL | VWR | 734-1869 |
| Dissection microscope | N/A | N/A |
| GentleMACS C tubes | Miltenyi Biotec | 130-093-237 |
| GentleMACS™ Dissociator | Miltenyi Biotec | 130-093-235 |
| Heating cage | N/A | N/A |
| Infusion Sets Microflex 25G -30 cm orange (x20) | Vygon | 240.05 |
| MS columns | Miltenyi Biotec | 130-042-201 |
| MACS MultiStand | Miltenyi Biotec | 130-042-303 |
| MiniMACS™ Separator | Miltenyi Biotec | 130-090-312 |
| Multipurpose- and Micro-centrifuge | N/A | N/A |
| Perfusion pump: Perfusor® fm (MFC) | B. Braun Malaysia | N/A |
| Silk Suture Thread, Size 5/0 (for lung insufflation) | Fine Science Tools | 18020-50 |
| Surgical Scalpel Blade No 10 | Swann-Morton | 0201 |
| Syringe, 1 mL | N/A | N/A |
| Syringe Pump Harvard Apparatus | Harvard Apparatus | PHD 22/2000 |
| Tweezers, forceps, scissors | N/A | N/A |
| Water bath or incubator adjusted to 37°C | N/A | N/A |
Materials and equipment
Media and buffers
The following media/buffers are required.
Anesthesia (∼15 min)
| Reagent | Final concentration | Amount |
|---|---|---|
| Ketamin (Nimatek®, Eurovet, Bladel, The Netherlands) | 3/8 of Vtot | 187.5 μL |
| Xylazin 2% (XYL-M®, VMD NV, Arendonk, Belgium) | 1/8 of Vtot | 62.5 μL |
| Saline (0.9% w/v NaCl, Mini-Plasco #0819-094, Belgium) | 1/2 of Vtot | 250 μL |
| Total | N/A | 500 μL |
Anesthesia can be stored at 4°C for up to 30 days. Here, we described the Total volume, Vtot required = number of mice × estimated weight of one mouse (20 g) × 4 (+ dead volume). For example, for six mice (with an average weight of 20 g) the required total volume is 500 μL.
RPMI 1640 (full) (∼30 min)
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI 1640 complete tissue culture medium (Thermo Fisher Scientific, #21875-034) | N/A | 440 mL |
| L-glutamine (Thermo Fisher Scientific, #25030149) | 2 mM | 5 mL |
| Penicillin/Streptomycin (Thermo Fisher Scientific, #15140122) | 1% | 5 mL |
| Fetal bovine serum (FBS) (Merck-Biochrom, #S0415) | 10% | 50 mL |
| Total | N/A | 500 mL |
RPMI 1640 (full) can be stored at 2°C–8°C in the dark for one month.
Control vehicle (∼3 h + stirring for 14–20 h)
| Reagent | Final concentration | Amount |
|---|---|---|
| NaCl | 1.8% | 1.8 g |
| Tween 80 (Sigma-Aldrich, #P1754) | 0.4% | 0.4 mL |
| Benzyl alcohol (Sigma-Aldrich, #402834) | 0.9% | 0.9 mL |
| Carboxymethylcellulose sodium (Sigma-Aldrich, #C9481) | 0.5% | 0.5 g |
| Distilled water | N/A | 100 mL |
| Total | N/A | 101.3 mL |
Control vehicle can be stored for up to one month at 4°C. Each mouse will be injected with 200 μL. Due to its viscosity, part of the solution will get lost during treatment. Therefore, it is recommended to prepare 30%–40% more. For example, if you have 20 mice that you have to treat for 7 days, you need to prepare at least 40 mL of control vehicle.
Sunitinib suspension (∼2 h)
| Reagent | Final concentration | Amount |
|---|---|---|
| Control vehicle | N/A | 30 mL |
| Sunitinib, Malate Salt (LC laboratories, #S-8803) | 5.2 mg/mL | 156 mg |
| Total | N/A | 30 mL |
The sunitinib suspension is stable for 1–2 weeks when stored at 4°C in the dark. Do not freeze the drug suspension. For a typical experiment we dose at 40 mg/kg/day. Taking into account that the weight of the active drug is less than the salt weight (the molecular weight of the L-malate salt of sunitinib is 532.58, while the molecular weight of the active drug is 398.5), a 10 mg/mL solution is equivalent to a 7.5 mg/mL active drug solution. Therefore, assuming that the bodyweight of the mice is 20 grams (range of 19–21 grams) we add sunitinib to the control vehicle at 5.2 mg of sunitinib per 1 mL of control vehicle in order to dose at 40 mg/kg/day with 200 μL per mouse per day).
CRITICAL:Prepare the sunitinib suspension under a fume hood to avoid health hazards.
MACS buffer (∼30 min)
| Reagent | Final concentration | Amount |
|---|---|---|
| BSA (Sigma-Aldrich, #A2058) | 0.5% | 2.5 g |
| EDTA (filtered) (Sigma-Aldrich, #ED2P-500G) | 2 mM | 0.2 mL (from a 5 M stock solution, prepared according to the manufacturer’s instructions) |
| DPBS (Thermo Fisher Scientific, #14190-094) | N/A | 499.8 mL |
| Total | N/A | 500 mL |
MACS buffer can be prepared one day in advance and stored for up to three days at 4°C.
Digestion buffer medium (Supplemented KnockOutTM DMEM-medium) (∼30 min)
| Reagent | Final concentration | Amount |
|---|---|---|
| KnockOut™ DMEM (Thermo Fisher Scientific, #10829018) | N/A | 477 mL |
| Penicillin/Streptomycin (Thermo Fisher Scientific, #15140122) | 1% (v/v) | 5 mL |
| Antibiotic-Antimycotic, 100X solution (Thermo Fisher Scientific, #15240062) | 2× | 10 mL |
| Sodium Pyruvate, 100 mM solution (Thermo Fisher Scientific, #11360070) | 1 mM | 5 mL |
| MEM Non-Essential Amino Acids Solution, 100X solution (MEM-NEAA) (Thermo Fisher Scientific, #11140035) | 1× | 5 mL |
| Endothelial Cell Growth Factor supplements (ECGS/ Heparin) (PromoCell, #C-30120) | 1× | 2 mL (one vial) |
| Total | N/A | 504 mL |
Digestion buffer medium (Supplemented KnockOutTM DMEM-medium) can be stored at 4°C, up to three months in a sterile manner. Freshly add the digestion enzymes (DNase and Collagenases) to the digestion medium before the start of the isolation procedures.
Lung digestion buffer (∼45 min)
| Reagent | Final concentration | Amount |
|---|---|---|
| Supplemented KnockOutTM DMEM-medium | N/A | 29.55 mL |
| Collagenase II (Thermo Fisher Scientific, #17101-015) | 0.1% (w/v) | 30 mg |
| Collagenase IV (Worthington, #LS004188) | 0.25% (w/v) | 75 mg |
| DNase I (Sigma-Aldrich, #D4527-10KU) | 15 μg/mL | 450 μL (75 μL per 5 mL buffer) Dissolve 1 mg DNase I in 1 mL PBS. Store in 75 μL aliquots at −20°C.) |
| Total | N/A | 30 mL |
Prepare lung digestion buffer immediately before isolation and store at 4°C until needed. It is recommended to use papier calque to weigh Collagenase type II and type IV, because powder sticks to the plastic. 5 mL of lung digestion buffer is sufficient for lungs from 1 adult mouse.
TBS (TRIS Buffered SALINE) (10×) (∼15 min)
| Reagent | Final concentration | Amount |
|---|---|---|
| TRIS-HCl (1 M) pH 7.5 | 0.1 M | 200 mL |
| NaCl (5 M) | 1.5 M | 600 mL |
| Thimerosal (Sodium Ethylmercurithiosalicylate) | 0.04% (w/v) | 800 mg |
| Triton X-100 | 1% (v/v) | 20 mL |
| Adjust to 2 L with distilled water | N/A | 1.18 L |
| Total | N/A | 2 L |
TBS can be stored at 20°C–22°C for 24 months. Use within one week once it has been diluted to 1×.
TNB (TRIS-NaCl-blocking buffer) (∼15 min)
| Reagent | Final concentration | Amount |
|---|---|---|
| TRIS-HCl (1 M) pH 7.5 | 0.1 M | 100 mL |
| NaCl (5 M) | 0.15 M | 30 mL |
| Blocking reagent from amplification kits | 0.5% | 5 g |
| Adjust to 1 L with distilled water | N/A | 870 mL |
| Total | N/A | 1 L |
To dissolve the blocking reagent, heat TNB blocking buffer to 60°C for 1 h with stirring. Aliquot and store TNB blocking buffer at –20°C for up to one month. Discard any unused blocking buffer which has been stored for more than 24 h at 20°C–22°C.
TNT (TRIS-NaCl-TRITON buffer) (∼15 min)
| Reagent | Final concentration | Amount |
|---|---|---|
| TRIS-HCl (1 M) pH 7.5 | 0.1 M | 100 mL |
| NaCl (5 M) | 0.15 M | 30 mL |
| Triton X-100 | 0.05% | 0.5 mL |
| Adjust to 1 L with distilled water | N/A | 869.5 mL |
| Total | N/A | 1 L |
TNT can be stored at 20°C–22°C for 1–2 weeks.
Alternatives: equipment and reagents
-
•
To perform transcardial perfusion, a perfusion pump Perfusor® fm (MFC) - B. Braun Malaysia or Harvard Apparatus PHD 22/2000 was used. Any other perfusion pump that allows the setting described in the protocols can be used.
-
•
Non-column based magnetic isolations methods are available (e.g., from STEMCELL Technologies) that could be optimized and used as alternatives for depletion and enrichment purposes.
-
•
Digestion efficiency may depend on the LOT number and/or the vendor of the enzymes used. Therefore, we recommend to test all newly purchased reagents before performing final experiments.
-
•
Applying sterile lab-practice and working in a laminar flow hood may be necessary depending on further downstream use of the isolated metastases-derived cells or ECs. For example, if the cells will be kept in culture, all reagents should be used under a laminar flow to avoid contamination and to keep them sterile. Alternatively, for end-stage experiments (e.g., sequencing, nucleic acid isolation, cell staining, Western blotting) the sterility of the purified cells might be less important.
-
•
A heating cage was used to improve vein visibility before tail vein injection. As an alternative for the heating cage, infrared heat lamps can be used.
-
•
GentleMACS™ Dissociator (Miltenyi Biotec, Cat#130-093-235), required for the digestion of lung tissue can be replaced by the gentleMACS™ Octo Dissociator (Miltenyi Biotec, Cat#130-095-937). The use of other mechanical tissue dissociation methods (i.e., razor blades or scalpel blades) was not tested and optimized within this protocol. As such, this could be optional, but would first require further optimization as the expected outcome cannot be guaranteed.
-
•
In this protocol, we used 40 μm cell strainers from Sigma-Aldrich (#CLS431750-50EA). Alternative 40 μm cell strainers from other vendors can be used.
-
•
We suggest to use the following antibodies: PDPN, (DSHB, Cat#8.1.1, 1/500); CD31, (BD Biosciences, Cat#550274, 1/50); ESM1, (R&D, Cat#AF1999, 1/50). However, alternative antibodies with similar properties (clone and antigen of the antibody), or with different clonality can also be used. If alternative antibodies will be used, we recommend optimizing the staining and antibody titration beforehand to obtain the most accurate results. Please take into consideration that the staining efficiency might also depend on the clonality, LOT number and/or vendor of the antibody.
-
•
Hoechst staining was used to visualize the nuclei. Alternatively, DAPI can also be used.
-
•
To anesthetize the mice, we used a mixture of Ketamine (Eurovet) and Xylazine (VMD) according to the local ethical committee guidelines. However, this can be replaced by alternative anesthetics that have been approved by the local ethical committee.
-
•
For fixation purposes, ROTI® HISTOFIX 4% (Carl Roth, #P087.1) was used. Alternatively, self-made paraformaldehyde 4%, or pre-made paraformaldehyde 4% solutions from other vendors can be used.
Step-by-step method details
Set-up of a RENCA and 4T1 mouse model with sunitinib treatment
Timing: ∼3–6 weeks
The following protocol describes the generation of sunitinib-induced (RENCA) and spontaneous (4T1) lung metastasis models of vessel co-option.
-
1.
Set-up of mouse model of renal cancer experimental lung metastasis (RENCA model) and of breast cancer experimental lung metastasis (4T1 model).
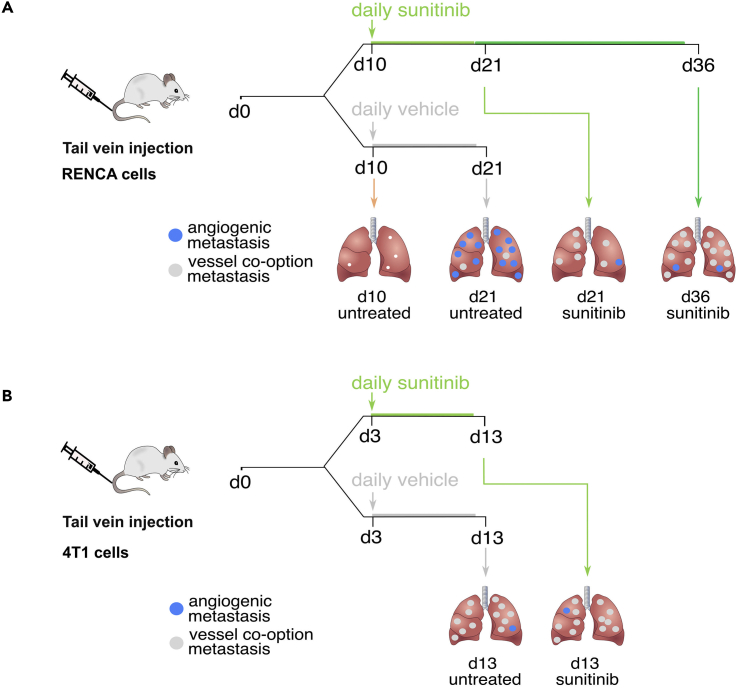
Figure 1A shows a detailed scheme of the RENCA model set-up and treatment schedule. Figure 1B shows a detailed scheme of the 4T1 model set-up and treatment schedule. The RENCA and the 4T1 model are based on the models described by Bridgeman et al. (Bridgeman et al., 2017).-
a.Turn on the heating cage.
-
b.When RENCA /4T1 cells reach 70% confluency after at least 2 passages, prepare them for tail vein injection:
-
i.Wash the cells 1× with phosphate buffered saline (PBS).
-
ii.Detach the cells with trypsin (1 mL for a cell culture flask T-75).
-
iii.Add 9 mL RPMI 1640 (full), resuspend and transfer to a 50 mL conical tube.
-
iv.Count the cells with a counting chamber.
-
v.Transfer the required volume of the cell suspension (2 × 105 cells per mouse) in a new 50 mL conical tube.
-
vi.Spin down 4 min at 145 g.
-
vii.Aspirate the supernatant, add 5 mL PBS and resuspend thoroughly.
-
viii.Spin down 4 min at 145 g.
-
ix.Resuspend with PBS (100 μL/mouse).
-
x.Place on ice.
-
i.
-
c.Place the mice (8- to 10-week-old female immunocompetent BALB/c mice) in the heating cage for a few min.
-
d.On day 0, inject RENCA / 4T1 cells (2 × 105 cells in 100 μL PBS) into the tail vein of the mice using a 29G needle.Note: The number of RENCA/4T1 cells to be injected was carefully titrated in advance in order to keep the mice alive without the risk of them becoming sick before the end of the experimental period.
-
a.
-
2.
Preparation of control vehicle (prepare two days before the start of the treatment).
Here we describe how to prepare 100 mL of control vehicle. Of note, the amount of control vehicle should be adjusted according to the number of mice included in the experiment.-
a.Fill a 100 mL glass bottle with 80 mL of reverse osmosis (RO) distilled water.
-
b.Add a stirring bar to the 80 mL of RO distilled water and place on a stirring plate at 20°C–22°C.
-
c.Dissolve 1.8 g of NaCl in the 80 mL of RO distilled water.
-
d.Add 0.4 mL of Tween 80. This will take about 10 min to dissolve as it is viscous.
-
e.Add 0.9 mL of benzyl alcohol and leave it for 5 min.
-
f.Bring the solution to pH 6.
 CRITICAL:We recommend to gradually and slowly adjust the pH by adding 50–100 μL of 0.5 M HCl or 0.5 M NaOH (as appropriate) at a time. The use of stronger acid / base might cause the pH to change too drastically because the control vehicle does not contain buffering compounds (like TRIS, etc.).
CRITICAL:We recommend to gradually and slowly adjust the pH by adding 50–100 μL of 0.5 M HCl or 0.5 M NaOH (as appropriate) at a time. The use of stronger acid / base might cause the pH to change too drastically because the control vehicle does not contain buffering compounds (like TRIS, etc.).
-
g.Filter sterilize the solution into a clean bottle.
-
h.Add a stirring bar and place back on the stirring plate. Slowly add 0.5 g of carboxymethylcellulose sodium and leave to stir for 14–20 h at 20°C–22°C.Note: Carboxymethylcellulose sodium takes a long time to dissolve.
-
i.The next day, the carboxymethylcellulose sodium will have mostly dissolved. It does not dissolve completely, so the control vehicle is always slightly cloudy.
-
j.Add another 20 mL of RO distilled water.
-
k.Store this control vehicle for up to one month at 4°C.
-
a.
-
3.Preparation of sunitinib treatment (prepare one day before the start of the treatment).
-
a.For oral dosing, add sunitinib malate powder (LC laboratories) to the control vehicle and vortex vigorously to create a suspension.
-
i.Allocate the mice randomly to the treatment condition (control vehicle or sunitinib treatment), balanced for body weight (RENCA on Day 9; 4T1 on Day 2).
-
ii.Calculate the average weight (in g) per treatment group, the number of treatment days and the number of mice and use this average weight to calculate the amount of sunitinib needed to get 40 mg/kg/day. We recommend administering a gavage volume of 200 μL per mouse (Serwer et al., 2010; Teuwen et al., 2021). Add the calculated amount of sunitinib to the prepared control vehicle into a 50 mL tube and vortex vigorously until the sunitinib has formed a mixed suspension.
-
i.
-
a.
Note: Sunitinib takes time to dissolve. Preheat the control vehicle to 37°C prior to addition of sunitinib powder and vortex vigorously for several rounds.
Note: Fresh stocks of sunitinib suspension should be prepared weekly (the drug suspension is stable for 1–2 weeks), and stored at 4°C in the dark (never freeze the drug suspension). The suspension has a bright yellow/orange color. The drug has a tendency to settle quickly, so before dosing each mouse, the drug should be vortexed or drawn up and down several times with the syringe/gavage needle.
-
4.Treatment schedule (start treatment on day 10 (RENCA); on day 3 (4T1)).
-
a.For the RENCA model, start treatment from day 10 after injection of RENCA cells and continue until day 20 or day 35. Sacrifice mice on day 21 or day 36 of the experiment and process lungs immediately. Include the following predetermined timepoints (see Figure 1A ‘Treatment schedule’):
-
i.d21 untreated (control vehicle).
-
ii.d21 treated (sunitinib).
-
iii.d36 treated (sunitinib).
-
i.
-
b.For the 4T1 model, start treatment from day 3 after injection of 4T1 cells and continue for 10 days. Sacrifice mice on day 13 of the experiment and process lungs immediately. Include the following predetermined timepoints (see Figure 1B ‘Treatment schedule’):
-
i.d13 untreated (control vehicle).
-
ii.d13 treated (sunitinib).
-
i.
-
a.
Note: Check the mice daily for signs of obvious discomfort or illness necessitating euthanasia to adhere to animal welfare guidelines in the event that humane endpoints have been reached. For example, monitor breathing speed/pattern, general behavior, bodyweight and other signs according to local/national animal welfare and monitoring guidelines.
Figure 1.
Set-up of RENCA model and 4T1 model
(A) Set-up of RENCA model and treatment schedule.
(B) Set-up of 4T1 model and treatment schedule. This figure is taken from Teuwen et al. (Teuwen et al., 2021).
Metastases-derived cell and endothelial cell single-cell isolation from lung tissues from RENCA and 4T1 mouse model with sunitinib treatment
Timing: ∼3–5.5 h
The following protocol describes the isolation of metastases-derived cells and ECs from murine lung tissue. The goal of this protocol is to retrieve high purity single cells. The lung EC isolation protocol mainly overlaps with the lung EC isolation section of the ‘STAR protocols for endothelial cell isolation from mouse tissues: brain, choroid, lung and muscle’ as recently published (Conchinha et al., 2021). The steps that are different from the previously published STAR protocol are the addition of a section on collection of metastases and the absence of a FACS purification step.
-
5.Collection of metastases. (Timing: 2 h)
-
a.Anesthetize mice with an intraperitoneal injection of ketamine (100 mg/kg body weight) and xylazine (10 mg/kg body weight).Timepoints of sacrifice are indicated above: on day 21 or on day 36 for the RENCA model and on day 13 for the 4T1 model.
-
b.Open the thoracic cavity.Note: No perfusion is needed when tumor ECs are isolated. When ECs from healthy mice are isolated, perform transcardial perfusion (using a butterfly needle) via the right ventricle with 5 mL ice-cold PBS at a perfusion rate of 120 mL/h for 5 min to rinse away the blood and cut the lungs into small pieces before placing them into the gentleMACS C tube.
-
c.Dissect both lungs and place them in a petri-dish on ice.
-
d.Micro-dissect (at least) 60 metastases per mouse under a dissection microscope using fine point tweezers. A hematoxylin and eosin (H&E) stained image of lung metastases is included in Figure 2.
-
e.Place the dissected tissue immediately in a gentleMACS C tube (Miltenyi Biotec, Cat#130-093-237) containing 5 mL of lung digestion buffer and keep the tube and the digestion buffer on ice.
 CRITICAL: Make sure the tube is closed tightly.
CRITICAL: Make sure the tube is closed tightly.
-
a.
-
6.
Digestion of metastases and isolation of metastases-derived cells (including cancer cells, immune cells, perivascular cells). (Timing: 1 h)
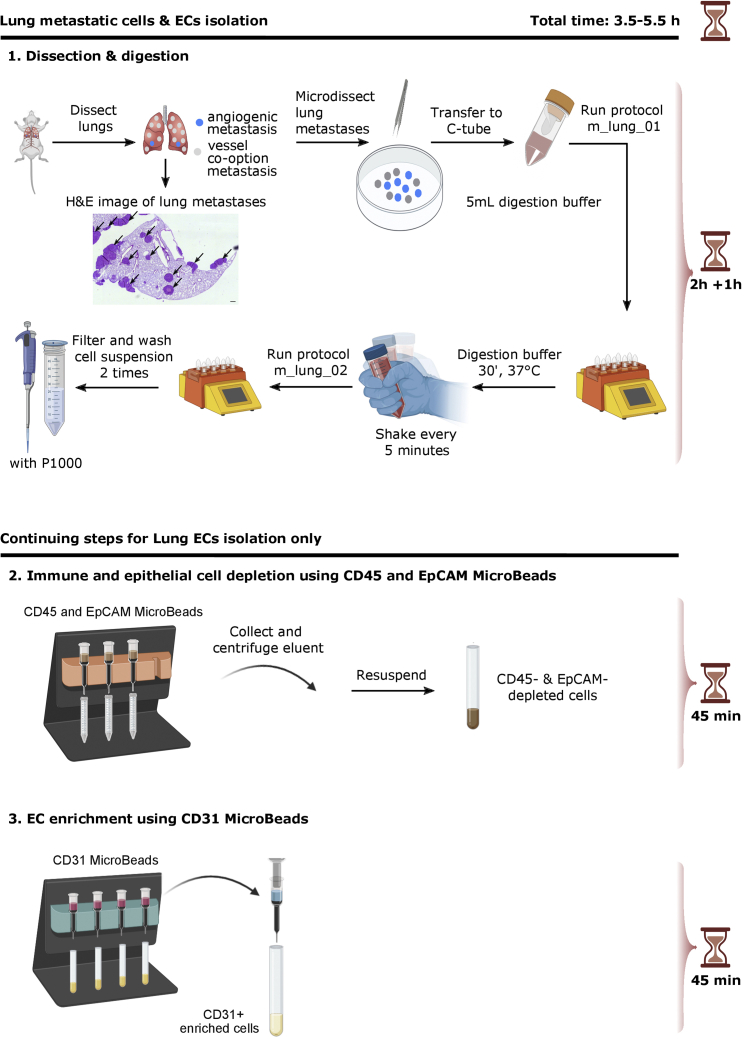
Figure 2 shows a detailed scheme of the isolation of cells derived from lung metastases. Digestion of lung tissue. (Timing: 1 h)-
a.Place the tubes into a gentleMACSTM Dissociator (Miltenyi Biotec).
-
b.Run the m_lung_01 protocol (pre-programmed by manufacturer).
-
c.Incubate for 30 min at 37°C.Note: Shake the tube vigorously by hand every 5 min for faster tissue dissociation.Note: After 20 min of incubation time, prepare 50 mL conical tubes with 40 μm cell strainers (blue), get the MACS buffer out of the fridge and prepare 15 mL conical tubes.
-
d.Place the tubes into a gentleMACSTM Dissociator (Miltenyi Biotec) and run the m_lung_02 protocol (pre-programmed by manufacturer).
-
e.Filter the cell suspension through a 40 μm cell strainer (blue; Sigma-Aldrich) into a 50 mL conical tube, rinse the strainer with 5 mL MACS buffer and transfer the suspension to a 15 mL conical tube.Note: use a pipette P1000 for this step.
-
f.Centrifuge the cell suspension at 250 g for 5 min and aspirate the supernatant.
-
g.Place the tubes on ice, resuspend the cell pellet in 100 μL of MACS buffer and 100 μL of RBC lysis buffer.
-
h.Leave on ice for 1 min.
-
i.Place in a 37°C water bath for 2 min.
-
j.Dilute the cell suspension with 10 mL MACS buffer, pipet up and down with a pipette P1000.
-
k.Centrifuge the cell suspension at 250 g for 5 min and aspirate the supernatant.Note: Expect a viability between 85%-94%. Keep cells on ice whenever possible.
-
a.
-
7.
Isolation of endothelial cells. (Timing: 2.5 h)
Figure 2 shows a detailed scheme of lung ECs isolation from lung metastases (also applicable to healthy lung).-
a.Digestion of lung metastases. (Timing: 1 h)
-
i.Place the tubes into a gentleMACSTM Dissociator (Miltenyi Biotec).
-
ii.Run the m_lung_01 protocol (pre-programmed by manufacturer).
-
iii.Incubate for 30 min at 37°C.Note: Shake the tube vigorously by hand every 5 min for better tissue dissociation.Note: After 20 min of incubation time, prepare 50 mL conical tubes with 40 μm cell strainers (blue), place MACS buffer on ice, and prepare 15 mL conical tubes.
-
iv.Place the tubes into a gentleMACSTM Dissociator (Miltenyi Biotec) and run the m_lung_02 protocol (pre-programmed by manufacturer).
-
v.Filter the cell suspension through a 40 μm cell strainer (blue; Sigma-Aldrich) into a 50 mL conical tube, rinse the strainer with 5 mL MACS buffer and transfer the suspension to a 15 mL conical tube.Note: use a pipette P1000 for this step.
-
vi.Centrifuge the cell suspension at 250 g for 5 min and aspirate the supernatant.
-
vii.Resuspend the cell pellet in 5 mL MACS buffer, pipet up and down with a pipette P1000, count the cells.
-
viii.Centrifuge the cell suspension at 250 g for 5 min and aspirate the supernatant. Proceed with the pellet to step b-i.
-
i.
-
b.Depletion of immune and epithelial cells using CD45 and CD326 (EpCAM) MicroBeads. (Timing: 45 min)
-
i.Add 80 μL MACS buffer, 10 μL CD45 MicroBeads and 10 μL CD326 (EpCAM) MicroBeads to the cell pellet.Note: We suggest to resuspend the cells in 80 μL of MACS buffer as indicated above. Adapt the amount of MicroBeads according to the cell number and follow the manufacturer's instructions, e.g. 10 μL of the beads for up to 1 x 107 cells; for larger numbers of cells, scale up the amount accordingly (https://www.miltenyibiotec.com/NL-en/products/cd45-microbeads-mouse.html?countryRedirected=1#gref; https://www.miltenyibiotec.com/US-en/products/cd326-epcam-microbeads-mouse.html#gref).
-
ii.Mix and incubate for 15 min at 4°C.Note: Process the samples quickly to avoid non-specific binding of MicroBeads at 20°C–22°C.
-
iii.Wash the cells by adding 3 mL of MACS buffer and centrifuge at 250 g for 5 min at 4°C.
-
iv.Aspirate the supernatant completely and resuspend the pellet in 0.5 mL of MACS buffer.
-
v.Place an MS column in the magnetic field of the suitable MACS Separator and rinse with 0.5 mL MACS buffer.
-
vi.Apply the cell suspension from step iv onto the column through a 40 μm cell strainer (blue) and collect unlabeled cells that flow through in 15 mL conical tube (this is the fraction that contains ECs).
-
vii.Perform three washing steps with 0.5 mL MACS buffer each.
-
viii.Centrifuge the flow-through (CD45 & CD326 (EpCAM) negative fraction) at 250 g for 5 min at 4°C.
-
i.
-
c.Enrichment of ECs using CD31 murine MicroBeads. (Timing: 45 min)
-
i.Add 90 μL MACS buffer and 10 μL CD31 MicroBeads to the cell pellet. Mix and incubate for 15 min at 4°C.
-
ii.Wash the cells by adding 3 mL of MACS buffer and centrifuge at 250 g for 5 min at 4°C.
-
iii.Aspirate the supernatant completely and resuspend the pellet in 0.5 mL of MACS buffer.
-
iv.Place an MS column in the magnetic field of the suitable MACS Separator and rinse the column with 0.5 mL MACS buffer.
-
v.Apply the cell suspension onto the column and collect the unlabeled cells that flow-through in a 15 mL conical tube.
-
vi.Perform three washing steps with 0.5 mL MACS buffer each, adding the MACS buffer each time once the column reservoir is empty.
-
vii.Remove the column from the separator, place it on a 15 mL conical tube, pipet 1 mL MACS buffer onto the column. Immediately flush out the magnetically labeled cells (CD31+) by firmly pushing the plunger into the column and centrifuge this fraction at 250 g for 5 min at 4°C. The EC-enriched suspension can be used for bulk or scRNA-seq or other downstream applications.Note: Expect a viability between 87%-94%. Keep cells on ice whenever possible.
-
i.
-
a.
Figure 2.
Isolation of metastases-derived cells and EC-s from mouse lung metastases
Scheme illustrating isolation of cells from metastases (1) and subsequent isolation of ECs from lung metastases (2, 3). For EC isolation, a MACS immune & epithelial cell depletion (2) and EC enrichment (3) was performed. The included H&E image shows lung metastases from the RENCA model (Arrows indicate metastases; scale bar: 250 μm). This figure is adapted from Conchinha et al. (Conchinha et al., 2021).
Distinguishing vessel co-option from angiogenesis in lung metastases by immunostaining
Timing: ∼2 days
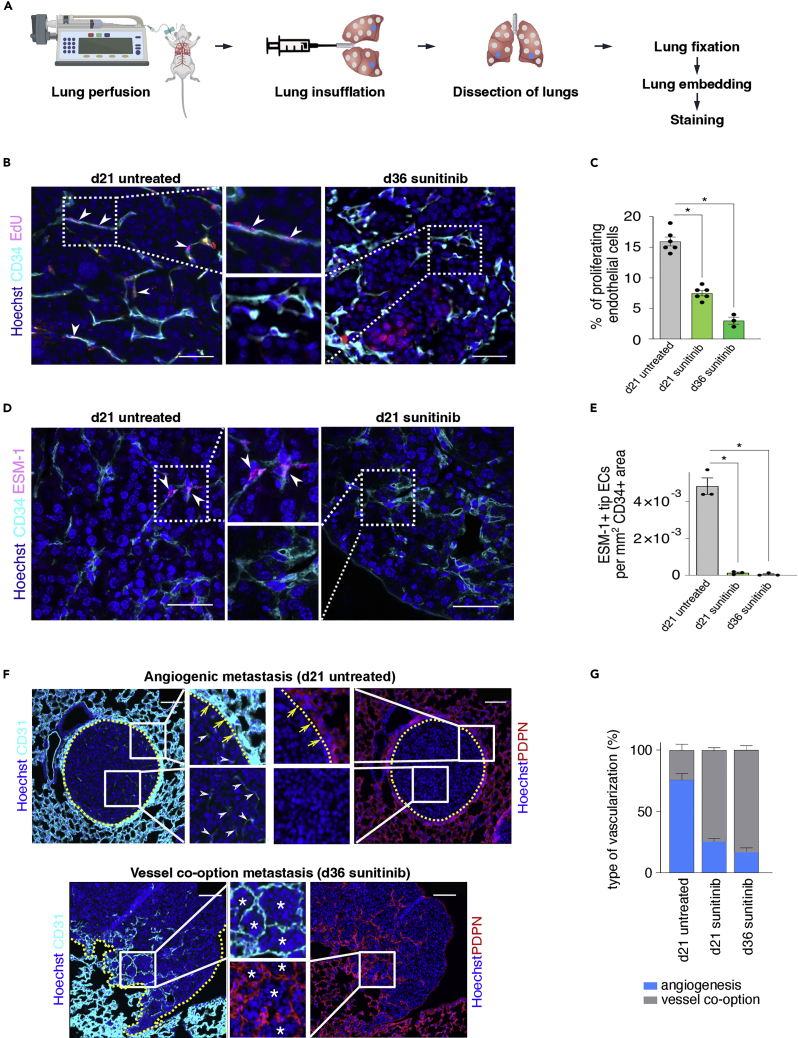
Here, we explain how to perform lung insufflation (Figure 3A) for immunohistochemistry and how to execute EdU (proliferation marker) or ESM1 (tip cell marker)/CD34 (EC marker) staining and CD31 (EC marker)-podoplanin (pneumocyte marker) staining (Figures 3B–3G), which are used to score the metastases as ‘angiogenic’ or ‘vessel co-option’. Figure 3B shows that ‘d36 sunitinib treated’ metastases (right panel, vessel co-option metastasis) contain less proliferating ECs (EdU-positive) than ‘d21 untreated’ metastases (left panel, angiogenic metastasis), as quantified in Figure 3C. Figure 3D shows that ‘d21 sunitinib treated’ metastases (right panel, vessel co-option metastasis) contain less tip ECs (ESM1-positive) when compared to ‘d21 untreated’ metastases (left panel, angiogenic metastasis), as quantified in Figure 3E. Furthermore, Figure 3F shows representative images of metastases with angiogenic sprouting and metastases with vessel co-option. Metastases relying on angiogenic sprouting grow as a sphere compressing the surrounding lung parenchyma and they show vessel sprouting within the metastasis, while cancer cells of metastases relying on vessel co-option are invading the surrounding lung tissue in an irregular and infiltrative manner and preserve the epithelial and vascular structure. In Figure 3G, the quantification of both vascularization types in the RENCA model is shown for the indicated conditions.
-
8.Lung insufflation for immunohistochemistry (injection of 500 μL PFA 4% (ROTI® HISTOFIX 4%) into trachea).
-
a.Anesthetize the mice with an intraperitoneal injection of ketamine (100 mg/kg body weight) and xylazine (10 mg/kg body weight).
-
b.Open the abdominal cavity, cut the diaphragm, and open the thoracic cavity by cutting the ribs along the midclavicular line towards the clavicula, while being careful not to touch the lungs.
-
c.Perform transcardial perfusion via the right ventricle.
-
i.Place the needle into the right ventricle.
-
ii.Start perfusion for 5 min with NaCl 0.9% at a perfusion rate of 2 mL/min.Note: Make a tiny cut after 1 min in the left atrium and continue with the perfusion.
-
iii.Start perfusion for 5 min with paraformaldehyde 4% (PFA4%) at a perfusion rate of 2 mL/min.
-
i.
-
d.Place a silk suture thread under the teeth and fix with tape to stabilize the head.
-
e.Dissect the neck area and pull the salivary glands apart.
-
f.Clear the trachea externally by removing tissue on the sides of the trachea by blunt dissection with tweezers and carefully removing the transparent layer on top with scissors.
-
g.Pull a silk suture thread under the trachea and make a loose double overhand knot.
-
h.Prepare a 1 mL syringe with 500 μL PFA 4% with a 24G needle and slightly bend the needle.
-
i.Insert the needle in the trachea, cranially from the knot.
-
j.Inject (not too slow – not too fast) and look at the expansion of the lungs.
-
k.Immediately after the injection tighten the knot and make two extra knots.
-
l.Dissect the lungs.
-
m.Wash the lungs in PBS.
-
n.Leave the lungs for 14–20 h in PFA 4%.
-
o.After 24 h, wash the lungs three times with PBS, remove the silk suture thread and prepare the lungs for embedding.
-
p.Embed the lungs in paraffin.
-
a.
-
9.CD31/podoplanin staining.
-
a.Deparaffinize the tissue sections and rehydrate for 5 min with distilled water.
-
b.Trypsinize the tissue sections for 7 min (1/80 in CaCl2 0.1%).
-
c.Wash 5 min with TBS (TRIS Buffered Saline).
-
d.To block endogenous peroxidase activity, incubate for 20 min with MeOH/H2O2 (200 mL methanol plus 600 μL H2O2 30%; Sigma-Aldrich 8.22287).
-
e.Wash 3 × 5 min with TBS.
-
f.Apply protein block (Goat serum) 1/5 in TNB (TRIS-NaCl-blocking Buffer) for 45 min to block nonspecific background staining.
-
g.Apply primary antibody Hamster anti-Podoplanin (DSHB 8.1.1-c) 1/500 in TNB.
-
h.Wash 3 × 5 min with TNT (TRIS-NaCl-TRITON Buffer).
-
i.Apply secondary antibody Goat anti-Hamster-B (Vector Laboratories, BA-9100) 1/200 in TNB for 45 min.
-
j.Wash 3 × 5 min with TNT.
-
k.Incubate for 30 min with Streptavidin-Peroxidase 1/100 in TNB.
-
l.Wash 3 × 5 min with TNT.
-
m.Incubate for 8 min with Cy3 1/50 in amplification diluent.Note: The amplification diluent is part of the TSA Fluorescence Systems KIT (containing Fluorescein Tyramide/Cyanine 3 Tyramide/Cyanine 5 Tyramide, Streptavidin-Peroxidase and blocking Reagent).
-
n.Wash 3 × 5 min with TNT.
-
o.Wash 5 min with TBS.
-
p.To block endogenous peroxidase activity, incubate for 20 min with MeOH/H2O2.
-
q.Wash 3 × 5 min with TBS.
-
r.Apply protein block (Donkey serum) 1/5 in TNB for 45 min to block nonspecific background staining.
-
s.Apply primary antibody Rat anti-CD31 (BD 550274) antibody 1/50 in TNB.
-
t.Wash 3 × 5 min with TNT.
-
u.Apply secondary antibody Donkey anti-Rat IgG, biotin-labeled (Jackson ImmunoResearch, 712-065-153) 1/200 in TNB for 45 min.
-
v.Wash 3 × 5 min with TNT.
-
w.Incubate for 30 min with Streptavidin-Peroxidase 1/100 in TNB.
-
x.Wash 3 × 5 min with TNT.
-
y.Incubate for 8 min with CY5 1/50 in amplification diluent.
-
z.Wash 3 × 5 min with TNT.
-
aa.Incubate for 30 min with Hoechst 1/500 in TNB.
-
bb.Wash 3 × 5 min with TNT.
-
cc.Add Mount Prolong Gold medium (without DAPI) to cover the section.
-
a.
-
10.ESM1/CD34-staining.
-
a.Deparaffinize the tissue sections and rehydrate for 5 min with distilled water.
-
b.Perform 20 min Antigen Retrieval (in Target Retrieval Solution pH6, Dako S1699, diluted to 1X in milliQ water and preheated to ca 100°C; the preheated solution should become milky in appearance) in the 100°C oven followed by 20 min cool down.
-
c.Wash 5 min with TBS.
-
d.Apply protein block (Donkey serum) 1/5 in TNB for 45 min to block nonspecific background staining.
-
e.Apply primary antibody Goat anti-ESM-1 (R&D AF1999) 1/50 in TNB.
-
f.Wash 3 × 5 min with TNT.
-
g.Add Donkey anti-goat-alexa 488 (Life Technologies, A-11055) 1/200 in TNB.
-
h.Wash 3 × 5 min with TNT.
-
i.Proceed to step j of section 11 for the subsequent CD34 staining.
-
a.
-
11.EdU/CD34-staining.
-
a.Prepare stock solutions according to the manufacturers’ protocol (website: https://www.thermofisher.com/be/en/home/references/protocols/cell-and-tissue-analysis/protocols/click-it-edu-imaging-protocol.html?ef_id=CjwKCAjw2bmLBhBREiwAZ6ugo38i0vhRD36_ne8gKIcEjFcRav5pyEegVUK8xqEBXqKounS_y1CgmxoCpq4QAvD_BwE:G:s&s_kwcid=AL!3652!3!447.
-
i.Bring vials to 20°C–22°C before opening.
-
ii.Add 2 mL DMSO (Component C) or an aqueous solution to Component A to make a 10 mM EdU stock solution. Vortex and store at –20°C.
-
iii.Prepare 1× Click-iT® EdU reaction buffer by adding 36 mL of deionized water to Component D. Store at 2°–8°C.
-
iv.Prepare Alexa Fluor® azide solution by adding 70 μL DMSO (Component C) to Component B. Mix well. Store at –20°C. When stored as directed, this working solution is stable for up to 1 year.
-
v.Prepare 10× Click-iT® EdU buffer additive (Component F) by adding 2 mL deionized water to the vial (Component F) and mix until fully dissolved. Store at –20°C. When stored as directed, this stock solution is stable for up to 1 year. If the solution develops a brown color, it has degraded and should be discarded.
-
vi.Dilute Hoechst® 33342 (Component G) 1:2,000 in PBS to obtain a 1× solution.
-
i.
-
b.Inject 40 μL (5 mg/kg for 20 g mouse) of the EdU solution (10 mM; 0.1 mg in 40 μL) intraperitoneally, 2 h before sacrificing mice for histology. Dissect and process the lungs as described in section 8.Note: Protect the EdU solution from light (turn off light in the laminar flow hood and put tinfoil around each vial).
-
c.Deparaffinize the tissue sections and rehydrate for 5 min with distilled water and rinse for 5 min with TBS.
-
d.Perform 20 min Antigen Retrieval in the oven followed by 20 min cool down (see section 10b).
-
e.Wash 5 min with TBS.
-
f.Rinse 15 min with PBS + 0.1% Triton X-100 at 20°C–22°C.
-
g.Make 1× Click-iT® EdU buffer additive by diluting the 10× solution prepared above 1:10 in deionized water. Use this solution within 8 h.
-
h.Apply the Click-iT® reaction cocktail at 4°C:Note: Add the ingredients in the order as listed.For a total volume of 500 μL:430 μL 1× Click-iT® EdU reaction buffer.+ 20 μL CuSO4.+ 1.2 μL Alexa Fluor® azide – 555.+ 50 μL 1× Click-iT® EdU buffer additive.
-
i.Rinse 2 × 15 min with PBS + 0.1% Triton X-100 at 20°C–22°C.
-
j.To block endogenous peroxidase activity, incubate for 20 min with MeOH/H2O2.
-
k.Wash 3 × 5 min with TBS.
-
l.Apply protein block (Donkey serum) 1/5 in TNB for 45 min to block nonspecific background staining.
-
m.Apply Rat anti-CD34 (BD 553731) 1/25 in TNB.
-
n.Wash 3 × 5 min with TNT.
-
o.Apply Donkey anti-Rat IgG, biotin-labeled (Jackson ImmunoResearch, 712-065-153) 1/200 in TNB for 45 min.
-
p.Wash 3 × 5 min with TNT.
-
q.Incubate for 30 min with Streptavidin-Peroxidase 1/100 in TNB.
-
r.Wash 3 × 5 min with TNT.
-
s.Incubate for 8 min with Cy5 Tyramide 1/50 in amplification diluent.
-
t.Wash 3 × 5 min with TNT.
-
u.Incubate for 30 min with Hoechst 1/500 in PBS.
-
v.Wash 3 × 5 min with TNT.
-
w.Add Mount Prolong Gold medium (without DAPI).
-
a.
Figure 3.
Distinguishing vessel co-option from angiogenic metastases
(A) Workflow for immunostaining of murine lung tissue.
(B) Representative images of renal cancer lung metastases, stained for EdU, CD34, and Hoechst (nuclei). Middle images are magnifications of respective boxed areas. Arrowheads, proliferating ECs; scale bar 50 μm.
(C) Quantification of the percentage of proliferating ECs in (B). Data are means ± SEM; n = 3–6. ∗p < 0.05 by one-way ANOVA with Tukey’s multiple comparisons test.
(D) Representative images of renal cancer lung metastases stained for ESM-1, CD34, and Hoechst (nuclei). Middle images are magnifications of the respective boxed areas. Arrowheads, tip ECs; scale bar 50 μm.
(E) Quantification of ESM-1+ ECs per CD34+ area in (D). Data are means ± SEM; n = 3. ∗p < 0.05 by one-way ANOVA with Tukey’s multiple comparisons test.
(F) Representative micrographs of angiogenic (top) and vessel co-option (bottom) renal cancer lung metastases sections, immunostained for CD31 (ECs) and PDPN (pneumocytes) on adjacent sections. Nuclei are counterstained with Hoechst. Images in the middle are magnifications of the respective boxed areas. Note how the metastasis, relying on angiogenic vessel sprouting, grows as a sphere that compresses the surrounding lung parenchyma and into which CD31low vessels sprout (inset), while the cancer cells of the metastasis, relying on vessel co-option, invade the surrounding lung tissue in an irregular/infiltrative manner with preservation of the epithelial and vascular structure. Dotted yellow line: border between metastatic tissue and surrounding lung tissue; white arrowheads: angiogenic vessels, yellow arrows: border between the metastasis and the compressed surrounding lung tissue; asterisks, co-opting cancer cells; scale bar, 100 μm.
(G) Quantification of vascularization type in the RENCA model in the indicated conditions. Data are mean ± SEM; n=3–5. This figure is adapted from Teuwen et al. (Teuwen et al., 2021).
Expected outcomes
The above protocols describe how to establish the RENCA and 4T1 model, how to isolate high purity single cells from the lung and how to perform immunostainings to discriminate vessel co-option from angiogenesis.
After setting-up the model, metastases can be analyzed typically at day 10, 21 and 36 in the RENCA and at day 13 in the 4T1 model.
For isolation of metastases-derived cells (including cancer cells, immune cells, perivascular cells) and ECs, mechanical and enzymatic digestion was used. Of note, for EC isolation, magnetic bead sorting was performed to deplete immune cells (CD45)/epithelial cells (EpCAM) and to enrich for ECs with CD31 MicroBeads. Expected viability ranges from 85%-94% for lung cells derived from metastases and from 87%-94% for lung ECs. The expected yield for normal ECs is approximately 650.000 cells per mouse and for metastases-derived ECs approximately 2.000.000 cells per mouse (these values are an average of six experiments). The purity for normal ECs and tumor ECs is 81% and 95%, respectively. Purity was checked by FACS immediately after MACS enrichment.
After isolation, these cells/tissues can be used for different assays. For example, high purity single cells can be further processed for single-cell RNA sequencing (scRNAseq) (not addressed in this protocol but described in more detail by Teuwen et al. (Teuwen et al., 2021)). To prepare cells for scRNAseq, isolated single-cell suspensions of freshly isolated metastases-derived cells and ECs can be resuspended in PBS containing 0.4% UltraPure BSA (50 mg/mL; Thermo Fisher Scientific), filtered over 40 μm cell strainers on ice, and converted to barcoded scRNA-seq libraries using the Chromium Single Cell 3′ Library, Gel Bead & Multiplex Kit and Chip Kit (10× Genomics). Besides scRNA-seq, isolated cells can also be used for bulk RNA-seq or for cell culture, etc.
Limitations
We acknowledge the following limitations related to the above described protocols. First, the protocols were established for metastates-derived cell and EC isolation from lungs using 8-10 week-old BALB/c mice. Therefore, adjustments might be needed if metastases-derived cells and/or ECs are isolated from a different organ or a different developmental stage. In addition, no other strain than BALB/c mice can be used to establish these tumor models because both the 4T1 cell line and RENCA cell line are derived from a spontaneous malignancy that occurred in BALB/c mice (Hrushesky and Murphy, 1973; Pulaski and Ostrand-Rosenberg, 2001; Sobczuk et al., 2020). Second, adjustments might be needed depending on the specific downstream use of the isolated cells (for example in vitro cell culture). Third, the results obtained after immunostaining are dependent on a proper perfusion and insufflation of the lungs and on the embedding/storage of the lung tissue. Fourth, the dilution of the antibodies might vary between different lots and vendors/manufacturers. Therefore, we recommend to first optimize these parameters for each antibody before use.
Troubleshooting
Problem 1
Correct number of RENCA/4T1 cells to inject in order to keep mice alive without the risk of them becoming sick before the end of the experimental period: It is important to carefully titrate the number of cells to be injected prior to the start of an experiment in order to ensure both tumor take and that mice do not meet a humane endpoint prior to 10 days post-injection in the vehicle group (steps 1b, 1d). Second, it has to be taken into account that results in terms of the tumor models may vary slightly from lab-to-lab and that mice might therefore have to be sacrificed due to signs of distress before 10 days, or that tumors might fail to grow within 10 days.
Potential solution
Perform cell number titration experiments: The amount of 2 × 105 cells per mouse should be considered as an estimate, and should be adapted in case necessary. We recommend to perform titration experiments with different cell numbers to make sure that the mice develop a tumor but also that the mice do not meet a humane endpoint before the end of the experimental period.
Problem 2
Preparation of control vehicle: Bring control vehicle solution to pH 6: Be aware that this step might be tricky to perform because there is no buffering compound (like TRIS, etc.) present in the control vehicle (step 2f).
Potential solution
To bring the control vehicle solution to pH 6: We recommend to add 50–100 microliters of 0.5 M HCl or 0.5 M NaOH (as appropriate) at a time and wait patiently as the pH changes very slowly. If you use stronger acid / base, the pH may change too rapidly to get this right.
Problem 3
Preparation of sunitinib: solubility: Sunitinib (malate powder) is difficult to dissolve into the control vehicle solution (step 3a).
Potential solution
To dissolve sunitinib in the control vehicle solution: We recommend to preheat the control vehicle to 37°C, vortex for several rounds and place the sunitinib solution in a 37°C water bath in between vortexing. The drug has a tendency to settle quickly, so before dosing each mouse, the drug should be vortexed or drawn up and down several times with the syringe/gavage needle.
Problem 4
Low viability of isolated metastases-derived cells or ECs: Low viability of cells derived from metastases or ECs can be caused by an extended isolation time, an extended digestion time and/or might be dependent on the number of manipulations on the cells (steps 5e, 6a–e, 7a(i-v), 7b, 7c).
Potential solution
To avoid low viability of isolated metastases-derived cells or ECs: It is recommended to process the samples as fast as possible in order to reduce the isolation time. Furthermore, during the isolation procedure it is recommended to store cell solutions at 4°C or on ice. A crucial step for cell viability is the enzymatic digestion. Therefore, if the cell viability is low, we advise to adjust the time of the enzymatic digestion step.
Problem 5
Low number of isolated metastases-derived cells: A low number of isolated cells derived from metastases may occur due to:
-
•
Underdigestion or overdigestion of the dissected tissue (steps 5e, 6a–e).
Low number of isolated ECs: A low number of isolated ECs may occur due to:
-
•
Underdigestion or overdigestion of the dissected tissue (steps 5e, 7a (i–v)).
-
•
Poor enrichment with CD31 MicroBeads (step 7c).
Potential solution
To avoid low numbers of isolated metastases-derived cells or isolated ECs, we recommend to optimize the different mechanical digestion steps (vigorous shaking by hand, pipetting, cutting of metastatic tissue with scalpel blade). By adjusting these steps, the maximum possible number of cells can be obtained after isolation. For the ECs in particular, we recommend to optimize the CD31 MicroBeads volume (increase volume to avoid loss of ECs during magnetic sorting) and antibody concentration (increase concentration to avoid loss of ECs during cell sorting) according to the manufacturer’s recommendations. Alternatively, other non-column based magnetic isolation technologies can be used for positive and negative selection (e.g., from STEMCELL Technologies).
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Peter Carmeliet (peter.carmeliet@kuleuven.be).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
L.-A.T. and P.B.V. are supported by Fonds Oncologie Augustinus-Koning Boudewijnstichting and GZA Ziekenhuizen, and A. Cuypers and L.P.M.H.d.R. by the Fonds Wetenschappelijk Onderzoek (FWO); A.R.R. is currently a full-time employee of AstraZeneca; V.L.B. is a principal laboratory research scientist at the Francis Crick Institute; J.K. is supported by AIAS-CO-FUND II: GA: MSCA: 754513, Lundbeckfonden: R307-2018-3667, Carlsberg Fonden: CF19-0687, Kræftens Bekæmpelse: R302-A17296, A.P. Møller Fonden: 20-L-0317, Riisfort Fonden, and Steno Diabetes Center Aarhus (SDCA); and P.C. is supported by grants from Methusalem funding (Flemish Government), the Fund for Scientific Research-Flanders (FWO-Vlaanderen), the European Research Council ERC Advanced Research Grant EU- ERC74307, and the NNF Laureate Research Grant from Novo Nordisk Foundation (Denmark).
Author contributions
Conceptualization and Direction: P.C.; investigation: A. Cuypers, L.-A.T., L.P.M.H.d.R., A.R.C., J.K., A.B., S.V., A. Carton, and A.M.; writing: A. Cuypers, L.-A.T., and A.R.R.; supervision: L.-A.T., L.P.M.H.d.R., V.L.B., G.E., M.D., and P.C.; funding acquisition: P.B.V. and P.C.
Declaration of interests
The authors declare no competing interests.
Data and code availability
The published article by Teuwen et al. (Teuwen et al., 2021) includes all datasets generated or analyzed during this study.
References
- Bridgeman V.L., Vermeulen P.B., Foo S., Bilecz A., Daley F., Kostaras E., Nathan M.R., Wan E., Frentzas S., Schweiger T., et al. Vessel co-option is common in human lung metastases and mediates resistance to anti-angiogenic therapy in preclinical lung metastasis models. J. Pathol. 2017;241:362–374. doi: 10.1002/path.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conchinha N.V., Sokol L., Teuwen L.A., Veys K., Dumas S.J., Meta E., García-Caballero M., Geldhof V., Chen R., Treps L., et al. Protocols for endothelial cell isolation from mouse tissues: brain, choroid, lung, and muscle. STAR Protoc. 2021;2:100508. doi: 10.1016/j.xpro.2021.100508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrushesky W.J., Murphy G.P. Investigation of a new renal tumor model. J. Surg. Res. 1973;15:327–336. doi: 10.1016/0022-4804(73)90096-6. [DOI] [PubMed] [Google Scholar]
- Kuczynski E.A., Vermeulen P.B., Pezzella F., Kerbel R.S., Reynolds A.R. Vessel co-option in cancer. Nat. Rev. Clin. Oncol. 2019;16:469–493. doi: 10.1038/s41571-019-0181-9. [DOI] [PubMed] [Google Scholar]
- Pulaski B.A., Ostrand-Rosenberg S. Mouse 4T1 breast tumor model. Curr. Protoc. Immunol. 2001;Chapter 20:Unit 20.2. doi: 10.1002/0471142735.im2002s39. [DOI] [PubMed] [Google Scholar]
- Serwer L., Hashizume R., Ozawa T., James C.D. Systemic and local drug delivery for treating diseases of the central nervous system in rodent models. J. Vis. Exp. 2010:1992. doi: 10.3791/1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczuk P., Brodziak A., Khan M.I., Chhabra S., Fiedorowicz M., Wełniak-Kamińska M., Synoradzki K., Bartnik E., Cudnoch-Jędrzejewska A., Czarnecka A.M. Choosing the right animal model for renal cancer research. Transl. Oncol. 2020;13:100745. doi: 10.1016/j.tranon.2020.100745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuwen L.A., De Rooij L.P.M.H., Cuypers A., Rohlenova K., Dumas S.J., García-Caballero M., Meta E., Amersfoort J., Taverna F., Becker L.M., et al. Tumor vessel co-option probed by single-cell analysis. Cell Rep. 2021;35:109253. doi: 10.1016/j.celrep.2021.109253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The published article by Teuwen et al. (Teuwen et al., 2021) includes all datasets generated or analyzed during this study.