Abstract
Advanced prostate cancer (aPC) in Black men was reported to present with aggressive features and to be associated with poor prognosis. Herein, we compared the cell-free DNA (cfDNA) genomic landscape of aPC in Black vs White men. Patients (pts) with aPC from 6 academic institutions and available cfDNA comprehensive genomic profiling (CGP) were included. Association between mutated genes and race was evaluated using Barnard’s test and a Probabilistic Graphical Model (PGM) machine learning approach. Analysis included 743 aPC pts (217 Black, 526 White) with available cfDNA CGP. The frequency of alterations in the androgen receptor gene was significantly higher in Black vs White men (55.3% vs 35% respectively, P < .001). Additionally, alterations in EGFR, MYC, FGFR1, and CTNNB1 were present at higher frequencies in Black men. PGM analysis and Barnard’s test were concordant. Findings from the largest cohort of Black men with aPC undergoing cfDNA CGP may guide further drug development in these men.
Keywords: prostate cancer, Black, White, cell-free DNA, comprehensive genomic profiling
This article compares the cell-free DNA (cfDNA) genomic landscape of men with advanced prostate cancer with a focus on differences based on race.
According to the United States Surveillance, Epidemiology and End Results Program (SEER), Black men have a higher incidence of prostate cancer and mortality rates compared with those of non-AA (175.8 vs 104.1 and 37.4 vs 17.9 per 100,000 individuals) respectively.1 Paradoxically, recent reports indicate that Black men with advanced prostate cancer (aPC) may respond better to systemic therapies and have better survival outcomes compared with White men.2,3 We hypothesize that this disconnect between the disease presentation and response to therapy may be explained by differences in the underlying tumor genomic landscape as assessed by liquid biopsy.
Toward this end, a retrospective analysis of the comprehensive genomic profiles (CGP) of cell-free DNA (cfDNA) extracted from plasma using the Guardant360 test (GuardantHealth) from patients with aPC managed at 6 academic institutions (Winship Cancer Institute of Emory University, Barbara Ann Karmanos Cancer Institute, Medical University of South Carolina, University of Alabama, Tulane University School of Medicine; Huntsman Cancer Institute at the University of Utah) is reported.
The first available cfDNA panel results were included in this analysis and the pairwise association between mutated genes and race was investigated by Barnard’s test and adjusted by Benjamini-Hochberg’s False Discovery Rate (BH-FDR). An independent Probabilistic Graphical Model (PGM) machine learning approach further explored the association between race and the landscape of altered genes.4
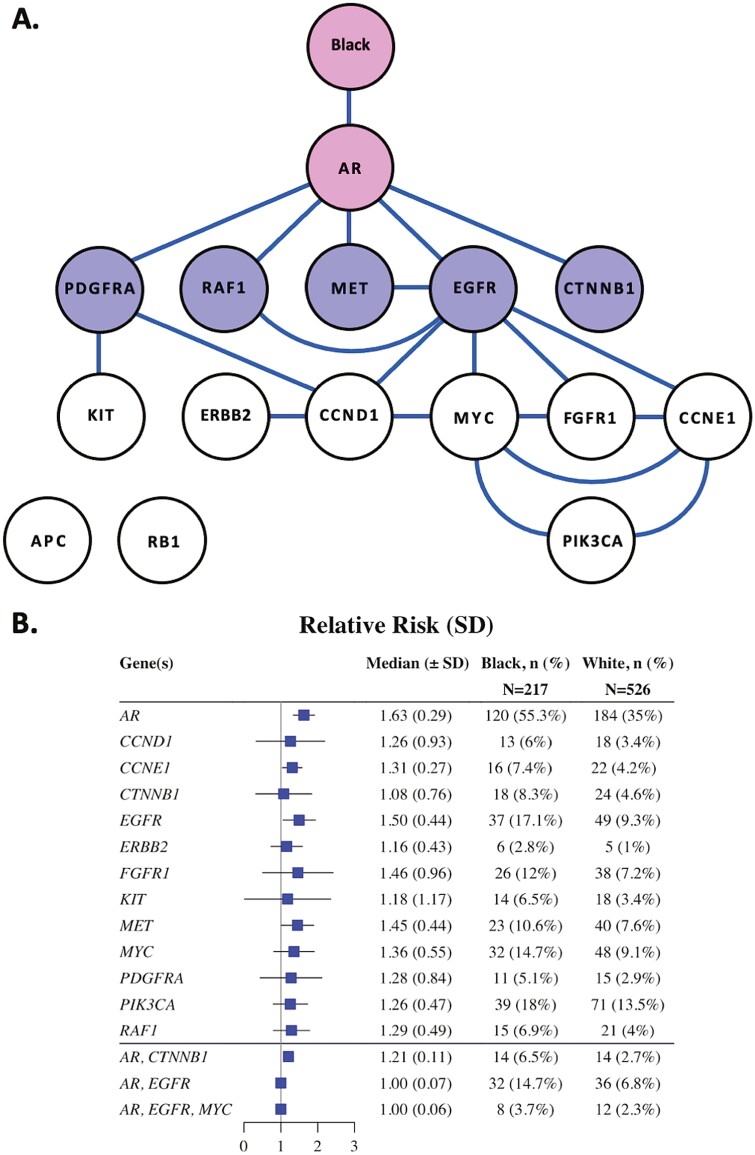
A total of 743 patients, 217 Black men (29%) and 526 (71%) White men with aPC who had undergone tumor genomic profiling by cfDNA CGP were available and were included in this study. Multiple genomic aberrations were enriched in Black patients (Table 1); these were also detected by PGM (Fig. 1). Pathogenic genomic alterations were found in 92% Black men and 83% of White men. Black men had a greater median number of alterations (n = 3) compared with White men (n = 2). The genomic landscape by race is shown in Supplementary Fig. S1. Targetable alterations of interest included EGFR, PIK3CA, and FGFR1. A full list of genomic alterations is available in the supplementary figure, with some patients having more than one alteration per gene.
Table 1.
Number of patients harboring a pathogenic alteration in the top 15 genes found in Black or White men based on BH-FDR.
| Affected gene | Black (N = 217) | White (N = 526) | P-value | BH-FDR | ||
|---|---|---|---|---|---|---|
| Patients | Frequency | Patients | Frequency | |||
| AR | 120 | 55.30 | 184 | 34.98 | <.001 | <0.001 |
| EGFR | 37 | 17.05 | 49 | 9.32 | .003 | 0.070 |
| MYC | 32 | 14.75 | 48 | 9.13 | .025 | 0.375 |
| FGFR1 | 26 | 11.98 | 38 | 7.22 | .036 | 0.375 |
| CTNNB1 | 18 | 8.29 | 24 | 4.56 | .047 | 0.375 |
| KIT | 14 | 6.45 | 18 | 3.42 | .065 | 0.375 |
| RB1 | 13 | 5.99 | 16 | 3.04 | .061 | 0.375 |
| ERBB2 | 6 | 2.76 | 5 | 0.95 | .063 | 0.375 |
| SMAD4 | 3 | 1.38 | 1 | 0.19 | .047 | 0.375 |
| CCNE1 | 16 | 7.37 | 22 | 4.18 | .073 | 0.381 |
| PIK3CA | 39 | 17.97 | 71 | 13.50 | .125 | 0.383 |
| APC | 17 | 7.83 | 27 | 5.13 | .184 | 0.383 |
| RAF1 | 15 | 6.91 | 21 | 3.99 | .094 | 0.383 |
| CCND1 | 13 | 5.99 | 18 | 3.42 | .120 | 0.383 |
| PDGFRA | 11 | 5.07 | 15 | 2.85 | .137 | 0.383 |
Figure 1.
Conditional risk landscape visualization. (A) Probabilistic graphical model representing the association between the genomic alterations and race in this cohort (N = 743). Each node represents a mutated gene, each edge indicates a direct dependence between mutated genes. (B) Forest plot indicating the relative risk of having an alteration in a specific gene and Black or White race. Moreover, investigation of more complex relationships such as those between race and secondarily connected genes (AR, CTNNB1 and AR, EGFR) is presented in lower half of the forest plot. Pink shading indicates that the gene has a direct connection to race; purple shaded nodes are for genes with a secondary connection to race.
To our knowledge, this is the largest dataset of genomic profiling of cfDNA in Black men with aPC reported to date. We found a significantly higher frequency of AR gene alterations in Black men compared to White men. In addition, alterations in the EGFR and MYC genes, as well as WNT pathway genes CTNNB1 and APC, were present in greater frequency in Black men. While these genomic alterations have been associated with poorer clinical outcomes in Black men, they are targetable using novel agents as a monotherapy or in combination with AR-targeting agents.5 The analysis of the available data by PGMs complements traditional statistical testing of pairwise comparisons. In particular, the PGM indicates that an Black individual has a 1.63-fold greater risk of having a pathogenic AR variant. Moreover, this risk ratio is little influenced by conditional dependencies between AR and other genes, suggesting that for this dataset, AR is the primary genetic alteration differentiating Black men from White men with prostate cancer.
Many of altered genes identified in this work have been shown to be inter-related in metastatic prostate cancer. For example, amplification of the oncogenic transcription factor C-MYC is commonly observed in prostate cancer in tumors expressing high levels of AR and antagonizes the expression of the AR transcriptional program.6 Alterations in WNT pathway and crosstalk with AR signaling have been reported in patients with prostate cancer and are associated with progression to androgen insensitive tumor growth and poor prognosis. Our PGM confirm this strong genomic co-dependence (Fig. 1).
In 2014, Dovey et al7 summarized the key molecular-specific characteristics of prostate cancer oncogenesis in Black vs White males. They found that defective AR signaling, telomerase shortening (elevated c-MYC expression), epigenetic differences affecting signaling and epithelial-mesenchymal transition pathways (such as PI3K signaling pathway), and deficient WNT signaling pathway mutations were key molecular characteristic in Black men. Furthermore, Dovey et al highlight that a distinguishing feature in the genomic landscapes between Black and White men is the higher frequency of a pathogenically mutated AR gene. Our results support this finding, albeit with a significantly greater number of AR alterations in Black men. However, previous work by our group and others has found a higher frequency of AR alterations in patients with mCRPC compared with patients with mCSPC.4 Of note, a report by Sivakumar et al suggested that Black men receive CGP later in their treatment course and are less likely to have access to the latest therapeutic options.8 In fact, previous reports indicate that if Black men have equal access to the standard-of-care treatments, their outcomes may be similar or better to the outcomes of White men with prostate cancer.3,9 Overall, the major distinction between the 2 cohorts presented here is the overall prevalence of AR alterations, which are higher in Black patients. In this cohort, the higher frequency of AR alterations in Black men could reflect a higher proportion of heavily treated patients, as also noted by Stopsack et al in their recent review of cancer genomes by race.10
The limitations of this project include the lack of clinical annotation such as the disease state, tumor volume, and treatment exposure (including the use of AR axis therapies). We also restrict this analysis to DNA testing and we lack a serial assessment of the genomic landscape changes, which would offer us additional insights into the exposure to different treatments during the course of the disease. Finally, the limited size of the cohort, number of alterations, and limited number of genes on the panel may have resulted in failure to detect weaker associations between genes, mutations, and race.
Future research that includes a multi-omics approach and socioeconomic disparities may help elucidate how these contribute to the poor outcomes seen in Black men with prostate cancer. The analysis of the available data may also be improved by exploring the utility of PGMs for investigations of conditional dependencies among variants and genes that influence outcomes. This approach complements traditional statistical testing of pairwise comparisons.
Supplementary Material
Contributor Information
Raquel Zimmerman, University of Utah School of Medicine, Salt Lake City, UT, USA; Department of Human Genetics, University of Utah, Salt Lake City, UT, USA.
Mehmet A Bilen, Department of Hematology and Medical Oncology, Emory University School of Medicine, Atlanta, GA, USA; Winship Cancer Institute of Emory University, Atlanta, GA, USA.
Elisabeth I Heath, Department of Hematology and Medical Oncology, Emory University School of Medicine, Atlanta, GA, USA.
Lakshminarayanan Nandagopal, University of Alabama at Birmingham, Birmingham, AL, USA.
Umang Swami, Huntsman Cancer Institute, University of Utah, Salt Lake City, UT, USA.
Adam Kessel, Huntsman Cancer Institute, University of Utah, Salt Lake City, UT, USA.
Ellen Jaeger, Deming Department of Medicine, Section of Hematology/Oncology, Tulane University Medical School, New Orleans, LA, USA.
Sergiusz Wesolowski, Department of Human Genetics, University of Utah, Salt Lake City, UT, USA.
Edgar J Hernanadez, Department of Human Genetics, University of Utah, Salt Lake City, UT, USA.
Jonathan Chipman, University of Utah School of Medicine, Salt Lake City, UT, USA.
Alleda Mack, Karmanos Cancer Institute, Department of Oncology, Wayne State University School of Medicine, Detroit, MI, USA.
Deepak Ravindranathan, Department of Hematology and Medical Oncology, Emory University School of Medicine, Atlanta, GA, USA.
Benjamin L Maughan, University of Utah School of Medicine, Salt Lake City, UT, USA.
Roberto Nussenzveig, Huntsman Cancer Institute, University of Utah, Salt Lake City, UT, USA.
Mark Yandell, Department of Human Genetics, University of Utah, Salt Lake City, UT, USA.
Manish Kohli, Huntsman Cancer Institute, University of Utah, Salt Lake City, UT, USA.
Michael B Lilly, Medical University of South Carolina Hollings Cancer Center, Charleston, SC, USA.
A Oliver Sartor, Deming Department of Medicine, Section of Hematology/Oncology, Tulane University Medical School, New Orleans, LA, USA.
Neeraj Agarwal, Huntsman Cancer Institute, University of Utah, Salt Lake City, UT, USA.
Pedro C Barata, Deming Department of Medicine, Section of Hematology/Oncology, Tulane University Medical School, New Orleans, LA, USA.
Conflict of Interest
Mehmet A. Bilen: Exelixis, Bayer, BMS, Eisai, Pfizer, AstraZeneca, Janssen, Calithera Biosciences, Genomic Health, Nektar, EMD Serono, SeaGen, Sanofi (C/A, SAB), Merck, Xencor, Bayer, Bristol-Myers Squibb, Genentech/Roche, SeaGen, Incyte, Nektar, AstraZeneca, Tricon Pharmaceuticals, Genome & Company, AAA, Peloton Therapeutics, Pfizer (RF [institutional]); Elisabeth I. Heath: Astellas Pharma (C/A), Astellas Pharma, Arvinas, AstraZeneca, BioXcel Therapeutics, Bristol-Myers Squibb, Calibr, Calithera Biosciences Inc., Caris Life Sciences. Corcept Therapeutics, Corvis Pharmaceuticals, Daiichi Sankyo Inc., Eisai Inc., Exelixis, Five Prime Therapeutics, Fortis Therapeutics, GlaxoSmithKline, Gilead Sciences Inc., Harpoon Therapeutics, Hoffman-La Roche, Infinity Pharmaceuticals, iTeos Therapeutics, Janssen Research & Development LLC, Merck Sharp & Dohme, Merck, Mirati Therapeutics, Modra Pharmaceuticals, Oncolys BioPharma, Peloton Therapeutics Inc., Pfizer, Pharmacyclics LLC, POINT Biopharma, Seattle Genetics (RF), Bayer, Sanofi, Seattle Genetics (H), Bayer, Sanofi, Astellas Pharma, Caris Life Sciences, Seattle Genetics (travel expenses), Janssen Research & Development LLC (Steering Committee), Bayer, Sanofi (SAB); Umang Swami: Astellas, Seattle Genetics (C/A), Janssen, Astellas/Seattle Genetics (RF [institutional]); Mark Yandell: Backdrop Health Inc. (a Founder of Backdrop Health Inc., which has a commercial license to some of the software used in the analyses); Michael B. Lilly: Bayer Pharmaceuticals (RF); A. Oliver Sartor: Advanced Accelerator Applications (AAA), Astellas, AstraZeneca, Bayer, Blue Earth Diagnostics, Inc., Bavarian Nordic, Bristol Myers Squibb, Clarity Pharmaceuticals, Clovis, Constellation, Dendreon, EMD Serono, Fusion, Isotopen Technologien Meunchen, Janssen, Merck, Moyvant, Myriad, Noria Therapeutics, Inc., Novartis, Noxopharm, Progenics, POINT Biopharma, Pfizer, Sanofi, Tenebio, Telix, Theragnostics (C/A), Advanced Accelerator Applications, Amgen, AstraZeneca, Bayer, Constellation, Endocyte, Invitae, Janssen, Lantheus, Merck, Progenics, Tenebio (RF); Neeraj Agarwal: Astellas, AstraZeneca, Aveo, Bayer, Bristol Myers Squibb, Calithera, Clovis, Eisai, Eli Lilly, EMD Serono, Exelixis, Foundation Medicine, Genentech, Gilead, Janssen, Merck, MEI Pharma, Nektar, Novartis, Pfizer, Pharmacyclics, Seattle Genetics (C/A), Astellas, AstraZeneca, Bavarian Nordic, Bayer, Bristol Myers Squibb, Calithera, Celldex, Clovis, Eisai, Eli Lilly, EMD Serono, Exelixis, Genentech, Gilead, Glaxo Smith Kline, Immunomedics, Janssen, Medivation, Merck, Nektar, New Link Genetics, Novartis, Pfizer, Prometheus, Rexahn, Roche, Sanofi, Seattle Genetics, Takeda, Tracon (RF [institutional]). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board.
Data Availability
The data underlying this article will be shared on reasonable request to the corresponding author.
References
- 1. SEER cancer facts: prostate cancer. https://seer.cancer.gov/statfacts/html/prost.html. Accessed May 5, 2021.
- 2. Sartor O, Armstrong AJ, Ahaghotu C, et al. Survival of African-American and Caucasian men after sipuleucel-T immunotherapy: outcomes from the PROCEED registry. Prostate Cancer Prostatic Dis. 2020;23(23):517-526. 10.1038/s41391-020-0213-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. George DJ, Heath EI, Sartor AO, et al. Abi Race: a prospective, multicenter study of black (B) and white (W) patients (pts) with metastatic castrate resistant prostate cancer (mCRPC) treated with abiraterone acetate and prednisone (AAP). J Clin Oncol. 2018;36 (Suppl_18):LBA5009LBA50. 10.1200/jco.2018.36.18_suppl.lba5009. [DOI] [Google Scholar]
- 4. Lin E, Hahn AW, Nussenzveig RH, et al. Identification of somatic gene signatures in circulating cell-free DNA associated with disease progression in metastatic prostate cancer by a Novel Machine Learning Platform. Oncologist. 2021 Sep;26(9):751-760. 10.1002/onco.13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de Bono J, Mateo J, Fizazi K, et al. Olaparib for Metastatic Castration-Resistant Prostate Cancer. N Engl J Med 2020;382:2091-2102. [DOI] [PubMed] [Google Scholar]
- 6. Barfeld SJ, Urbanucci A, Itkonen HM, et al. c-Myc antagonises the transcriptional activity of the androgen receptor in prostate cancer affecting key gene networks. EBioMedicine 2017;18:83-93. 10.1016/j.ebiom.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dovey ZS, Nair SS, Chakravarty D, Tewari AK.. Racial disparity in prostate cancer in the African American population with actionable ideas and novel immunotherapies. Cancer Reports 2021;4(5):e1341. 10.1002/cnr2.1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sivakumar S, Lee JK, Moore JA, et al. Ancestral characterization of the genomic landscape, comprehensive genomic profiling utilization, and treatment patterns may inform disparities in advanced prostate cancer: a large-scale analysis. JCO. 2021;39(Suppl_15):5003-5003. 10.1200/JCO.2021.39.15_suppl.5003. [DOI] [Google Scholar]
- 9. Halabi S, Dutta S, Tangen CM, et al. Overall survival of Black and White men with metastatic castration-resistant prostate cancer treated with docetaxel. J Clin Oncol. 2019;37(5):403-410. 10.1200/JCO.18.01279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stopsack KH, Nandakumar S, Arora K, et al. Differences in prostate cancer genomes by self-reported race: contributions of genetic ancestry, modifiable cancer risk factors, and clinical factors. Clin Cancer Res. 2022;28(2):318-326. https://doi-org.ezproxy.lib.utah.edu/10.1158/1078-0432.CCR-21-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.